Abstract
We reported previously that HIV-1 the Tat-interacting protein of 110 kDa (TIP110; P110(NRB)/SART3/p110) is important in regulation of hematopoiesis, and in maintaining pluripotent factor (NANOG, OCT4, and SOX2) expression in and pluripotency of human embryonic stem cells (hESCs). Here we show that TIP110 expression in hESCs line H9 and embryonal carcinoma cell line NT-2 is regulated by C-MYC expression in ESCs through an E box present in the TIP110 promoter region. Through up- and down- modulation of expression, TIP110 induces OCT4 exon 1a inclusion and exon 1b skipping in our OCT4 minigene model. Thus, TIP110 expression in ESCs regulates alternative splicing of OCT4, an event required for pluripotency of ESCs.
Introduction
Embryonic stem cells (ESCs), derived from the inner cell mass of blastocyst-stage embryos, can self-renew and differentiate along 3 germ layer lineages [1,2]. The undifferentiated state is maintained by actions of transcription factors (TF). The POU domain TF OCT4 (also known as OCT3, OCT3/4, and POU5F1), located on chromosome 6p21.3, is a core component controlling self-renewal and maintenance of the undifferentiated state of ESCs [3,4]. OCT4 is also an essential factor for generation of induced pluripotent stem (iPS) cells [5]. The OCT4 gene can potentially encode 2 different spliced variants, OCT4A and OCT4B. Both isoforms share identical POU DNA-binding and C-terminal trans-activation domains, but they differ at their N termini. Alternative splicing of OCT4 is important in that OCT4A is highly expressed in pluripotent ESCs that self-renew and maintain the undifferentiated state of early pluripotent cells, while OCT4B can be detected in various nonpluripotent cell types and cannot sustain self-renewal and pluripotency of ESCs [6,7]. Takeda et al. first characterized human OCT4 cDNA, and amplified OCT4A and OCT4B cDNA sequences [8]. They analyzed the OCT4 sequence of exon–intron organization and identified OCT4A with exon 1 of the 447 base pair and OCT4B with exon 1b of the 344 base pair, as well as detailed splice donor and splice acceptor sites [8]. However, it is not known how OCT4A and OCT4B isoform splicing is regulated.
Although the pluripotent state of ESCs is controlled through certain levels of core TFs, such as OCT4, SOX2, and NANOG, CMYC is also essential for maintaining a pluripotent state [9]. The oncoprotein C-MYC is a master regulator for cell proliferation, is virtually undetectable in quiescent cells, and its expression is rapidly induced as cells enter the G1 phase of the cell cycle in response to stimulation. Expression of C-MYC is transient and directly related to the proliferative potential of cells, and subsequently, the abundance of C-MYC decreases gradually to a low steady-state level, where it remains for as long as the cells continue to proliferate [10,11]. C-MYC forms a heterodimer with the bHLH/Zip protein Max that binds to the E-box motif (cacgtg) containing genes to activate its target gene transcription [12].
We hypothesized that TIP110 expression is modulated through C-MYC and herein demonstrate that the oncogenic TF C-MYC upregulates transcription of the RNA binding protein TIP110 through interaction with the TIP110 E-box in the TIP110 promoter, thus ensuring high-level Tip110 expression in proliferating hESCs. We further show that TIP110 regulates OCT4 alternative splicing in hESCs.
Materials and Methods
Cell culture and cell transfections
The hESC line, H9, was cultured in the knockout Dulbecco's modified Eagle's medium (DMEM): F12, supplied with 20% serum replacement (KSR; Invitrogen) and the basic fibroblast growth factor (bFGF; Invitrogen) on mouse embryonic feeders mitotically inactivated with mitomycin C as described [13]. The human embryonal carcinoma cell line NTero-2 cl.D1(NT-2: CRL-1973) was purchased from American Tissue Culture Collection (ATCC) and maintained in the DMEM supplemented with 10% fetal calf serum at 37°C in 5% CO2. Human cord blood was collected and used according to institutional guidelines. CD34 cells were purified within 24 h of collection using immunomagnetic selection (Miltenyi Biotec). CD34+ cells (>93% pure) were cultured in 10% FBS with the cytokine combination of: 100 ng/mL of stem cell factor, 100 ng/mL FLT3 ligand, and 20 ng/mL of thrombopoietin (SFT; R&D Systems). Detailed purification and culture of human cord blood CD34+ cells were as described [14]. The K562 cell line was purchased from American Tissue Culture Collection (ATCC, CCL-243) and maintained in the RPMI medium supplemented with 10% fetal calf serum at 37°C in 5% CO2. NT-2 cells were transfected by using Lipofectamine 2000 (Invitrogen). H9 cells were transfected by using the Amaxa Human Stem cell Nucleofector Kit (Lonza, Cat. No. VPH-5002) and Lipofectamine LTX and Plus reagent (invitrogen, Cat No. 15338).
