Abstract
Understanding the determinants of protein stability remains one of protein science's greatest challenges. There are still no computational solutions that calculate the stability effects of even point mutations with sufficient reliability for practical use. Amino acid substitutions rarely increase the stability of native proteins; hence, large libraries and high-throughput screens or selections are needed to stabilize proteins using directed evolution. Consensus mutations have proven effective for increasing stability, but these mutations are successful only about half the time. We set out to understand why some consensus mutations fail to stabilize, and what criteria might be useful to predict stabilization more accurately. Overall, consensus mutations at more conserved positions were more likely to be stabilizing in our model, triosephosphate isomerase (TIM) from Saccharomyces cerevisiae. However, positions coupled to other sites were more likely not to stabilize upon mutation. Destabilizing mutations could be removed both by removing sites with high statistical correlations to other positions and by removing nearly invariant positions at which “hidden correlations” can occur. Application of these rules resulted in identification of stabilizing mutations in 9 out of 10 positions, and amalgamation of all predicted stabilizing positions resulted in the most stable yeast TIM variant we produced (+8 °C). In contrast, a multimutant with 14 mutations each found to stabilize TIM independently was destabilized by 2 °C. Our results are a practical extension to the consensus concept of protein stabilization, and they further suggest the importance of positional independence in the mechanism of consensus stabilization.
Keywords: protein engineering, consensus design, sequence correlation, protein stability, triosephosphate isomerase
Introduction
Most native proteins are only marginally stable, meaning the folded and unfolded states are generally separated by no more than 5–15 kcal mol−1.1,2 Many natural proteins are not stable enough for research, pharmaceutical, or industrial applications, and many disease pathologies arise from single mutations that destabilize proteins. For example, most of the “hot spot” mutations observed in the tumor suppressor p53 in cancer mutations are far from the DNA binding site and merely reduce the stability of the protein.3 However, the prediction of protein stability remains one of the most difficult problems in protein biochemistry, due to inadequate performance of potential functions, difficulty in sampling backbone motion, lack of knowledge of the unfolded state, and the challenge of modeling entropic effects.4–6 A systematic analysis of the performance of 11 stability prediction algorithms by Khan and Vihinen7 concluded that Dmutant8 and FoldX9 were among the most reliable, but even these were only about 60% accurate in correctly predicting qualitatively if mutations were stabilizing or destabilizing. For example, for FoldX, the standard deviation of the difference between the experimental and calculated ΔΔG values for a mutation is 0.5– 1.0 kcal mol−1 (depending on the implementation and elimination of outliers), but the mean experimental ΔΔG values are about 2.5 kcal mol−1.9 Part of the challenge in understanding protein stability is that its measurement, by calorimetry or spectroscopic observation of thermal or chemical denaturation, is slow and labor and material intensive. In general, library-based strategies to improve protein stability have been very successful, but these require library construction, an appropriate screen, and/or some rational design.10–13 These types of experiments demonstrate that few mutations to natural proteins are stabilizing, on the scale of 1% or less.
Advances in DNA sequencing technologies have provided a wealth of genomic data that can be readily translated into protein sequences. Many families of proteins now have hundreds to thousands of known sequences, allowing one to interrogate the determinants of protein fitness statistically. One such approach, consensus design, or the replacement of an amino acid with the most common amino acid in a multiple sequence alignment (MSA), has been shown to increase the stability of antibodies as well as other proteins.14–18 For example, Steipe et al. engineered 10 consensus mutations in the Vκ domain of murine antibody McPC603.14 Enhanced stability was observed in six variants, three were neutral, and only one was less stable than wild-type McPC603. This and other studies show that consensus mutations stabilize proteins about 50% of the time, which is dramatically better than random mutagenesis. Consensus design has also been applied to full consensus repeats such as the tetratricopeptide repeats and ankyrins, in addition to whole enzymes including the fungal phytases and, recently, triosephosphate isomerase (TIM).19–25 In general, these full-consensus proteins are dramatically more stable than the proteins from which their sequences arise. A consensus fungal phytase was 15–22 °C more stable than its parental sequences, and previously constructed consensus TIM variants cannot be fully melted at 95 °C.22,25 Recently, the concept of ancestral design, replacing an amino acid with one froma common ancestor in phylogeny, has seen similar results for stabilization.26–29 The Yamagishi laboratory individually replaced 12 residues with ancestral amino acids in 3-isopropylmalate dehydrogenase and found that half of the mutations improved stability.26
We wished to understand why consensus mutations are only stabilizing about half the time and, ideally, to predict which half would be stabilizing. For one thing, we hypothesized that positions that are highly variable (i.e., not conserved) are not likely to be stabilized by the consensus mutation, since those sites contain relatively little information. For another, we hypothesized that consensus mutations in sites that are strongly coupled to other positions might result in destabilization, at least without some kind of compensatory mutation. For example, one can imagine that mutation of a residue in a buried polar interaction to a consensus hydrophobic residue would be destabilizing unless the partner polar amino acid was also mutated. To test these ideas, we used the well-studied TIM from Saccharomyces cerevisiae as a host for a large number of consensus mutations, and we examined the effects on thermal stability for different levels of sequence conservation and correlation, as well as structural properties such as surface exposure and secondary structure.
TIM is the archetypical member of the (β/α)8-barrel fold family, which is seen in more than 10% of all natural enzymes.30,31 TIM catalyzes the isomerization between dihydroxyacetone phosphate (DHAP) and glyceraldehyde-3-phosphate (GAP) in glycolysis; therefore, it is present in nearly every organism and amenable to statistical analysis. The enzyme, a homodimer in most species, has been characterized in detail from several organisms including Escherichia coli, S. cerevisiae, Trypanosoma brucei, and Homo sapiens.32–38 The active-site residues of (β/α)8-barrel proteins are typically found on the surface loops connecting the β-strand core to the α-helical surface, as are those in TIM (e.g., K12, H95, and E165 in yeast TIM). Other loops are critical for function, including loop 3, which is interdigitated into the other monomer, and loop 6, the opening and closing motion of which is coordinated with catalytic activity. Despite their ubiquity and apparently modular nature, loop swapping and other TIM-barrel engineering have proven more difficult than expected.39 The mutability of TIM has been studied in the Harbury laboratory. Silverman et al. found that many single conservative mutations (e.g., Glu to Asp) of yeast TIM were tolerated, but libraries of conservative mutations resulted in only 1 in 1010 active variants, suggesting the importance of coupling between those mutations.40
We present the characterization of single consensus mutations made in a large number of sites in S. cerevisiae TIM (S.c. TIM). We demonstrate that, in general, higher levels of conservation lead to stabilization, but that both the most highly conserved sites and the most highly correlated sites are less likely to be stabilizing, due to coupling effects including “hidden correlations.” Application of the resulting algorithm allows one to identify stabilizing mutations in TIM with high reliability (9 of 10 tested were stabilizing). Furthermore, while aggregation of all the mutations found to be individually stabilizing actually resulted in net destabilization, aggregation of all of the mutations predicted to be stabilizing by our algorithm resulted in dramatic thermostabilization.
Results
re-S.c. TIM
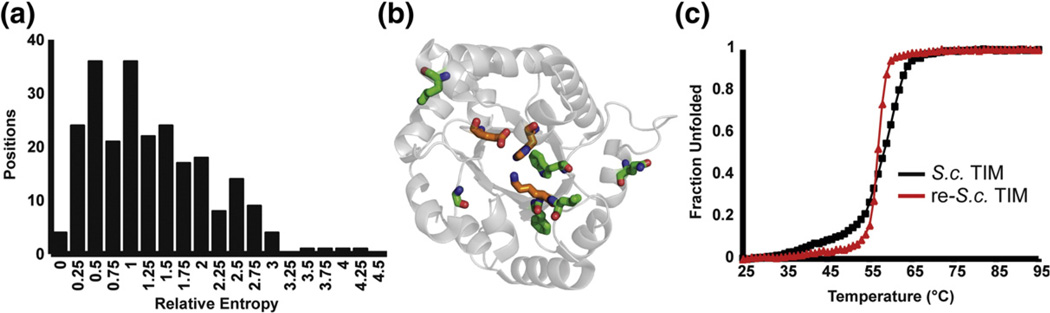
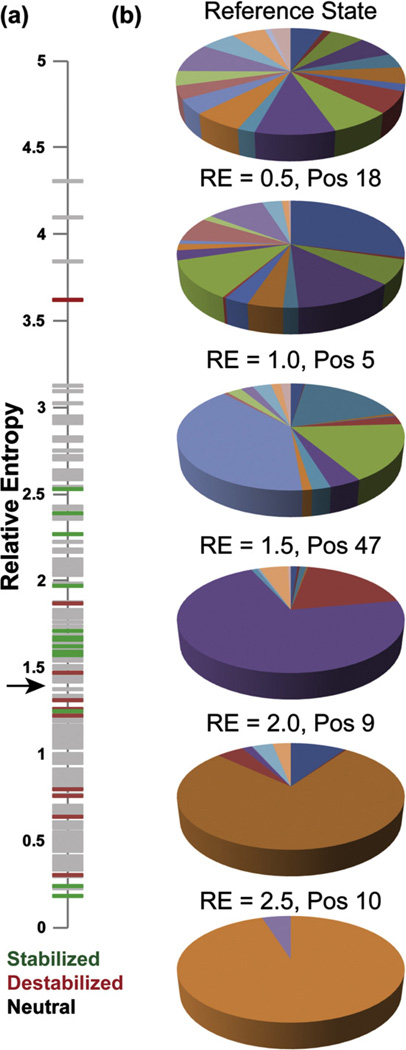
We hypothesized that highly conserved positions imply greater importance in defining the family, and therefore consensus mutations at these positions might result in greater thermostabilization. To quantify the extent of conservation, we calculated the relative entropy between the distribution of amino acids in a neutral reference state and the distribution in each position in the MSA of TIM. Relative entropy is an easily calculated information theoretic estimate of the log of the probability of observing a given distribution if one expects a reference distribution.41,42 The reference state was taken from the codon usage in the yeast genome,43 which approximates equal usage with slight deviations from codon bias and chemical constraints of the amino acids. Positions 31 and 126 in TIM were the least and most conserved, respectively, with relative entropies of 0.31 and 4.31, and the average relative entropy was 1.42 (Fig. 1a). We chose to simultaneously mutate the six most conserved positions in wild-type S.c. TIM that were not already consensus amino acids. This yielded the TIM variant re-S.c. TIM (F11W L13M Q82M W90Y K134R A212V).
