Abstract
Adenoviruses have attracted much attention as probes to study biological processes such as DNA replication, transcription, splicing, and cellular transformation. More recently these viruses have been used as gene-transfer vectors and oncolytic agents. On the other hand, adenoviruses are notorious pathogens in people with compromised immune functions. This article will briefly summarize the basic replication strategy of adenoviruses and the key proteins involved and will deal with the new developments since 2006. In addition, we will cover the development of antivirals that interfere with human adenovirus (HAdV) replication and the impact of HAdV on human disease.
An adenovirus-infected cell produces about one million copies of viral DNA within 40 h. This efficient replication mechanism is a desirable drug target, given the morbidity associated with adenovirus infections.
Members of the family Adenoviridae are nonenveloped icosahedral viruses that replicate in the nucleus. In recent years, new DNA amplification techniques and sequencing efforts have led to the identification of many new human and nonhuman adenoviruses (AdV). The human adenoviruses (HAdV) are grouped as seven species (formerly called “subgroups”), HAdV-A–G, in the Mastadenovirus genus. Within the species, distinct “types” (formerly called “serotypes”) are recognized based on the absence of serological cross-neutralization. With the advent of high-throughput sequencing of new virus isolates, a debate sparked around the question how sequence data should be used to define types and species (Aoki et al. 2011; Seto et al. 2011).
The successful propagation in tissue culture of small DNA tumor viruses like adenoviruses, polyoma virus, and SV40 opened up the field of studies on the replication and transcription of the genomes of these viruses in the 1960s and 1970s (Tooze 1980; Yaniv 2009). These studies yielded many of the basic principles of mammalian DNA replication. AdV DNA replication was reviewed in detail in Hay (1966), Van der Vliet (1996), and Van der Vliet and Hoeben (2006). The mechanisms of AdV DNA replication have now been firmly established, and many of the roles of the key actors in the process have been elucidated in detail. During the last decade, the advancement in acquiring insight in the molecular processes involved in AdV DNA replication slowed down as the interest of many groups shifted from the study of viral DNA replication to the study of cellular processes.
A DETAILED MODEL OF ADENOVIRUS REPLICATION
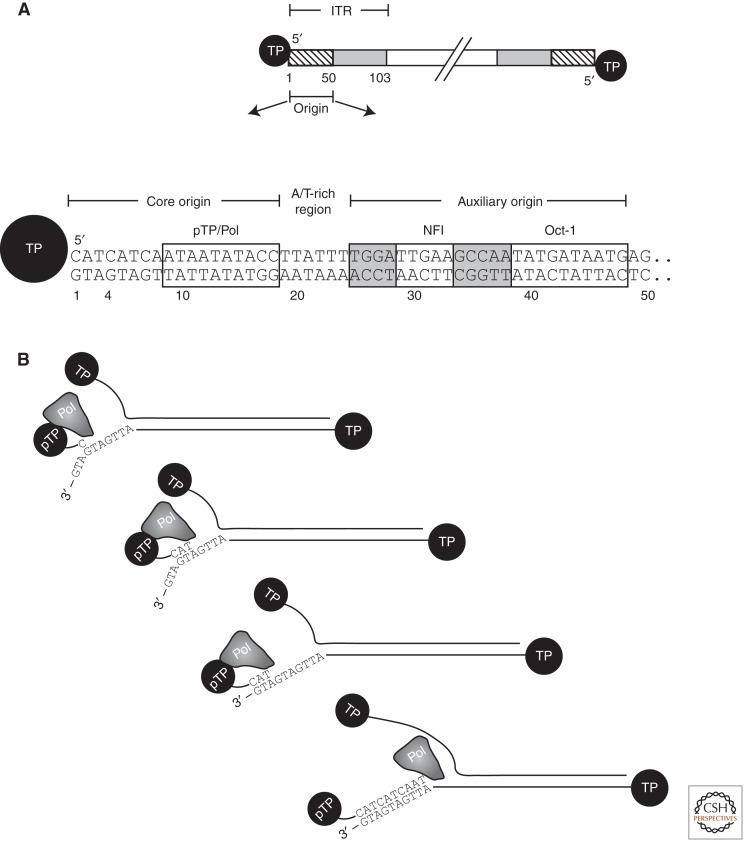
HAdV contain a linear double-stranded genome of about 36 kb with inverted terminal repetitions of ∼100 bp. Both 5′ termini are covalently attached to a 55-kDa terminal protein (TP). AdV DNA replication is a very efficient process. Within 40 h, an infected cell produces approximately one million copies of viral DNA. Much information on the replication mechanism came from an in vitro system that replicates HAdV-5 and -2 DNA with purified proteins. Two identical origins of replication are located within the inverted terminal repeats, covering ∼1–50 bp. The terminal 18 bp form the minimal origin and the remainder act as the auxiliary origin (Fig. 1A). Protein-primed DNA synthesis starts by covalent addition of a dCMP residue to an 80-kDa precursor of the TP (pTP). Replication requires three viral proteins encoded by E2 genes: pTP, AdV DNA polymerase (AdV Pol), and the DNA-binding protein (DBP). Two cellular transcription factors, NFI and Oct-1, bind the auxiliary origin and thereby enhance initiation several hundredfold. In the final stages of replication, pTP is cleaved by a viral protease to TP, resulting in progeny DNA, which is subsequently packaged in virions.
Figure 1.
(A) Structure of the adenovirus type 5 origin and regions required for interaction with viral and cellular proteins. (B) Schematic representation of the initiation of DNA replication illustrating how the nucleotides 4–6 are used as a template for the formation of the pTP-CAT complex. The complex subsequently displaces to pair with nucleotides 1–3 of the template stand, after which the elongation continues.
