Abstract
microRNAs (miRNAs) and small interfering RNAs from plants are 2′-O-methylated at their 3′ termini. Although miRNAs from animals are not methylated, two studies show that the recently identified mammalian Piwi-interacting RNAs carry a 2′-O-methyl group on the 3′ terminal ribose.
Small RNAs of 20–30 nucleotides (nt) have emerged as universal regulators of gene expression and genome stability in fungi, plants and animals. The three currently known types of small RNAs — miRNAs, small interfering RNAs (siRNAs) and Piwi-interacting RNAs (piRNAs) — are unified by their common structural features, such as 5′-phosphate and 3′-hydroxyl groups, and their interaction with Argonaute proteins. On pages 347 and 349 of this issue, studies from two groups reveal 2′-O-methylation of piRNAs1,2, a modification that has implications for the metabolism of this type of small RNA.
miRNAs and siRNAs are 21- to 24-nt RNAs derived from hairpin RNAs and long double-stranded RNAs, respectively3,4. Dicer processes the precursor RNAs into small-RNA duplexes5. One strand of the small-RNA duplex is incorporated into a protein complex containing an Argonaute protein and guides the Argonaute-mediated endonucleolytic cleavage of target messenger RNAs during RNA interference5,6. miRNAs are also known to cause translational inhibition of their target mRNAs. Endogenous siRNAs from Schizosaccharomyces pombe and plants can guide histone H3 Lys9 methylation and DNA methylation to confer transcriptional regulation7–9.
piRNAs, 26- to 30-nt RNAs found in spermatogenic cells in mammals, constitute a distinct class of small RNAs10–14. The overwhelming enrichment for uridine as the 5′ nucleotide and the clustering of numerous piRNAs along one strand at some loci are evidence against a biogenesis pathway involving Dicer. Whereas miRNAs and siRNAs are associated with the Argonaute subfamily of Argonaute proteins, piRNAs are bound by the Piwi subfamily of proteins. piRNAs seem to share features with the Drosophila melanogaster germline-specific repeat-associated small interfering RNAs (rasiRNAs), which ensure genome stability by silencing transposons15. Like piRNAs, rasiRNAs are longer (24–29 nt) than miRNAs and siRNAs, are produced in a Dicer-independent manner and are bound by the Piwi subfamily of Argonaute proteins. A recent study uncovered an intriguing aspect of rasiRNA biogenesis: an existing rasiRNA leads to the production of a second rasiRNA from a target RNA through the slicer activity of an interacting protein of the Piwi subfamily of Argonautes16. The cleavage event defines the 5′ end of the new rasiRNA, but how the 3′ end is defined still remains to be determined.
What we do know about the 3′ ends of rasiRNAs is that the terminal ribose is modified. The 3′ terminal modification was inferred from the observation that Drosophila rasiRNAs are resistant to sodium periodate treatment followed by β-elimination15. Such chemical treatments result in an RNA shortened by one nucleotide. Now, two groups have demonstrated for the first time that piRNAs isolated from mouse testis, like Drosophila rasiRNAs, are resistant to such chemical treatments1,2. This suggests that piRNAs are also modified at their 3′ termini. The two groups defined the nature of this modification using different technical approaches. Kirino and Mourelatos1 isolated bulk piRNAs from mouse testes, ligated them to radioactive pCp at their 3′ termini and digested them with RNase T2. Two-dimensional thin layer chromatography of the radioactive products showed a migration pattern that perfectly matched 2′-O-methylated dinucleotides with a terminal cytidine. The presence of the dinucleotides was probably due to the inhibitory effect of the methyl group toward RNase T2. Ohara et al.2 digested bulk piRNAs isolated from mouse testes with RNase T2 and analyzed the resulting nucleosides by liquid chromatography and mass spectrometry (LC/MS). As only the 3′-terminal nucleotides were converted to nucleosides by RNase T2 (and all others to di- or monophosphonucleotides), the nucleosides represented the 3′ termini of piRNAs. LC revealed traces for all four 2′-O-methylnucleosides, three of which were confirmed by MS. Collision-induced dissociation confirmed that the methyl group was on the ribose. Although the studies from both groups could have benefited from the use of 3′-O-methylnucleoside or 3′-O-methylnucleotide standards to exclude the possibility of 3′-O-methylation, it is very likely that the methylation of piRNAs occurs on the 2′-hydroxyl of the 3′-terminal nucleotide.
