Abstract
Unlike prototypical receptor tyrosine kinases (RTKs), which are single-chain polypeptides, the insulin receptor (InsR) is a preformed, covalently linked tetramer with two extracellular α subunits and two membrane-spanning, tyrosine kinase-containing β subunits. A single molecule of insulin binds asymmetrically to the ectodomain, triggering a conformational change that is transmitted to the cytoplasmic kinase domains, which facilitates their trans-phosphorylation. As in prototypical RTKs, tyrosine phosphorylation in the juxtamembrane region of InsR creates recruitment sites for downstream signaling proteins (IRS [InsR substrate] proteins, Shc) containing a phosphotyrosine-binding (PTB) domain, and tyrosine phosphorylation in the kinase activation loop stimulates InsR’s catalytic activity. For InsR, phosphorylation of the activation loop, which contains three tyrosine residues, also creates docking sites for adaptor proteins (Grb10/14, SH2B2) that possess specialized Src homology-2 (SH2) domains, which are dimeric and engage two phosphotyrosines in the activation loop.
Many structural and mechanistic aspects of signaling by the insulin receptor are now known, yet a key question remains unanswered: How does insulin binding to the receptor ectodomain trigger phosphorylation of the cytoplasmic kinase domains?
Insulin is a highly potent anabolic hormone that is critical for tissue development and for glucose homeostasis (Taniguchi et al. 2006). Released from the β cells of the pancreas, insulin regulates glucose output from the liver and glucose uptake into (primarily) skeletal muscle and adipose tissue. In addition, insulin promotes the synthesis and storage of carbohydrates, lipids, and protein. Insulin’s actions are mediated by the insulin receptor (InsR), a plasma membrane-resident glycoprotein and member of the receptor tyrosine kinase (RTK) family. Other members of the InsR subfamily of RTKs include the insulinlike growth factor-1 receptor (IGF1R) and insulin receptor-related receptor, the latter of which has no known ligand. As an RTK, InsR is ligand-activated through mechanisms that are both prototypical and atypical of RTKs. These mechanisms will be the focus of this article.
InsR ARCHITECTURE
InsR comprises two α and two β subunits, forming an α2β2 heterotetramer (Fig. 1). The two α subunits are extracellular, whereas the two β subunits begin on the extracellular side of the membrane and then traverse the membrane into the cytoplasmic region. The β subunits contain the cytoplasmic tyrosine kinase domains. The α subunits are disulfide-bridged by at least two (possibly four) cysteine residues, and each α subunit is paired to one β subunit through a single disulfide bridge (Sparrow et al. 1997).
Figure 1.
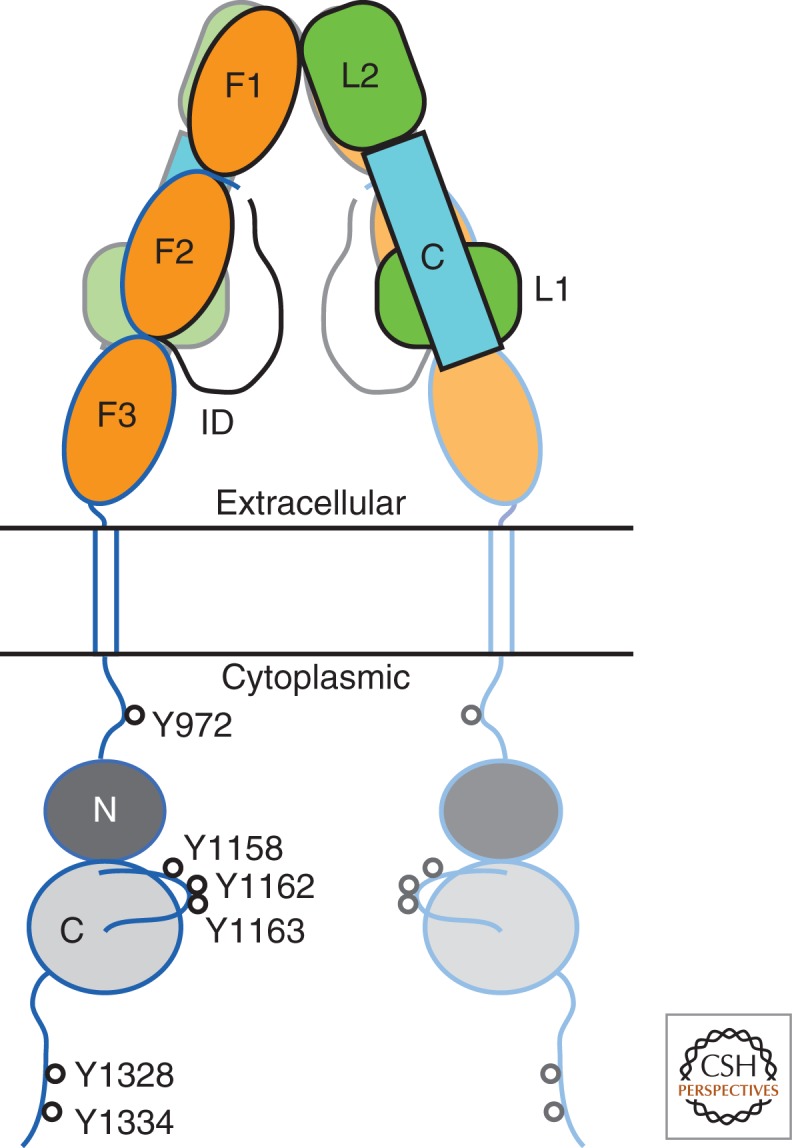
Structure-based schematic diagram of InsR. The α subunits (extracellular, denoted in black outline) comprise two leucine-rich domains (L1 and L2) with an intervening cysteine-rich domain (C), one intact fibronectin type III (FnIII) domain (F1), a partial FnIII domain (F2), and a long insert domain (ID) that contains the site of furin cleavage (L1-C-L2-F1-F2-ID). The β subunits (extracellular and cytoplasmic, denoted in blue outline) contain the completion of the second FnIII domain (F2), followed by an intact FnIII domain (F3), a transmembrane helix, and a cytoplasmic tyrosine kinase domain (N and C lobes). The second αβ half-receptor is shown semitransparent. The major sites of tyrosine autophosphorylation in the cytoplasmic domain are indicated (InsR-B isoform numbering). The molecular twofold axis is vertical, in the plane of the figure.
The α subunit consists of a leucine-rich domain followed by a cysteine-rich domain, a second leucine-rich domain, one complete fibronectin type III (FnIII) domain, a partial FnIII domain, and a long carboxy-terminal segment that, during maturation, is cleaved by furin to yield α and β subunits. The β subunit begins (after a short amino-terminal segment) with the completion of the second FnIII domain, followed by a third FnIII domain, which leads into the transmembrane segment. The transmembrane segment is believed to adopt an α-helical conformation.