Multiple interactions between the three viral and two cellular proteins and the origin orchestrate the formation of a preinitiation complex. This step is accompanied by subtle changes in the origin structure by DBP and by extensive origin bending, mainly induced by NFI and Oct-1. Initiation occurs at position 4 of the template (sequence 3′-G1TAG4TA) followed by formation of a pTP-trinucleotide intermediate, pTP-CAT (Fig. 1B). This pTP-CAT intermediate jumps back three bases and becomes paired with template residues 1–3 (King and Van der Vliet 1994). This jumping-back mechanism explains the presence of a short 3-bp (or sometimes 2- or 4-bp) repeat sequence in the first 10 bp of all AdV origins (De Jong et al. 2003). For replication to begin at an internal site, rather than at the genome termini, appears to be a common feature among protein-primed replication systems (see below). A specific amino acid in pTP, Ser580 in HAdV-5, is used as a primer for covalent binding of dCMP by AdV Pol, generating pTP-C (Desiderio and Kelly 1981; Smart and Stillman 1982). During, or shortly after the three-nucleotide jump, AdV Pol dissociates from pTP (King and van der Vliet 1994). This leads to an increased rate of polymerization and proofreading. Further disassembly of the preinitiation complex takes place when NFI dissociates early in initiation, whereas Oct-1 dissociates only when the recognition site has become single stranded on passing of the replication fork (van Leeuwen et al. 1997). The pTP-CAT intermediate is efficiently elongated by the combined action of AdV Pol and DBP, leading to the formation of a new duplex genome and the displacement of the nontemplate strand. The ITRs of displaced nontemplate strands may anneal together (either intra- or intermolecularly) to restore functional origins that could serve as substrates for new replication rounds. Newly generated Ad genome duplexes (formed either by replication or the annealing of complementary displaced single strands) may also enter subsequent replication rounds or be processed and encapsidated into virions. A schematic representation of the protein–protein interactions and the protein–template interactions can be found in an earlier version of this article (van der Vliet and Hoeben 2006).
Although the mechanism of adenovirus DNA replication is unique among the eukaryotic viruses, it bears many similarities with the protein-primed DNA replication of several bacteriophages such as the Bacillus subtilis bacteriophages ϕ29 and GA-1, the Escherichia coli bacteriophage PRD1, and the Streptococcus pneumoniae phage CP-1. Similar to AdV, these phages contain linear double-stranded DNA genomes, which are replicated by protein-primed DNA replication mechanisms. The replication leaves a TP covalently coupled to the viral DNA (Blanco et al. 1989; Salas 1991; Illana et al 1996). Also in the replication of these bacteriophages, sliding-back or jumping-back mechanisms are used in which the nucleotides other than the 3′ terminal nucleotides are used as a template for the 5′ terminal nucleotide that remains covalently coupled to the phage’s TP (Salas 1991; Méndez et al. 1992; Caldentey et al. 1993; Illana et al. 1996; Martín et al. 1996).
VIRAL REPLICATION PROTEINS
The Precursor TP
The precursor TP (pTP) is an 80-kDa protein that functions as the primer for initiation. pTP binds both single-strand DNA (ssDNA) and double-strand DNA (dsDNA), and the affinity of pTP for ssDNA might function in stabilizing AdV Pol on partially unwound origin DNA (de Jong et al. 2003). During initiation, AdV Pol covalently couples the first dCMP residue to Ser580 of pTP. The initiation of replication by pTP-binding dCMP is well conserved in AdV. Hence almost all adenoviruses characterized to date have a C residue at the 5′ end of their genome, suggesting an inherent preference of pTP to bind dCMP. The single exception to this rule has been found in the fowl adenovirus-A (FAdV-A). In the FAdV-A isolates Phelps and KUR, initiation starts with incorporation of a dGMP. Interestingly, the highly similar FAdV-A strain OTE is conventional and starts initiation by binding of dCMP (Rademaker et al. 2006). Studies with minireplicons showed that the pTP and Pol of CELO-OTE and CELO-Phelps do not have different specificities, but rather show relaxed template specificity. This allows these viruses to use 3′-GTAGTAG as well as 3′-CTACTAC as templates for replication initiation (Rademaker et al. 2006). This implies the efficient formation of pTP-GMP complexes. This is in contrast to HAdV-5, in which mutation of G4 in the template strand fully blocked replication initiation in vitro. For HAdV-5, even in the absence of any template DNA, pTP-C-Pol complexes are formed. The preference of HAdV-5 pTP to bind dCMP was reduced in the presence of manganese ions in the replication-initiation assay, suggesting that structural factors govern the template specificity (King and van der Vliet 1994).