Fig. 1.
re-S.c. TIM. (a) Histogram of relative entropy values for all 240 aligned positions in the TIM family. The mean RE is 1.42. (b) The six most conserved positions in S.c. TIM that are not consensus amino acids are shown in green sticks. The active-site residues are shown in orange on the 1YPI crystal structure. (c) Ellipticity at 222 nm is followed with increasing temperature. The wild type melts at 59.1 °C, but re-S.c. TIM melts at 57.0 °C.
The gene was assembled from synthetic oligonucleotides, cloned into an E. coli overexpression vector, and purified to near homogeneity by Ni-NTA chromatography before and after cleavage of an N-terminal hexahistidine tag with the capsid protein protease of tobacco etch virus. Purification of re-S.c. TIM yielded 5–10 mg L−1 of culture from the soluble fraction. The enzyme was assayed for activity under Vmax (saturating) conditions at 4 mM GAP (meaning, ~5× Km for wild-type S.c. TIM). The specific activity is within twofold of wild type (~104 µmol min−1 mg−1). Far-UV circular dichroism produced nearly identical spectra with broad minima spanning the 208-, 215-, and 222-nm peaks observed for mixed α/β proteins (Fig. 2a). The ellipticity at 222 nm was monitored with increasing temperature to compare relative stabilities (Fig. 1c). Both proteins maintain folded baselines until ~50 °C before unfolding through a single, cooperative transition. The wild-type enzyme remains half folded at 59.1 °C, but the engineered re-S.c. TIM unfolds with a T1/2 of 57.0 °C. (We say T1/2 here because all of the variants in this study unfold irreversibly, with precipitation upon continued heating in the unfolded state.) In contrast to our initial expectation, combining consensus mutations from the most conserved sites did not stabilize the protein.
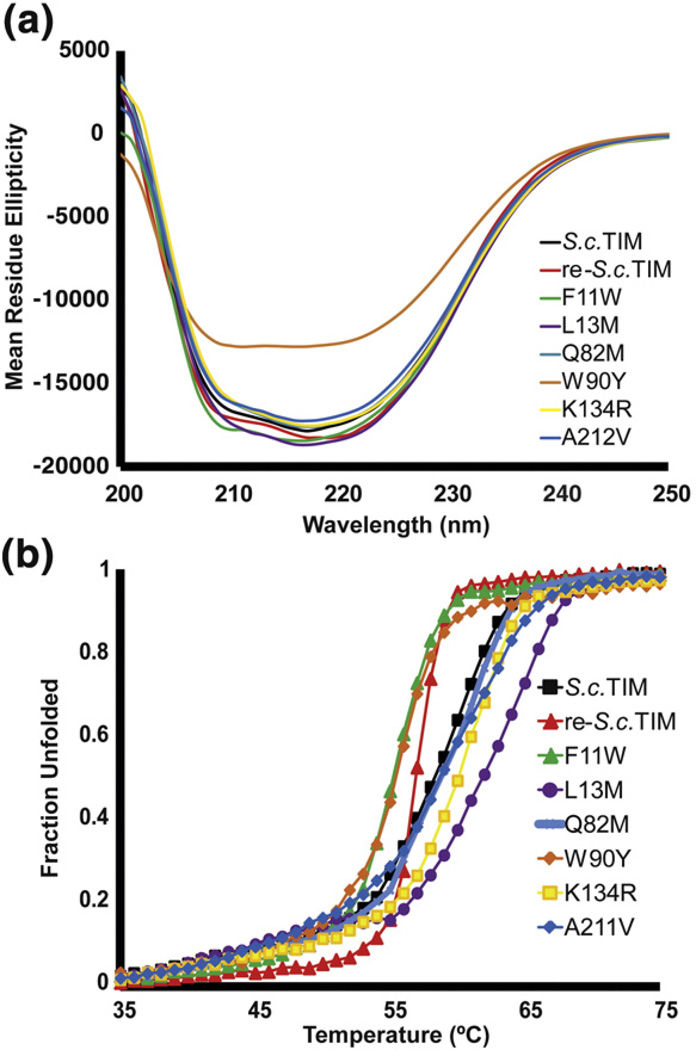
Fig. 2.
CD characterization of highly conserved mutations. (a) The circular dichroism spectra of wild type and consensus variants of TIM. All have similar ellipticity when normalized for concentration except W90Y, which may be partially unfolded. (b) The CD thermal melts indicate that four individual consensus variants are more stable than wild type, but the remaining two are less stable.
Individual consensus mutants
To determine why the six highly conserved mutations did not stabilize the protein, we constructed the mutations individually. All variants are within an order of magnitude of the wild-type activity. Five of the six variants share similarly shaped CD spectra with comparable mean residue ellipticities at 222 nm (Fig. 2a). W90Y is the only exception, which, taken together with the activity data, suggests that this variant may be partially unfolded. Thermal denaturation reveals that F11W and W90Y are destabilized, but L13M, Q82M, K134R, and A212V are more stable than wild type (Fig. 2b). re-S.c. TIM was 2 °C less stable than S.c. TIM, and the individual mutants ranged ±4 °C from the T1/2 of S.c. TIM. Surprisingly, mutation to even some of the most conserved residues in the MSA was destabilizing.
To further understand the consensus mutation phenomenon and its role in stabilization, we engineered and assayed a variety of consensus mutations with varying levels of conservation. There are 240 aligned positions in the TIM family. Of these positions, 43% of the positions deviate between S.c. TIM and the consensus sequence. Of these 103 positions, we chose to characterize 23 individual consensus mutations that vary in solvent exposure, secondary structure, conservation, and evolutionary substitution frequency.
The 23 variants were expressed and purified from BL21(DE3) E. coli in similar yield to S.c. TIM and re-S.c. TIM. The I20A, G122T, and F229A mutants did not express in sufficient quantities to characterize. Multiple codons and contexts were tested for I20A (GCG, GCT) and G122T (ACA, ACT, ACC, ACG) with similar results. Consequently, these three mutations were classified as destabilizing. The remaining 20 proteins were assayed for catalytic activity monitoring the turnover of GAP to DHAP. All the variants turned over substrate with specific activity values of ~103–104 µmol min−1 mg−1, which is on par with wild type. All variants displayed similar mean residue ellipticities at 222 nm(data not shown). The T1/2 for each variant was determined by CD thermal denaturation (Fig. 3a and b). The T1/2 values of 12 of the 23 mutants were greater than wild type, one was the same as wild type, and the remaining 10 exhibited a loss in stability—essentially the same as the 50% rate of stabilization previously seen for both consensus and ancestral mutations.
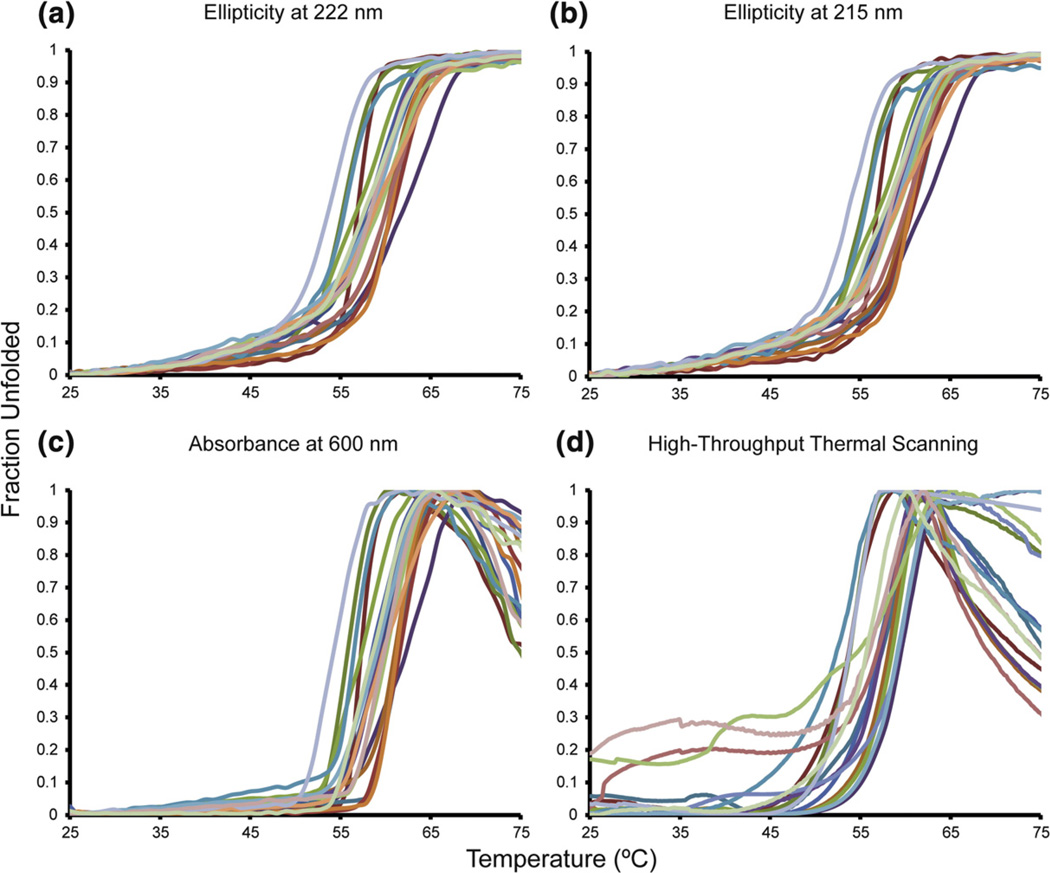
Fig. 3.
Thermal stabilities of consensus TIM variants. We monitored the loss of secondary structure with increasing temperature at 222 nm for helices (a) and 215 nm for sheets (b). (c) The optical density at 600 nm from aggregation reports similar two-state unfolding profiles as the CD thermal melts. (d) High-throughput thermal scanning was used to assay the melting temperatures based on hydrophobic dye binding. Note that the same colors are used for the same variants in (a)–(d).
The most destabilized variant in our data set was N213K at ΔT1/2=−5.1 °C, and the most stabilizing mutation was L13M at ΔT1/2=+4.0 °C. Since the entire data set differs by only 9.1 °C, we relied on several other thermal assays to accurately rank the relative stabilities of our variants: (1) We also observed thermal denaturation of the TIMs by 215 nm CD signal. (2) TIM thermal denaturation leads to aggregation and precipitation upon unfolding. We measured the T1/2 values from the scattering of light (optical density) at 600 nm. This method is essentially the same as what is referred to as differential static light scattering.44 (3) We previously showed that TIM stability differences could be accurately ascertained by high-throughput thermal scanning.10 The data from each of these methods are highly concordant, as shown in Figs. 3 and 4 and Supplemental Fig. 1. Taken together, we are able to accurately measure small differences in stability. (Throughout this report, unless specified, T1/2 values refer to those obtained by loss of ellipticity at 222 nm on thermal denaturation.)