The cytoplasmic region of the β subunit consists of a juxtamembrane region (∼30 residues; between the transmembrane helix and the tyrosine kinase domain), the tyrosine kinase domain (∼300 residues), and a carboxy-terminal region (∼70 residues). Tyrosine autophosphorylation sites have been mapped in all three regions (White et al. 1988; Kohanski 1993), with Y972 (+exon 11 numbering) in the juxtamembrane region and Y1158, Y1162, and Y1163 in the kinase domain having been shown to be functionally important. The juxtamembrane and carboxy-terminal regions are thought to be unstructured polypeptide segments, save for possible β turns in the juxtamembrane region near Y965 and Y972 (Backer et al. 1992).
InsR THREE-DIMENSIONAL STRUCTURE
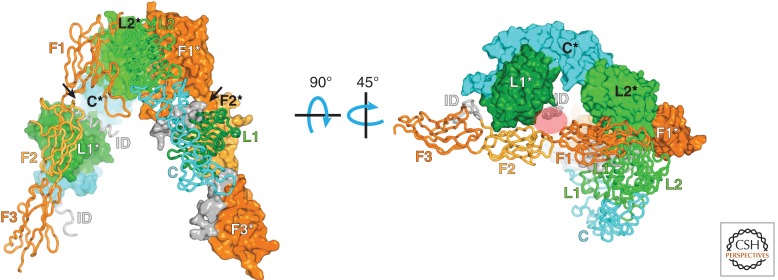
Because InsR is a large, heavily glycosylated membrane-spanning protein, structural studies have been confined thus far to crystallographic studies of either the soluble ectodomain or the soluble cytoplasmic kinase domain. A crystal structure of the full (disulfide-linked) dimeric ectodomain was reported in 2006 (McKern et al. 2006) and later re-refined (Smith et al. 2010). The structure shows an antiparallel “inverted V” arrangement (Fig. 2, left). Previous photolabeling cross-linking studies and alanine-scanning mutagenesis have implicated two regions on InsR in insulin binding (Whittaker et al. 2008). Site 1 is formed by the first leucine-rich domain in one α subunit and the carboxy-terminal segment of the other α subunit. Site 2 is thought to involve loops from the first and second FnIII domains of the other αβ half-receptor (Fig. 2, right).
Figure 2.
Structure of the ectodomain of InsR. In this disulfide-linked symmetric dimer (apo—no insulin) (McKern et al. 2006; Smith et al. 2010), one of the αβ half-receptors (in front) is shown as a Cα trace, and the other αβ half-receptor (in back) is shown with a molecular surface. The molecular twofold axis is vertical, in the plane of the figure. The two L domains are labeled L1 and L2, the cysteine-rich domain is labeled C, the FnIII domains are labeled F1–3, and the insert domain (end of the α subunit, beginning of the β subunit) is labeled ID. An asterisk in the label denotes a domain from the second half-receptor. The arrows point to the two (identical) insulin-binding sites (although binding of a single insulin molecule is sufficient for receptor activation). In the right panel, the structure has been rotated first by 90°, then by 45°, as indicated. The red oval shows where insulin is thought to bind (corresponds to the arrow on the left in the left panel).
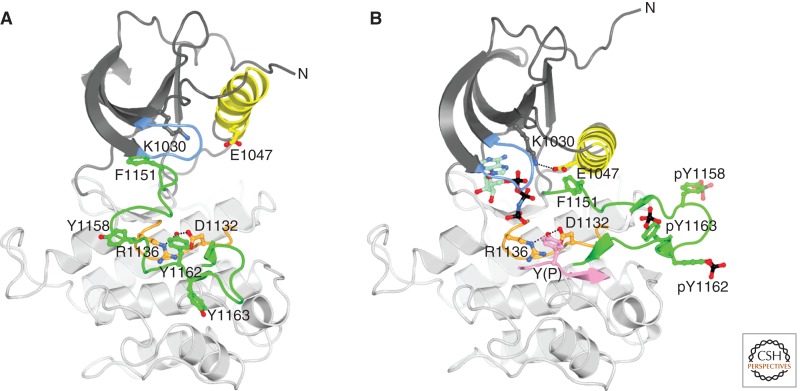
Crystal structures of the InsR tyrosine kinase domain (IRK) have been determined in several different phosphorylation states and with bound substrates (Mg-ATP and substrate peptide) (Hubbard et al. 1994; Hubbard 1997). IRK possesses a canonical protein-kinase architecture, with an amino-terminal lobe (N lobe) comprising a five-stranded β sheet and a single α helix (αC), and a C lobe that is mainly helical and contains most of the catalytic residues, in the so-called catalytic and activation loops (Fig. 3). One deviation in the C-lobe architecture relative to other RTKs is the presence of an additional α helix (αJ) at the carboxy-terminal end. (The function of this extra helix has not been established, but in the structure of a complex between IRK and the protein tyrosine phosphatase PTP1B (Li et al. 2005), this helix is part of the phosphatase binding site.) Tyrosine kinase activity is regulated by the phosphorylation state of the activation loop in the C lobe, which begins with the kinase-conserved 1150DFG motif and ends with a conserved proline (P1172). The IRK activation loop contains three sites of tyrosine autophosphorylation, Y1158, Y1162, and Y1163, which are phosphorylated in trans on insulin binding to the ectodomain.
Figure 3.
Structure of the tyrosine kinase domain of InsR. (A) Ribbon diagram of inactive, unphosphorylated IRK (Hubbard et al. 1994). The N lobe is colored dark gray except for αC and the nucleotide binding loop (blue), and the C lobe is colored light gray except for the catalytic loop (orange) and activation loop (green). Carbon atoms are colored according to their position in the structure, oxygen atoms are colored red, and nitrogen atoms are colored blue. Select side chains are shown in ball-and-stick representation. Hydrogen bonds between Y1162 (the “pseudosubstrate”) in the activation loop and D1132 and R1136 in the catalytic loop are shown as black dashed lines. The amino terminus is labeled (N) (the carboxyl terminus is hidden behind the structure). (B) Ribbon diagram of active, tris-phosphorylated IRK in complex with an ATP analog (AMPPNP) and a peptide substrate (Hubbard 1997). Same coloring as in (A) and, in addition, phosphorus atoms are colored black, carbon atoms of AMPPNP are colored pale green, and the peptide substrate is colored pink. F1151, at the beginning of the activation loop, is part of the DFG motif. Hydrogen bonds between the substrate tyrosine (Y[P]) and D1132 and R1136 are shown by black dashed lines as is the salt bridge between conserved residues K1030 (β3) and E1047 (αC).