The AdV DNA Polymerase
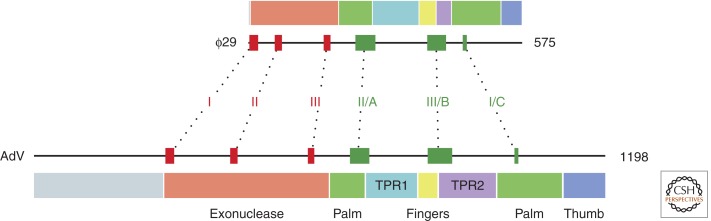
The AdV DNA polymerase (AdV Pol) belongs to the distinct group of family B (or α-like) DNA polymerases that uses a protein primer for the initiation of replication. The crystal structure of AdV Pol has not been determined, but sequence comparisons with polymerases of known structure as well as biochemical and mutagenesis studies have revealed AdV Pol’s general domain organization (Fig. 2) (Joung et al. 1991; Joung and Engler 1992; Knopf 1998; Liu et al. 2000; Brenkman et al. 2001, 2002; Liu et al. 2003). Besides consisting of the common polymerase subdomains palm, fingers, and thumb, which are associated with 5′–3′ polymerase activity, AdV Pol carries a 3′–5′ exonuclease domain that is necessary for its intrinsic proofreading ability (King et al. 1997; Brenkman et al. 2002; Liu et al. 2003). Furthermore, sequence comparisons have suggested that AdV Pol contains certain insertions, termed TPR1 and TPR2, which are specific for the protein-primed family B polymerases (Blasco et al. 1990; Dufour et al. 2000). Although no information is available on the roles of TPR1 and TPR2 in AdV Pol, studies on the bacteriophage ϕ29 provided some insights into their function. The crystal structure of the ϕ29 polymerase has been determined (Kamtekar et al. 2004, 2006; Berman et al. 2007). In the phage ϕ29 polymerase, TPR1 and TPR2 form discrete subdomains. Based on these structural data as well as biochemical studies on ϕ29 polymerase and its protein primer (Rodríguez et al. 2005), the TPR structures are known to be responsible for specific binding to the protein primer (TPR1) and/or suspected to contribute to ϕ29 Pol’s intrinsic strand-displacement capacity (TPR2) and/or high processivity (TPR1 and TPR2). They would contribute to the latter two functionalities by making part of the tunnels encircling, respectively, the downstream template strand and the upstream duplex product. These tunnels would topologically and functionally resemble, respectively, the helicases and sliding clamp proteins of replisomes. Interestingly, sequence comparisons with the other protein-primed family B polymerases indicate that the putative TPR2 domain of AdV Pol is rather large (Fig. 1) (Uil et al. 2011), which might suggest that it has an additional or different function.
Figure 2.
Representations of AdV and ϕ29 polymerases showing the locations of conserved sequence motifs and structural domains. The domain organization shown for ϕ29 Pol was described previously by Kamtekar et al. (2004) and is based on structural data. The domain organization depicted for AdV Pol is inferred from that of ϕ29 Pol and is based on the relative location of motifs shared among (many of) the DNA polymerases. Indicated for both polymerases are the most highly conserved of these sequence motifs within the exonuclease domain (“Exo” motifs I, II, and III) and the polymerase region (“Pol” motifs I, II, and III, also known as, respectively, motifs C, A, and B) (Wong et al. 1988; Bernad et al. 1989; Delarue et al. 1990). All representations are to scale.
Previously, AdV Pol function has mostly been studied by biochemical assays using purified proteins or cell extracts (Liu et al. 2003). Recent work, however, gave insight into the nucleotide selection and proofreading functions of AdV Pol in the context of full-fledged viral replication. In a study that aimed to make mutator AdV polymerases that could serve in viral directed evolution procedures, AdV Pol mutants were made that carried single-amino-acid substitutions in either the nucleotide binding pocket or the exonuclease active site (Uil et al. 2011). Using a polymerase complementation approach (in which a polymerase-defective virus is replicated on cells stably expressing the AdV Pol), these mutants were tested for their ability to support productive adenovirus replication. Furthermore, they were tested for mutator activity by performing deep sequencing on pools of replicated viruses.
Mutation of highly conserved residues within the nucleotide binding pocket generally had drastic effects on the ability to support Ad replication. For most of the mutants this would have likely been owing to a decrease in viral genome replication efficiency (rather than an excessive mutation rate). Also most of the exonuclease domain mutants tested were unable to support AdV replication. Interestingly, all substitutions of the most highly conserved exonuclease residues (those that contribute to the metal ion binding framework of the exonuclease active site) were found to be lethal for virus replication. Mutations applied to the homologs of these residues in other viral polymerases are known to effectively knock out 3′–5′ exonuclease activity and therewith proofreading activity (Bernad et al. 1989; Esteban et al. 1994; Abdus Sattar et al. 1996; Kuhn and Knopf 1996), but are not necessarily lethal in the context of productive virus replication, as has been found for RB69, HSV, and CMV (Hwang et al. 1997; Bebenek et al. 2001; Chou and Marousek 2008). Therefore, barring nonspecific structural effects, the lethality of these mutations for adenovirus might point to an additional role of these exonucleolytic residues. Perhaps related to this issue is that homologous mutations in ϕ29 polymerase result in a reduced velocity of DNA polymerization on dsDNA templates (Soengas et al. 1992; Esteban et al. 1994).
Of the viable Adv Pol mutants, several caused excess mutations during viral passaging, confirming the importance of the respective mutated motifs for incoming nucleotide selection and proofreading. For example, mutation of a nonhighly conserved residue located in a loop between two α helices of the fingers domain (D827A) led to a degree of mutator activity. As the concerning residue is not thought to directly contact the incoming nucleotide, it may be that its substitution indirectly affects nucleotide selection, for example, by changing the relative positions of the two α helices. The strongest mutator AdV Pols identified in the study were those of two mutants of the exonuclease motifs I and II (Fig. 1). These mutants, T286I and F421Y, carry substitutions of residues whose homologs in ϕ29 are known to play a role in primer terminus stabilization at the exonuclease active site during proofreading (de Vega et al. 1996, 1998). The mutator activities found for these mutants would support a similar role of these highly conserved residues in AdV Pol.
The AdV DBP
On infection the major viral replication protein produced is DBP. DBP binds dsDNA and ssDNA, the latter in a cooperative fashion, and it changes the DNA structure drastically (Kanellopoulos et al. 1995). DBP is involved in several steps during DNA replication. Besides protecting ssDNA against nucleases, DBP acts during initiation by stimulating binding of AdV Pol and NFI, presumably by slight changes in the configuration of origin DNA (Hay 1966; van Breukelen et al. 2003; van der Vliet and Hoeben 2006). Moreover, DBP lowers the Km for coupling of the first nucleotide to pTP. During elongation, DBP enhances the rate and processivity of AdV DNA polymerase and changes its sensitivity to inhibitors. DBP, which has a helix destabilizing activity, has an essential role in DNA unwinding, which in vitro is an ATP-independent process not requiring a helicase. DBP drives template unwinding by formation of DBP multimers.
