Abstract
Introduction
Glioblastoma multiforme (GBM; World Health Organization astrocytoma grade IV) is the most frequent and most malignant primary brain tumor in adults. Despite multimodal therapy, all such tumors practically recur during the course of therapy, causing a median survival of only 14.6 months in patients with newly diagnosed GBM. The present study was aimed at examining the expression of the DNA repair protein AlkB homolog 2 (ALKBH2) in human GBM and determining whether it could promote resistance to temozolomide chemotherapy.
Methods
ALKBH2 expression in GBM cell lines and in human GBM was determined by quantitative real-time PCR (qRT-PCR) and gene expression analysis, respectively. Drug sensitivity was assessed in GBM cells overexpressing ALKBH2 and in cells in which ALKBH2 expression was silenced by small-interfering (si)RNA. ALKBH2 expression following activation of the p53 pathway was examined by western blotting and qRT-PCR.
Results
ALKBH2 was abundantly expressed in established GBM cell lines and human GBM, and temozolomide exposure increased cellular ALKBH2 expression levels. Overexpression of ALKBH2 in the U87 and U251 GBM cell lines enhanced resistance to the methylating agents temozolomide and methyl methanesulfonate but not to the nonmethylating agent doxorubicin. Conversely, siRNA-mediated knockdown of ALKBH2 increased sensitivity of GBM cells to temozolomide and methyl methanesulfonate but not to doxorubicin or cisplatin. Nongenotoxic activation of the p53 pathway by the selective murine double minute 2 antagonist nutlin-3 caused a significant decrease in cellular ALKBH2 transcription levels.
Conclusion
Our findings identify ALKBH2 as a novel mediator of temozolomide resistance in human GBM cells. Furthermore, we place ALKBH2 into a new cellular context by showing its regulation by the p53 pathway.
Keywords: DNA repair, glioblastoma, ALKBH2, p53, temozolomide
Glioblastoma multiforme (GBM; World Health Organization astrocytoma grade IV) is the most common primary brain tumor in adults. It represents a highly invasive, proliferative, and aggressive cancer type for which no cure is presently available. Resistance to postoperative radio- and chemotherapy is the main cause of treatment failure in patients with GBM. In patients with newly diagnosed disease, the addition of concomitant and adjuvant temozolomide chemotherapy to postoperative fractionated radiotherapy has recently been shown to increase median survival as well as 2- and 5-year survival rates compared with radiotherapy alone.1,2 During the course of therapy, however, radio- and chemoresistance typically become evident through local tumor recurrence. As a result, only 1 in 4 GBM patients survives 2 years from diagnosis despite aggressive multimodal treatment.1 To improve this poor prognosis, there is a critical need to uncover the molecular basis for the low sensitivity of GBM to chemotherapy.
Temozolomide is a prodrug spontaneously converted at physiological pH to its active metabolite 5-(3-methyltriazen-1-yl)imidazole-4-carboxamide, which creates cytotoxic methyl adducts in nitrogen and oxygen atoms in DNA bases.3 However, several independent DNA repair mechanisms, which normally safeguard genome integrity, can facilitate drug resistance and cancer cell survival by removing chemotherapy-induced DNA adducts. For instance, O6-methylguanine adducts are substrates for the ubiquitous DNA repair protein O6-methylguanine-DNA methyltransferase (MGMT), whereas the frequent N3-methyladenine and N7-methylguanine adducts are repaired by the base excision repair system in cooperation with poly(ADP-ribose) polymerase–1.4,5 Thus, the anticancer efficacy of temozolomide is highly influenced by cellular DNA repair capacity. Antagonizing specific DNA repair proteins in GBM may prove a promising therapeutic strategy to increase temozolomide cytotoxicity and prolong patient survival.
The DNA repair protein human AlkB homolog 2 (ALKBH2) is an oxidative demethylase capable of directly reversing the N1-methyladenine and N3-methylcytosine base lesions in double- and single-stranded DNA.6 Although these lesions are considered weakly mutagenic, they are thought to be highly cytotoxic due to their ability to disturb DNA replication. ALKBH2 is solely expressed in the cell nucleus, where it localizes mainly to replication foci during the S phase of the cell cycle through interaction with proliferating cell nuclear antigen (PCNA) via an ALKBH2 PCNA-interacting motif.6,7 It has therefore been suggested that ALKBH2 may have a role in DNA repair close to the replication fork.8 Embryonal fibroblasts from gene-targeted mice deficient in ALKBH2 have been shown to be incapable of repairing N1-methyladenine in DNA after methyl methanesulfonate treatment.9 As a result, ALKBH2-deficient cells were more sensitive to methyl methansesulfonate exposure than wild type (wt) cells, indicating that ALKBH2 has a functionally important role in the cellular defense against methylating agents. Since N1-methyladenine and N3-methylcytosine are thought to be induced predominantly during the S phase of the cell cycle, the cytotoxic effect of these lesions could be expected to be substantial if left unrepaired in rapid proliferating cells such as cancer cells. However, it is currently unknown to what extent ALKBH2 is expressed in GBM cells and human GBM and whether it can promote resistance to the clinically potent drug temozolomide.