Constructs and primers
pCSC.Tip110GFP and pCSC.c-mycGFP, as well as RNAi for TIP110 and C-MYC were as described [15]. The OCT4 minigene was constructed by cloning human OCT4 genomic DNA (Genebank: Z11900) from 60-1130, named minigene 1, which includes OCT4 exon 1a, and from 3864 to 4710, named minigene 2, which includes exon 1b/2. Minigene1 primers were Oct4-60 5′-taccgagctcggatctaacagggcacagt-3′ and Oct4-1130 5′-ctggactagtggatcctcaccggcagtt-3′; minigene 2 primers were Oct4-3864 5′-ccgtcgacaggtgttctcgaggccagggtctc-3′ and Oct4-4710 5′-ccctcgagccagtgatggaagcaatgga-3. Minigene1 was inserted into pcDNA3.1 (Invitrogen; Cat. No. K4800-01). Minigene 2 was cloned into the clonning site right after minigene 1 on the same construct.
RNA isolation, semiquantitative and real-time polymerase chain reaction analysis
Total cellular RNAs were isolated using an Invitrogen TRizol RNA isolation kit according to the manufacturer's instructions. Before RNA precipitation, RNA is extracted one more time with Acid-pheno:chloroform (PH:4.5, with IAA 125:24:1) to remove traces of DNA. One to 20 nanograms of total RNA were used for reverse transcription and polymerase chain reaction (PCR) in a one-step semiquantitative reverse transcription polymerase chain reaction (RT-PCR) reaction (Roche, Cat.No. 11939832001). Primers for OCT4a, OCT4b, C-MYC, and the internal control β-actin gene have been described [15,16]. The same c-myc primers were applied for one tube RT-PCR and q-PCR, which could detect both endogenous and transfected c-myc gene. The one tube RT-PCR was set at 50°C for 30 min and 95°C for 2 min for reverse transcription, followed by a total of 30 cycles of PCR. Five ng total RNA is used for detection OCT4 from the OCT minigene-transfected NT-2 or hESC cells, while 20 ng total RNA is used for detection of endogenous OCT4 genes in NT-2 cells and hESCs. Aliquots of PCR products were analyzed on a 1.2% agarose gel. For quantitative real-time PCR, reverse transcription was performed by using a Superscript III kit with random hexamer primers. Q-PCR was performed by using SYBR Green PCR Master Mix reagents (Invitrogen).
His-tagged Tip110 protein purification and western blot
6× his-tagged TIP110 expression plasmid was transfected into 293T cells. The cells were harvested at day 3 after transfection. After washing with cold PBS, the cells were resuspended in a washing buffer (50 mM Tris.HCl, pH 8.0, 30 mM NaCl, 10% glycerol, 1% NP-400) with protease inhibitors and placed on ice for 20 min. The resuspended cells were sonicated, and then centrifuged for 15 min at 4°C. The supernatant was incubated with the Ni-beads column (Qiagen, Cat: 31014). The Tip110 protein was eluted with 250 mM imidazole in 1× Buffer E (20 mM Hepes-HCl, pH 7.9, 100 mM KCl, 0.2 mM EDTA, 10% glycerol, and 1 mM DTT) and frozen in aliquots at −80°C. The Tip110 antibody used for western blot has been described before [15]; the antibody used for western Blot analysis is anti-c-myc (Santa Cruz; Cat: SC-789)
In vitro splicing assay
The OCT4 minigene, which contains a T7 promoter was in vitro transcribed into pre-mRNA by using a 5× MEGAscript+ T7 kit (Ambion; Cat: AMB 1334-5) at 30°C for 3 h. After transcription, the DNA template was digested by adding DNase I from the kit. In vitro splicing was performed in 40 μL splicing reactions, which contained a 4 μL 10× SP buffer (5 mM ATP, 0.2 M creatine phosphate, 16 mM MgCl2), a 20 μL NT-2 cell extract, and a 5-μL pre-mRNA probe. A 20 μL reaction was taken out right after the reaction was set up as input, and the other 20 μL reaction was incubated at 30°C for 3 h. RNA from both the input and splicing reactions were extracted from Trizol and resuspended in a 50 μL buffer. One microliter out of the 50 μL total RNA was used for quantitative RT-PCR.