Other Viral Proteins
In addition to the viral proteins directly involved in DNA replication, HAdV encodes several proteins that affect cellular processes that are involved in the down-regulation of cellular antiviral responses, stimulate cell-cycle progression, or inhibit the DNA-damage responses. These factors facilitate and promote efficient viral DNA replication in the cells. Many such viral factors and processes have been reviewed recently (Giberson et al. 2012).
Cellular Factors Optimize Initiation
The pTP-Pol complex binds weakly to the origin and can catalyze only minimal initiation. In vitro replication assays facilitated the identification of nuclear factors (NF) that enhance AdV replication considerably, up to the levels observed in vivo. NFI has been identified to be identical to CTF-1, a cellular transcription factor, NFII is a topoisomerase type 1 required for the synthesis of genome-length molecules, and NFIII is identical to the transcription factor Oct-1. NFI and Oct-1 form part of the preinitiation complex and both interact with AdV DNA and with one of the viral replication proteins (van der Vliet 1996; de Jong and van der Vliet 1999; van der Vliet and Hoeben 2006).
NFI encompasses a family of cellular transcription factors that also enhances initiation of AdV DNA replication up to 60-fold. NFI binds to position 25–38 in the origin of HAdV-5. Mutation of this sequence reduces replication of the virus in infected cells. The efficiency of initiation is highly dependent on the AdV Pol concentration, suggesting that AdV Pol is recruited to the origin by an interaction with NFI. A second way in which NFI stimulates initiation is by changing the origin structure. NFI bends the origin 60° as shown by scanning force microscopy (Mysiak et al. 2004a). The bending is dependent on the conserved AT-rich sequence directly preceding the core recognition site of NFI.
Oct-1 belongs to the family of octamer-binding transcription factors containing a POU DNA-binding domain. The POU domain has a bipartite structure with two subdomains, POU specific (POUs) and POU homeodomain (POUhd), that both contribute to DNA binding and to the stimulation of initiation. Oct-1 binds next to the NFI recognition sequence at origin position 39–47.
Whereas NFI interacts with AdV Pol, Oct-1 interacts with pTP and tethers it to the origin as shown by a reduced dissociation rate of the pTP-Pol-origin interaction (van Leeuwen et al. 1997). Like in the case of NFI, stimulation of initiation depends on the structural changes in the origin induced by Oct-1. Scanning force microscopy showed a 42°-bend angle in origin DNA on binding the Oct-1 POU domain, confirming earlier biochemical data. Simultaneous binding of NFI and Oct-1 induces an 82° bend indicating that both bends are oriented toward each other (Mysiak et al. 2004b). Because both NFI and Oct-1 act synergistically to enhance initiation, this strengthens the notion that origin bending is essential for initiation, presumably by facilitating the optimal assembly of a preinitiation complex.
In conclusion, the two cellular transcription factors stimulate initiation in two different ways: recruitment of the viral replication proteins as well as bending the origin. Combined with the role of DBP in modifying the origin structure and facilitating the kinetics of initiation, a picture arises in which all proteins act in a concerted fashion, contributing to optimal assembly of a preinitiation complex as well as to the kinetics of the actual initiation and elongation reactions.
Human Adenoviruses and Disease
Although in immune-competent persons most HAdV infections occur subclinically, AdV infections are related to a wide range of malignancies (reviewed by Horwitz 2001; Lenaerts et al. 2008). The gastrointestinal disease caused by enteric AdV (i.e., HAdV-F) is considered a major contributing factor in childhood diarrhea in underdeveloped and high-population density areas. In addition, the HAdV are the most common infectious cause of ocular disease worldwide. AdV are isolated frequently from patients with a compromised immune system (Kojaoghlanian et al. 2003; Walls et al. 2003; Lenaerts et al. 2008). They are a frequent cause of morbidity and mortality after allogeneic stem cell transplantation (Symeonidis et al. 2007). Remarkably, the incidence of HAdV infections in pediatric stem cell recipients is rising sharply (Bruno et al. 2003; van Tol et al. 2005). Another group that is at risk is HIV-infected individuals.
Despite these disease associations, HAdV-derived vectors have been very popular as gene-transfer vehicles in experimental gene therapy. Initially, replication-deficient vectors were used. In these vectors, a deletion of the E1 genes blocks expression of the viral genes and replication, and provides the space needed to insert a gene of interest. Although the gene transfer is efficient, immune-competent hosts mount an immune response against viral and transgene antigens (reviewed by Bangari and Mittal 2006). This leads to the elimination of virally transduced cells in immune-competent hosts. In the newer generations of AdV vectors the residual AdV genes were removed (reviewed by Seiler et al. 2007). The propensity of the E1-deleted HAdV vectors to induce potent vector and transgene product-specific immune responses led to their use as live-virus vaccine carriers (Liu et al. 2009). In recent years, HAdV attracted a renewed interest as oncolytic agents in cancer therapy. Although the use of replication-competent HAdV in cancer therapy has been explored already more than 50 years ago (Kelly and Russell 2007), the current approaches use modifications in the vector genomes that limit viral replication to tumor cells. In this respect, the vectors HAdV-5 mutant “ONYX-015” and HAdV-5 Δ24 can serve as archetypes. The AdV ONYX-015 virus has a deletion in the region coding for the E1B 55 kDa protein. As a result, the virus is incapable of down-regulating the p53 activity. The HAdV Δ24 virus carries a deletion in the viral E1A gene that encompasses the region responsible for binding the RB protein. As a result, the Δ24 E1A protein cannot bind RB to relieve RB repression of E2F transcription factors. As a result, replication of HAdV-5 Δ24 viruses can only occur in tumor cells in which RB is inactivated (Berk 2005). RB and p53 are two critical transcription regulatory proteins and the pathways in which they operate are disturbed in nearly all human cancers (Polager and Ginsberg 2009). This should lead to preferential replication in tumor cells with alterations in the p53 pathway. These viruses have been evaluated in several clinical studies for a wide variety of tumor types with promising results (reviewed by Jiang et al. 2009). In 2006, the AdV “H101,” which is very similar to AdV ONYX-015, was registered in China for clinical use.