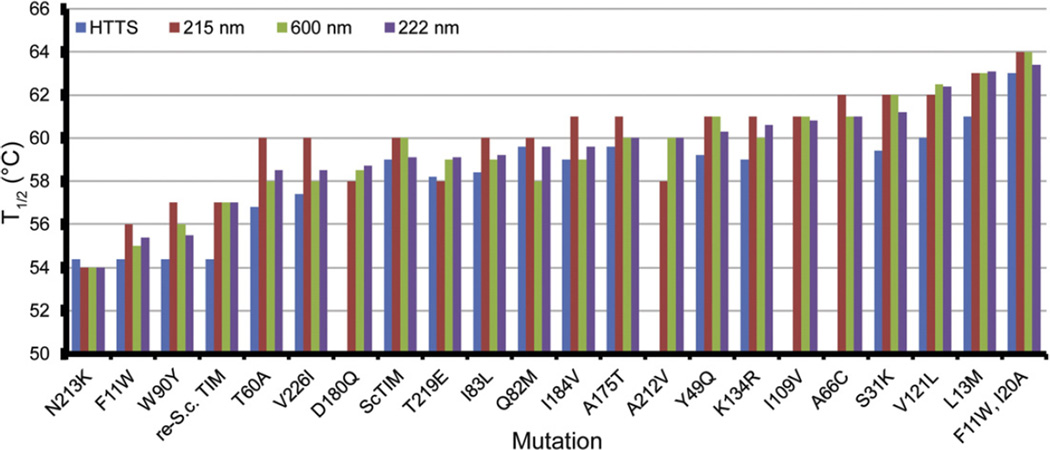
Fig. 4.
Concordance of stability assays. The variants are arranged by the T1/2 values derived from CD thermal denaturation at 222 nm. Data were not collected by high-throughput thermal scanning for A66C, I109V, D180Q, and A212V.
Structural nature of consensus mutations
For each of the consensus mutations, we analyzed the physical and chemical properties of the amino acids, their context within the folded protein, and their sequence statistics (Table 1). On average, residues in TIM are 17% solvent exposed and our consensus mutants average 13% ranging from 0% to 60%. In our data set, there is no correlation between solvent-accessible surface area and T1/2 (R2=0.20, see Supplemental Fig. 2). Of the 7 loop mutants, 5 were stabilized; 6 of the 10 helical mutations were stabilizing; and only 1 of the 6 β-sheet mutants was more stable. The Harbury laboratory has previously shown that the β-strand core of TIM is highly sensitive to mutations.40 BLOSUM scores are based on the mutational propensities between amino acids as calculated across phylogeny and are consequently a way of quantifying how conservative a mutation is.45 Here, 6 out of 10 common substitutions (positive BLOSUM) were more stable, and 6 out of 13 rarer substitutions (zero to negative BLOSUM) were more stable. Therefore, except that few positions in β-strands result in stabilization, the general structural properties of the mutations were not predictive of stabilization.
Table 1.
Characterization of mutants
| T1/2 | ΔT1/2 | Spec. act. | Corr.h | FoldX (ΔΔG)i | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Mutant | REa | (°C)b | (°C)c | Predictiond | (µmol min−1 mg−1)e | Solvent (%)f | BLOSUMg | 2° | (>0.5?) | (kcal mol−1) |
| S.c. TIM | NA | 59.1 | — | 1.0±0.1×104 | NA | NA | NA | NA | ||
| re-S.c. TIM | NA | 57.0 | −2.1 | 5±1×103 | NA | NA | NA | NA | ||
| F11W | 3.62 | 55.4 | −3.7 | 5±1×103 | 0.7 | 1 | β-Sheet | No | 5.3 | |
| L13M | 2.53 | 63.1 | +4.0 | + | 8.5±0.4×103 | 59.6 | 2 | Loop | No | −0.4 |
| Q82M | 2.39 | 59.6 | +0.5 | + | 1.1±0.4×104 | 36.8 | 0 | α-Helix | No | −0.5 |
| A212V | 2.27 | 60.0 | +0.9 | + | 8.6±0.5×103 | 3.1 | −2 | Loop | No | −2.4 |
| K134R | 1.97 | 60.6 | +1.5 | + | 6.0±0.8×103 | 21.7 | 2 | α-Helix | No | +0.2 |
| W90Y | 1.87 | 55.5 | −3.6 | 2.5±0.8×103 | 8.3 | 2 | β-Sheet | Yes | +0.8 | |
| V123P | 1.76 | DNEj | DNE | DNE | 0.0 | −2 | β-Sheet | Yes | +4.0 | |
| V226I | 1.71 | 58.5 | −0.6 | + | 9±2×103 | 1.3 | 1 | Loop | No | −0.6 |
| I109V | 1.67 | 60.8 | +1.7 | + | 8±1×103 | 0.5 | 1 | α-Helix | No | +0.7 |
| A66C | 1.66 | 61.0 | +1.9 | + | 6.6±0.5×103 | 0.9 | 0 | Loop | No | −0.1 |
| I83L | 1.62 | 59.2 | +0.1 | + | 5.1±0.6×103 | 1.0 | 2 | α-Helix | No | +0.5 |
| V121L | 1.59 | 62.4 | +3.3 | + | 3.9±0.6×103 | 1.6 | 3 | Loop | No | −0.2 |
| I184V | 1.57 | 59.6 | +0.5 | + | 3.9±0.2×103 | 7.2 | 1 | α-Helix | No | +1.9 |
| D180Q | 1.47 | 58.7 | −0.4 | 8.0±0.3×103 | 17.3 | 0 | α-Helix | Yes | +0.5 | |
| Ave. RE | 1.42 | |||||||||
| N213K | 1.31 | 54.0 | −5.1 | 5±1×103 | 0.0 | 0 | α-Helix | No | +1.1 | |
| F229A | 1.26 | DNE | DNE | DNE | 0.1 | −2 | β-Sheet | Yes | +4.2 | |
| A175T | 1.25 | 60.0 | +0.9 | 5±2×103 | 38.7 | −1 | Loop | No | −0.3 | |
| T219E | 1.22 | 59.1 | 0.0 | 6.0±0.8×103 | 25.4 | 0 | α-Helix | No | −0.5 | |
| I20A | 0.81 | DNE | DNE | DNE | 0.0 | −1 | α-Helix | Yes | +3.0 | |
| G122T | 0.64 | DNE | DNE | DNE | 4.1 | 1 | β-Sheet | Yes | +8.5 | |
| T60A | 0.30 | 58.5 | −0.6 | 6.5±0.8×103 | 14.2 | −1 | Loop | Yes | −0.5 | |
| Y49Q | 0.24 | 60.3 | +1.2 | 6.0±0.4×103 | 17.8 | −1 | α-Helix | Yes | +2.0 | |
| S31K | 0.18 | 61.2 | +2.1 | 8±2×103 | −42.6 | 0 | β-Sheet | Yes | −0.5 | |
| Compensatories | ||||||||||
| F11W, I20A | NA | 63.4 | +4.3 | 5.2±0.8×103 | NA | NA | NA | NA | +2.8 | |
| W90Y, G122T | NA | DNE | DNE | DNE | NA | NA | NA | NA | +3.0 | |
| W90Y, G122T, V123P | NA | DNE | DNE | DNE | NA | NA | NA | NA | +6.6 | |
| algoTIM mutations | ||||||||||
| I127V | 1.68 | ND | ND | + | ND | 1.6 | 1 | β-Sheet | Low | |
| I40V | 1.63 | ND | ND | + | ND | 0.0 | 1 | β-Sheet | Low | |
| V162I | 1.60 | ND | ND | + | ND | 0.2 | 1 | β-Sheet | Low | |
| K135E | 1.54 | ND | ND | + | ND | 50.1 | 1 | α-Helix | Low | |
| N78I | 1.49 | ND | ND | + | ND | 21.6 | −3 | Loop | Low | |
| algoTIM | NA | 67.2 | +8.2 | + | 1.0±0.8×104 | NA | NA | NA | NA | |
| comboTIM | NA | 55.0 | −4.1 | 2.9±0.2×103 | NA | NA | NA | NA | ||
Relative entropy between the positional distribution and the yeast neutral reference state.
Derived from CD thermal data at 222 nm.
T1/2–59.1 °C, where 59.1 °C is the T1/2 of S.c. TIM.
(+) indicates mutations that are expected to increase the stability of S.c. TIM based on conservation and correlation filters.
The specific activity for the turnover of GAP to DHAP. The enzymes were assayed at 4 mM GAP, which corresponds to > 5× Km of S.c. TIM.
The solvent exposure of each residue in Protein Data Bank entry 1YPI was calculated in MOLMOL version 2K.2.0 with a 1.4 Å sphere.
The Henikoff score is a quantitative value to describe the conservativeness of a mutation based on phylogenetic analysis. Here, neutral mutations are given a score of 0, common mutations score positive, and rare mutations are scored as negative values.
We consider a site to be highly correlated if its maximal mutual information score is greater than 0.5.
The ΔΔG values were calculated for each mutant using FoldX (see Materials and Methods). Here, a destabilized and a stabilized mutation have positive and negative values, respectively.
Did not express.
Sequence statistics and stability of tested mutants
Our initial results from re-S.c. TIM and its individual constituents suggested that high positional conservation alone is not more predictive of which consensus mutations will be stabilizing. We reexamined this in light of the full set of 23 mutations. Mutations at sites that are more conserved than average (relative entropy greater than 1.42) yielded more stable mutants in 9 of 14 consensus variants, while only 3 of 9 consensus mutations at weakly conserved positions were stabilizing (Fig. 5a). Overall, limiting consensus mutations to sites with more than average conservation improves the chances of making stabilizing mutations from about 1-in-2 to about 2-in-3, even though some mutations at highly conserved positions were destabilizing. We also found that the stabilization was not significantly related to the degree to which the consensus residue was more common at the given position than the residue found in yeast TIM (Supplemental Fig. 3). The R2 for this metric (ln fcons/fwt) versus T1/2 is about the same as for relative entropy alone to T1/2 (0.07 versus 0.03) over all the expressing mutants tested here.
Fig. 5.
Filtering by conservation. (a) All positions in TIM have been plotted against their relative entropies from the neutral reference state. All sites are shown in gray, and stabilizing and destabilizing consensus mutations are shown in green and red, respectively. Note that the stable mutations aggregate above the black arrow, which indicates the mean relative entropy of 1.42. (b) Amino acid distributions for the yeast neutral reference state and positions with relative entropy values of 0.5, 1.0, 1.5, 2.0, and 2.5 are shown.