InsR ACTIVATION
As described above, InsR is a preformed disulfide-linked dimer, and there is compelling evidence that trans-phosphorylation/activation occurs within the dimer rather than through higher-order oligomerization (Frattali et al. 1992). Although InsR is a symmetric αβ homodimer (or α2β2 heterotetramer), the active signaling complex is thought to be a single molecule of insulin (which circulates in the bloodstream in a hexameric form) bound asymmetrically to InsR with subnanomolar affinity, and thus the conformational change is induced in (or mainly in) only one αβ half-receptor. Higher concentrations of insulin display negative cooperativity for binding to the two equivalent sites on InsR (De Meyts 2008). Even though insulin is not present in the available crystal structure of the ectodomain (McKern et al. 2006), sites 1 and 2 are already closely apposed (Fig. 2, right), suggesting that the conformational changes induced by insulin binding will be relatively subtle (Renteria et al. 2008; Ward and Lawrence 2012).
In the basal (noninsulin) state, trans-phosphorylation of the kinase domains is limited by an unknown mechanism, and insulin binding to the ectodomain induces a structural transition within the receptor that facilitates trans-phosphorylation of the cytoplasmic kinase domains. Two scenarios can be envisioned for the basal state. In the first, the kinase domains are spatially separated and thus cannot undergo efficient trans-phosphorylation. In the second, the kinase domains are already within “striking distance,” but are arranged in such a way as to prevent trans-phosphorylation (perhaps as an inhibitory dimer). A partially activating point mutation (Y984A) in the juxtamembrane region of InsR is more consistent with the latter mechanism (Li et al. 2003).
cis-INHIBITION AND trans-ACTIVATION IN THE InsR KINASE DOMAIN
As observed in the first crystal structure of IRK (Hubbard et al. 1994), the unphosphorylated (basal state) activation loop, in the absence of Mg-ATP, adopts an autoinhibitory configuration in which Y1162, the second of the three activation-loop tyrosines, is bound in the kinase active site, hydrogen-bonded to D1132 and R1136 in the catalytic loop (Fig. 3A). In this configuration, although Y1162 is positioned for phosphoryl transfer, the amino-terminal end of the activation loop (1150DFG motif) occludes the ATP-binding site, preventing phosphorylation of Y1162 in cis. That is, because of the finite length of the activation loop, it is not sterically possible to bind simultaneously ATP in its binding cleft and Y1162 in the active site in cis. This cis-inhibitory/pseudosubstrate (Y1162) conformation first viewed in IRK has now been observed in several other RTKs, including IGF1R (Munshi et al. 2002), TrkA-B (Artim et al. 2012; Bertrand et al. 2012), MuSK (Till et al. 2002), and Ror2 (Artim et al. 2012). The activation loops of all of these RTKs have in common three (phosphorylatable) tyrosines in a YXXDYY motif (where X is any residue), in which the second tyrosine is bound in the active site as a pseudosubstrate. The activation loops of ALK (anaplastic lymphoma kinase) and Met also contain three tyrosines in the same positions, but lack the aspartic acid preceding the second tyrosine (a key residue in stabilizing the pseudosubstrate conformation of InsR [Till et al. 2001]), and their conformations in the unphosphorylated state differ (Wang et al. 2006; Lee et al. 2010). In the presence of cellular (millimolar) concentrations of Mg-ATP, the unphosphorylated activation loop probably adopts multiple conformations, including the pseudosubstrate conformation, as well as conformations in which Mg-ATP is bound and the active site is available for trans-phosphorylation of a tyrosine from the neighboring kinase domain.
It was shown many years ago that Y1162 was the first of the three tyrosines in the IRK activation loop to undergo autophosphorylation, followed by Y1158 and Y1163 (Wei et al. 1995). Several years ago, a crystal structure of the IGF1R kinase domain (IGF1RK) was determined in which the tyrosine equivalent to Y1162, Y1135, was bound in the active site of a symmetry-related kinase domain (in trans) (Wu et al. 2008a). This configuration was made possible through the tight binding of an ATP-competitive small-molecule inhibitor, which forced the activation loop out of its pseudosubstrate configuration. This structure provides a snapshot of the first trans-phosphorylation reaction in IGF1RK as well as in IRK (100% sequence conservation in the activation loop), and afforded rationalization of a mutation in the activation loop of InsR, R1164Q, which causes loss of InsR autophosphorylation (Formisano et al. 1993). The structure indicates that R1137 (R1164 in InsR) and E1132 (E1159 in InsR) in the IGF1R activation loop form a salt bridge that is probably important for positioning Y1135 (Y1162 in InsR) in the active site for trans-phosphorylation.
On trans-phosphorylation of the IRK activation loop on Y1158, Y1162, and Y1163, the loop is stabilized in a conformation that is optimized for binding of substrates (Mg-ATP, tyrosine) and for catalysis (Fig. 3B). Of the three phosphotyrosines, pY1163 is the most important one for loop stabilization, corresponding structurally to the single phosphotyrosine in the activation loop of Src-family tyrosine kinases (Yamaguchi and Hendrickson 1996). Although the unphosphorylated activation loops of RTKs display (in crystal structures) a number of inactive conformations, the active (usually phosphorylated) conformations are remarkably similar (Huse and Kuriyan 2002). Although the other two phosphotyrosines in the InsR activation loop, pY1158 and pY1162, may contribute to stabilization, they are also directly involved in binding of downstream signaling proteins (discussed below).
RECRUITMENT OF DOWNSTREAM SIGNALING PROTEINS TO THE ACTIVATED InsR
On insulin-stimulated activation/autophosphorylation of InsR, several proteins are recruited to the receptor for downstream signal propagation, including the IRS proteins, Shc, SH2B1 (formerly SH2-B), and SH2B2 (formerly APS). These proteins contain either a phosphotyrosine-binding (PTB) domain (IRS proteins, Shc) or an Src homology-2 (SH2) domain (SH2B1-2). The PTB domains of the IRS proteins and Shc bind to the juxtamembrane autophosphorylation site pY972, a canonical PTB-domain binding site (NPXpY). The SH2 domains of SH2B1–2 and of Grb10 and Grb14 (negative regulators described below) bind to the triply phosphorylated IRK activation loop.
One unusual feature of pY972 in the InsR juxtamembrane region is that it is a low-affinity binding site for the PTB domain of IRS1, arguably the most important substrate (along with IRS2) of InsR. A phosphopeptide representing the native pY972 sequence binds to the isolated IRS1 PTB domain with a dissociation constant (Kd) of 87 µm (Farooq et al. 1999), whereas most physiological PTB domain–phosphopeptide interactions are characterized by Kd values in the low micromolar range. The residue that precedes Y972 is E971, and an E971A substitution dramatically lowers the Kd to 2 µm (Farooq et al. 1999). The high Kd for binding to the native NPEpY sequence can be rationalized from the IRS1 PTB-domain structure (Eck et al. 1996). Owing to an extended carboxy-terminal helix, the pocket on the PTB domain for the residue preceding pY972 (E971) is shallow and suboptimal for glutamic acid. In fact, a phosphopeptide harboring E971A was used for cocrystallization.
