Abstract
Nuclear receptors are activated by lipid-soluble signals (e.g., steroid hormones) that cross the plasma membrane. Once activated, most function as transcription factors to control gene expression for numerous biological processes.
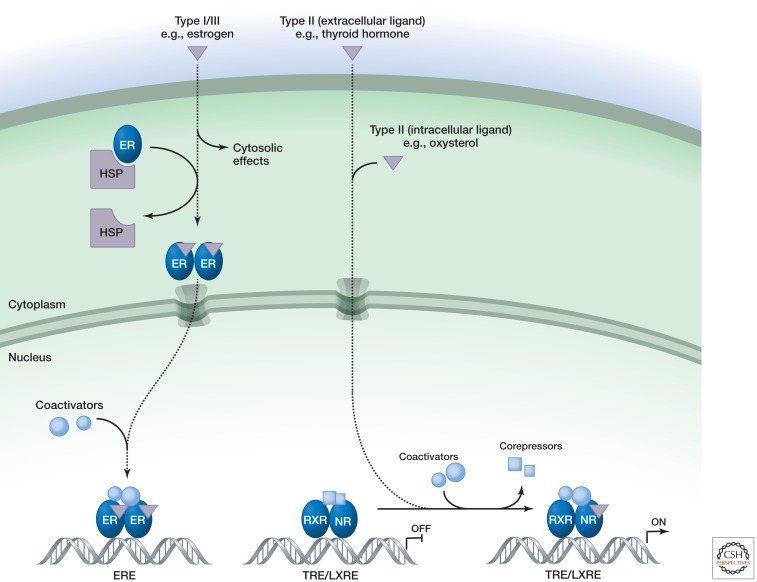
Nuclear receptors are a family of ligand-regulated transcription factors that are activated by steroid hormones, such as estrogen and progesterone, and various other lipid-soluble signals, including retinoic acid, oxysterols, and thyroid hormone (Mangelsdorf et al. 1995). Unlike most intercellular messengers, the ligands can cross the plasma membrane and directly interact with nuclear receptors inside the cell (Fig. 1), rather than having to act via cell surface receptors. Once activated, nuclear receptors directly regulate transcription of genes that control a wide variety of biological processes, including cell proliferation, development, metabolism, and reproduction. Although nuclear receptors primarily function as transcription factors, some have also been found to regulate cellular functions within the cytoplasm. For example, estrogens act through the estrogen receptor in the cytoplasm of endothelial cells to rapidly activate signaling pathways that control vascular tone and endothelial cell migration (Wu et al. 2011).
Figure 1.
Nuclear receptor signaling.
Studies of the salivary glands of insect larva in the 1960s first indicated that steroid hormones regulate transcription. Subsequent work showed that estrogen can selectively activate the genes encoding egg-white and yolk proteins, leading to the cloning of the estrogen, glucocorticoid, and thyroid hormone receptors in the 1980s (Hollenberg et al. 1985; Green et al. 1986; Miesfeld et al. 1986; Sap et al. 1986; Weinberger et al. 1986). We now know that 48 nuclear receptors are encoded in the human genome (Mangelsdorf et al. 1995). In many cases, ligands for these have been identified, but several “orphan receptors” remain (Burris et al. 2012). Whether all of these have bona fide ligands is unclear, because some nuclear receptors can act in the absence of a ligand (Table 1).
Table 1.
Common nuclear receptors and their ligands
| Receptor | Abbreviation | Ligand |
|---|---|---|
| Androgen receptor | AR | Testosterone |
| Estrogen receptor | ER | Estrogen |
| Estrogen-related receptor | ERR | ? |
| Glucocorticoid receptor | GR | Cortisol |
| Mineralocorticoid receptor | MR | Aldosterone |
| Progesterone receptor | PR | Progesterone |
| Retinoic acid receptor | RAR | Retinoic acid |
| Retinoid orphan receptor | ROR | ? |
| Retinoic acid-related receptor | RXR | Rexinoids |
| Liver X receptor | LXR | Oxysterols |
| Peroxisome proliferator-activated receptor γ | PPARγ | Fatty acid metabolites |
| Thyroid hormone receptor | TR | Thyroid hormone |
| Vitamin D3 receptor | VDR | Vitamin D3 |
Nuclear receptors share a common structure, comprising a highly variable amino-terminal domain that includes several distinct transactivation regions (the A/B domain; also referred to as AF1 for activation function 1), a central conserved DNA-binding domain that includes two Zn fingers (the C domain), a short region responsible for nuclear localization (the D domain), and a large fairly well-conserved carboxy-terminal ligand-binding domain (the E domain, or LBD) that also contributes to interactions of the subset of nuclear receptors that form heterodimers (Mangelsdorf et al. 1995). Some also possess a highly variable carboxy-terminal tail (the F domain) that in most cases has unknown functions.