Chromatin immunoprecipitation assay
Chromatin immunoprecipitation (ChIP) assays were performed by using a ChIP assay kit (Upstate Biotechnology). Briefly, Hela cells or NT2 cells in 10-cm dishes were plated at 60% confluency and allowed to grow until 80% confluency over 40 h. Cells (107) were crosslinked and lysed. Chromatin was sheared to 300- to 900-bp fragments. Normalized inputs were incubated with the 4-μg anti-c-Myc antibody (Santa Cruz; Cat: SC-789) at 4°C overnight. IgG (Rabbit anti-mouse IgG, (MP Biomedical Cat#55436) was used as a negative control. The RNA polymerase II antibody (the RNA polymerase II 8WG monoclonal antibody, Covance Cat#MMS-126R) was used as a positive control (primers: 5′-agatgaaaccgttgtccaaact-3′ and 5′-aggttacggcagtttgtctctc-3′). Immunoprecipitated DNA was amplified by PCR using TIP110 promoter primers, which span the −412- to −250-bp region flanking the putative E-box (5′-) in the human TIP110 promoter (primers: 5′- gcgtattaatgtaatttact-3′ and 5′-atccatttcccagattcctc-3′).
Preparing nuclear extract and DNA electrophoretic mobility shift assays
Electrophoretic mobility shift assays (EMSAs) were performed with the ChIP assay kit (Upstate Biotechnology) using the protocol recommended by the manufacturer. Briefly, Hela cells and NT2 cells were cultured and nuclear extracts were prepared as described in the protocol. Double-stranded E-box probes (5′-aaccggaaaactcacgtgtatttaacgtct-3′ and 5′-agacgttaaatacacgtgagttttccggtt-3′) or E-box mutant probes (5′-aaccggaaaactcccgggtatttaacgtct-3′ and 5′-agacgttaaatacccgggagttttccggtt-3′) were labeled with biotin at the 3′ end. The DNA binding reaction was performed by incubation of the nuclear extract protein (20 μg) with a biotin-labeled probe for 30 min at room temperature. The nonbiotin-labeled probe or the C-MYC antibody were used for competition. Complexes were analyzed by electrophoresis through a 6% acrylamide nondenaturing gel. Biotin-labeled probes were transferred to Amersham Hibond-N+ (GE Healthcare) and detected by using the LightShift Chemiluninescent EMSA kit (Thermo Scientific) according to the manufacturer's instructions.
Results and Discussion
C-MYC upregulates TIP110 transcription and protein expression
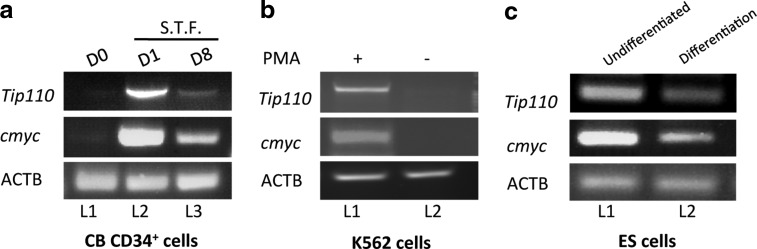
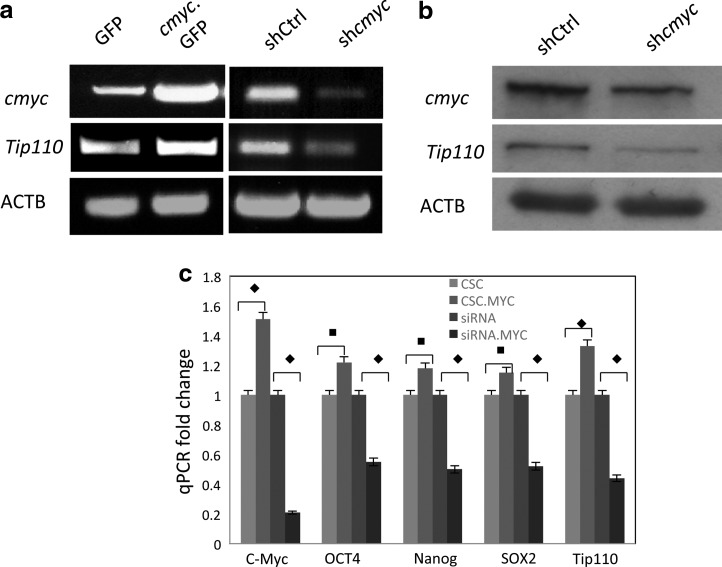
We previously reported that TIP110 was expressed in hematopoietic stem cells and hESC [13,15]. Since C-MYC was shown to activate the TIP110 promoter by using a luciferase reporter system [15], we tested whether C-MYC expression correlated with TIP110 expression. We analyzed C-MYC and TIP110 expression from cord blood CD34+ cells at different stages. The TIP110 expression pattern was similar to that of C-MYC (Fig. 1a). C-MYC and TIP110 are not expressed in quiescent CD34+ cells (Fig.1a, L1), but are expressed in cytokine-stimulated undifferentiated CD34+ cells (Fig.1a, L2) as we previously reported [15], with decreased expression during