Our second initial hypothesis was that some consensus mutations would fail to stabilize because of coupling. One potential way to predict coupling is from statistical correlation of positions in an MSA. Here, statistical correlation was determined from the mutual information between the amino acid distributions at each pair of positions (Fig. 6). Mutual information is the relative entropy between the observed pairwise distribution and the joint distribution calculated from the positional frequencies. As an illustration, if position i is amino acid a 25% of the time, and position j is amino acid b 10% of the time, then we randomly expect a–b pairs in 2.5% of sequences. The degree to which we see more (or fewer) a–b pairs than this increases the information in one distribution about the other distribution and implies correlation (or anticorrelation).
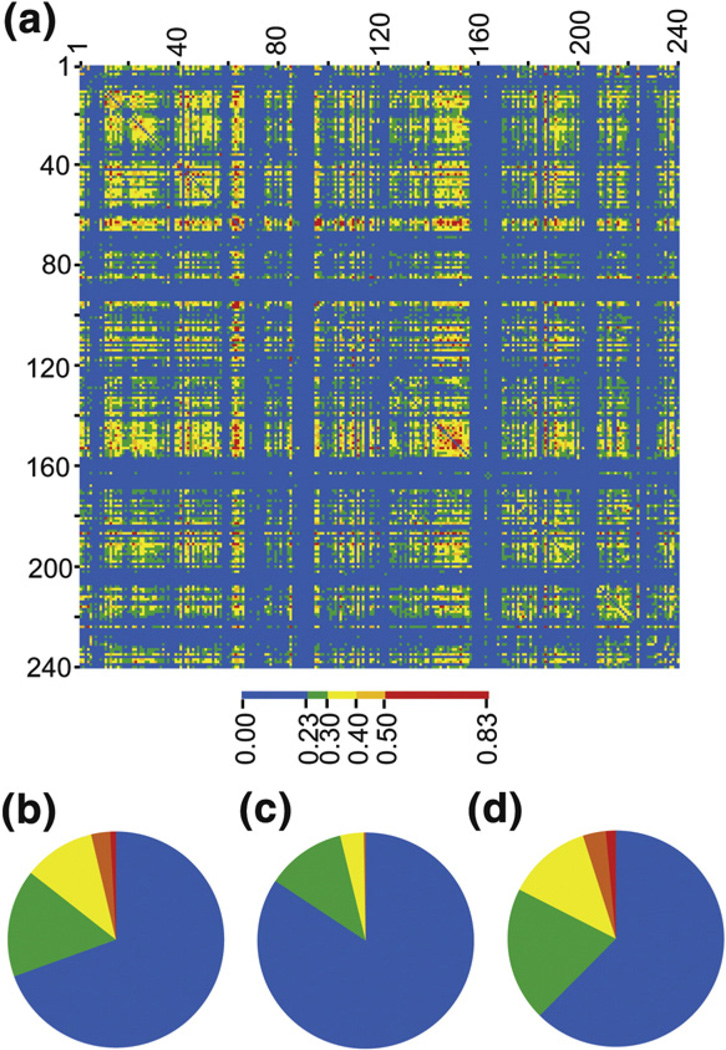
Fig. 6.
Mutual information and protein stability. (a) The mutual information matrix for all 240 positions in TIM is shown. The matrix is symmetric (x–y is the same as y–x), and there is no meaning to the self-correlations (x–x), which were not calculated. (b) The distribution of mutual information scores is shown for the entire matrix. Here, approximately 30% of all pairwise correlations are above the noise threshold of 0.23. The distribution of mutual information scores are shown for stabilizing mutations (c) and destabilizing mutations (d). Note that there is a significantly higher fraction of strong correlations at the positions that lead to a loss in stability.
By this calculation, only a small number of positions are seen to interact strongly (Fig. 6a). This was also observed in WW and PDZ domains using a related metric for sequence correlation (SCA).46,47 Of the 14 positions with above-average conservation, the 5 that were destabilized (Fig. 6d) upon mutation to the consensus residue are more highly correlated to other positions than the nine that were stabilized (Fig. 6c). Three of the five less stable variants are hubs for interaction networks, with detectable correlations to multiple positions. To estimate the importance of mutual information scores, we generated mock alignments in which the positional distributions were maintained but amino acids were scrambled between the sequences and then recalculated the mutual information scores. Detectable here means above this “noise” threshold (MI=0.23). The strengths of several correlations to the destablized conserved positions are significant. Pairwise correlations between positions 90–157 (MI=0.72), 90–123 (MI=0.63), and 180– 229 (MI=0.51) are all within the top 0.4% of the 28,680 possible unique correlations for 240 positions. A full analysis of the statistical interactions in the TIM family is beyond the scope of this article and will be presented separately (V.D., B.J.S., and T.J.M., manuscript in preparation).
Two variants with above-average conservation and little sequence correlation, F11W (−3.7 °C) and V266I (−0.6 °C), were destabilizing. Position 11 is so conserved that it is almost invariant in the TIM family—it is Trp in 595 out of 719 sequences in our MSA. Coupling to very highly conserved positions cannot be detected by sequence correlation because if position 11 is nearly always Trp, then i-11 pairs will always be a-Trp, and no additional information occurs in the pairwise distribution than in what would be expected at random. However, it is still possible that a highly conserved position could be physically coupled to another position, in the sense that mutation of the conserved positions might require a compensatory mutation at a second position to rescue stability or function. These types of hidden correlations can only occur at the most conserved sites, and so they can be eliminated from sites for potential consensus mutations by putting an upper limit on conservation (e.g., a relative entropy greater than 3), in addition to the lower limit already described.
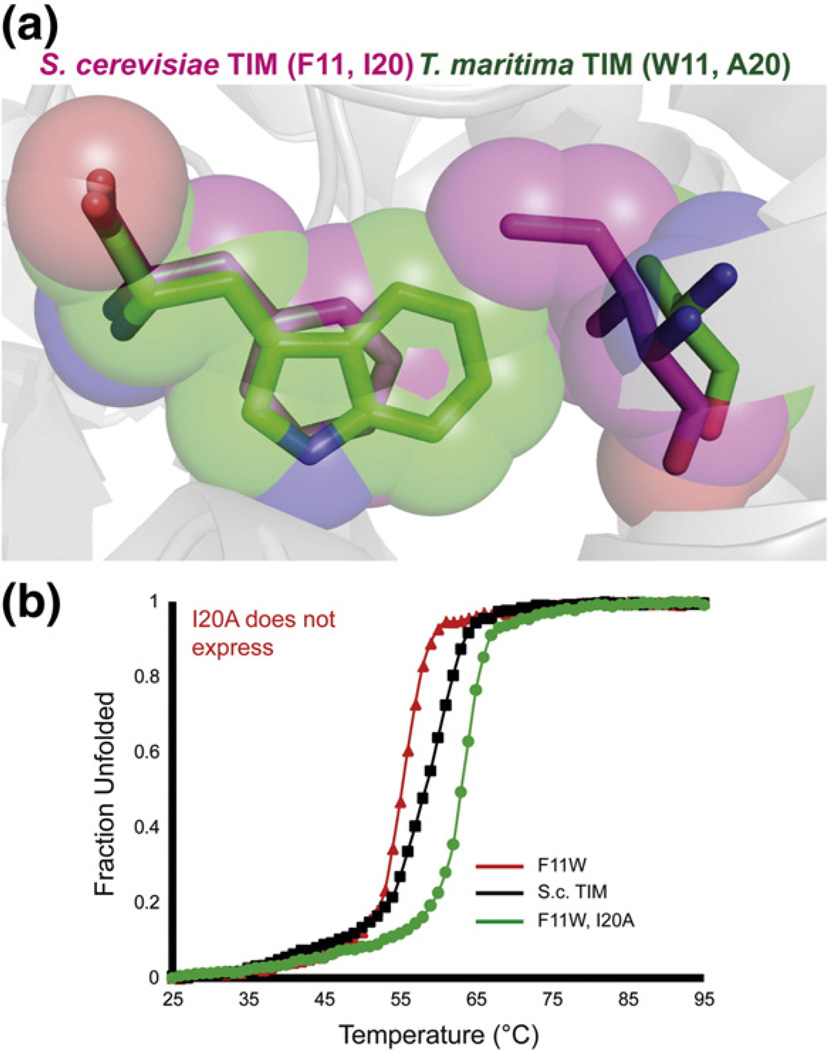
To explore the role of both statistical correlations and hidden correlations further, we attempted to design compensatory mutants for consensus variants F11W and W90Y. We analyzed the MSA and the crystal structures of TIMs with these different amino acids. As noted, yeast TIM is one of very few with Phe at position 11; TIM from most organisms, such as Thermotoga maritima, have a Trp at this position. It appears that position 20, in van der Waals contact with F11, is a larger amino acid, Ile, than is typically seen in this position (Ala in T. maritama). Alignment of the crystal structures from S. cerevisiae and T. maritima shows that the F11W mutation would sterically clash with the Ile in position 20 (Fig. 7a). The compensatory mutant F11W I20A in yeast TIM was 4.3 °C more stable than yeast TIM. In the context of F11W, the I20A mutation netted 8 °C of thermal stabilization. The I20A mutant alone did not express, perhaps because of destabilization due to under-packing against Phe11. Thus, positions 11 and 20 are coupled in TIM, although this could not be detected by correlation statistics.
Fig. 7.
A hidden correlation between positions 11 and 20. (a) The crystal structures of S.c. TIM and T. maritima TIM [Protein Data Bank entries 1YPI (pink) and 1B9B (green)] are aligned and residues 11 and 20 are highlighted. The F11W mutation may have introduced a steric clash resulting in destabilization. (b) CD thermal denaturation of F11W and F11W I20A. I20A alone did not express in appreciable quantities.
W90Y was a second mutation where above-average conservation did not yield consensus thermostabilization. Mutual information shows that this position is a hub of statistical interactions. We attempted to “correct” the strongest broken correlation, 90–122, by mutating the glycine at position 122 to the larger threonine. Thr is the consensus amino acid at position 122 and co-evolves with Tyr at position 90. The G122T substitution did not express in the context of S.c. TIM or W90Y S.c. TIM with codons ACA or ACC. Mutual information analysis also suggested that position 123 co-evolved with 90 and 122. A V123P consensus mutation was also constructed but did not express in any scaffold (V123P, W90Y/V123P, W90Y/V123P/G122T). There are 16 residues that cage the aromatic ring at position 90, half of which directly pack against the side chain. All are consensus amino acids except 122 and 123, at which we tested possible substitutions. G122R is a known human mutation that leads to thermolability.34 The hub-like nature of position 90 makes it difficult to engineer compensatory mutations without disrupting other possible interactions.