In this work, we report that ALKBH2 is highly expressed in GBM cells and human GBM compared with nontumoral human brain (NHB) and that temozolomide exposure may further increase transcription levels of ALKBH2. Moreover, we show that overexpression of ALKBH2 enhances cellular resistance to temozolomide and methyl methanesulfonate but has no effect on cellular sensitivity to nonmethylating agents. We show that silencing of ALKBH2 activity increases cellular sensitivity to both temozolomide and methyl methanesulfonate, suggesting that a high level of ALKBH2 expression could be an important mediator of chemoresistance in human GBM. We also show that nongenotoxic activation of the p53 pathway leads to a substantial downregulation of ALKBH2 at both mRNA and protein levels in GBM cells.
Materials and Methods
Cell Culture and Reagents
The human GBM cell lines A172 (Massachusetts Institute of Technology), CCF-STTG1 (Cleveland Clinic Foundation), D37 (Duke University Medical Center), GaMg (University of Bergen), HF66 (Henry Ford Hospital), LN18, LN229 (Centre Hospitalier Universitaire Vaudois, Switzerland), T98 (Stanford University), U87, U118, and U251 (University of Uppsala) were cultured in Dulbecco's modified Eagle's medium (Sigma) supplemented with 10% fetal bovine serum, l-glutamine, nonessential amino acids, and antibiotics (100 U/mL penicillin, 100 U/mL streptomycin, and 5 × 10–3 mg/mL plasmocin). All cell lines were maintained at 37°C in a 5% CO2-humidified atmosphere. The authentication of the cell lines was confirmed by short tandem repeat genomic profiling. Methyl methanesulfonate was purchased from Sigma. Temozolomide (Tocris Bioscience) was dissolved in dimethyl sulfoxide (DMSO) to obtain a 100 mM stock solution. Doxorubicin (Pharmacia & Upjohn) was dissolved in sterile water to obtain a 1 mM stock solution. The proteasome inhibitor MG132 (Calbiochem) and the murine double minute 2 (MDM2) antagonist nutlin-3 (Calbiochem) were dissolved in DMSO to obtain 42 mM stock solutions. All stock solutions were kept as small aliquots in a freezer at –20°C and further diluted in growth medium before addition to each cell culture experiment.
Total RNA Extraction and Quantitative Real-time PCR
Total RNA was isolated using the RNeasy Mini Kit (Qiagen). Human brain reference total RNA was purchased from Ambion. Synthesis of cDNA and quantitative real-time PCR (qRT-PCR) were performed as described previously.10 The following primer pairs were used: ALKBH2, forward 5′-GCTGGGCTGACCTACACATT-3′ and reverse 5′-TGCCGGAAGACAAAGTCTCT-3′; MGMT, forward 5′-CTGGAGCTGTCTGGTTGTGA-3′ and reverse 5′-CTGGTGAACGACTCTTGCTG-3′; 18S, forward 5′-CGGCTACCACATCCAAGGAA-3′ and reverse 5′-GCTGGAATTACCGCGGCT-3′.
Generation of Stable ALKBH2-Overexpressing GBM Cell Lines
ALKBH2 cDNA expressing pDEST26 vector was purchased from imaGenes. Plasmid DNA was isolated with the Wizard Plus SV Minipreps DNA Purification System (Promega) as instructed by the manufacturer. We transfected 1 × 105 GBM cells with ∼2 µg plasmid DNA in a 6-well plate using FuGENE HD transfection reagent (Roche Applied Science) according to the manufacturer's instructions. Antibiotic selection of transfected cells was performed with 300 µg/mL geneticin (Invitrogen). Geneticin-resistant colonies were subsequently screened for established ALKBH2 overexpression by western blot.