At least one reason why glutamate precedes Y972 is that an acidic residue at the P-1 position (relative to the substrate tyrosine) decreases the substrate Km (Shoelson et al. 1992). Indeed, substitution of E971 with alanine in InsR results in loss of autophosphorylation of Y972 (SR Hubbard, unpubl.). That is, E971 is critical for autophosphorylation of Y972, which resides in a nonoptimal substrate sequence for InsR. Interestingly, the PTB domain of Shc binds to the native pY972 phosphopeptide (NPEpY) with a Kd of 4 µm (Farooq et al. 1999) (i.e., within the typical range of PTB domain–phosphopeptide-binding affinities). IRS1–2 contain a pleckstrin homology (PH) domain upstream of the PTB domain (Dhe-Paganon et al. 1999), which plays a major role in recruitment of IRS1–2 to InsR (Yenush et al. 1996). Dimerization of the PH-PTB domains of IRS1 may also facilitate binding to the dimeric InsR (SR Hubbard, unpubl.).
IRS1–2 contain numerous tyrosine phosphorylation sites in their carboxy-terminal regions, which reside in a YΦXM motif (where Φ is hydrophobic). These sites, once phosphorylated by InsR, serve as recruitment sites for the SH2 domains of phosphatidylinositol 3-kinase (PI3K) (Myers et al. 1992), which, on activation, leads to activation of Akt. Other tyrosine phosphorylation sites in IRS1–2 recruit the adapter protein Grb2 (which, through Sos, activates Ras) and the protein tyrosine phosphatase SHP2 (White 2002). IRS1–2 also possess numerous sites of serine/threonine phosphorylation that negatively regulate tyrosine phosphorylation, either in the course of normal negative feedback or in pathological insulin resistance (Pirola et al. 2004).
Previous yeast two-hybrid studies showed that a second InsR-interacting region (in addition to the PTB domain) existed in IRS2, but was lacking in IRS1, and this region was named the kinase regulatory-loop binding (KRLB) region (Sawka-Verhelle et al. 1996, 1997) or receptor binding domain-2 (RBD2) (He et al. 1996). These studies showed that the KRLB region binds to the kinase domain of InsR in a phosphorylation-dependent manner (He et al. 1996; Sawka-Verhelle et al. 1996). The KRLB region was coarsely mapped to residues 591–733, starting ∼300 residues carboxy terminal to the PTB domain. This region is predicted to lack secondary and tertiary structure (it contains a high proportion of glycine, serine, and proline residues), and mutagenesis studies identified two non-YΦXM tyrosines, Y624 and Y628, as critical residues in the KRLB–IRK interaction (Sawka-Verhelle et al. 1997).
The molecular basis for the interaction of the IRS2 KRLB region with InsR was elucidated through a cocrystal structure of a 15-residue peptide from the KRLB region (containing Y624 and Y628) bound to phosphorylated IRK (Wu et al. 2008b). The structure revealed that the KRLB region binds in the active site of IRK, with Y628 positioned for phosphorylation. Biochemical experiments showed that Y628 is phosphorylated by IRK, but with a Km (ATP) that is unusually high compared to that of a YΦXM substrate (1.6 mm vs. 40 µm) (Wu et al. 2008b). In addition, the phosphorylated Y628 peptide retains significant binding affinity in the IRK active site, which results in poor substrate turnover. Consequently, this segment of IRS2 inhibits phosphorylation by InsR of other tyrosine sites in IRS2. Importantly, Y628 in IRS2 was mapped as an insulin-stimulated phosphorylation site in cells (Schmelzle et al. 2006). A possible functional role for the KRLB region is to suppress tyrosine phosphorylation of IRS2, setting a threshold such that only metabolic pathways via PI3K are stimulated by insulin and not mitogenic pathways via Grb2/Ras; there are 10 potential PI3K recruitment sites in IRS2 versus a single Grb2 site.
InsR-interacting proteins that contain an SH2 domain (SH2B2, Grb10/14) bind to the phosphorylated activation loop of InsR. The phosphorylated activation loop represents an atypical SH2-domain binding site, in that the loop is multiply phosphorylated and is stabilized in a turn-containing (rather than an extended) conformation. Indeed, these SH2 domains possess unique features that allow them to bind efficiently to the phosphorylated activation loop of InsR (Hu et al. 2003; Stein et al. 2003; Depetris et al. 2005). First, they are dimeric. Second, each protomer in the dimer binds to two phosphotyrosines in the activation loop: pY1158 canonically, through the invariant arginine (βB5) in the SH2 domain, and pY1163 noncanonically, through two lysine residues (βD1 and βD3). Third, they have evolved to bind poorly to canonical phosphotyrosine sequences (presumably, to reduce competition), those containing a pYXXΦ sequence, by disrupting the pY+3 (hydrophobic) binding pocket. Although other RTKs such as TrkA-C, MuSK, Met, Ror2, and ALK also contain three activation-loop tyrosines at the equivalent positions of Y1158, Y1162, and Y1163 in InsR, only in the case of Trks have SH2 domain-containing proteins (SH2B1 and SH2B2) been implicated in binding to the phosphorylated activation loop (Qian et al. 1998).
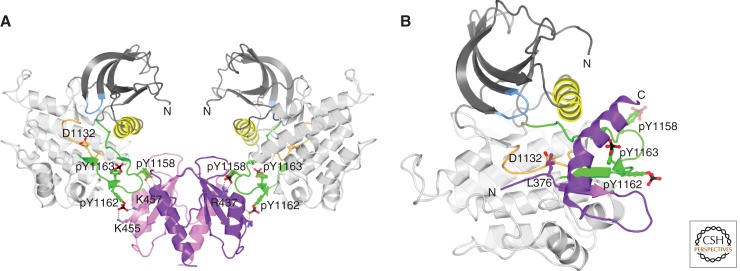
A crystal structure of the SH2 domain of SH2B2 bound to IRK (Hu et al. 2003) revealed that this SH2 domain possesses a noncanonical architecture, in which the carboxy-terminal half of the domain forms a long α helix rather than two β strands (βE and βF) and a shorter α helix (αB), as found in a typical SH2 domain. This structural rearrangement facilitates formation of a novel SH2-domain dimer, which interacts with the phosphorylated activation loops in the two kinase domains of InsR (Fig. 4A). Each protomer of the SH2-domain dimer coordinates two phosphotyrosines, the first (pY1158) in the canonical (arginine-containing) phosphate- binding pocket and the second (pY1162) by two lysines in βD. Binding of the SH2B2 SH2 domain does not perturb the conformation of the phosphorylated activation loop, but rather this mode of recruitment facilitates phosphorylation by InsR of a carboxy-terminal tyrosine (Y618) in SH2B2 (Liu et al. 2002). Of note, despite 79% sequence identity between the SH2 domains of SH2B1 and SH2B2, the SH2B1 SH2 domain is monomeric, which switches its binding preference from the phosphorylated activation loop of IRK to pY813 of Jak2, a conventional SH2-domain ligand with a pY+3 hydrophobic residue (Hu and Hubbard 2006).
Figure 4.