further differentiation of CD34+ cells (Fig. 1a, L3). Thus, C-MYC expression correlates with TIP110 expression in cord blood CD34+ cells. When the human leukemia cell line K562 was induced to differentiate toward the megakaryocyte lineage by PMA, expression of both TIP110 and C-MYC was reduced (Fig. 1b). To test whether C-MYC and TIP110 show the same expression pattern in hESCs, ESCs were cultured for maintenance of pluripotency and after differentiation. qRT-PCR showed that TIP110 as well as C-MYC were expressed in hESCs (Fig. 1c, L1). hESCs were removed from feeder layers and bEGF for 3 days, to allow ESC differentiation. C-MYC expression levels were dramatically reduced (by 67%), along with a large reduction in expression of TIP110 (85%) (Fig. 1c, L2), suggesting that TIP110 might be under the control of C-MYC. To examine direct involvement of C-MYC over Tip110 expression, we exogenously overexpressed C-MYC, or silenced C-MYC expression in NT-2 cells to test whether C-MYC regulates TIP110 expression. There was a clear increase of TIP110 expression with overexpression of C-MYC, and a reduction of TIP110 expression with knockdown of C-MYC expression in NT-2 cells at both the mRNA (Fig. 2a) and protein (Fig. 2b) level. After overexpressing of C-MYC or silencing C-MYC expression in NT-2 cells, we analyzed the mRNA levels of stem cell markers for pluripotency, including OCT4, SOX2, and NANOG by qPCR. The results showed that the expression levels of OCT4, SOX2, and NANOG are increased with overexpression of C-MYC; the expression levels are reduced with silencing C-MYC expression (Fig. 2c).
FIG. 1.
TIP110 and C-MYC share similar expression patterns. Semiquantitative RT-polymerase chain reaction (PCR) of: (a) Freshly isolated CD34+ cells (L1), CD34+ cells cultured in the Iscove's modified Dulbecco's medium (IMDM) with fetal bovine serum, stem cell factor, thrombopoietin, and Fms-related tyrosine kinase 3 ligand (FL) for 24 h (L2), or cells continued in culture for 8 days (L3); (b) K562 cells cultured in the RPMI medium (L1) or included with 20 nM phorbol 12-myristate 13-acetate for 5 days (L2). (c) ESC line (H9) cultured to yield undifferentiated human embryonic stem cells (hESCs) (L1) or differentiated hESC offspring under feeder-free conditions by withdrawing basic fibroblast growth factor in a medium for 3 days (L2). β-actin was used as a loading control. Data shown are from 1 of at least 3 reproducible experiments.
FIG. 2.
C-MYC regulates TIP110 expression. NT-2 cells were transduced with c-myc expression lentivirus and cells were harvested 3 days after cell transduction for analysis of TIP110 expression via semiquantitative RT-PCR (a), western blot (b). C-MYC, TIP110, OCT4, SOX2, and NANOG mRNA expression was analyzed through qPCR (c). β-actin was used to normalize q-PCR results. Data in panel c are expressed as mean±SD of 3 independent experiments. ♦P<0.001; ■P<0.05 for the compared pair.