AdV Antivirals
There is a considerable morbidity and mortality associated with HAdV infections. This would clearly be limited by the availability of effective antiviral drugs. In addition, the increased clinical use of HAdV-derived replication-competent vectors makes the availability of effective antivirals against HAdV infections desirable. To date, there is no FDA-approved antiviral for the treatment of HAdV infections available. Several phosphonyl acyclic nucleotide analogs have activity against HAdV. Cidofovir, (S)-9-(3-hydroxy-2-phosphonomethoxy propyl)cytosine dehydrate, is among the most potent of antivirals. It has been registered by the FDA for treatment of human cytomegalovirus-induced retinitis in AIDS patients. Mechanistically, cidofovir mimics the monophosphate form of deoxycytosine and targets the AdV Pol directly. This has been elegantly confirmed by selecting cidofovir-resistant HAdV-5 mutants. All these mutations are located in the AdV Pol-coding region (Kinchington et al. 2002). Cidofovir inhibits all HAdV types tested (Kinchington et al. 2005). This is in contrast to, for instance, Ribavirin, which inhibits replication in all HAdV-C isolates, but had variable activity in isolates of HAdV-A, B, and D (Morfin et al. 2009).
So far the clinical efficacy of cidofovir has been promising but not unequivocal (reviewed by Ljungman 2004; Lenaerts et al. 2008). Current efforts are concentrated on optimizing the formulation of cidofovir and the development of new derivatives (Toth et al. 2008; Paolino et al. 2011; Ruiz et al. 2011). In addition, high-throughput screening for inhibitors of viral replication can identify new compounds that function as broad-spectrum antiviral agents. Already a compound was identified that is a potent inhibitor of dihydroorotate dehydrogenase, which is a key enzyme of the de novo pyrimidine synthesis pathway. This compound inhibits the replication of several viruses, including AdV (Hoffmann et al. 2011).
In parallel, immunotherapy protocols are developed involving infusion of AdV-specific T lymphocytes. The feasibility of this approach has been established in patients undergoing hematopoietic stem cell transplantation (HSCT) (Feuchtinger et al. 2006). This approach may be facilitated by the cross-reactivity of Ad-specific T cells (Heemskerk et al. 2003; Leen et al. 2006; Veltrop-Duits et al. 2006). Although promising, the immunotherapy approach is very labor intensive and expensive, restricting its application. Therefore it will remain a major challenge to develop effective antivirals for treatment for AdV infections that have activity against the wide diversity of HAdV serotypes. The detailed insight in key aspects of AdV DNA replication may facilitate rational development of new AdV-specific antivirals.
Footnotes
Editors: Stephen D. Bell, Marcel Méchali, and Melvin L. DePamphilis
Additional Perspectives on DNA Replication available at www.cshperspectives.org
REFERENCES
- Abdus Sattar AK, Lin TC, Jones C, Konigsberg WH 1996. Functional consequences and exonuclease kinetic parameters of point mutations in bacteriophage T4 DNA polymerase. Biochemistry 35: 16621–16629 [DOI] [PubMed] [Google Scholar]
- Aoki K, Benko M, Davison AJ, Echavarria M, Erdman DD, Harrach B, Kajon AE, Schnurr D, Wadell G 2011. Toward an integrated human adenovirus designation system that utilizes molecular and serological data and serves both clinical and fundamental virology. J Virol 85: 5703–5704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bangari DS, Mittal SK 2006. Current strategies and future directions for eluding adenoviral vector immunity. Curr Gene Ther 6: 215–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bebenek A, Dressman HK, Carver GT, Ng S, Petrov V, Yang G, Konigsberg WH, Karam JD, Drake JW 2001. Interacting fidelity defects in the replicative DNA polymerase of bacteriophage RB69. J Biol Chem 276: 10387–10397 [DOI] [PubMed] [Google Scholar]
- Berk AJ 2005. Recent lessons in gene expression, cell cycle control, and cell biology from adenovirus. Oncogene 24: 7673–7685 [DOI] [PubMed] [Google Scholar]
- Berman AJ, Kamtekar S, Goodman JL, Lázaro JM, de Vega M, Blanco L, Salas M, Steitz TA 2007. Structures of ϕ29 DNA polymerase complexed with substrate: The mechanism of translocation in B-family polymerases. EMBO J 26: 3494–3505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernad A, Blanco L, Lázaro JM, Martín G, Salas M 1989. A conserved 3′→5′ exonuclease active site in prokaryotic and eukaryotic DNA polymerases. Cell 59: 219–228 [DOI] [PubMed] [Google Scholar]
- Blanco L, Bernad A, Lázaro JM, Martin G, Garmendia C, Salas M 1989. Highly efficient DNA synthesis by the phage φ29 DNA polymerase. Symmetrical mode of DNA replication. J Biol Chem 264: 8935–8940 [PubMed] [Google Scholar]
- Blasco MA, Blanco L, Parés E, Salas M, Bernad A 1990. Structural and functional analysis of temperature-sensitive mutants of the phage ϕ29 DNA polymerase. Nucleic Acids Res 18: 4763–4770 [PMC free article] [PubMed] [Google Scholar]
- Brenkman AB, Heideman MR, Truniger V, Salas M, Van der Vliet PC 2001. The (I/Y)XGG motif of adenovirus DNA polymerase affects template DNA binding and the transition from initiation to elongation. J Biol Chem 276: 29846–29853 [DOI] [PubMed] [Google Scholar]