The receptors can exist as monomers, homodimers, or heterodimers and recognize DNA sequences termed hormone response elements (HREs) derived from pairs of sequences with the consensus RGGTCA (R is a purine). They can be grouped into four subtypes based on their mode of action. Type I receptors, such as the androgen receptor, the estrogen receptor, and the progesterone receptor, are anchored in the cytoplasm by chaperone proteins (e.g., HSP90) (Echeverria and Picard 2010). Ligand binding frees the receptor from the chaperone, allowing homodimerization, exposure of the nuclear localization sequence, and entry into the nucleus (Fig. 1). Once in the nucleus, the ligand–receptor complex associates with transcriptional coactivators that facilitate binding to and activation of target genes (Glass and Rosenfeld 2000; Bulynko and O’Malley 2011). Recent genome-wide location analysis indicates that most nuclear-receptor binding sites in the genome are located in enhancer elements that are far away from the transcriptional start site, as first documented for the estrogen receptor (Carroll et al. 2006). Studies of the glucocorticoid receptor suggest that the ligand-bound receptor rapidly exchanges with its binding sites and that increases and decreases in receptor activity follow changes in the concentration of endogenous glucocorticoids.
Type II receptors, such as the thyroid hormone receptor and the retinoic acid receptor, in contrast, reside in the nucleus bound to their specific DNA response elements even in the absence of ligand. They generally form heterodimers with the retinoid X receptor (RXR) and in the absence of ligand exert active repressive functions through interactions with NCoR and SMRT corepressor complexes (Chen and Evans 1995; Horlein et al. 1995) that are associated with histone deacetylases (HDACs) (Watson et al. 2012). Binding of ligand to the LBD leads to dissociation of corepressors and their replacement with coactivator complexes. Coactivator complexes typically contain proteins with enzymatic functions, including histone acetyltransferases, that help open up chromatin and facilitate activation of target genes (Glass and Rosenfeld 2000). Note that several type II receptors bind to ligands produced in the same cell (e.g., LXR responses to oxysterols), which allows cell-autonomous feedback regulation.
Type III receptors function similarly to type I receptors except that the organization of the HRE differs (it is a direct repeat rather than inverted) and type IV receptors instead bind as monomers to half-site HREs (Mangelsdorf et al. 1995).
Ligands allosterically control the interactions of nuclear receptors with coactivators and corepressors by influencing the conformation of a short helix, referred to as AF2 (activation function 2), at the carboxy-terminal end of the LBD (Glass and Rosenfeld 2000). In the absence of ligand, the AF2 helix is in an open conformation that enables binding of corepressors to type II receptors. Upon agonist binding, the AF2 helix adopts a conformation in which it forms one side of a charge clamp that grips the ends of a short helix of consensus sequence LxxLL present in coactivator proteins that interact directly with the LBD (Nolte et al. 1998). Selective modulation of nuclear receptor activities can be achieved by synthetic ligands that differentially alter the AF2 conformation (Glass and Rosenfeld 2000). For example, the estrogen receptor modulator tamoxifen prevents AF2 from adopting a charge-clamp conformation, thereby blocking AF2-dependent transcriptional activity.
The functions of nuclear receptors can also be modulated by posttranslational modifications that include phosphorylation, ubiquitylation, and SUMOylation (Berrabah et al. 2011; Treuter and Venteclef 2011; Lee and Lee 2012). Phosphorylation can activate some nuclear receptors independently of ligand binding and function as the major mechanism regulating activities of orphan receptors (Berrabah et al. 2011). Receptor ubiquitylation can occur in response to ligand binding and may contribute to termination of hormonal signaling (Lee and Lee 2012). SUMOylation typically reduces the activation function of nuclear receptors and/or promotes repressor activity (Treuter and Venteclef 2011).