How are piRNAs 2′-O-methylated? At present, we can only speculate on the basis of our knowledge of 2′-O-methylation of other types of RNAs. In plants, miRNAs and siRNAs carry a 3′-terminal 2′-O-methyl group introduced by the methyltransferase protein HEN1 (refs. 17,18). HEN1 is a 942-residue protein with an N-terminal double-stranded RNA–recognition motif (dsRM) and a C-terminal methyltransferase domain (Fig. 1a). HEN1 is highly specific for small-RNA duplexes. It is unable to act on single-stranded small RNAs and requires free 2′- and 3′-hydroxyl groups on the 3′-terminal nucleotide and a 2-nt 3′ overhang; both of these are characteristic of Dicer products (Fig. 1b). Intriguingly, proteins that show approximately 40%–50% similarity to a C-terminal region in HEN1, but lack the dsRM, are present in animal genomes (Fig. 1a). One model of piRNA methylation is that single-stranded piRNAs are methylated by HEN1 homologs after the 3′ ends are generated (Fig. 2a). The lack of the dsRM in the HEN1 homologs is consistent with the Dicer-independent biogenesis of rasiRNAs and piRNAs.
Figure 1. miRNAs and siRNAs are methylated by HEN1 in Arabidopsis thaliana.
(a) HEN1 homologs in other organisms (NCBI Protein accession codes indicated). HEN1 and its homolog in rice are similar in size and in the entire length of their sequences. The animal homologs are similar to only the C-terminal region (oval) of HEN1. This region shows sequence similarity to methyltransferases. (b) Methylation as an intermediate step in small-RNA biogenesis in Arabidopsis. The precursors to miRNAs and siRNAs are processed by Dicer-like (DCL) proteins into duplexes of small RNAs, which serve as the substrates of the methyltransferase HEN1. The methylated small RNAs are then incorporated into a protein complex containing an Argonaute protein.
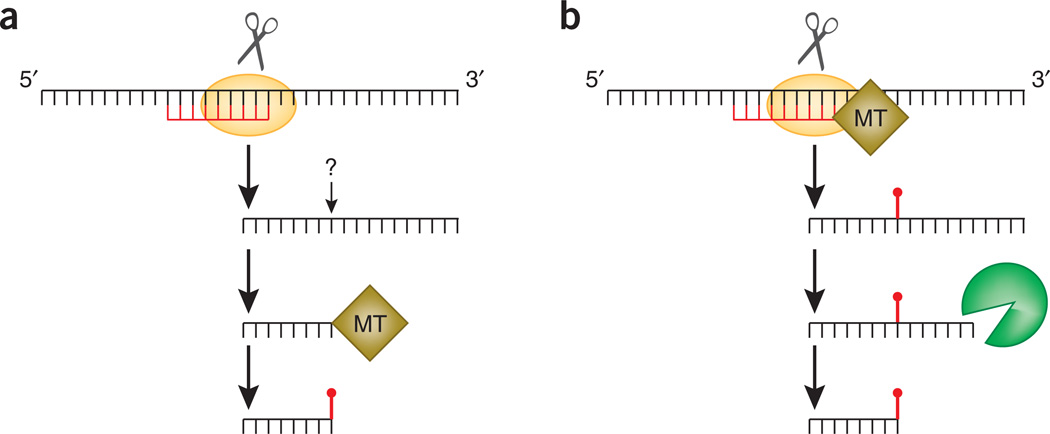
Figure 2. Two models of rasiRNA and piRNA methylation.
(a) In model 1, a rasiRNA guides the cleavage of a target RNA to define the 5′ end of a future rasiRNA. The 3′ end of the rasiRNA is generated by an unknown mechanism. The single-stranded rasiRNA is then methylated by a methyltransferase (MT). (b) In model 2, a rasiRNA guides the cleavage of a target RNA to define the 5′ end of a future rasiRNA and recruits an MT to methylate the target RNA at a nucleotide a certain distance away. The methyl group defines the 3′ end of the future rasiRNA.
Another model of 2′-O-methylation of piRNAs can be proposed on the basis of what is known about 2′-O-methylation of ribosomal RNAs. In eukaryotes, rRNAs harbor tens of internal 2′-O-methylated nucleotides. The methylation at most of the sites is guided by small nucleolar RNAs (snoRNAs)19. In addition to their conserved box C and D motifs, snoRNAs harbor a 10- to 21-nt antisense element that base-pairs perfectly with rRNA sequences. A methyltransferase is guided to a region of an rRNA by a snoRNA to methylate the rRNA at the nucleotide positioned 5 nt upstream of the D box of the snoRNA. Given that methylation of an internal nucleotide of an RNA can be guided by another RNA in trans, another model of piRNA and rasiRNA methylation can be envisioned by incorporating recent findings by Gunawardane et al.16. In this model (Fig. 2b), the binding of an existing rasiRNA to a target RNA not only induces the cleavage of the target RNA within the complementary region (to define the 5′ end of the new rasiRNA) but also recruits a methyltransferase to methylate the target RNA at a nucleotide positioned 16–20 nt away from the 5′ end of the complementary rasiRNA. This methyl group would define the 3′ end of the future rasiRNA by impeding the activity of a 3′→5′ exonuclease that acts on the precursor.