Protein Extraction and Western Blot Analysis
Cells were rinsed in phosphate buffered saline, lysed in a buffer (pH 7.2) containing 20 mM 3-(N-morpholino)propanesulfonic acid, 5 mM EDTA, 2 mM ethylene glycol tetraacetic acid, 30 mM NaF, 0.5% Triton-X, 40 mM β-glycerophosphate, 20 mM Na-pyrophosphate, 1 mM Na-orthovanadate, 3 mM benzamidine, 5 µM pepstatin, 10 µM leupeptin, and 1 mM phenylmethylsulfonyl fluoride and repeatedly sonicated using Sonics Vibra Cell (Cole-Parmer Instruments). Total protein lysates were centrifuged at 10 000 g for 15 min, and the resulting supernatant was used. Total protein extracts were electrophoresed using NuPage Bis-Tris 4%–12% precast gels (Invitrogen). The remaining procedure was carried out as described previously.10 The following primary antibodies were used: mouse monoclonal anti-ALKBH2 (A8228; Sigma), rabbit polyclonal anti–β-actin (ab8227; Abcam), mouse monoclonal anti-MDM2 (OP46; Calbiochem), mouse monoclonal anti-p21 (556431; BD Pharmingen), and rabbit polyclonal anti–p53 upregulated modulator of apoptosis (PUMA) (#4976; Cell Signaling Technology). The secondary antibodies goat anti-mouse immunoglobulin (Ig)G–horseradish peroxidase (HRP) (sc-2005) and goat anti-rabbit IgG-HRP (IM0831) were from Santa Cruz Biotechnology and Beckman Coulter, respectively.
Cell Viability and Clonogenicity Assays
We seeded 3 × 103 cells in 100 µL growth medium in each well of a 96-well plate. The growth medium was removed the following day and replaced with new medium containing 500–2000 μM temozolomide, 100–400 μM methyl methanesulfonate, or 0.5–3 μM doxorubicin. Control cells received drug vehicle only. After 96 h, 20 µL of CellTiter 96 AQueous One Solution Cell Proliferation Assay (MTS assay, Promega) was added to each well for 1 hr before absorbance at 490 nm was recorded on a 96-well plate reader. The absorbance for wells containing only growth medium was subtracted to adjust for background absorbance. Each drug concentration was assayed in quadruplicate, and each experiment was repeated at least twice. Clonogenic survival assays were performed by seeding 300 cells in each well in 6-well plates. Cells were treated with 1500 μM temozolomide, 300 μM methyl methanesulfonate, or 2 μM doxorubicin for 24h before the growth medium was replaced with fresh drug-free medium, and cells were left to proliferate for 10 days. Colonies were then fixed with 6% glutaraldehyde, stained with 0.5% crystal violet, and counted. Only colonies counting more than 50 cells were included.
Flow Cytometric Analyses
DNA synthesis as a measure of cellular proliferation was determined by assessing incorporation of the thymidine analog 5-ethynyl-2′-deoxyuridine (Edu) into genomic DNA using the Click-IT Edu Alexa Fluor 647 flow cytometry kit (Invitrogen). Cells were labeled with 10 µM Edu for 1h, fixed in 4% paraformaldehyde, and stained according to the manufacturer's instructions. In addition, cells were stained with propidium iodide to determine DNA content and cell cycle phase distribution. Cells were analyzed on a C6 flow cytometer (Accuri Cytometers), and the acquired data were analyzed using FlowJo software version 7.6.3 (Tree Star).
ALKBH2-Targeted Small-interfering RNA Transfection
Cells were seeded to 50%–60% confluency in a 6-well plate. Small-interfering (si)RNA transfections were performed the following day under serum-free conditions using Oligofectamine Reagent (Invitrogen) according to the manufacturer's instructions with minor modifications. Cells were harvested 72h after transfection, and ALKBH2 knockdown was verified by western blotting. Three distinct siRNA sequences (Ambion) against ALKBH2 were used. A negative control siRNA was included in all transfection experiments. All siRNAs were used at a final concentration of 20 nM. The sequences were: siRNA#1, sense 5′-GAAUCUGACUUUUCGUAAAtt-3′ and antisense 5′-UUUACGAAAAGUCAGAUUCac-3′; siRNA#2, sense 5′-GUCUUCCCGUGAGAAAGAAtt-3′ and antisense 5′-UUCUUUCUCACGGGAAGACtg-3′; siRNA#3, sense 5′-GCACCGAGAUGAUGAAAGAtt-3′ and antisense 5′-UCUUUCAUCAUCUCGGUGCtc-3′.
Statistical Analyses
Data are presented as mean ± SEMs of 3 independent experiments. Statistical analyses were carried out using a two-tailed Student's t-test assuming equal variances. P < .05 was considered statistically significant.