- Brenkman AB, Breure EC, Van der Vliet PC 2002. Molecular architecture of adenovirus DNA polymerase and location of the protein primer. J Virol 76: 8200–8207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruno B, Gooley T, Hackman RC, Davis C, Corey L, Boeckh M 2003. Adenovirus infection in hematopoietic stem cell transplantation: Effect of ganciclovir and impact on survival. Biol Blood Marrow Transplant 9: 341–352 [DOI] [PubMed] [Google Scholar]
- Caldentey J, Blanco L, Bamford DH, Salas M 1993. In vitro replication of bacteriophage PRD1 DNA. Characterization of the protein-primed initiation site. Nucleic Acids Res 21: 3725–3730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou S, Marousek GI 2008. Accelerated evolution of maribavir resistance in a cytomegalovirus exonuclease domain II mutant. J Virol 82: 246–253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jong RN, Van der Vliet PC 1999. Mechanism of DNA replication in eukaryotic cells: Cellular host factors stimulating adenovirus DNA replication. Gene 236: 1–12 [DOI] [PubMed] [Google Scholar]
- de Jong RN, Van der Vliet PC, Brenkman AB 2003. Adenovirus DNA replication: Protein priming, jumping back and the role of the DNA binding protein DBP. Curr Top Microbiol Immunol 272: 187–211 [DOI] [PubMed] [Google Scholar]
- de Vega M, Lazaro JM, Salas M, Blanco L 1996. Primer-terminus stabilization at the 3′–5′ exonuclease active site of ϕ29 DNA polymerase. Involvement of two amino acid residues highly conserved in proofreading DNA polymerases. EMBO J 15: 1182–1192 [PMC free article] [PubMed] [Google Scholar]
- de Vega M, Lázaro JM, Salas M, Blanco L 1998. Mutational analysis of ϕ29 DNA polymerase residues acting as ssDNA ligands for 3′–5′ exonucleolysis. J Mol Biol 279: 807–822 [DOI] [PubMed] [Google Scholar]
- Delarue M, Poch O, Tordo N, Moras D, Argos P 1990. An attempt to unify the structure of polymerases. Protein Eng 3: 461–467 [DOI] [PubMed] [Google Scholar]
- Desiderio SV, Kelly TJ Jr 1981. Structure of the linkage between adenovirus DNA and the 55,000 molecular weight terminal protein. J Mol Biol 145: 319–337 [DOI] [PubMed] [Google Scholar]
- Dufour E, Méndez J, Lázaro JM, de Vega M, Blanco L, Salas M 2000. An aspartic acid residue in TPR-1, a specific region of protein-priming DNA polymerases, is required for the functional interaction with primer terminal protein. J Mol Biol 304: 289–300 [DOI] [PubMed] [Google Scholar]
- Esteban JA, Soengas MS, Salas M, Blanco L 1994. 3′→5′ Exonuclease active site of ϕ29 DNA polymerase. Evidence favoring a metal ion-assisted reaction mechanism. J Biol Chem 269: 31946–31954 [PubMed] [Google Scholar]
- Feuchtinger T, Matthes-Martin S, Richard C, Lion T, Fuhrer M, Hamprecht K, Handgretinger R, Peters C, Schuster FR, Beck R, et al. 2006. Safe adoptive transfer of virus-specific T-cell immunity for the treatment of systemic adenovirus infection after allogeneic stem cell transplantation. Br J Haematol 134: 64–76 [DOI] [PubMed] [Google Scholar]
- Giberson AN, Davidson AR, Parks RJ 2012. Chromatin structure of adenovirus DNA throughout infection. Nucleic Acids Res 40: 2369–2376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay RT 1966. Adenovirus DNA replication. In DNA replication in eukaryotic cells (ed. DePamphilis ML), pp. 699–721 Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Heemskerk B, Veltrop-Duits LA, Van Vreeswijk T, Ten Dam MM, Heidt S, Toes RE, Van Tol MJ, Schilham MW 2003. Extensive cross-reactivity of CD4+ adenovirus-specific T cells: Implications for immunotherapy and gene therapy. J Virol 77: 6562–6566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann HH, Kunz A, Simon VA, Palese P, Shaw ML 2011. Broad-spectrum antiviral that interferes with de novo pyrimidine biosynthesis. Proc Natl Acad Sci 108: 5777–5782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horwitz MS 2001. Adenoviruses, Philadelphia. In Field’s virology (ed. Knipe DM, Howley PM, Griffin DE, Lamb RA, Martin MA, Roizman B, Strauss SE), pp. 2301–2325 Lippincott, Williams & Wilkins, Philadelphia, PA [Google Scholar]
- Hwang YT, Liu BY, Coen DM, Hwang CB 1997. Effects of mutations in the Exo III motif of the herpes simplex virus DNA polymerase gene on enzyme activities, viral replication, and replication fidelity. J Virol 71: 7791–7798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Illana B, Blanco L, Salas M 1996. Functional characterization of the genes coding for the terminal protein and DNA polymerase from bacteriophage GA-1. Evidence for a sliding-back mechanism during protein-primed GA-1 DNA replication. J Mol Biol 264: 453–464 [DOI] [PubMed] [Google Scholar]
- Jiang H, Gomez-Manzano C, Lang FF, Alemany R, Fueyo J 2009. Oncolytic adenovirus: Preclinical and clinical studies in patients with human malignant gliomas. Curr Gene Ther 9: 422–427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joung I, Engler JA 1992. Mutations in two cysteine-histidine-rich clusters in adenovirus type 2 DNA polymerase affect DNA binding. J Virol 66: 5788–5796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joung I, Horwitz MS, Engler JA 1991. Mutagenesis of conserved region I in the DNA polymerase from human adenovirus serotype 2. Virology 184: 235–241 [DOI] [PubMed] [Google Scholar]