If the three criteria described here are taken together—above-average conservation (here, relative entropy greater then 1.42), elimination of the most coupled sites (here, maximal mutual information less than 0.50), and elimination of nearly invariant sites with possible hidden correlations (here, relative entropy greater than 3)—then 15 consensus mutations would be predicted to be stabilizing. We tested 10 of those individually, and 9 were stabilizing.
Multimutants
After the failure of the original re-S.c. TIM variant, we wanted to test whether a super-stable mutant of yeast TIM could be made by amalgamating predicted stabilizing mutations. Of the 240 aligned positions in the TIM alignment, 103 are not consensus. Only 19 positions have relative entropies between 1.42 and 3.00, and four of those positions (C41A, W90Y, V123P, and D180Q) have high maximal mutation information values as well as large numbers of significant correlations (see Supplemental Fig. 4). As a result, we designed algoTIM, which includes 15 consensus mutations (L13M I40V A66C N78I Q82M I83L I109V V121L I127V K134R K135E V162I I184V A212V V226I). These mutations include nine stabilizing mutations, one destabilizing mutation, and five uncharacterized mutations. In addition, we characterized a second TIM we named comboTIM that simply combines all stabilizing mutations characterized in this study (F11W L13M I20A S31K Y49Q A66C Q82M I83L I109V V121L K134R A175T I184V A212V). Note that this variant contains the stabilizing F11W I20A pair, and does not contain the destabilizing V226I mutation or any of the mutations we removed from algoTIM due to high correlations.
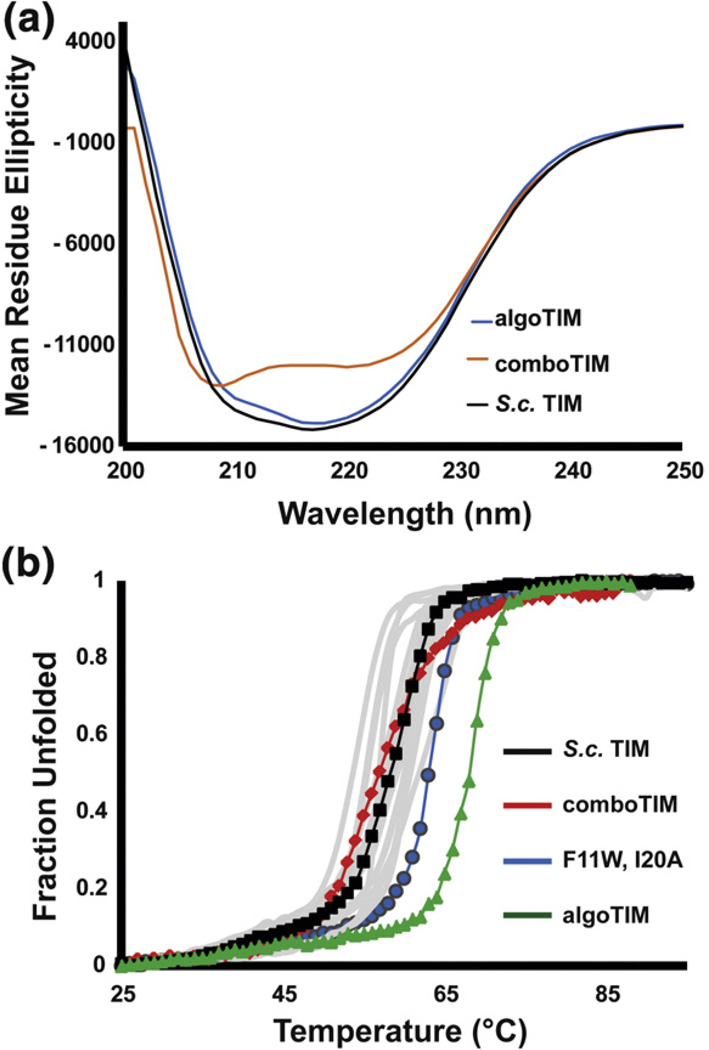
The algorithmic multimutant algoTIM melted with a T1/2 of 67.2 °C, nearly 10 °C greater than S.c. TIM and an additional 4 °C more stable than any variant previously characterized (Fig. 8). In stark contrast, comboTIM was destabilized (T1/2=56.7 °C) from wild type despite harboring 14 known stabilizing mutations.
Fig. 8.
Characterization of algoTIM and comboTIM. (a) The CD wavelength scans of wild-type S.c. TIM and algoTIM are nearly identical. comboTIM shows less ellipticity at 222 nm and has its deepest minima at 205 nm, suggesting some random coil. (b) The CD thermal melts monitored at 222 nm are shown for all characterized proteins in gray, with comboTIM, S.c. TIM, the F11W I20A mutant, and algoTIM highlighted.
Discussion
A number of lines of evidence show that about 50% of consensus mutations are stabilizing. We set out to understand how to identify which half are stabilizing and the basis for that distinction. Our two fundamental hypotheses were that consensus mutations at weakly conserved positions would be less likely to stabilize, and that mutations at positions that are coupled to other sites might destabilize more frequently. We originally tried to simply amalgamate the consensus mutations at the six most conserved sites that were not already the consensus residue in yeast TIM, but this actually resulted in slight destabilization. Dissection of the re-S.c. TIM multimutant into its constituent mutations showed that two of the mutations, F11W and W90Y, were destabilizing. Position 90 is both strongly correlated to several other positions and correlated at least weakly to a large number of positions. Our attempts to generate compensatory mutants for the W90Y mutation illustrate how difficult it can be to mutate highly correlated positions. All but two positions around 90 are already consensus residues, but mutation of those two positions to consensus residues, G122T and V123P, alone, in combination with W90Y, or altogether, resulted in no expression. Consequently, we suggest removal of highly correlated positions from the set of stabilizing mutations to test.
F11W represents a different and more subtle kind of coupling. Position 11 is Trp in virtually every TIM, and we initially were quite surprised that F11W was destabilizing in yeast TIM. In retrospect, it seems reasonable that in order for yeast TIM to have a mutation at the highly conserved W11 seen in most TIMs, something else might also have mutated in response. That turns out to be the case. Mutation of adjacent position 20 from the larger Ile to the smaller (consensus) Ala seen in most TIMs apparently compensates for the larger Trp in position 11; moreover, I20A alone (i.e., in the context of F11) results in no expression. The F11W I20A dual mutant is the most stable simple mutant we engineered here, which suggests that consensus mutations at the most conserved positions can have a big payoff, but with some peril. Namely, we cannot statistically detect correlations to invariant positions. If two residues are highly conserved in a protein, it is impossible to say if they are conserved together or separately, unless the single mutants reduce fitness and a double mutant rescues it. We think of this as a kind of hidden correlation that, like the statistical correlations we can detect, are best to avoid to maximize the number of stabilizing mutations.
When we look at the entire group of 23 consensus mutations made here, the fraction of mutations that stabilize versus destabilize or abrogate expression is better in the more-conserved half of positions (two thirds are stabilizing, versus half overall). There is little pattern to which mutations stabilize otherwise. Few stabilizing mutations were found in β-strands, but many stabilizing mutations were solvent exposed or in loops, where we might not expect stabilization and certainly cannot meaningfully predict it computationally. Even fairly non-conservative mutations, like Y49Q, were often stabilizing.
We analyzed the consensus mutations with the computational protein stability predictor FoldX (Table 1).9 There was essentially no correlation between the predicted ΔΔG values and T1/2 values (see Supplemental Fig. 2b). FoldX is able to identify which mutations are stabilizing (i.e., the sign of ΔΔG agrees with the sign of ΔT1/2) in about 60% of cases, but this is about the fraction of consensus mutations that are stabilizing overall. FoldX did predict large destabilizations (3–8.5 kcal mol−1) for the six mutants that did not express, which is valuable information for the protein engineer. It is important to note that while both ΔΔG values and ΔT1/2 values report relative stabilities, they are not the same thermodynamically, and the irreversible thermal denaturations here are not under equilibrium conditions. It is possible that some variants have, for example, decreased thermal stability but a greater free energy difference between the folded and unfolded states.
One interesting note about the consensus mutations explored here is that, except for the variants that do not express, all of the variants, including algoTIM with 15 mutations, have extremely high catalytic activity. None is reduced even an order of magnitude, and wild-type TIM is among the most efficient enzymes known. This is not because TIM is especially mutable. It is a highly tuned enzyme that works by exquisite positioning of catalytic residues with coordinated loop dynamics in the catalytic cycle. Harbury found that vanishingly few variants with multiple conservative mutations were active.48 Unlike most mutations designed by humans and computers, consensus mutations have been tested for fitness by nature in a variety of contexts. When we choose to replace an amino acid with the most common one in an MSA, there is greater confidence in the maintenance of function. Interestingly, Hilvert recently reported that “consensus” mutations in libraries of chorismate mutase from directed evolution were also stabilizing, but the consensus variants ranged significantly in activity (from 2-fold higher to 30-fold lower).49
Three multimutants were constructed for this study: re-S.c. TIM, comboTIM, and algoTIM. The sum of the ΔT1/2 values for the six re-S.c. TIM mutants is −0.6 °C, but re-S.c. TIM is actually destabilized about 2.1 °C. More strikingly, comboTIM is made up of the stabilized F11WI20Amutant and 12 additional mutations all found to be stabilizing. The sum of those ΔT1/2 values is +22.9 °C, but the protein is actually destabilized by 2.5 °C. In contrast, amalgamating the 15 residues suggested by our conservation–correlation algorithm results in 8.2 °C of stabilization, the most we saw in this study. This variant includes one mutation we know to be destabilizing and five that we did not test separately. We suggest that, besides helping to identify which consensus mutations will be stabilizing, removal of coupled positions also increases the additivity of the mutations. comboTIM includes several residues with below-average relative entropy that are enriched in statistical interactions to other sites (Fig. 9b and d). Although each of these mutations is stabilizing in the context of wild type, coupling among the 16 mutated positions negates additive gains in T1/2 (Fig. 9b and d). In contrast, the positions in algoTIM were selected for independence (Fig. 9a and c).