Results
ALKBH2 Is Abundantly Expressed in Established GBM Cell Lines and Human GBM
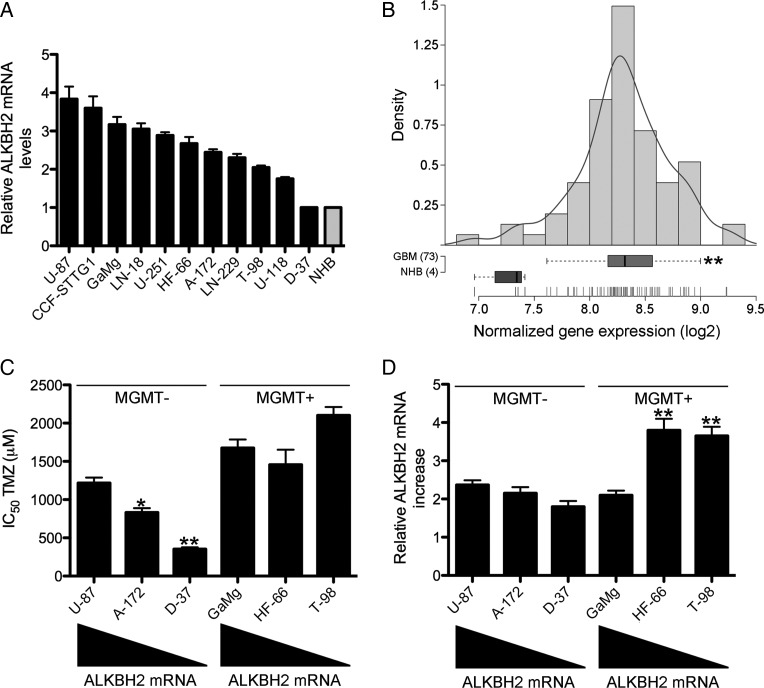
ALKBH2 is ubiquitously expressed in a wide range of normal human tissues, with peak levels in the testis and pancreas.11 However, to what extent ALKBH2 is expressed in human cancer cells in general is at present largely elusive. To determine whether GBM cells express ALKBH2, we initially screened a panel of 11 established GBM cell lines using qRT-PCR. As shown in Fig. 1A, apart from the D37 cell line, ALKBH2 was abundantly expressed, with mRNA expression levels 2- to 4-fold higher compared with NHB. We subsequently analyzed ALKBH2 mRNA expression levels in an independent recently published gene expression data set consisting of 4 NHB samples and 73 newly diagnosed GBM.12 Interestingly, ALKBH2 expression was significantly upregulated in human GBM by ∼2-fold compared with NHB (Fig. 1B; P = .001). Thus, our findings identify increased ALKBH2 expression as a novel feature of established GBM cell lines and human GBM.
Fig. 1.
ALKBH2 expression in GBM cells and human GBM. (A) ALKBH2 mRNA expression in a panel of 11 DNA fingerprinted GBM cell lines compared with NHB determined by qRT-PCR. (B) ALKBH2 gene expression (Affymetrix probe 225625_at) in 4 NHB samples and in 73 newly diagnosed GBM obtained from a previously published data set.12 Recurrent GBM was excluded in the analysis. **P < .01. (C) Correlation between ALKBH2 mRNA expression levels and temozolomide sensitivity in MGMT-deficient (U87, A172, and D37) and MGMT-proficient (GaMg, HF66, and T98) GBM cell lines. *P < .05, **P < .01. (D) Levels of ALKBH2 induction in GBM cell lines with distinct baseline ALKBH2 expression and MGMT status were quantified by qRT-PCR following 24h of temozolomide (TMZ) exposure with each cell line's corresponding half-maximal inhibitory concentration (IC50) dosage. The increase in ALKBH2 mRNA in cells receiving temozolomide was calculated relative to corresponding control cells treated with drug vehicle (DMSO) only. **P < .01.
Because ALKBH2 mRNA expression covered a continuum from high to low in our panel of GBM cell lines, we next examined whether there was any correlation between ALKBH2 expression levels and temozolomide sensitivity. We selected the 3 cell lines U87, A172, and D37 to represent high, intermediate, and low levels, respectively, of ALKBH2 mRNA expression. None of these cell lines expressed any MGMT as determined by qRT-PCR (data not shown). Interestingly, we observed an inverse relationship between ALKBH2 expression levels and temozolomide sensitivity in these cell lines (Fig. 1C). In contrast, there was no correlation between ALKBH2 mRNA expression levels and temozolomide sensitivity in the MGMT-proficient cell lines GaMg, HF66, and T98. Taken together, these data suggest that ALKBH2 mRNA expression levels inversely correlate with temozolomide sensitivity in MGMT-deficient GBM cell lines. Because cellular exposure to chemotherapeutic drugs may provoke a DNA damage response, we treated the 6 different GBM cell lines in Fig. 1C with their respective half-maximal inhibitory concentration (IC50) dosage of temozolomide for 24h and quantified levels of ALKBH2 induction by qRT-PCR. As shown in Fig. 1D, ALKBH2 levels increased ∼2-fold in all the cell lines following 24h of temozolomide exposure. However, we did not observe any significant association between the induced levels of ALKBH2 and the corresponding baseline expression levels, but ALKBH2 expression was most induced in the MGMT-proficient cell lines HF66 and T98. In contrast, MGMT expression levels did not significantly change in GaMg, HF66, or T98 cells during temozolomide exposure, as determined by qRT-PCR (data not shown).