Structures of InsR-interacting proteins. (A) Ribbon diagram of the dimeric SH2 domain of SH2B2 bound to two IRK (InsR tyrosine kinase domain) molecules (Hu et al. 2003). The two SH2-domain protomers are colored pink and purple. Tris-phosphorylated IRK is colored as in Figure 3B. The molecular twofold axis is vertical. D1132 (catalytic loop), pY1158, pY1162, and pY1163 (activation loop) of IRK are shown in ball-and-stick representation, as are R437, K455, and K457 of SH2B2. Salt bridges are shown by black dashed lines. (B) Ribbon diagram of the BPS region of Grb14 bound to tris-phosphorylated IRK (Depetris et al. 2005). Grb14 BPS is colored purple and the pseudosubstrate residue L376 is shown in ball-and-stick representation. The amino and carboxyl termini of the BPS region are labeled N and C.
RECRUITMENT OF NEGATIVE REGULATORS TO THE ACTIVATED InsR
In addition to recruitment to InsR of positive regulators of insulin signaling, several proteins are recruited to the activated InsR to attenuate signaling, including the adapter proteins Grb10 (Smith et al. 2007) and Grb14 (Cooney et al. 2004) and the protein tyrosine phosphatase PTP1B (Elchebly et al. 1999; Klaman et al. 2000). As mentioned above, Grb10/14 contains a carboxy-terminal SH2 domain that binds directly to the phosphorylated activation loop of InsR. In addition to the SH2 domain, Grb10/14 possess several other signaling modules, including a Ras-associating (RA) domain, a PH domain, and a ∼45-residue region known as BPS (between PH and SH2) (He et al. 1998) or PIR (phosphorylated insulin receptor-interacting region) (Kasus-Jacobi et al. 1998), which is unique to this adapter family.
Biochemical studies showed that the BPS region of Grb10/14 is capable of directly inhibiting the catalytic activity of InsR (Stein et al. 2001; Bereziat et al. 2002). A crystal structure of the Grb14 BPS region in complex with phosphorylated IRK revealed at least one mechanism by which Grb14 inhibits signaling by InsR (Depetris et al. 2005). The amino-terminal portion of the BPS region binds in the substrate-binding groove in the C lobe of IRK, with a leucine (L376) inserted into the active site instead of a tyrosine (Fig. 4B). Hydrophobic residues at the +1, +3, and +5 positions relative to L376 mimic binding of an optimal IRK substrate (YΦXMXΦ). Thus, this segment of the BPS region functions as a pseudosubstrate inhibitor of IRK.
Carboxy terminal to the pseudosubstrate segment, the Grb14 BPS region adopts a 16-residue α helix whose residues make interactions with the phosphorylated activation loop. These interactions fortify the BPS–IRK interaction and provide specificity; the BPS region of Grb14/10 inhibits only InsR and the highly related IGF1R. The BPS α helix ends near the kinase N lobe, which positions the carboxy-terminal SH2 domain for interaction with pY1158 and pY1162 in the activation loop (Depetris et al. 2005). The SH2 domain of Grb14/10 is dimeric (Stein et al. 2003; Depetris et al. 2005), like the SH2B2 SH2 domain (Hu et al. 2003), but each protomer possesses a prototypical SH2-domain architecture. Binding of canonical phosphotyrosine sequences (with P+3 hydrophobic residues) are selected against by a valine in the BG loop (glycine in Src and Abl), which effectively seals off the P+3 binding pocket (Stein et al. 2003).
In recent studies, two groups have identified Grb10 as a downstream phosphorylation target of mTORC1 (Hsu et al. 2011; Yu et al. 2011). Grb10 provides the “missing link” in a negative-feedback system involving insulin/IGF1, PI3K/Akt, and mTORC1, in which activation of mTORC1 leads to inhibition of PI3K/Akt signaling. Grb10 negatively regulates PI3K/Akt signaling by binding to and inhibiting the catalytic activity of the insulin/IGF1 receptor (as described above for Grb14). Phosphorylation of Grb10 by mTORC1 was shown to increase the stability of Grb10 protein levels in cells (Hsu et al. 2011; Yu et al. 2011), which provides one mechanism by which mTORC1-mediated phosphorylation of Grb10 potentiates PI3K/Akt inhibition. Another possible mechanism is that phosphorylation of Grb10 increases the interaction between Grb10 and IRK/IGF1RK and thus the Grb10 inhibitory effect. The mTORC1-mediated serine/threonine sites in Grb10 have been mapped (Hsu et al. 2011; Yu et al. 2011), and several are located in the BPS region and the BPS-SH2 linker, regions that interact (BPS) or potentially interact (BPS-SH2 linker) directly with IRK/IGF1K. In particular, S428 is four residues upstream of the leucine pseudosubstrate in the Grb10 BPS region (see Fig. 4B), and because IRK/IGF1RK prefers tyrosine substrates with acidic residues in this region (Shoelson et al. 1992), it is plausible that pS428 (now acidic) increases Grb10 binding affinity.
PTP1B is capable of dephosphorylating numerous RTKs in vitro and in cells in which it is overexpressed, yet PTP1B-null mice are of normal size (and not tumor-prone) and show two major phenotypes: insulin hypersensitivity and resistance to high-fat-diet-induced weight gain (Elchebly et al. 1999; Klaman et al. 2000). The two main targets of PTP1B’s action are thought to be InsR and the cytoplasmic tyrosine kinase Jak2 (Myers et al. 2001). A crystal structure of PTP1B with a bound phosphopeptide representing the activation loop of IRK indicated that specificity for InsR may be dictated in part by the twin phosphotyrosines pY1162/1163, in the context (E/D)pYpY(K/R) (Salmeen et al. 2000). Jak2 also contains twin phosphotyrosines (pY1007/1008) in this sequence motif in its activation loop, but other RTKs do as well, including IGF1R, fibroblast growth factor receptors, Trks, MuSK, and Mer. Therefore, additional determinants of PTP1B substrate specificity must exist.
A crystal structure of a complex between PTP1B and phosphorylated IRK was determined (Li et al. 2005), which revealed an unusual mode of interaction. Rather than binding to the phosphorylated activation loop, PTP1B was bound to the “backside” of IRK. This binding mode was clearly facilitated by high concentrations of ammonium sulfate, a common precipitating agent for protein crystallization. Although this binding mode between PTP1B and InsR remains to be corroborated in cell-based assays, the specificity of the interaction is intriguing: even the highly related IGF1R would not be predicted to engage PTP1B in this manner. Furthermore, two tyrosines in PTP1B (Y152, Y153) that are found in the PTP1B–IRK interface were shown (before the structure was solved) to be important in the interaction between PTP1B and InsR in cells (Bandyopadhyay et al. 1997; Dadke et al. 2000). It thus remains an intriguing hypothesis that the mode of binding observed in the PTP1B–IRK crystal structure represents a recruitment interaction to localize PTP1B to InsR. No crystal structure of the catalytic mode of interaction between PTP1B and phosphorylated IRK has been reported. Presumably, this structure would reveal additional specificity determinants not highlighted by the PTP1B-phosphopeptide (IRK activation loop) structure (Salmeen et al. 2000).