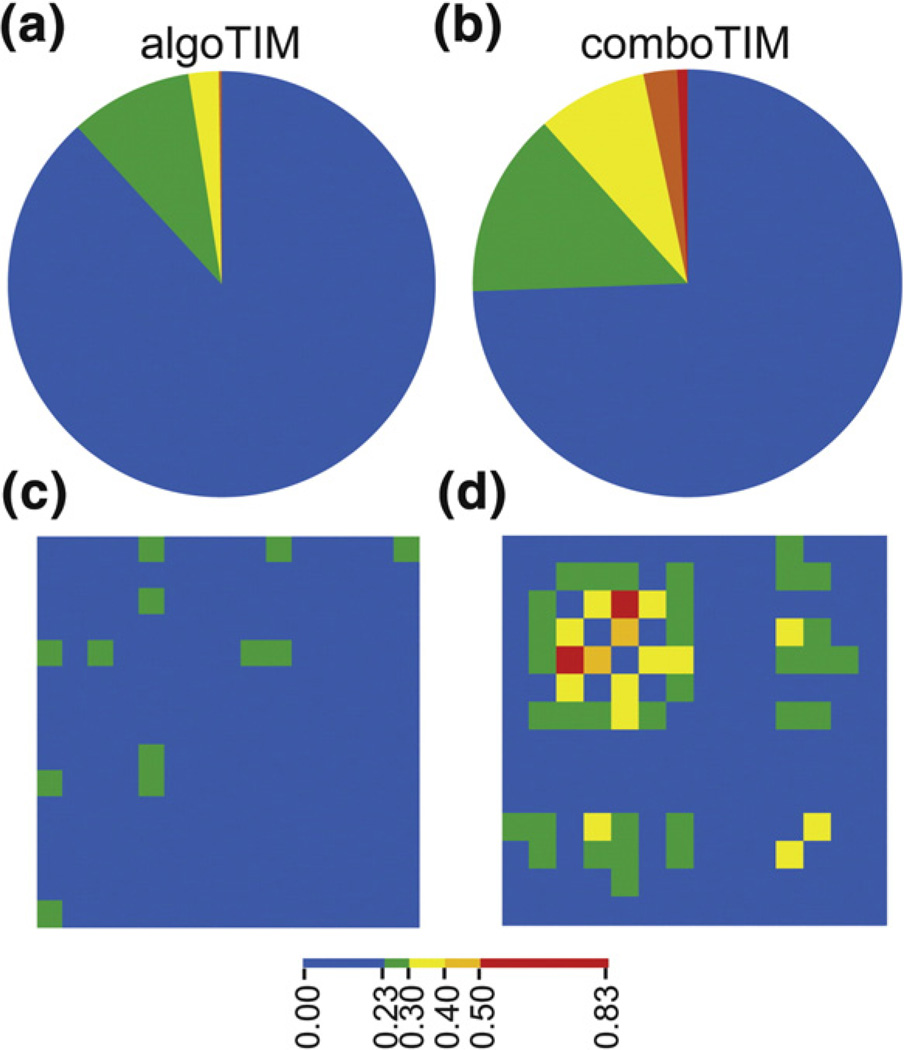
Fig. 9.
Mutual information for comboTIM and algoTIM mutation sites. The positions of mutation for comboTIM and algoTIM have been isolated from the mutual information matrix of all pairwise interactions. (a) The positions of mutation in algoTIM have virtually no strong (red, orange) correlations to other sites in the protein. (b) In contrast, the 14 positions of mutation in comboTIM have many strong correlations with other positions within TIM. (c) The 15 mutations in algoTIM are assembled into a matrix with the correlations displayed as a heat map. The positions of mutation are not correlated to each other. (d) The 14 mutations in comboTIM are assembled into a matrix with the correlations displayed as a heat map. Although these mutations were stabilizing independently, there are many strong correlations between sites of mutation in comboTIM, perhaps leading to nonadditive effects.
We examined three other studies in the literature on stabilization by consensus mutations in light of our suggested metrics. Steipe et al. made 10 consensus mutations to the Vκ domain of the McPC603 antibody.14 Based on our analysis of the current Kabat database, three of those mutations (F32Y, Q79E, and N90Q) are at positions with greater than average conservation, and two of them (32 and 90) are stabilizing. The destabilizing mutation has the highest maximum mutual information value (0.39 versus 0.23 and 0.25), but it is somewhat below the top 1 percentile of all values (0.48). The next three most conserved positions, all just below the mean relative entropy here (1.97), were all stabilizing, and only one of those had a high mutual information score (0.42). The remaining four less conserved positions resulted in three neutral and one stabilizing mutation.
We also looked at a number of mutations made to the human p53 core domain by Nikolova et al.50 Eleven of those mutations were consensus mutations based on our analysis of the p53 family from Pfam. Only two of those mutations were at positions with higher than average conservation; both had low maximal mutual information values, and both were stabilizing. The next two most conserved positions just below mean relative entropy (here, 1.91) both had low correlation scores and were both stabilizing. Of the remaining seven mutations at less conserved sites, three were stabilizing and four were destabilizing.
Watanabe et al. made 12 “ancestral” mutations to Thermus thermophilus 3-isopropylmalate dehydrogenase, of which 10 were also consensus mutations.26 Six of those mutations have higher than average conservation but all with relative entropy values below 3, and four of those were stabilizing. The four mutants at less conserved positions were neutral or destabilized. Three of the six more conserved positions have at least one significant interaction (maximum mutual information above the top 1 percentile), of which two were stabilizing and one was destabilizing. In total, our algorithm would have suggested three of these mutations for stabilization, and two of the three would have stabilized. None of these studies provides a sufficiently large number of mutations that meet the criteria of our algorithm to verify the effectiveness better than our current study, but they do suggest overall that consensus mutations at more conserved sites are more likely to be stabilizing and that removal of coupled sites discards some of the destabilizing mutations at more conserved positions.
We cannot definitively say from this work what exact quantitative standards should be applied for the conceptual filters proposed here. We chose to make a relative entropy of 3 from the yeast codon usage reference state our upper limit on conservation (for removing hidden correlations). If the most common amino acid at a position is Leu, a relative entropy of 2.27 corresponds to 99% conservation, but if the most common amino acid is Trp, it is 4.46—because Trp is used less overall, and so it would be more improbable for it to dominate a site. In practice, values above 2.5 or even 2.0 represent very highly biased positions, and it would take a much larger data set to quantitatively set this limit. Likewise, it is not clear if the mean relative entropy score is the optimal lower limit on conservation, and, further, this value might change substantially for proteins enriched in rarer amino acids, but until a much larger data set is available, the top half of conservation scores is a reasonable place to look. It is much more difficult to articulate a quantitative criterion for “too correlated.” The residues we removed here had both very high maximal mutual information values (in the top 1%) and also had a large number of significant correlations overall. It is unclear if one of these criteria is more important than the other. Again, until much more data are available, removing positions with the top 1% of mutual information scores is practical.
Another difficulty in consensus design is that there is significant bias to the sequence databases with respect to taxonomic distribution. We chose to use a database of all TIM sequences with sequence repeats and fragments removed, mainly to simplify correlation calculations. One could also eliminate not just identical sequences, but even those that were highly similar. Alternatively, one could limit the database to only those variants that are phylogenetic neighbors, such as those from all eukaryotes or all fungi in this case, so that the “average” sequence would not be too far evolved from the host sequence. For example, at the two problematic mutations F11W and W90Y, F11 is the consensus residue among the fungal variants (but not among all eukaryotic variants) and W90 is the consensus residue in both fungi and all eukaryotes. However, other stabilizing mutations are discarded using these limited databases (e.g., Q82 is most common in fungi and I109 is most common in eukaryotes). We speculate that much of the information gained from limiting the taxonomic distribution is also represented in the correlation criteria. Furthermore, it is difficult to know how much to limit the database (what “neighborhood” is appropriate). Adjusting the composition of the MSA might be a fruitful avenue for future investigation.
Finally, it is worth noting that while this work offers some explanation for why some consensus mutations are not stabilizing, it does not tell us why consensus mutations are stabilizing in general. Several groups have articulated the notion that adding consensus mutations to a protein generates a superposition of stabilizing interactions, only a fraction of which are necessary in any one protein to achieve sufficient stability for fitness. That necessarily implies that the effects of consensus mutations are mostly additive, which virtually must be true for fully consensus enzymes with sequences far from any natural variant to be stable and active, as they sometimes are. Here, we see that the consensus mutations that are most likely to stabilize are the ones that are the most independent, which is consistent with the importance of additivity for consensus stabilization. Still, given the multitude of evolutionary pressures for fitness besides adequate stability (activity, solubility, folding rate, etc.), it is remarkable that so many consensus mutations stabilize proteins.
In summary, we have demonstrated that consensus mutations at more conserved sites were more likely to stabilize yeast TIM and that removal of mutations at nearly invariant and highly correlated positions increased the likelihood of stabilization. These mutations could be amalgamated into a highly stable multimutant, probably in part because of their independence. The high activity of all resulting proteins suggests that application of this algorithm to proteins even for which little is known about the structure or mechanism is a promising way to rapidly generate stable proteins for research and applied uses. At least in the case of TIM, our method improves the identification of stabilizing mutations from ~50% to ~90%, which is of great practical use to the protein engineer.
Materials and Methods
Databases
The MSA of TIM was produced from the hidden Markov model alignment of 781 full-length, nonredundant sequences downloaded from Pfam (v22.0). All partial sequences shorter than 205 aa were first removed from the full 1239 sequence TIM alignment leaving 888 sequences. An additional 107 sequences were removed as repeats leaving the 781 studied sequences. This is the same curated TIM database we described previously.25
The full MSAs of the isocitrate/isopropylmalate dehydrogenase family (PF00180) and p53 DNA binding domain (PF00870) were downloaded from Pfam (v26.0). For the dehydrogenases, the database was curated by keeping sequences that were 300 to 400 aa long. Only the 343 positions corresponding to the 3-isopropylmalate dehydrogenase from thermos thermophiles (LEU3_THETH/2-344) were used for the analysis. The final MSA had 4710 sequences and 343 positions. For p53, the database was curated by removing all sequences shorter than 150 aa in length. Positions with less that 60% occupancy were also discarded. The final MSA had 235 sequences and 195 positions. Sequences of the light chain of the kappa class were downloaded from the Kabat sequence database KabatMan v2.29†. This database containing 3387 sequences and 123 positions was used for the consensus analysis. Positions with occupancy of less than 60% were discarded from the database, leaving an MSA with 3387 sequences and 107 positions for the correlation analysis.
Consensus calculations
The extent of conservation for each position was determined by calculating the relative entropy where yeast codon usage serves as the neutral reference state. Relative entropy is determined from Eq. (1),
| (1) |
where px is the frequency for amino acid x in a given position and fx is the frequency for amino acid x in the neutral reference state. S. cerevisiae amino acid codon usage is as follows: A—5.6%, C—1.3%, D—5.8%, E—6.5%, F—4.4%, G—5.1%, H—2.1%, I—6.5%, K—7.3%, L—9.5%, M—2.1%, N—6.1%, P—4.3%, Q—3.9%, R—4.4%, S—8.9%, T—5.9%, V—5.7%, W—1.0%, Y—3.4%.43 By this calculation, high relative entropies quantitatively describe more biased (conserved) positions. A position that resembles the neutral reference state will have a relative entropy near zero.