ALKBH2 Overexpression Promotes Temozolomide Resistance in GBM Cells
To investigate the potential functional significance of ALKBH2 upregulation in GBM cells, we stably overexpressed ALKBH2 in the U87 and U251 GBM cell lines. Two independent clones overexpressing ALKBH2 were selected from each cell line for subsequent experiments (Fig. 2A). Because the cytotoxicity of chemotherapeutic drugs is highly dependent on the cellular proliferation rate, we first examined whether ALKBH2 overexpression influenced cell proliferation by measuring Edu incorporation in the ALKBH2-overexpressing clones and in their respective geneticin-resistant controls. As shown in Supplementary Fig. 1A and B, no significant difference in the size of the S-phase population was observed between ALKBH2-overexpressing cells and their corresponding control cells in either cell line, indicating that elevated ALKBH2 levels do not alter cell proliferation. We then treated the cell lines with increasing doses of temozolomide, methyl methanesulfonate, or doxorubicin for 96h before assessing drug-induced cytotoxicity using the MTS cell proliferation assay. ALKBH2-overexpressing U87 and U251 cells displayed significantly increased resistance to both temozolomide and methyl methanesulfonate chemotherapy compared with corresponding control cells (Fig. 2B and C). In contrast, ALKBH2 overexpression did not increase cellular resistance to doxorubicin treatment (Fig. 2B and C), suggesting that increased ALKBH2 activity mediates drug resistance predominantly to methylating agents. To further substantiate these findings, we performed clonogenic survival assays using ALKBH2-overexpressing U251 cells and their corresponding geniticin-resistant controls. After temozolomide or methyl methanesulfonate treatment, ALKBH2-overexpressing cells displayed significantly higher levels of clonogenic survival compared with control cells, confirming that increased ALKBH2 expression protects against methylating agent–induced cytotoxicity (Fig. 2D). Taken together, these data demonstrate that ALKBH2 overexpression does not alter cell proliferation but increases resistance to temozolomide and methyl methanesulfonate in GBM cells.
Fig. 2.
ALKBH2 overexpression increases resistance to methylating agents. (A) Clones stably overexpressing ALKBH2 were established by transfection of U87 and U251 cells with a His-tagged plasmid encoding ALKBH2. The upper band in the western blot represents the His-tagged plasmid, whereas the lower band represents endogenous ALKBH2 expression. (B) U87 and (C) U251 ALKBH2-overexpressing clones and their respective controls were treated with temozolomide (TMZ), methyl methanesulfonate (MMS), or doxorubicin (DOX) for 96h before drug-induced cytotoxicity was determined using an MTS assay. Cell viability (%) was calculated relative to corresponding control cells receiving drug vehicle only. The results shown were obtained using U87 clone #1 and U251 clone #1. Equally significant data were obtained using U87 clone #2 and U251 clone #2 (data not shown). *P < .05, **P < .01. (E) Colony formation assay using U251 ALKBH2-overexpressing cells and their respective controls. *P < .05, **P < .01.
Suppression of ALKBH2 Activity by SiRNA Transfection Sensitizes GBM Cells to Temozolomide
To further demonstrate the functional role of ALKBH2 in temozolomide resistance, we next silenced ALKBH2 expression in the human GBM cell lines GaMg (MGMT proficient) and U87 (MGMT deficient). ALKBH2 protein expression was efficiently suppressed when these cells were transfected with ALKBH2-targeted siRNAs, while a nontargeting siRNA yielded no effect on ALKBH2 expression levels (Fig. 3A). In contrast, siRNA-mediated suppression of ALKBH2 had no impact on cellular MGMT expression levels (Supplementary Fig. 2). Cells were treated with temozolomide, methyl methanesulfonate, or doxorubicin for 72h after siRNA transfection, and cell viability was determined using the MTS cell proliferation assay. Knockdown of ALKBH2 using either sequence #1 or sequence #3 significantly sensitized GaMg and U87 cells to temozolomide and methyl methanesulfonate chemotherapy compared with control transfectants (Fig. 3B and C). In contrast, ALKBH2 silencing had no effect on cellular response to either doxorubicin (Fig. 3B and C) or the nonmethylating DNA-damaging agent cisplatin (data not shown), further substantiating ALKBH2 as a mediator of methylating-agent resistance. These results suggest that ALKBH2 expression may compromise temozolomide-induced cytotoxicity and that silencing of ALKBH2 expression accordingly increases cellular temozolomide susceptibility.
Fig. 3.