Because Grb14 and Grb10 engage all three activation-loop phosphotyrosines of InsR via their SH2 domains and BPS regions (Depetris et al. 2005), these negative regulators would be predicted to antagonize PTP1B-catalyzed dephosphorylation of InsR. This indeed appears to be the case, at least for Grb14, based on both in vitro and cell-based experiments (Bereziat et al. 2002; Nouaille et al. 2006). It is conceivable that, in cells in which Grb14 is present (muscle and liver), Grb14 acts initially to attenuate InsR signaling at the plasma membrane and, on internalization of InsR, PTP1B (tethered to the endoplasmic reticulum) outcompetes Grb14 for binding and dephosphorylates InsR.
CONCLUDING REMARKS
Since the cloning of human InsR in the mid-1980s (Ebina et al. 1985; Ullrich et al. 1985), a vast amount of knowledge has been gained on the biochemical, structural, and functional properties of this crucial RTK in human biology. The key mechanistic questions that remain to be answered are (i) What prevents the kinase domains from trans-phosphorylating in the absence of insulin (i.e., how is the basal state maintained?) (ii) What is the insulin-induced conformational change in the ectodomain? (iii) How is this conformational change transduced to the kinase domains to facilitate trans-phosphorylation? (iv) On activation, do the kinase domains function in tandem or individually in the phosphorylation of downstream substrates? The prevalence of type II diabetes in developed countries is increasing at an alarming rate. One therapeutic strategy (out of many) for treating this disease is to design or screen for agents that directly potentiate the signaling output from InsR. Our current knowledge and that gained from answering these mechanistic questions should prove invaluable in the quest for such therapeutic agents. A recent crystal structure reveals how insulin binds to its primary site on InsR (Menting et al. 2013).
ACKNOWLEDGMENTS
Past support for InsR research is acknowledged from the National Institutes of Health (DK052916).
Footnotes
Editors: Joseph Schlessinger and Mark A. Lemmon
Additional Perspectives on Signaling by Receptor Tyrosine Kinases available at www.cshperspectives.org
REFERENCES
- Artim SC, Mendrola JM, Lemmon MA 2012. Assessing the range of kinase autoinhibition mechanisms in the insulin receptor family. Biochem J 448: 213–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backer JM, Shoelson SE, Weiss MA, Hua QX, Cheatham RB, Haring E, Cahill DC, White MF 1992. The insulin receptor juxtamembrane region contains two independent tyrosine/β-turn internalization signals. J Cell Biol 118: 831–839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandyopadhyay D, Kusari A, Kenner KA, Liu F, Chernoff J, Gustafson TA, Kusari J 1997. Protein-tyrosine phosphatase 1B complexes with the insulin receptor in vivo and is tyrosine-phosphorylated in the presence of insulin. J Biol Chem 272: 1639–1645 [DOI] [PubMed] [Google Scholar]
- Bereziat V, Kasus-Jacobi A, Perdereau D, Cariou B, Girard J, Burnol AF 2002. Inhibition of insulin receptor catalytic activity by the molecular adapter Grb14. J Biol Chem 277: 4845–4852 [DOI] [PubMed] [Google Scholar]
- Bertrand T, Kothe M, Liu J, Dupuy A, Rak A, Berne PF, Davis S, Gladysheva T, Valtre C, Crenne JY, et al. 2012. The crystal structures of TrkA and TrkB suggest key regions for achieving selective inhibition. J Mol Biol 423: 439–453 [DOI] [PubMed] [Google Scholar]
- Cooney GJ, Lyons RJ, Crew AJ, Jensen TE, Molero JC, Mitchell CJ, Biden TJ, Ormandy CJ, James DE, Daly RJ 2004. Improved glucose homeostasis and enhanced insulin signalling in Grb14-deficient mice. EMBO J 23: 582–593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dadke S, Kusari J, Chernoff J 2000. Down-regulation of insulin signaling by protein-tyrosine phosphatase 1B is mediated by an N-terminal binding region. J Biol Chem 275: 23642–23647 [DOI] [PubMed] [Google Scholar]
- De Meyts P 2008. The insulin receptor: A prototype for dimeric, allosteric membrane receptors? Trends Biochem Sci 33: 376–384 [DOI] [PubMed] [Google Scholar]
- Depetris RS, Hu J, Gimpelevich I, Holt LJ, Hubbard SR 2005. Structural basis for inhibition of the insulin receptor by the adaptor protein Grb14. Mol Cell 20: 325–333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhe-Paganon S, Ottinger EA, Nolte RT, Eck MJ, Shoelson SE 1999. Crystal structure of the pleckstrin homology-phosphotyrosine binding (PH-PTB) targeting region of insulin receptor substrate 1. Proc Natl Acad Sci 96: 8378–8383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebina Y, Ellis L, Jarnagin K, Edery M, Graf L, Clauser E, Ou JH, Masiarz F, Kan YW, Goldfine ID, et al. 1985. The human insulin receptor cDNA: The structural basis for hormone-activated transmembrane signalling. Cell 40: 747–758 [DOI] [PubMed] [Google Scholar]
- Eck MJ, Dhe-Paganon S, Trub T, Nolte RT, Shoelson SE 1996. Structure of the IRS-1 PTB domain bound to the juxtamembrane region of the insulin receptor. Cell 85: 695–705 [DOI] [PubMed] [Google Scholar]
- Elchebly M, Payette P, Michaliszyn E, Cromlish W, Collins S, Loy AL, Normandin D, Cheng A, Himms-Hagen J, Chan CC, et al. 1999. Increased insulin sensitivity and obesity resistance in mice lacking the protein tyrosine phosphatase-1B gene. Science 283: 1544–1548 [DOI] [PubMed] [Google Scholar]
- Farooq A, Plotnikova O, Zeng L, Zhou MM 1999. Phosphotyrosine binding domains of Shc and insulin receptor substrate 1 recognize the NPXpY motif in a thermodynamically distinct manner. J Biol Chem 274: 6114–6121 [DOI] [PubMed] [Google Scholar]
- Formisano P, Sohn KJ, Miele C, Di Finizio B, Petruzziello A, Riccardi G, Beguinot L, Beguinot F 1993. Mutation in a conserved motif next to the insulin receptor key autophosphorylation sites de-regulates kinase activity and impairs insulin action. J Biol Chem 268: 5241–5248 [PubMed] [Google Scholar]
- Frattali AL, Treadway JL, Pessin JE 1992. Transmembrane signaling by the human insulin receptor kinase. J Biol Chem 267: 19521–19528 [PubMed] [Google Scholar]