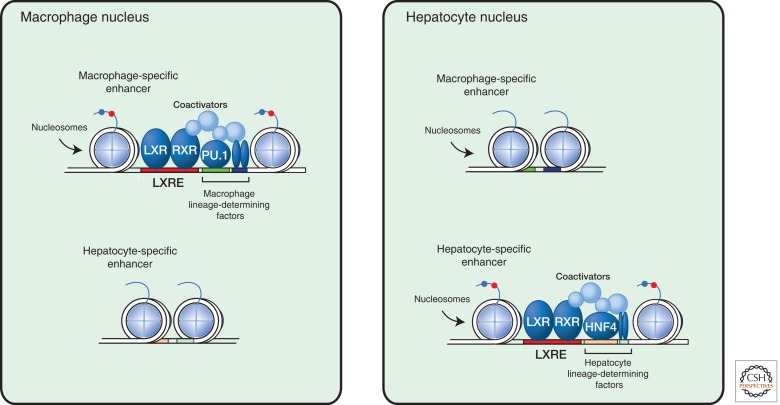
A characteristic feature of nuclear receptors with respect to their integrative roles in development and homeostasis is their ability to regulate different genes in different cell types. For example, estrogen receptors regulate different sets of genes in the brain, breast, and uterus that contribute to the distinct functions of those organs. Recent studies indicate that tissue-specific responses are a consequence of binding of nuclear receptors to enhancer elements that are selected in a cell-specific manner. Cell-specific enhancer selection is conferred by the key lineage-determining factors for each cell type, which interact in a collaborative manner to generate open regions of chromatin that provide access points for signal-dependent transcription factors (Fig. 2) (Heinz et al. 2010). In the case of LXRs, for example, macrophage-specific binding sites are established by interactions between macrophage-lineage-determining factors that include PU.1 and AP-1, whereas in liver (Heinz et al. 2010) LXR-binding sites occur in association with the hepatocyte-lineage-determining factors HNF4 and C/EBPα (Boergesen et al. 2012). In each case, a complex multistep process involving numerous coactivator proteins is involved in building a functional enhancer, and the tissue-specific responses can be further tailored by expression of distinct coactivator/corepressor complexes (Fig. 2) (Bulynko and O’Malley 2011).
Figure 2.
Tissue-specific nuclear receptor signaling in hepatocytes versus macrophages.
Given the wide variety of processes controlled by nuclear receptors, their dysregulation can contribute to numerous diseases, including cancer, diabetes, and infertility. However, because they bind to small molecules, they represent promising therapeutic targets for which selective agonists and antagonists can be engineered (Burris et al. 2012). Tamoxifen, for example, is an estrogen receptor antagonist currently used to treat breast cancer, and thiazolidinediones that target peroxisome proliferator-activated receptor γ (PPARγ) are used to treat type 2 diabetes. Because nuclear receptors regulate many genes in many tissues, synthetic ligands usually show beneficial therapeutic effects and unwanted side effects that limit clinical use. Major goals in the nuclear receptor field therefore include attaining a better understanding of the mechanisms underlying their actions in specific cell types and ways in which to selectively modulate their activities (Burris et al. 2012).