Silencing of ALKBH2 expression increases cellular susceptibility to methylating agents. (A) GaMg and U87 cells were transfected with 3 distinct ALKBH2-targeted siRNA sequences, and ALKBH2 knockdown was verified by western blotting 72h after transfection. A negative control siRNA was included in all experiments. (B) GaMg and (C) U87 cells were transfected with siRNA sequence #1 or #3 or a negative control and treated with temozolomide (TMZ), methyl methanesulfonate (MMS), or doxorubicin (DOX) for 72h before drug-induced cytotoxicity was assessed using an MTS assay. Cell viability (%) was calculated relative to corresponding control cells receiving drug vehicle only. *P < .05, **P < .01.
Nongenotoxic Activation of the p53 Pathway Downregulates ALKBH2 Expression Levels
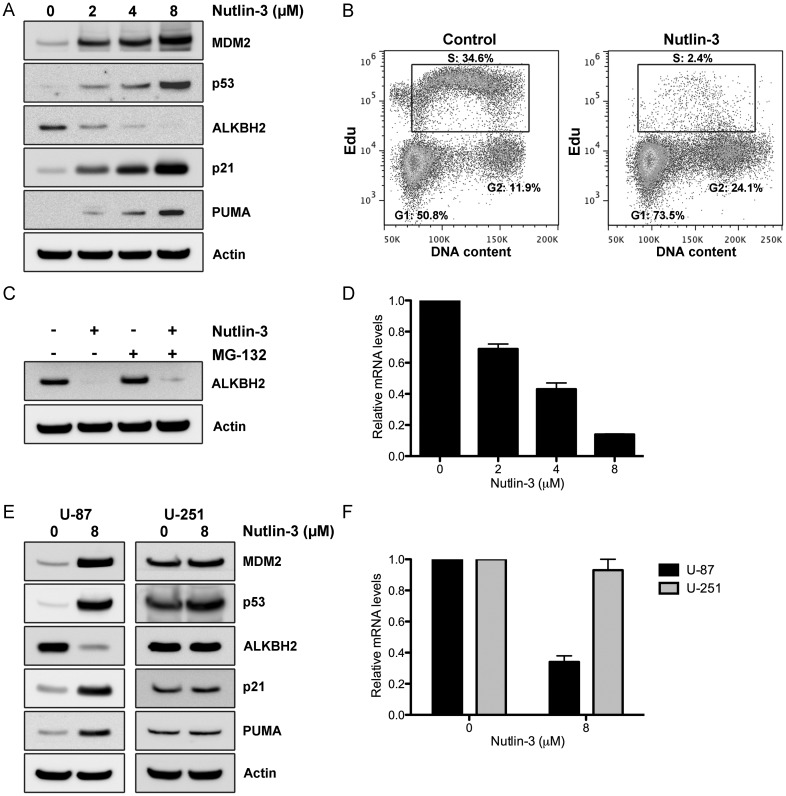
The tumor suppressor p53 acts as a sequence-specific transcription factor that regulates the expression of a broad range of genes (encoding both proteins and microRNAs) involved in diverse cellular processes, including DNA repair. Having shown that ALKBH2 could contribute to temozolomide resistance, we subsequently examined whether activation of the p53 pathway would alter the expression levels of ALKBH2 in GBM cells. The CCF-STTG1 GBM cell line harbors an amplification of the MDM2 gene on chromosome 12 that results in functional abrogation of the p53 pathway.13 Sequencing of the TP53 gene in CCF-STTG1 cells did not reveal any mutations (data not shown), which is consistent with the notion that MDM2 amplification and p53 mutation are mutually exclusive events.14 To investigate the effect of p53 activation on cellular ALKBH2 levels, we treated CCF-STTG1 cells with increasing concentrations of the selective MDM2 antagonist nutlin-3 for 24h. Nutlin-3 effectively disrupts the p53/MDM2 interaction, stabilizes p53, and activates the p53 pathway without the need for upstream signaling events such as DNA damage.15 In agreement with activation of a functional p53 pathway, nutlin-3 treatment led to increased protein levels of p53 and its bona fide targets MDM2, p21, and PUMA in a dose-dependent manner (Fig. 4A). In addition, nutlin-3 induced a prominent cell cycle arrest in the G1 and G2/M phases, which is consistent with previous observations (Fig. 4B).16 Interestingly, ALKBH2 levels diminished accordingly during nutlin-3 exposure, suggesting that increased p53 signaling may suppress ALKBH2 (Fig. 4A and C). CCF-STTG1 cells were then cotreated with nutlin-3 and the proteasome inhibitor MG132 to investigate whether decreased ALKBH2 levels were due to increased protein degradation. However, as shown in Fig. 4C, MG132 exposure failed to significantly restore ALKBH2 protein levels during nutlin-3 therapy. To see whether p53 activation decreased ALKBH2 at the transcriptional level, we quantified ALKBH2 mRNA using qRT-PCR following nutlin-3 treatment in CCF-STTG1 cells. We found that ALKBH2 mRNA levels were increasingly repressed with increasing nutlin-3 concentration (Fig. 4D), indicating that decreased ALKBH2 levels were due to transcriptional downregulation rather than increased protein degradation. To further validate these findings, we treated U87 and U251 GBM cells, which express wt and mutant p53, respectively, with nutlin-3 for 24h. In agreement with our results in the CCF-STTG1 cell line, nutlin-3 treatment increased the protein levels of p53 and its transcriptional targets MDM2, p21, and PUMA in U87 cells but not in U251 cells (Fig. 4E). Importantly, ALKBH2 expression levels after nutlin-3 exposure declined exclusively in U87 cells (Fig. 4E), indicating that wt-p53 expression is an absolute requirement for nutlin-mediated repression of ALKBH2. Moreover, ALKBH2 mRNA levels were significantly decreased in U87 but not in U251 cells after nutlin-3 therapy (Fig. 4F). Taken together, these data indicate that nongenotoxic activation of wt-p53 by the MDM2 antagonist nutlin-3 represses ALKBH2 transcription levels in human GBM cells.