- Kamtekar S, Berman AJ, Wang J, Lázaro JM, de Vega M, Blanco L, Salas M, Steitz TA 2004. Insights into strand displacement and processivity from the crystal structure of the protein-primed DNA polymerase of bacteriophage ϕ29. Mol Cell 16: 609–618 [DOI] [PubMed] [Google Scholar]
- Kamtekar S, Berman AJ, Wang J, Lázaro JM, de Vega M, Blanco L, Salas M, Steitz TA 2006. The ϕ29 DNA polymerase:protein-primer structure suggests a model for the initiation to elongation transition. EMBO J 25: 1335–1343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanellopoulos PN, Van der Zandt H, Tsernoglou D, Van der Vliet PC, Tucker PA 1995. Crystallization and preliminary x-ray crystallographic studies on the adenovirus ssDNA binding protein in complex with ssDNA. J Struct Biol 115: 113–116 [DOI] [PubMed] [Google Scholar]
- Kelly E, Russell SJ 2007. History of oncolytic viruses: Genesis to genetic engineering. Mol Ther 15: 651–659 [DOI] [PubMed] [Google Scholar]
- Kinchington PR, Araullo-Cruz T, Vergnes JP, Yates K, Gordon YJ 2002. Sequence changes in the human adenovirus type 5 DNA polymerase associated with resistance to the broad spectrum antiviral cidofovir. Antiviral Res 56: 73–84 [DOI] [PubMed] [Google Scholar]
- Kinchington PR, Romanowski EG, Jerold GY 2005. Prospects for adenovirus antivirals. J Antimicrob Chemother 55: 424–429 [DOI] [PubMed] [Google Scholar]
- King AJ, Van der Vliet PC 1994. A precursor terminal protein-trinucleotide intermediate during initiation of adenovirus DNA replication: Regeneration of molecular ends in vitro by a jumping back mechanism. EMBO J 13: 5786–5792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King AJ, Teertstra WR, Van der Vliet PC 1997. Dissociation of the protein primer and DNA polymerase after initiation of adenovirus DNA replication. J Biol Chem 272: 24617–24623 [DOI] [PubMed] [Google Scholar]
- Knopf CW 1998. Evolution of viral DNA-dependent DNA polymerases. Virus Genes 16: 47–58 [DOI] [PubMed] [Google Scholar]
- Kojaoghlanian T, Flomenberg P, Horwitz MS 2003. The impact of adenovirus infection on the immunocompromised host. Rev Med Virol 13: 155–171 [DOI] [PubMed] [Google Scholar]
- Kühn FJ, Knopf CW 1996. Herpes simplex virus type 1 DNA polymerase. Mutational analysis of the 3′-5′-exonuclease domain. J Biol Chem 271: 29245–29254 [DOI] [PubMed] [Google Scholar]
- Leen AM, Bollard CM, Myers GD, Rooney CM 2006. Adenoviral infections in hematopoietic stem cell transplantation. Biol Blood Marrow Transplant 12: 243–251 [DOI] [PubMed] [Google Scholar]
- Lenaerts L, De Clercq E, Naesens L 2008. Clinical features and treatment of adenovirus infections. Rev Med Virol 18: 357–374 [DOI] [PubMed] [Google Scholar]
- Liu H, Naismith JH, Hay RT 2000. Identification of conserved residues contributing to the activities of adenovirus DNA polymerase. J Virol 74: 11681–11689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Naismith JH, Hay RT 2003. Adenovirus DNA replication. Curr Top Microbiol Immunol 272: 131–164 [DOI] [PubMed] [Google Scholar]
- Liu J, O’Brien KL, Lynch DM, Simmons NL, La PA, Riggs AM, Abbink P, Coffey RT, Grandpre LE, Seaman MS, et al. 2009. Immune control of an SIV challenge by a T-cell-based vaccine in rhesus monkeys. Nature 457: 87–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ljungman P 2004. Treatment of adenovirus infections in the immunocompromised host. Eur J Clin Microbiol Infect Dis 23: 583–588 [DOI] [PubMed] [Google Scholar]
- Martín AC, Blanco L, García P, Salas M, Méndez J 1996. In vitro initiation of pneumococcal phage Cp-1 DNA replication occurs at the third 3′ nucleotide of the linear template: A stepwise sliding-back mechanism. J Mol Biol 260: 369–377 [DOI] [PubMed] [Google Scholar]
- Méndez J, Blanco L, Esteban JA, Bernad A, Salas M 1992. Initiation of φ29 DNA replication occurs at the second 3′ nucleotide of the linear template: A sliding-back mechanism for protein-primed DNA replication. Proc Natl Acad Sci 89: 9579–9583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morfin F, Dupuis-Girod S, Frobert E, Mundweiler S, Carrington D, Sedlacek P, Bierings M, Cetkovsky P, Kroes AC, Van Tol MJ, et al. 2009. Differential susceptibility of adenovirus clinical isolates to cidofovir and ribavirin is not related to species alone. Antivir Ther 14: 55–61 [PubMed] [Google Scholar]
- Mysiak ME, Bleijenberg MH, Wyman C, Holthuizen PE, Van der Vliet PC 2004a. Bending of adenovirus origin DNA by nuclear factor I as shown by scanning force microscopy is required for optimal DNA replication. J Virol 78: 1928–1935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mysiak ME, Wyman C, Holthuizen PE, Van der Vliet PC 2004b. NFI and Oct-1 bend the Ad5 origin in the same direction leading to optimal DNA replication. Nucleic Acids Res 32: 6218–6225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paolino K, Sande J, Perez E, Loechelt B, Jantausch B, Painter W, Anderson M, Tippin T, Lanier ER, Fry T, et al. 2011. Eradication of disseminated adenovirus infection in a pediatric hematopoietic stem cell transplantation recipient using the novel antiviral agent CMX001. J Clin Virol 50: 167–170 [DOI] [PubMed] [Google Scholar]