- He W, Craparo A, Zhu Y, O’Neill TJ, Wang L-M, Pierce J, Gustafson TA 1996. Interaction of insulin receptor substrate-2 (IRS-2) with the insulin and insulin-like growth factor I receptors. J Biol Chem 271: 11641–11645 [DOI] [PubMed] [Google Scholar]
- He W, Rose DW, Olefsky JM, Gustafson TA 1998. Grb10 interacts differentially with the insulin receptor, insulin-like growth factor I receptor, and epidermal growth factor receptor via the Grb10 Src homology 2 (SH2) domain and a second novel domain located between the pleckstrin homology and SH2 domains. J Biol Chem 273: 6860–6867 [DOI] [PubMed] [Google Scholar]
- Hsu PP, Kang SA, Rameseder J, Zhang Y, Ottina KA, Lim D, Peterson TR, Choi Y, Gray NS, Yaffe MB, et al. 2011. The mTOR-regulated phosphoproteome reveals a mechanism of mTORC1-mediated inhibition of growth factor signaling. Science 332: 1317–1322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J, Hubbard SR 2006. Structural basis for phosphotyrosine recognition by the Src homology-2 domains of the adapter proteins SH2-B and APS. J Mol Biol 361: 69–79 [DOI] [PubMed] [Google Scholar]
- Hu J, Liu J, Ghirlando R, Saltiel AR, Hubbard SR 2003. Structural basis for recruitment of the adaptor protein APS to the activated insulin receptor. Mol Cell 12: 1379–1389 [DOI] [PubMed] [Google Scholar]
- Hubbard SR 1997. Crystal structure of the activated insulin receptor tyrosine kinase in complex with peptide substrate and ATP analog. EMBO J 16: 5572–5581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard SR, Wei L, Ellis L, Hendrickson WA 1994. Crystal structure of the tyrosine kinase domain of the human insulin receptor. Nature 372: 746–754 [DOI] [PubMed] [Google Scholar]
- Huse M, Kuriyan J 2002. The conformational plasticity of protein kinases. Cell 109: 275–282 [DOI] [PubMed] [Google Scholar]
- Kasus-Jacobi A, Perdereau D, Auzan C, Clauser E, Van Obberghen E, Mauvais-Jarvis F, Girard J, Burnol AF 1998. Identification of the rat adapter Grb14 as an inhibitor of insulin actions. J Biol Chem 273: 26026–26035 [DOI] [PubMed] [Google Scholar]
- Klaman LD, Boss O, Peroni OD, Kim JK, Martino JL, Zabolotny JM, Moghal N, Lubkin M, Kim YB, Sharpe AH, et al. 2000. Increased energy expenditure, decreased adiposity, and tissue-specific insulin sensitivity in protein-tyrosine phosphatase 1B-deficient mice. Mol Cell Biol 20: 5479–5489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohanski RA 1993. Insulin receptor autophosphorylation. II. Determination of autophosphorylation sites by chemical sequence analysis and identification of the juxtamembrane sites. Biochemistry 32: 5773–5780 [DOI] [PubMed] [Google Scholar]
- Lee CC, Jia Y, Li N, Sun X, Ng K, Ambing E, Gao MY, Hua S, Chen C, Kim S, et al. 2010. Crystal structure of the ALK (anaplastic lymphoma kinase) catalytic domain. Biochem J 430: 425–437 [DOI] [PubMed] [Google Scholar]
- Li S, Covino ND, Stein EG, Till JH, Hubbard SR 2003. Structural and biochemical evidence for an autoinhibitory role for tyrosine 984 in the juxtamembrane region of the insulin receptor. J Biol Chem 278: 26007–26014 [DOI] [PubMed] [Google Scholar]
- Li S, Depetris RS, Barford D, Chernoff J, Hubbard SR 2005. Crystal structure of a complex between protein tyrosine phosphatase 1B and the insulin receptor tyrosine kinase. Structure 13: 1643–1651 [DOI] [PubMed] [Google Scholar]
- Liu J, Kimura A, Baumann CA, Saltiel AR 2002. APS facilitates c-Cbl tyrosine phosphorylation and GLUT4 translocation in response to insulin in 3T3-L1 adipocytes. Mol Cell Biol 22: 3599–3609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKern NM, Lawrence MC, Streltsov VA, Lou MZ, Adams TE, Lovrecz GO, Elleman TC, Richards KM, Bentley JD, Pilling PA, et al. 2006. Structure of the insulin receptor ectodomain reveals a folded-over conformation. Nature 443: 218–221 [DOI] [PubMed] [Google Scholar]
- Menting JG, Whittaker J, Margetts MB, Whittaker LJ, Kong GK-W, Smith BJ, Watson CJ, Žáková L, Kletvíková E, Jiráček J, et al. 2013. How insulin engages its primary binding site on the insulin receptor. Nature 493: 241–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munshi S, Kornienko M, Hall DL, Reid JC, Waxman L, Stirdivant SM, Darke PL, Kuo LC 2002. Crystal structure of the Apo, unactivated insulin-like growth factor-1 receptor kinase. Implication for inhibitor specificity. J Biol Chem 277: 38797–38802 [DOI] [PubMed] [Google Scholar]
- Myers MG Jr, Backer JM, Sun XJ, Shoelson S, Hu P, Schlessinger J, Yoakim M, Schaffhausen B, White MF 1992. IRS-1 activates phosphatidylinositol 3'-kinase by associating with src homology 2 domains of p85. Proc Natl Acad Sci 89: 10350–10354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers MP, Andersen JN, Cheng A, Tremblay ML, Horvath CM, Parisien JP, Salmeen A, Barford D, Tonks NK 2001. TYK2 and JAK2 are substrates of protein-tyrosine phosphatase 1B. J Biol Chem 276: 47771–47774 [DOI] [PubMed] [Google Scholar]
- Nouaille S, Blanquart C, Zilberfarb V, Boute N, Perdereau D, Burnol AF, Issad T 2006. Interaction between the insulin receptor and Grb14: A dynamic study in living cells using BRET. Biochem Pharmacol 72: 1355–1366 [DOI] [PubMed] [Google Scholar]
- Pirola L, Johnston AM, Van Obberghen E 2004. Modulation of insulin action. Diabetologia 47: 170–184 [DOI] [PubMed] [Google Scholar]
- Qian X, Riccio A, Zhang Y, Ginty DD 1998. Identification and characterization of novel substrates of Trk receptors in developing neurons. Neuron 21: 1017–1029 [DOI] [PubMed] [Google Scholar]
- Renteria ME, Gandhi NS, Vinuesa P, Helmerhorst E, Mancera RL 2008. A comparative structural bioinformatics analysis of the insulin receptor family ectodomain based on phylogenetic information. PloS ONE 3: e3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmeen A, Andersen JN, Myers MP, Tonks NK, Barford D 2000. Molecular basis for the dephosphorylation of the activation segment of the insulin receptor by protein tyrosine phosphatase 1B. Mol Cell 6: 1401–1412 [DOI] [PubMed] [Google Scholar]