Tip110 expression transactivated by C-MYC is through putative E-box binding sites located in the Tip110 promoter
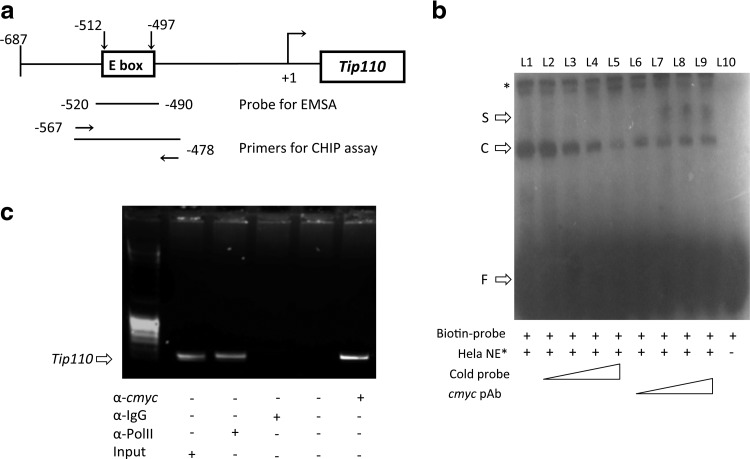
Next, we searched for the elements through which C-MYC could regulate TIP110 expression in the TIP110 promoter region. Genomatix programs were applied to predict binding sites of potential TFs within the TIP110 promoter region (−687 to +1 bp). A putative E-box (CACGTG), a binding site for C-MYC TFs, was identified in the promoter of TIP110 (Fig. 3a). To determine whether C-MYC could bind to this E box region of the TIP110 promoter, we first performed EMSA experiments to identify C-MYC interactions with this specific E box on the TIP110 promoter (Fig. 3b). Associations between the E box probe and nuclear extract from NT-2 cells were tested. One single complex was formed from the gel shift result (L1). The complex formation is competed by including the same, but unlabeled E box probe (L2–L5). The shifted bands were supershifted by including the C-MYC antibody in the binding reactions (L6–L9). When we mutate the E box sequence to destroy the E box, while still keeping other transcription sites intact, the association between the mutated E box probe and TIP110 is no longer apparent (data not shown).
FIG. 3.
C-MYC association in vivo/vitro with E box element in the TIP110 promoter. (a) Schematic localization of specific primers/probes used for chromatin immunoprecipitation (ChIP) and electrophoretic mobility shift assays (EMSA); (b) EMSA was performed with biotin-labeled probes as indicated in (a). 20 μg of nuclear extracts obtained from NT-2 cells transfected with C-MYC expression vector were used for the gel shift assays. Unlabeled probes were used in excess from 10× to 100× more for competition (lane 3–6). (c) CHIP assay is performed on NT-2 hESCs overexpressing C-MYC. Input DNA (L1); RNA polymerase II antibody as a positive control (L2); Rabbit IgG as negative control (L3), no antibody added as negative control (L4) and C-MYC antibody for bound or not bound fraction (L5) were amplified with the primers indicated in (a). *nonspecific binding; S: supershift; C: cmyc/probe complex; F: free probe.
Next, we performed CHIP assay to determine if C-MYC localized to this TIP110 E box region in NT-2 cells. Using ChIP analysis, we found that this putative binding site E box was bound by the endogenous C-MYC protein (Fig. 3c), confirming that the C-MYC protein interacts with the E box located in the TIP110 promoter.