Correlation calculations
Mutual information was used to calculate the pairwise statistical interactions between positions in the MSA. The mutual information is determined from Eq. (2),
| (2) |
with px, frequency of amino acid residue x at position i; py, frequency of amino acid residue y at position j; and px,y, frequency of co-occurrence of amino acid residue x at position i and amino acid residue y at position j. High mutual information scores correspond to highly correlated (or anticorrelated) distributions.
Cloning
The wild-type S.c. TIM construct was previously cloned and characterized (see Supplemental Information for exact sequence, linkers, and cleavage sites).25 The consensus variant genes were assembled by PCR of S.c. TIM with overlapping primers containing the desired mutations. re- S.c. TIM was assembled from synthetic oligonucleotides via PCR reassembly.51 comboTIM and algoTIM were ordered as full-length genes from Genewiz (South Plainfield, NJ). Full-length genes were digested with restriction enzymes NcoI and BamHI before ligation into pHLIC, a T7 overexpression plasmid constructed in our laboratory. All clones were confirmed by analytical restriction digests and DNA sequencing at Genewiz.
Protein expression and purification
Vectors harboring the TIM genes were transformed into BL21(DE3) E. coli for T7 overexpression. Liquid cultures of 1 L 2× YT were grown at 37 °C and induced with 0.1 mM IPTG at OD600=~0.75. The cultures were grown at 37 °C for 3–4 h postinduction. Proteins were purified as 6× His fusions using Ni-NTA chromatography.25 The 6× His tag was freed from the N-terminus by tobacco etch virus protease yielding the native protein, and the solution was subjected to Ni-NTA chromatography again to remove the 6× His tag and 6× His-tobacco etch virus protease. Protein concentration and purity were determined by A280 and SDS-PAGE. Extinction coefficients at 280 nm were calculated using Scripps Protein Calculator v3.3 (e.g., 24,750 M−1 cm−1 for S.c. TIM).
Circular dichroism
Circular dichroism spectra and melts were obtained on a Jasco J-815 spectrometer at 14 µM protein in 100 mM potassium phosphate, pH 8, and 300 mM NaCl. Wavelength scans were collected in triplicate from 195 to 275 nm with 2-s integration time at 100 nm min−1 scanning speed. Data collected with HT voltage greater than 600 V were discarded. Thermal denaturation was monitored by observing the loss of ellipticity at 222 nm or 215 nm (see Results). Data were collected in 1 °C steps with 6-s temperature equilibration, 1 °C min−1 ramping, and 2-s integration. All scans and thermal melts were exported and plotted in Microsoft Excel 2007. The T1/2 values were calculated by fitting the equation described by Koepf52 to the data:
| (3) |
where ΔHm is the enthalpy change at the unfolding transition, T1/2 is the temperature in Kelvin at which half the protein is unfolded, T is the temperature in Kelvin, R is the universal gas constant, mn is the slope of the pretransition baseline, yn is the intercept of the pretransition baseline, md is the slope of the posttransition baseline, and yd is the intercept of the posttransition baseline.
The Jasco J-815 spectrometer also recorded the absorbance at 600 nm with increasing temperatures. The T1/2 value for each variant was calculated as the position on the curve with the greatest slope. The slopes at each temperature were calculated from a 5° window around each point.
High-throughput thermal scanning
The thermal denaturation of TIM variants was performed at 25 µM protein with 5× SYPRO Orange dye (the absolute concentration of the dye is not disclosed by Invitrogen). The melts were assayed using a Bio-Rad C1000 thermal cycler with a ramp rate of 1 °C min−1 at 0.2 °C intervals. The data were exported into Microsoft Excel 2007. The T1/2 was calculated as the temperature with the maximum slope as determined from a 5° window around each point.
Activity assay
The kinetic activities of all variants were determined by the method described by Plaut and Knowles using the background subtraction technique of John Richard.32,53 Detailed procedures are described in previously published work on TIM.25 Single-point kinetics were performed in triplicate at 4 mM L-GAP (~5× Km of S.c. TIM) and 30 pM enzyme.
FoldX calculations
The FoldX algorithm was downloaded as a YASARA add-in.54 The S. cerevisiae crystal structure, 1YPI, was “repaired” and saved before in silico mutagenesis. The stability change was calculated from the average of three runs using the FoldX parameters: pH 7, 298 K, ionic strength of 500, and van der Waals design 2. Neighboring residues were allowed to move during the energy minimization.
Structural calculations
Residue solvent exposure was calculated with MOLMOL.55 To determine the conservative nature of mutations, we used the BLOSUM62 matrix.45
Supplementary Material
Acknowledgements
B.J.S. was a National Institutes of Health Chemistry- Biology Interface Program Fellow and Ohio State Presidential Fellow. T.N. was an Ohio State Arts and Sciences College Research Scholar and Dean's Undergraduate Research Fund awardee. M.T. was a University Summer Research Intern. D.M. and S.R. were visiting summer students from Cornell University and Kenyon College, respectively. We thank Nicholas Callahan and Deepamali Perera for helpful conversations and suggestions. This work was supported by The Ohio State University and NIH grant R01 GM083114 to TJM.
Abbreviations used
- DHAP
dihydroxyacetone phosphate
- GAP
glyceraldehyde-3-phosphate
- MSA
multiple sequence alignment
- S.c. TIM
Saccharomyces cerevisiae TIM
- TIM
triosephosphate isomerase
Footnotes
Supplementary Data
Supplementary data to this article can be found online at doi:10.1016/j.jmb.2012.04.025
References
- 1.Dill KA. Dominant forces in protein folding. Biochemistry. 1990;29:7133–7155. doi: 10.1021/bi00483a001. [DOI] [PubMed] [Google Scholar]
- 2.Rose GD, Wolfenden R. Hydrogen bonding, hydrophobicity, packing, and protein folding. Annu. Rev. Biophys. Biomol. Struct. 1993;22:381–415. doi: 10.1146/annurev.bb.22.060193.002121. [DOI] [PubMed] [Google Scholar]
- 3.Joerger AC, Ang HC, Fersht AR. Structural basis for understanding oncogenic p53 mutations and designing rescue drugs. Proc. Natl Acad. Sci. USA. 2006;103:15056–15061. doi: 10.1073/pnas.0607286103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Magliery TJ, Lavinder JJ, Sullivan BJ. Protein stability by number: high-throughput and statistical approaches to one of protein science's most difficult problems. Curr. Opin. Chem. Biol. 2011;15:443–451. doi: 10.1016/j.cbpa.2011.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cordes MH, Davidson AR, Sauer RT. Sequence space, folding and protein design. Curr. Opin. Struct. Biol. 1996;6:3–10. doi: 10.1016/s0959-440x(96)80088-1. [DOI] [PubMed] [Google Scholar]
- 6.Richards FM. Protein stability: still and unsolved problem. Cell. Mol. Life Sci. 1997;53:790–802. doi: 10.1007/s000180050100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Khan S, Vihinen M. Performance of protein stability predictors. Hum. Mutat. 2010;31:675–684. doi: 10.1002/humu.21242. [DOI] [PubMed] [Google Scholar]
- 8.Zhou H, Zhang C, Liu S, Zhou Y. Web-based toolkits for topology prediction of transmembrane helical proteins, fold recognition, structure and binding scoring, folding-kinetics analysis and comparative analysis of domain combinations. Nucleic Acids Res. 2005;33:W193–W197. doi: 10.1093/nar/gki360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schymkowitz J, Borg J, Stricher F, Nys R, Rousseau F, Serrano L. The FoldX web server: an online force field. Nucleic Acids Res. 2005;33:W382–W388. doi: 10.1093/nar/gki387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lavinder JJ, Hari SB, Sullivan BJ, Magliery TJ. High-throughput thermal scanning: a general, rapid dye-binding thermal shift screen for protein engineering. J. Am. Chem. Soc. 2009;131:3794–3795. doi: 10.1021/ja8049063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Arnold FH. Combinatorial and computational challenges for biocatalyst design. Nature. 2001;409:253–257. doi: 10.1038/35051731. [DOI] [PubMed] [Google Scholar]
- 12.Edgell MH, Sims DA, Pielak GJ, Yi F. High-precision, high-throughput stability determinations facilitated by robotics and a semiautomated titrating fluorometer. Biochemistry. 2003;42:7587–7593. doi: 10.1021/bi034063g. [DOI] [PubMed] [Google Scholar]
- 13.Aucamp JP, Cosme AM, Lye GJ, Dalby PA. High-throughput measurement of protein stability in microtiter plates. Biotechnol. Bioeng. 2005;89:599–607. doi: 10.1002/bit.20397. [DOI] [PubMed] [Google Scholar]
- 14.Steipe B, Schiller B, Pluckthun A, Steinbacher S. Sequence statistics reliably predict stabilizing mutations in a protein domain. J. Mol. Biol. 1994;240:188–192. doi: 10.1006/jmbi.1994.1434. [DOI] [PubMed] [Google Scholar]
- 15.Ohage E, Steipe B. Intrabody construction and expressionIThe critical role of VL domain stability. J. Mol. Biol. 1999;291:1119–1128. doi: 10.1006/jmbi.1999.3019. [DOI] [PubMed] [Google Scholar]
- 16.Knappik A, Ge L, Honegger A, Pack P, Fischer M, Wellnhofer G, et al. Fully synthetic human combinatorial antibody libraries (HuCAL) based on modular consensus frameworks and CDRs randomized with trinucleotides. J. Mol. Biol. 2000;296:57–86. doi: 10.1006/jmbi.1999.3444. [DOI] [PubMed] [Google Scholar]