What might be the function of piRNA methylation? If the second model (Fig. 2b) of piRNA methylation is correct, methylation has an active role in defining the 3′ ends of piRNAs. In addition, the 2′-O-methyl group on mature piRNAs could serve as a mark to discourage binding to the Argonaute subfamily of proteins and encourage binding to the Piwi subfamily of proteins. It is known that a terminal 2′-O-methyl group in an siRNA reduces the efficiency of binding to the PAZ domain of Argonaute proteins20. In plants, the lack of miRNA methylation in hen1 mutants results in exonucleolytic degradation of miRNAs and in the addition of short uridine-rich tails to the 3′ ends of miRNAs21. It is conceivable that 2′-O-methylation of piRNAs protects the terminal 3′-hydroxyl group from undesirable enzymatic activities.
The finding that piRNAs are 2′-O-methylated adds another type of RNA to the repertoire of 2′-O-methylated RNAs. Future research on how, where and why piRNAs are methylated will enrich our understanding of small-RNA biogenesis and function.
Footnotes
COMPETING INTERESTS STATEMENT
The author declares no competing financial interests.
References
- 1.Kirino Y, Mourelatos Z. Nat. Struct. Mol. Biol. 2007;14:347–348. doi: 10.1038/nsmb1218. [DOI] [PubMed] [Google Scholar]
- 2.Ohara T, et al. Nat. Struct. Mol. Biol. 2007;14:349–350. doi: 10.1038/nsmb1220. [DOI] [PubMed] [Google Scholar]
- 3.Du T, Zamore PD. Development. 2005;132:4645–4652. doi: 10.1242/dev.02070. [DOI] [PubMed] [Google Scholar]
- 4.Kim VN. Nat. Rev. Mol. Cell Biol. 2005;6:376–385. doi: 10.1038/nrm1644. [DOI] [PubMed] [Google Scholar]
- 5.Hammond SM. FEBS Lett. 2005;579:5822–5829. doi: 10.1016/j.febslet.2005.08.079. [DOI] [PubMed] [Google Scholar]
- 6.Tolia NH, Joshua-Tor L. Nat. Chem. Biol. 2007;3:36–43. doi: 10.1038/nchembio848. [DOI] [PubMed] [Google Scholar]
- 7.Hamilton A, Voinnet O, Chappell L, Baulcombe D. EMBO J. 2002;21:4671–4679. doi: 10.1093/emboj/cdf464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Volpe TA, et al. Science. 2002;297:1833–1837. doi: 10.1126/science.1074973. [DOI] [PubMed] [Google Scholar]
- 9.Xie Z, et al. PLoS Biol. 2004;2:E104. doi: 10.1371/journal.pbio.0020104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aravin A, et al. Nature. 2006;442:203–207. doi: 10.1038/nature04916. [DOI] [PubMed] [Google Scholar]
- 11.Girard A, Sachidanandam R, Hannon GJ, Carmell MA. Nature. 2006;442:199–202. doi: 10.1038/nature04917. [DOI] [PubMed] [Google Scholar]
- 12.Grivna ST, Beyret E, Wang Z, Lin H. Genes Dev. 2006;20:1709–1714. doi: 10.1101/gad.1434406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lau NC, et al. Science. 2006;313:363–367. doi: 10.1126/science.1130164. [DOI] [PubMed] [Google Scholar]
- 14.Watanabe T, et al. Genes Dev. 2006;20:1732–1743. doi: 10.1101/gad.1425706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vagin VV, et al. Science. 2006;313:320–324. doi: 10.1126/science.1129333. [DOI] [PubMed] [Google Scholar]
- 16.Gunawardane LS, et al. Science. 2007;315:1587–1590. doi: 10.1126/science.1140494. [DOI] [PubMed] [Google Scholar]
- 17.Yang Z, Ebright YW, Yu B, Chen X. Nucleic Acids Res. 2006;34:667–675. doi: 10.1093/nar/gkj474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yu B, et al. Science. 2005;307:932–935. doi: 10.1126/science.1107130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kiss T. EMBO J. 2001;20:3617–3622. doi: 10.1093/emboj/20.14.3617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ma JB, Ye K, Patel DJ. Nature. 2004;429:318–322. doi: 10.1038/nature02519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li J, Yang Z, Yu B, Liu J, Chen X. Curr. Biol. 2005;15:1501–1507. doi: 10.1016/j.cub.2005.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]