Tip110 regulates an alternative splicing of OCT4
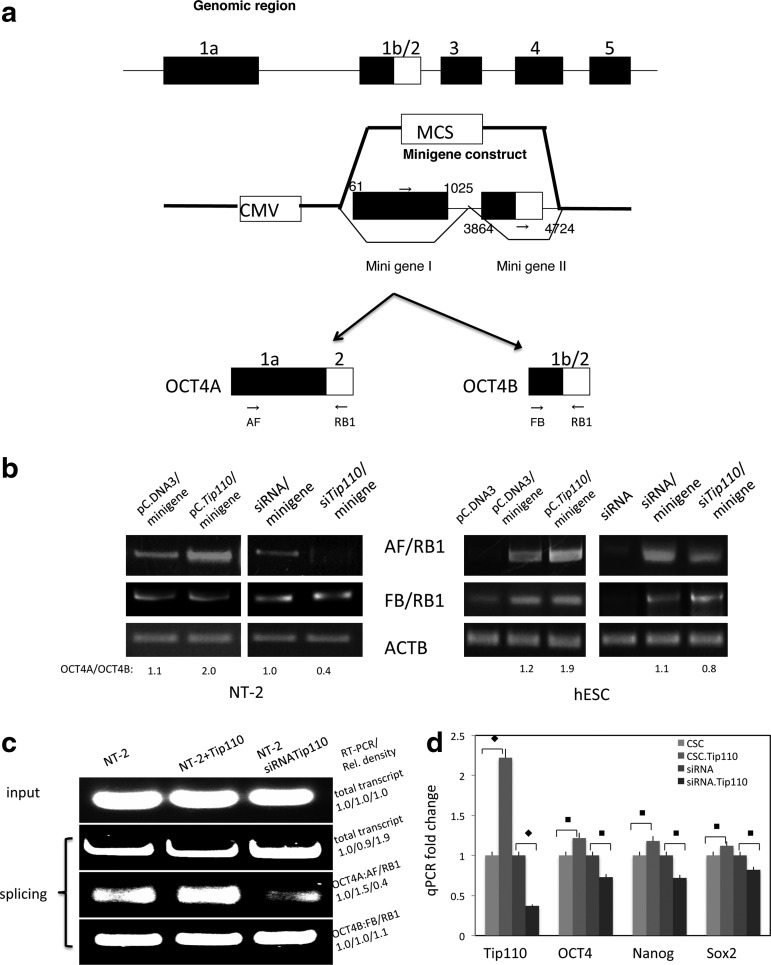
Studies of others [17] and ourselves (unpublished data) showed that TIP110 was involved in general splicing through interaction with U6 snRNA. The association of TIP110 with splicing machinery and splicing factors suggests TIP110 might have a regulatory role in alternative splicing. Our recent work [13] showed that TIP110 functions in maintaining pluripotent factor OCT4 expression in hESCs, and we had hypothesized that TIP110 could play a role in alternative splicing of OCT4. To test our hypothesis that TIP110 functions in OCT4 alternative splicing, we first cloned a minigene for OCT4 that includes exon 1a (minigene 1) and exon 1b/2 (minigene 2) (OCT4 cDNA, GenBank: Z11900.1). Minigene I starts from gene 60 to 980 and includes exon 1a and its splicing donor, minigene II that starts from gene 3864 to gene 4710, which includes exon 1b/2, exon 1b splicing donor and its acceptor, and exon 2 and exon 2 splicing acceptor (Fig. 4a). We inserted minigene I and minigene II sequentially to pC.DNA3 vector cloning sites, which contain a CMV promoter. The product of the OCT4 minigene undergoes 2 alternative splicing decisions to include either exon 1a or exon 2 (part of OCT4A product), or excluding exon 1a, but including exon 1b and exon 2 (part of the OCT4B product) (Fig. 4a). To test TIP110 function in OCT4 splicing, we cotransfected the Oct4 minigene with Tip110 expression or silencing constructs in NT-2 cells as well as hESCs. When the TIP110 gene is overexpressed, the OCT4 minigene is able to splice in the form of Oct4A; when TIP110 gene expression is downregulated, the OCT4 minigene is not able to splice in the form of OCT4A; while the OCT4B level is not changed (Fig. 4b). To validate the in vivo results that TIP110 expression is able to splice the OCT4A form, we designed an in vitro splicing reaction. The OCT4 minigene was in vitro transcribed into a pre-mRNA probe. The OCT4 pre-mRNA probe was incubated with the NT-2 cell extract, the same amount of the NT-2 cell extract with the TIP110 recombinant protein, and the same amount of TIP110 siRNA NT-2 cell extract in the splicing reaction for 3 h. Instead of running a denature RNA gel for detecting unprocessed OCT4 pre-mRNA, OCT4A and OCT4B isoforms due to the large sizes; total RNA was extracted at the end of the splicing reaction. 1/50 total RNA was used for quantitative RT-PCR to amplify unprocessed RNA, OCT4A, and OCT4B RNA. The results showed that TIP110 caused more spliced OCT4A, but not OCT4B; while siRNA for Tip110 caused less spliced OCT4A, but not OCT4B. Total transcript either decreased with TIP110 or increased without TIP110 presence accordingly (Fig. 4c). Both in vivo and in vitro results showed that TIP110 is an important factor for OCT4A splicing. There is a slight increase in the stem cell marker OCT4A, SOX2, and NANOG levels with overexpression TIP110; however, the stem cell marker levels decreased with silent TIP110 expression (Fig. 4d). This is consistent with our previous results from hESC [13]. We further tested whether expression of TIP110 could activate the OCT4 promoter when we cotransfected the OCT4 promoter-reporter gene and the TIP110 expression vector. We did not detect TIP110 activation of this OCT4 promoter under these conditions (data not shown). This suggests that TIP110 regulates OCT4A expression in hESCs through alternative splicing rather than by activation of this promoter.