Fig. 4.
Nongenotoxic activation of the p53 pathway by nutlin-3 downregulates ALKBH2 expression. (A) CCF-STTG1 cells were treated with the MDM2 antagonist nutlin-3 (0–8 µM) for 24h before cell lysates were collected for western blotting. (B) Cell cycle profiles of CCF-STTG1 cells treated with either drug vehicle (DMSO) alone or 8 µM nutlin-3 for 24h were determined by Edu labeling and flow cytometry. Cells in each phase were calculated from the flow cytograms and expressed as percentage of the total cell population. (C) Western blotting of CCF-STTG1 cells treated with drug vehicle, 8 µM nutlin-3, 25 µM MG132, or both for 24h. (D) ALKBH2 mRNA levels determined by qRT-PCR in CCF-STTG1 cells treated with 0–8 µM nutlin-3 for 24h. (E) U87 and U251 cells were treated with drug vehicle or 8 µM nutlin-3 for 24h before cell lysates were collected for western blotting. (F) ALKBH2 mRNA levels determined by qRT-PCR in U87 and U251 cells treated with drug vehicle or 8 µM nutlin-3 for 24h.
Discussion
Resistance to temozolomide chemotherapy is a major therapeutic challenge in the clinical management of GBM. Epigenetic silencing of the MGMT gene in tumor cells by promoter hypermethylation is currently considered the most reliable predictor of response to temozolomide in patients with newly diagnosed GBM. However, around half of all GBM patients harbor an unmethylated MGMT tumor promoter.17 As a result, these patients demonstrate less survival benefit from the addition of concomitant and adjuvant temozolomide chemotherapy to standard radiotherapy.18 Thus, there is a need to identify additional mediators of temozolomide resistance in human GBM and to decide whether these mechanisms may serve as novel therapeutic targets in concert with conventional modalities such as radio- and chemotherapy. Our data show that the DNA repair protein ALKBH2 is highly expressed in GBM cells and in human GBM compared with NHB and that temozolomide exposure may further increase ALKBH2 transcription levels. A similar finding has been reported in a small panel of pediatric brain tumors in which ALKBH2 mRNA expression was found to be enhanced compared with NHB samples.19 In contrast, ALKBH2 was frequently downregulated in biopsies from gastric carcinomas compared with adjacent non-neoplastic mucosa, suggesting that loss of ALKBH2 expression might contribute to malignant transformation of the gastric mucosa, perhaps due to impaired DNA repair function.20 These conflicting observations may imply that ALKBH2 is regulated differently in different tissues and their corresponding tumors, but further in-depth studies are needed to address this.
In this work, we have identified the DNA repair protein ALKBH2 as a novel mechanism of temozolomide resistance in human GBM cells. In contrast, altered ALKBH2 expression levels did not influence cellular response to the nonmethylating agents doxorubicin and cisplatin. This indicates that ALKBH2 acts as a mediator of methylating-agent resistance, which is in line with its enzymatic property as an oxidative demethylase. The previously reported crystal structure of the ALKBH2–double-stranded DNA complex provides the structural basis for designing small-molecule inhibitors of ALKBH2-mediated DNA repair.21 Our results showing the role of ALKBH2 in temozolomide resistance provide a rationale for the future development of ALKBH2 inhibitors in order to improve the efficacy of temozolomide chemotherapy in patients with GBM. To address whether increased ALKBH2 expression was associated with poor outcome in newly diagnosed GBM, we analyzed 2 independent recently published gene expression data sets in which 46 and 35 patients were treated with radiation and temozolomide, respectively.12,22 However, we could not detect any statistically significant association between ALKBH2 expression at time of diagnosis and outcome in either of the 2 cohorts (P = .26 and P = .18, respectively). This could be due to the low sample size or the fact that ALKBH2 expression at time of diagnosis does not necessarily reflect the expression levels achieved during the course of therapy, since we have shown that ALKBH2 is induced by temozolomide, whereas activation of wt-p53 can repress ALKBH2 transcription.