- Polager S, Ginsberg D 2009. p53 and E2f: Partners in life and death. Nat Rev Cancer 9: 738–748 [DOI] [PubMed] [Google Scholar]
- Rademaker HJ, Fallaux FJ, Van den Wollenberg DJ, De Jong RN, Van der Vliet PC, Hoeben RC 2006. Relaxed template specificity in fowl adenovirus 1 DNA replication initiation. J Gen Virol 87: 553–562 [DOI] [PubMed] [Google Scholar]
- Rodríguez I, Lázaro JM, Blanco L, Kamtekar S, Berman AJ, Wang J, Steitz TA, Salas M, de Vega M 2005. A specific subdomain in φ29 DNA polymerase confers both processivity and strand-displacement capacity. Proc Natl Acad Sci 102: 6407–6412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz J, Beadle JR, Buller RM, Schreiwer J, Prichard MN, Keith KA, Lewis KC, Hostetler KY 2011. Synthesis, metabolic stability and antiviral evaluation of various alkoxyalkyl esters of cidofovir and 9-(S)-[3-hydroxy-2-(phosphonomethoxy)propyl]adenine. Bioorg Med Chem 19: 2950–2958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salas M 1991. Protein-priming of DNA replication. Annu Rev Biochem 60: 39–71 [DOI] [PubMed] [Google Scholar]
- Seiler MP, Cerullo V, Lee B 2007. Immune response to helper dependent adenoviral mediated liver gene therapy: Challenges and prospects. Curr Gene Ther 7: 297–305 [DOI] [PubMed] [Google Scholar]
- Seto D, Chodosh J, Brister JR, Jones MS 2011. Using the whole-genome sequence to characterize and name human adenoviruses. J Virol 85: 5701–5702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smart JE, Stillman BW 1982. Adenovirus terminal protein precursor. Partial amino acid sequence and the site of covalent linkage to virus DNA. J Biol Chem 257: 13499–13506 [PubMed] [Google Scholar]
- Soengas MS, Esteban JA, Lázaro JM, Bernad A, Blasco MA, Salas M, Blanco L 1992. Site-directed mutagenesis at the Exo III motif of ϕ29 DNA polymerase; overlapping structural domains for the 3′–5′ exonuclease and strand-displacement activities. EMBO J 11: 4227–4237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symeonidis N, Jakubowski A, Pierre-Louis S, Jaffe D, Pamer E, Sepkowitz K, O’Reilly RJ, Papanicolaou GA 2007. Invasive adenoviral infections in T-cell-depleted allogeneic hematopoietic stem cell transplantation: High mortality in the era of cidofovir. Transpl Infect Dis 9: 108–113 [DOI] [PubMed] [Google Scholar]
- Tooze J 1980. DNA tumor viruses. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Toth K, Spencer JF, Dhar D, Sagartz JE, Buller RM, Painter GR, Wold WS 2008. Hexadecyloxypropyl-cidofovir, CMX001, prevents adenovirus-induced mortality in a permissive, immunosuppressed animal model. Proc Natl Acad Sci 105: 7293–7297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uil TG, Vellinga J, de Vrij J, Van den Hengel SK, Rabelink MJ, Cramer SJ, Eekels JJ, Ariyurek Y, Van Galen M, Hoeben RC 2011. Directed adenovirus evolution using engineered mutator viral polymerases. Nucleic Acids Res 39: e30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Breukelen B, Brenkman AB, Holthuizen PE, Van der Vliet PC 2003. Adenovirus type 5 DNA binding protein stimulates binding of DNA polymerase to the replication origin. J Virol 77: 915–922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Vliet PC 1996. Roles of transcription factors in DNA replication. In DNA replication in eukaryotic cells. (ed. DePamphilis ML), pp. 87–119 Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- van der Vliet PC, Hoeben RC 2006. Adenovirus. In DNA replication and human disease (ed. DePamphilis ML), pp. 645–661 Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- van Leeuwen HC, Rensen M, Van der Vliet PC 1997. The Oct-1 POU homeodomain stabilizes the adenovirus preinitiation complex via a direct interaction with the priming protein and is displaced when the replication fork passes. J Biol Chem 272: 3398–3405 [DOI] [PubMed] [Google Scholar]
- van Tol MJ, Claas EC, Heemskerk B, Veltrop-Duits LA, De Brouwer CS, van Vreeswijk T, Sombroek CC, Kroes AC, Beersma MF, de Klerk EP, et al. 2005. Adenovirus infection in children after allogeneic stem cell transplantation: Diagnosis, treatment and immunity. Bone Marrow Transplant 35 (Suppl 1): S73–S76 [DOI] [PubMed] [Google Scholar]
- Veltrop-Duits LA, Heemskerk B, Sombroek CC, van Vreeswijk T, Gubbels S, Toes RE, Melief CJ, Franken KL, Havenga M, van Tol MJ, et al. 2006. Human CD4+ T cells stimulated by conserved adenovirus 5 hexon peptides recognize cells infected with different species of human adenovirus. Eur J Immunol 36: 2410–2423 [DOI] [PubMed] [Google Scholar]
- Walls T, Shankar AG, Shingadia D 2003. Adenovirus: An increasingly important pathogen in paediatric bone marrow transplant patients. Lancet Infect Dis 3: 79–86 [DOI] [PubMed] [Google Scholar]
- Wong SW, Wahl AF, Yuan PM, Arai N, Pearson BE, Arai K, Korn D, Hunkapiller MW, Wang TS 1988. Human DNA polymerase α gene expression is cell proliferation dependent and its primary structure is similar to both prokaryotic and eukaryotic replicative DNA polymerases. EMBO J 7: 37–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaniv M 2009. Small DNA tumour viruses and their contributions to our understanding of transcription control. Virology 384: 369–374 [DOI] [PubMed] [Google Scholar]