FIG. 4.
Human OCT4 genomic schematic organization, OCT4 minigene construct, and RT-PCR analysis of OCT4A transcripts. (a) Two fragments of the OCT4 genomic clones spanning OCT exon 1 and exon 2 were amplified by PCR, then they are cloned into pCDNA3 vector multiple cloning sites (MCS), which could be spliced with either OCT4A and/or OCT4B. (b) Coexpression of plasmids for pC.DNA3 (control) and pC.DNA3TIP110; and shRNA (control) and shRNA.TIP110 with OCT4 minigene in NT-2 cells and H9 hESCs. Cells were harvested 48 h after transfection for OCT4A and OCT4B mRNA expression by RT-PCR. OCT4A/OCT4B ratio was calculated through densitometry reading. β-actin was used as a loading control. (c) OCT4 minigene was in vitro transcribed into pre-mRNA. The OCT4 minigene transcribed pre-mRNAs were incubated with NT-2 cell extract (L1), NT-2 cell extract plus recombinant TIP110 protein (L2), and NT-2 cell with siRNA for TIP110 extract in splicing buffer for 0 hrs (input) or 3 hrs reaction at 30°C. RNAs were extracted from the input for quantitative RT-PCR of total transcript; quantitative RT-PCR for total transcript, OCT4A, and OCT4B from splicing reactions. (d) The stem cell markers (OCT4, Nanog, and Sox2) were analyzed in NT-2 cells by qPCR with overexpressing TIP110 or silencing the expression. β-actin was used to normalize q-PCR results. Data in panel d are expressed as mean±SD of 3 independent experiments. ♦P<0.05; ■P<0.001 for the compared pair.
Expression of C-MYC is generally high during early embryonic development, where it is required for ESCs pluripotency and reprogramming in addition to proliferation. C-MYC expression is low or undetectable in differentiated adult tissues, which is consistent with the virtual absence of cell proliferation. In this study, we demonstrated that TIP110 expression in human CB CD34+ cells and hESCs is controlled by C-MYC expression. This explains expression of TIP110 in these cells, which ensures TIP110 functions in maintaining hematopoiesis and in maintaining pluripotent factor (NANOG, OCT4, and SOX2) expression in and pluripotency of hESCs.
We observed from our previous work that TIP110 expression in ESCs is necessary for OCT4A expression. OCT4A, which is an alternatively spliced isoform of the OCT4 gene, sustains ESC self-renewal. It is possible that other factors besides TIP110 might also be important in determining OCT4 alternative splicing, as our minigene could not produce the OCT4A isoform in Hela cells with TIP110 expression (data not shown). Through our current demonstration that Tip110 regulates expression of the OCT4A alternatively spliced isoform instead of the OCT4B isoform in our OCT4 minigene model, we have enhanced understanding of the contribution of TIP110 to pluripotency of hESCs.