- Sawka-Verhelle D, Tartare-Deckert S, White MF, Van Obberghen E 1996. Insulin receptor substrate-2 binds to the insulin receptor through its phosphotyrosine-binding domain and through a newly identified domain comprising amino acids 591–786. J Biol Chem 271: 5980–5983 [DOI] [PubMed] [Google Scholar]
- Sawka-Verhelle D, Baron V, Mothe I, Filloux C, White MF, Van Obberghen E 1997. Tyr624 and Tyr628 in insulin receptor substrate-2 mediate its association with the insulin receptor. J Biol Chem 272: 16414–16420 [DOI] [PubMed] [Google Scholar]
- Schmelzle K, Kane S, Gridley S, Lienhard GE, White FM 2006. Temporal dynamics of tyrosine phosphorylation in insulin signaling. Diabetes 55: 2171–2179 [DOI] [PubMed] [Google Scholar]
- Shoelson SE, Chatterjee S, Chaudhuri M, White MF 1992. YMXM motifs of IRS-1 define substrate specificity of the insulin receptor kinase. Proc Natl Acad Sci 89: 2027–2031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith FM, Holt LJ, Garfield AS, Charalambous M, Koumanov F, Perry M, Bazzani R, Sheardown SA, Hegarty BD, Lyons RJ, et al. 2007. Mice with a disruption of the imprinted Grb10 gene exhibit altered body composition, glucose homeostasis, and insulin signaling during postnatal life. Mol Cell Biol 27: 5871–5886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith BJ, Huang K, Kong G, Chan SJ, Nakagawa S, Menting JG, Hu SQ, Whittaker J, Steiner DF, Katsoyannis PG, et al. 2010. Structural resolution of a tandem hormone-binding element in the insulin receptor and its implications for design of peptide agonists. Proc Natl Acad Sci 107: 6771–6776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparrow LG, McKern NM, Gorman JJ, Strike PM, Robinson CP, Bentley JD, Ward CW 1997. The disulfide bonds in the C-terminal domains of the human insulin receptor ectodomain. J Biol Chem 272: 29460–29467 [DOI] [PubMed] [Google Scholar]
- Stein EG, Gustafson TA, Hubbard SR 2001. The BPS domain of Grb10 inhibits the catalytic activity of the insulin and IGF1 receptors. FEBS Lett 493: 106–111 [DOI] [PubMed] [Google Scholar]
- Stein EG, Ghirlando R, Hubbard SR 2003. Structural basis for dimerization of the Grb10 Src homology 2 domain. Implications for ligand specificity. J Biol Chem 278: 13257–13264 [DOI] [PubMed] [Google Scholar]
- Taniguchi CM, Emanuelli B, Kahn CR 2006. Critical nodes in signalling pathways: Insights into insulin action. Nat Rev Mol Cell Biol 7: 85–96 [DOI] [PubMed] [Google Scholar]
- Till JH, Ablooglu AJ, Frankel M, Bishop SM, Kohanski RA, Hubbard SR 2001. Crystallographic and solution studies of an activation loop mutant of the insulin receptor tyrosine kinase: Insights into kinase mechanism. J Biol Chem 276: 10049–10055 [DOI] [PubMed] [Google Scholar]
- Till JH, Becerra M, Watty W, Lu Y, Ma Y, Neubert TA, Burden SJ, Hubbard SR 2002. Crystal structure of the MuSK tyrosine kinase: Insights into receptor autoregulation. Structure 10: 1187–1196 [DOI] [PubMed] [Google Scholar]
- Ullrich A, Bell JR, Chen EY, Herrera R, Petruzzelli LM, Dull TJ, Gray A, Coussens L, Liao YC, Tsubokawa M, et al. 1985. Human insulin receptor and its relationship to the tyrosine kinase family of oncogenes. Nature 313: 756–761 [DOI] [PubMed] [Google Scholar]
- Wang W, Marimuthu A, Tsai J, Kumar A, Krupka HI, Zhang C, Powell B, Suzuki Y, Nguyen H, Tabrizizad M, et al. 2006. Structural characterization of autoinhibited c-Met kinase produced by coexpression in bacteria with phosphatase. Proc Natl Acad Sci 103: 3563–3568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward CW, Lawrence MC 2009. Ligand-induced activation of the insulin receptor: A multi-step process involving structural changes in both the ligand and the receptor. BioEssays 31: 422–434 [DOI] [PubMed] [Google Scholar]
- Ward CW, Lawrence MC 2012. Similar but different: Ligand-induced activation of the insulin and epidermal growth factor receptor families. Curr Opin Struct Biol 22: 360–366 [DOI] [PubMed] [Google Scholar]
- Wei L, Hubbard SR, Hendrickson WA, Ellis L 1995. Expression, characterization, and crystallization of the catalytic core of the human insulin receptor protein-tyrosine kinase domain. J Biol Chem 270: 8122–8130 [DOI] [PubMed] [Google Scholar]
- White MF 2002. IRS proteins and the common path to diabetes. Am J Physiol Endocrinol Metab 283: E413–E422 [DOI] [PubMed] [Google Scholar]
- White MF, Shoelson SE, Keutmann H, Kahn CR 1988. A cascade of tyrosine autophosphorylation in the β-subunit activates the phosphotransferase of the insulin receptor. J Biol Chem 263: 2969–2980 [PubMed] [Google Scholar]
- Whittaker L, Hao C, Fu W, Whittaker J 2008. High-affinity insulin binding: Insulin interacts with two receptor ligand binding sites. Biochemistry 47: 12900–12909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Li W, Craddock BP, Foreman KW, Mulvihill MJ, Ji QS, Miller WT, Hubbard SR 2008a. Small-molecule inhibition and activation-loop trans-phosphorylation of the IGF1 receptor. EMBO J 27: 1985–1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Tseng YD, Xu CF, Neubert TA, White MF, Hubbard SR 2008b. Structural and biochemical characterization of the KRLB region in insulin receptor substrate-2. Nat Struct Mol Biol 15: 251–258 [DOI] [PubMed] [Google Scholar]
- Yamaguchi H, Hendrickson WA 1996. Structural basis for activation of the human lymphocyte kinase Lck upon tyrosine phosphorylation. Nature 384: 484–489 [DOI] [PubMed] [Google Scholar]
- Yenush L, Makati KJ, Smith-Hall J, Ishibashi O, Myers MG Jr, White MF 1996. The pleckstrin homology domain is the principal link between the insulin receptor and IRS-1. J Biol Chem 271: 24300–24306 [DOI] [PubMed] [Google Scholar]
- Yu Y, Yoon SO, Poulogiannis G, Yang Q, Ma XM, Villen J, Kubica N, Hoffman GR, Cantley LC, Gygi SP, et al. 2011. Phosphoproteomic analysis identifies Grb10 as an mTORC1 substrate that negatively regulates insulin signaling. Science 332: 1322–1326 [DOI] [PMC free article] [PubMed] [Google Scholar]