- 17.Godoy-Ruiz R, Perez-Jimenez R, Ibarra-Molero B, Sanchez-Ruiz JM. A stability pattern of protein hydrophobic mutations that reflects evolutionary structural optimization. Biophys. J. 2005;89:3320–3331. doi: 10.1529/biophysj.105.067025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pey AL, Rodriguez-Larrea D, Bomke S, Dammers S, Godoy-Ruiz R, Garcia-Mira MM, Sanchez-Ruiz JM. Engineering proteins with tunable thermodynamic and kinetic stabilities. Proteins. 2008;71:165–174. doi: 10.1002/prot.21670. [DOI] [PubMed] [Google Scholar]
- 19.Main ER, Xiong Y, Cocco MJ, D'Andrea L, Regan L. Design of stable alpha-helical arrays from an idealized TPR motif. Structure. 2003;11:497–508. doi: 10.1016/s0969-2126(03)00076-5. [DOI] [PubMed] [Google Scholar]
- 20.Mosavi LK, Minor DL, Jr, Peng ZY. Consensus-derived structural determinants of the ankyrin repeat motif. Proc. Natl Acad. Sci. USA. 2002;99:16029–16034. doi: 10.1073/pnas.252537899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Binz HK, Stumpp MT, Forrer P, Amstutz P, Pluckthun A. Designing repeat proteins: well-expressed, soluble and stable proteins from combinatorial libraries of consensus ankyrin repeat proteins. J. Mol. Biol. 2003;332:489–503. doi: 10.1016/s0022-2836(03)00896-9. [DOI] [PubMed] [Google Scholar]
- 22.Lehmann M, Kostrewa D, Wyss M, Brugger R, D'Arcy A, Pasamontes L, van Loon AP. From DNA sequence to improved functionality: using protein sequence comparisons to rapidly design a thermostable consensus phytase. Protein Eng. 2000;13:49–57. doi: 10.1093/protein/13.1.49. [DOI] [PubMed] [Google Scholar]
- 23.Lehmann M, Loch C, Middendorf A, Studer D, Lassen SF, Pasamontes L, et al. The consensus concept for thermostability engineering of proteins: further proof of concept. Protein Eng. 2002;15:403–411. doi: 10.1093/protein/15.5.403. [DOI] [PubMed] [Google Scholar]
- 24.Lehmann M, Pasamontes L, Lassen SF, Wyss M. The consensus concept for thermostability engineering of proteins. Biochim. Biophys. Acta. 2000;1543:408–415. doi: 10.1016/s0167-4838(00)00238-7. [DOI] [PubMed] [Google Scholar]
- 25.Sullivan BJ, Durani V, Magliery TJ. Triosephosphate isomerase by consensus design: dramatic differences in physical properties and activity of related variants. J. Mol. Biol. 2011;413:195–208. doi: 10.1016/j.jmb.2011.08.001. [DOI] [PubMed] [Google Scholar]
- 26.Watanabe K, Ohkuri T, Yokobori S, Yamagishi A. Designing thermostable proteins: ancestral mutants of 3-isopropylmalate dehydrogenase designed by using a phylogenetic tree. J. Mol. Biol. 2006;355:664–674. doi: 10.1016/j.jmb.2005.10.011. [DOI] [PubMed] [Google Scholar]
- 27.Miyazaki J, Nakaya S, Suzuki T, Tamakoshi M, Oshima T, Yamagishi A. Ancestral residues stabilizing 3-isopropylmalate dehydrogenase of an extreme thermophile: experimental evidence supporting the thermophilic common ancestor hypothesis. J. Biochem. 2001;129:777–782. doi: 10.1093/oxfordjournals.jbchem.a002919. [DOI] [PubMed] [Google Scholar]
- 28.Shimizu H, Yokobori S, Ohkuri T, Yokogawa T, Nishikawa K, Yamagishi A. Extremely thermophilic translation system in the common ancestor commonote: ancestral mutants of glycyl-tRNA synthetase from the extreme thermophile Thermus thermophilus. J. Mol. Biol. 2007;369:1060–1069. doi: 10.1016/j.jmb.2007.04.001. [DOI] [PubMed] [Google Scholar]
- 29.Yamashiro K, Yokobori S, Koikeda S, Yamagishi A. Improvement of Bacillus circulans beta-amylase activity attained using the ancestral mutation method. Protein Eng. Des. Sel. 2010;23:519–528. doi: 10.1093/protein/gzq021. [DOI] [PubMed] [Google Scholar]
- 30.Alber T, Banner DW, Bloomer AC, Petsko GA, Phillips D, Rivers PS, Wilson IA. On the three-dimensional structure and catalytic mechanism of triose phosphate isomerase. Philos. Trans. R. Soc. Lond., B. 1981;293:159–171. doi: 10.1098/rstb.1981.0069. [DOI] [PubMed] [Google Scholar]
- 31.Nagano N, Orengo CA, Thornton JM. One fold with many functions: the evolutionary relationships between TIM barrel families based on their sequences, structures and functions. J. Mol. Biol. 2002;321:741–765. doi: 10.1016/s0022-2836(02)00649-6. [DOI] [PubMed] [Google Scholar]
- 32.Plaut B, Knowles JR. pH-dependence of the triose phosphate isomerase reaction. Biochem. J. 1972;129:311–320. doi: 10.1042/bj1290311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Noble ME, Zeelen JP, Wierenga RK, Mainfroid V, Goraj K, Gohimont AC, Martial JA. Structure of triosephosphate isomerase from Escherichia coli determined at 2.6 Å resolution. Acta Crystallogr. Sect. D: Biol. Crystallogr. 1993;49:403–417. doi: 10.1107/S0907444993002628. [DOI] [PubMed] [Google Scholar]
- 34.Mande SC, Mainfroid V, Kalk KH, Goraj K, Martial JA, Hol WG. Crystal structure of recombinant human triosephosphate isomerase at 2.8 Å resolution. Triosephosphate isomerase-related human genetic disorders and comparison with the trypanosomal enzyme. Protein Sci. 1994;3:810–821. doi: 10.1002/pro.5560030510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lolis E, Alber T, Davenport RC, Rose D, Hartman FC, Petsko GA. Structure of yeast triosephosphate isomerase at 1.9-Å resolution. Biochemistry. 1990;29:6609–6618. doi: 10.1021/bi00480a009. [DOI] [PubMed] [Google Scholar]
- 36.Wierenga RK, Kalk KH, Hol WG. Structure determination of the glycosomal triosephosphate isomerase from Trypanosoma brucei brucei at 2.4 Å resolution. J. Mol. Biol. 1987;198:109–121. doi: 10.1016/0022-2836(87)90461-x. [DOI] [PubMed] [Google Scholar]
- 37.Lambeir AM, Opperdoes FR, Wierenga RK. Kinetic properties of triose-phosphate isomerase from Trypanosoma brucei brucei. A comparison with the rabbit muscle and yeast enzymes. Eur. J. Biochem. 1987;168:69–74. doi: 10.1111/j.1432-1033.1987.tb13388.x. [DOI] [PubMed] [Google Scholar]
- 38.Dabrowska A, Kamrowska I, Baranowski T. Purification, crystallization and properties of triosephosphate isomerase from human skeletal muscle. Acta Biochim. Pol. 1978;25:247–256. [PubMed] [Google Scholar]
- 39.Gerlt JA, Raushel FM. Evolution of function in (beta/alpha)8-barrel enzymes. Curr. Opin. Chem. Biol. 2003;7:252–264. doi: 10.1016/s1367-5931(03)00019-x. [DOI] [PubMed] [Google Scholar]
- 40.Silverman JA, Balakrishnan R, Harbury PB. Reverse engineering the (beta/alpha)8 barrel fold. Proc. Natl Acad. Sci. USA. 2001;98:3092–3097. doi: 10.1073/pnas.041613598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cover TM, Thomas JA. Elements of Information Theory. 2nd edit. Hoboken, NJ: John Wiley & Sons, Inc.; 2006. [Google Scholar]
- 42.Magliery TJ, Regan L. Sequence variation in ligand binding sites in proteins. BMC Bioinformatics. 2005;6:240. doi: 10.1186/1471-2105-6-240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kanehisa M, Goto S, Kawashima S, Nakaya A. The KEGG databases at GenomeNet. Nucleic Acids Res. 2002;30:42–46. doi: 10.1093/nar/30.1.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Senisterra GA, Finerty PJ., Jr High throughput methods of assessing protein stability and aggregation. Mol. Biosyst. 2009;5:217–223. doi: 10.1039/b814377c. [DOI] [PubMed] [Google Scholar]
- 45.Henikoff S, Henikoff JG. Amino acid substitution matrices from protein blocks. Proc. Natl Acad. Sci. USA. 1992;89:10915–10919. doi: 10.1073/pnas.89.22.10915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Russ WP, Lowery DM, Mishra P, Yaffe MB, Ranganathan R. Natural-like function in artificial WW domains. Nature. 2005;437:579–583. doi: 10.1038/nature03990. [DOI] [PubMed] [Google Scholar]
- 47.Socolich M, Lockless SW, Russ WP, Lee H, Gardner KH, Ranganathan R. Evolutionary information for specifying a protein fold. Nature. 2005;437:512–518. doi: 10.1038/nature03991. [DOI] [PubMed] [Google Scholar]
- 48.Silverman JA, Balakrishnan R, Harbury PB. Reverse engineering the (beta/alpha )8 barrel fold. Proc. Natl Acad. Sci. USA. 2001;98:3092–3097. doi: 10.1073/pnas.041613598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jackel C, Bloom JD, Kast P, Arnold FH, Hilvert D. Consensus protein design without phylogenetic bias. J. Mol. Biol. 2010;399:541–546. doi: 10.1016/j.jmb.2010.04.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nikolova PV, Henckel J, Lane DP, Fersht AR. Semirational design of active tumor suppressor p53 DNA binding domain with enhanced stability. Proc. Natl Acad. Sci. USA. 1998;95:14675–14680. doi: 10.1073/pnas.95.25.14675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stemmer WP, Crameri A, Ha KD, Brennan TM, Heyneker HL. Single-step assembly of a gene and entire plasmid from large numbers of oligodeoxyribonucleotides. Gene. 1995;164:49–53. doi: 10.1016/0378-1119(95)00511-4. [DOI] [PubMed] [Google Scholar]
- 52.Koepf EK, Petrassi HM, Sudol M, Kelly JW. WW: an isolated three-stranded antiparallel beta-sheet domain that unfolds and refolds reversibly; evidence for a structured hydrophobic cluster in urea and GdnHCl and a disordered thermal unfolded state. Protein Sci. 1999;8:841–853. doi: 10.1110/ps.8.4.841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Go MK, Koudelka A, Amyes TL, Richard JP. Role of Lys-12 in catalysis by triosephosphate isomerase: a two-part substrate approach. Biochemistry. 2010;49:5377–5389. doi: 10.1021/bi100538b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Van Durme J, Delgado J, Stricher F, Serrano L, Schymkowitz J, Rousseau F. A graphical interface for the FoldX forcefield. Bioinformatics. 2011;27:1711–1712. doi: 10.1093/bioinformatics/btr254. [DOI] [PubMed] [Google Scholar]
- 55.Koradi R, Billeter M, Wuthrich K. MOLMOL: a program for display and analysis of macromolecular structures. J. Mol. Graphics. 1996;14:51–55. 29–32. doi: 10.1016/0263-7855(96)00009-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.