Acknowledgments
These studies were supported by Public Health Service Grants from the National Institutes of Health: R01 HL 056416, R01 HL 67384, and P01 DK090948 to H.E.B.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Rossant J. Stem cells and lineage development in the mammalian blastocyst. Reprod Fertil Dev. 2007;19:111–118. doi: 10.1071/rd06125. [DOI] [PubMed] [Google Scholar]
- 2.Yamanaka Y. Ralston A. Stephenson RO. Rossant J. Cell and molecular regulation of the mouse blastocyst. Dev Dyn. 2006;235:2301–2314. doi: 10.1002/dvdy.20844. [DOI] [PubMed] [Google Scholar]
- 3.Nichols J. Zevnik B. Anastassiadis K, et al. Formation of pluripotent stem cells in the mammalian embryo depends on the POU transcription factor Oct4. Cell. 1998;95:379–391. doi: 10.1016/s0092-8674(00)81769-9. [DOI] [PubMed] [Google Scholar]
- 4.Boyer LA. Lee TI. Cole MF, et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122:947–956. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Takahashi K. Tanabe K. Ohnuki M. Narita M. Ichisaka T. Tomoda K. Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 6.Cauffman G. Liebaers I. Van Steirteghem A. Van de Veld H. POU5F1 isoforms show different expression patterns in human embryonic stem cells and preimplantation embryos. Stem Cells. 2006;24:2685–2691. doi: 10.1634/stemcells.2005-0611. [DOI] [PubMed] [Google Scholar]
- 7.Zeng F. Baldwin DA. Schultz RM. Transcript profiling during preimplantation mouse development. Dev Biol. 2004;272:483–496. doi: 10.1016/j.ydbio.2004.05.018. [DOI] [PubMed] [Google Scholar]
- 8.Takeda J. Seino S. Bell GI. Human Oct3 gene family: cDNA sequences, alternative splicing, gene organization, chromosomal location, and expression at low levels in adult tissues. Nucleic Acid Res. 1992;20:4613–4620. doi: 10.1093/nar/20.17.4613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cartwright P. McLean C. Sheppard A. Rivett D. Jones K. Dalton S. LIF/STAT3 controls ES cell self-renewal and pluripotency by a Myc-dependent mechanism. Development. 2005;132:885–896. doi: 10.1242/dev.01670. [DOI] [PubMed] [Google Scholar]
- 10.Shaulian E. Karin M. AP-1 as a regulator of cell life and death. Nat Cell Biol. 2002;4:E131–E136. doi: 10.1038/ncb0502-e131. [DOI] [PubMed] [Google Scholar]
- 11.Pelengaris S. Khan M. Evan G. c-MYC: more than just a matter of life and death. Nat Rev Cancer. 2002;2:764–776. doi: 10.1038/nrc904. [DOI] [PubMed] [Google Scholar]
- 12.Yada M. Hatakeyama S. Kamura T. Nishiyama M. Tsunematsu R. Imaki H. Ishida N. Okumura F. Nakayama K. Nkayama K. Phosphorylation-dependent degradation of c-Myc is mediated by the F-box protein Fbw7. EMBO J. 2004;23:2116–2125. doi: 10.1038/sj.emboj.7600217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu Y. Lee MR. Timani K. He JJ. Broxmeyer HE. Tip110 maintains expression of pluripotent factors in and pluripotency of human embryonic stem cells. Stem Cells Dev. 2012;21:829–833. doi: 10.1089/scd.2011.0512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu Y. Hangoc G. Campbell TB. Broxmeyer HE. Identification of parameters required for efficient lentiviral vector transduction and engraftment of human cord blood CD34(+) NOD/SCID- repopulating cells. Exp Hematol. 2008;36:947–956. doi: 10.1016/j.exphem.2008.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu Y. Timani K. Mantel C. Fan Y. Hangoc G. Cooper S. He JJ. Broxmeyer HE. TIP110/p110nrb/SART3/p110 regulation of hematopoiesis through CMYC. Blood. 2011;117:5643–5651. doi: 10.1182/blood-2010-12-325332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Atlasi Y. Mowla SJ. Ziaee SA. Gokhale PJ. Andrews PW. OCT4 spliced variants are differentially expressed in human pluripotent and nonpluripotent cells. Stem Cells. 2008;26:3068–3074. doi: 10.1634/stemcells.2008-0530. [DOI] [PubMed] [Google Scholar]
- 17.Bell M. Schreiner S. Damianov A. Reddy R. Bindereif A. p110, a novel human U6 snRNP protein and U4/U6 snRNP recycling factor. EMBO J. 2002;21:2724–2735. doi: 10.1093/emboj/21.11.2724. [DOI] [PMC free article] [PubMed] [Google Scholar]