Footnotes
Editors: Lewis Cantley, Tony Hunter, Richard Sever, and Jeremy Thorner
Additional Perspectives on Signal Transduction available at www.cshperspectives.org
REFERENCES
- Berrabah W, Aumercier P, Lefebvre P, Staels B 2011. Control of nuclear receptor activities in metabolism by post-translational modifications. FEBS Lett 585: 1640–1650 [DOI] [PubMed] [Google Scholar]
- Boergesen M, Pedersen TA, Gross B, van Heeringen SJ, Hagenbeek D, Bindesboll C, Caron S, Lalloyer F, Steffensen KR, Nebb HI, et al. 2012. Genome-wide profiling of liver X receptor, retinoid X receptor, and peroxisome proliferator-activated receptor α in mouse liver reveals extensive sharing of binding sites. Mol Cell Biol 32: 852–867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulynko YA, O’Malley BW 2011. Nuclear receptor coactivators: Structural and functional biochemistry. Biochemistry 50: 313–328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burris TP, Busby SA, Griffin PR 2012. Targeting orphan nuclear receptors for treatment of metabolic diseases and autoimmunity. Chem Biol 19: 51–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll JS, Meyer CA, Song J, Li W, Geistlinger TR, Eeckhoute J, Brodsky AS, Keeton EK, Fertuck KC, Hall GF, et al. 2006. Genome-wide analysis of estrogen receptor binding sites. Nat Genet 38: 1289–1297 [DOI] [PubMed] [Google Scholar]
- Chen JD, Evans RM 1995. A transcriptional co-repressor that interacts with nuclear hormone receptors. Nature 377: 454–457 [DOI] [PubMed] [Google Scholar]
- Echeverria PC, Picard D 2010. Molecular chaperones, essential partners of steroid hormone receptors for activity and mobility. Biochim Biophys Acta 1803: 641–649 [DOI] [PubMed] [Google Scholar]
- Glass CK, Rosenfeld MG 2000. The coregulator exchange in transcriptional functions of nuclear receptors. Genes Dev 14: 121–141 [PubMed] [Google Scholar]
- Green S, Walter P, Kumar V, Krust A, Bornert JM, Argos P, Chambon P 1986. Human oestrogen receptor cDNA: Sequence, expression and homology to v-erb-A. Nature 320: 134–139 [DOI] [PubMed] [Google Scholar]
- Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK 2010. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell 38: 576–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollenberg SM, Weinberger C, Ong ES, Cerelli G, Oro A, Lebo R, Thompson EB, Rosenfeld MG, Evans RM 1985. Primary structure and expression of a functional human glucocorticoid receptor cDNA. Nature 318: 635–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horlein AJ, Naar AM, Heinzel T, Torchia J, Gloss B, Kurokawa R, Ryan A, Kamei Y, Soderstrom M, Glass CK, et al. 1995. Ligand-independent repression by the thyroid hormone receptor mediated by a nuclear receptor co-repressor. Nature 377: 397–404 [DOI] [PubMed] [Google Scholar]
- Lee JH, Lee MJ 2012. Emerging roles of the ubiquitin–proteasome system in the steroid receptor signaling. Arch Pharm Res 35: 397–407 [DOI] [PubMed] [Google Scholar]
- Mangelsdorf DJ, Thummel C, Beato M, Herrlich P, Schutz G, Umesono K, Blumberg B, Kastner P, Mark M, Chambon P, et al. 1995. The nuclear receptor superfamily: The second decade. Cell 83: 835–839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miesfeld R, Rusconi S, Godowski PJ, Maler BA, Okret S, Wikstrom AC, Gustafsson JA, Yamamoto KR 1986. Genetic complementation of a glucocorticoid receptor deficiency by expression of cloned receptor cDNA. Cell 46: 389–399 [DOI] [PubMed] [Google Scholar]
- Nolte RT, Wisely GB, Westin S, Cobb JE, Lambert MH, Kurokawa R, Rosenfeld MG, Willson TM, Glass CK, Milburn MV 1998. Ligand binding and co-activator assembly of the peroxisome proliferator-activated receptor-γ. Nature 395: 137–143 [DOI] [PubMed] [Google Scholar]
- Sap J, Munoz A, Damm K, Goldberg Y, Ghysdael J, Leutz A, Beug H, Vennstrom B 1986. The c-erb-A protein is a high-affinity receptor for thyroid hormone. Nature 324: 635–640 [DOI] [PubMed] [Google Scholar]
- Treuter E, Venteclef N 2011. Transcriptional control of metabolic and inflammatory pathways by nuclear receptor SUMOylation. Biochim Biophys Acta 1812: 909–918 [DOI] [PubMed] [Google Scholar]
- Watson PJ, Fairall L, Schwabe JW 2012. Nuclear hormone receptor co-repressors: Structure and function. Mol Cell Endocrinol 348: 440–449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberger C, Thompson CC, Ong ES, Lebo R, Gruol DJ, Evans RM 1986. The c-erb-A gene encodes a thyroid hormone receptor. Nature 324: 641–646 [DOI] [PubMed] [Google Scholar]
- Wu Q, Chambliss K, Umetani M, Mineo C, Shaul PW 2011. Non-nuclear estrogen receptor signaling in the endothelium. J Biol Chem 286: 14737–14743 [DOI] [PMC free article] [PubMed] [Google Scholar]