At present, 9 human AlkB homologs have been identified, and their biological properties as well as their potential roles in cancer are rapidly beginning to be uncovered. Lentivirus-mediated overexpression of ALKBH2 in gastric cancer cell lines has recently been reported to cause decreased proliferation, increased apoptosis, and cell cycle arrest in the G1 phase due to increased p16 and p21 levels.20 Moreover, ALKBH2 knockdown in the overexpressing cells accelerated cellular proliferation compared with control transfectants, indicating that loss of ALKBH2 expression promotes gastric cancer cell growth. ALKBH2 expression has previously been implicated in resistance to photodynamic therapy (PDT) in U87 GBM cells.23 SiRNA-mediated knockdown of ALKBH2 was accordingly reported to increase the efficacy of PDT, making the investigators speculate that PDT could induce DNA base lesions that could be substrates for ALKBH2-mediated repair. However, whether this is the exact mechanism behind these findings remains elusive. Among the other ALKBH proteins, ALKBH3, which differs from ALKBH2 in its cellular localization as well as in its substrate specificity, is highly expressed in prostate, lung, and rectal carcinomas.24–26 In addition, ALKBH8 has been implicated in the progression of urothelial carcinomas.27 Our results demonstrating the role of ALKBH2 in temozolomide resistance in human GBM cells add further insight into the continuously expanding knowledge of the ALKBH family of proteins.
The tumor suppressor p53 is a sequence-specific transcription factor that upon cellular stress such as DNA damage, oncogene activation, and hypoxia transactivates a broad range of genes whose products are involved in DNA repair, cell cycle arrest, cellular senescence, and apoptosis. In addition, p53 influences other cellular processes such as cellular metabolism,28 stem cell development,29 and microRNA regulation.30 In normal, unstressed cells, p53 activity is kept tightly suppressed by the MDM2 oncoprotein, which in turn is transcriptionally upregulated by p53 itself, thereby establishing a negative feedback loop. We found that ALKBH2 downregulation following p53 activation occurred at the transcriptional level, indicating that an active p53 pathway may suppress ALKBH2 transcription levels in human GBM cells. Interestingly, p53 has been known to transcriptionally repress, through both direct and indirect mechanisms, a large number of genes involved in different cellular processes such as apoptosis, cell cycle and cell growth, cell fate and development, DNA replication and repair, and metabolism, among others.31 Increased ALKBH2 levels following DNA damage have previously been correlated with increased binding of p53 to the ALKBH2 gene promoter, indicating that p53 may stimulate ALKBH2 transcription levels.23 Taken together with our findings, this might suggest that p53 can regulate ALKBH2 transcription differently depending on the cellular context, that is, whether genotoxic damage is present or not. Further studies are, however, needed to establish this as well as the exact mechanism behind p53-mediated transcriptional repression of ALKBH2. Because the majority of GBM cases express wt-p53,14 it is conceivable that nutlin-mediated p53 activation in these tumors could increase temozolomide sensitivity not only by increasing the transcription of pro-apoptotic genes such as PUMA and bcl-2–associated X protein, but also by downregulating proteins such as ALKBH2 that would otherwise promote temozolomide resistance.
In conclusion, we have shown that ALKBH2 is highly expressed in GBM cells and in human GBM. Moreover, this study is, to our best knowledge, the first to implicate the DNA repair protein ALKBH2 in GBM drug resistance. Thus, our findings provide a rationale for antagonizing ALKBH2 activity—for instance, through small-molecule inhibitors—in order to increase temozolomide efficacy in GBM cells. Future studies should explore larger gene expression data sets to determine whether ALKBH2 expression levels in GBM have any clinical prognostic and/or predictive value. Moreover, the ALKBH2 expression levels in other temozolomide-resistant cancers, such as malignant melanomas, and the potential therapeutic significance of p53-mediated hABH2 repression should be explored.
Supplementary Material
Funding
This work has been supported by the Norwegian Cancer Society, the Norwegian Research Council, Innovest AS, Helse Vest, Haukeland University Hospital, and the Bergen Medical Research Fund.
Supplementary Material
Acknowledgments
The authors would like to thank Pierre Brady for assistance in gene expression analysis and Tove Johansen and Marianne Enger for technical support.
Conflict of interest statement. None declared.
References
- 1.Stupp R, Mason WP, van den Bent MJ, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352(10):987–996. doi: 10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- 2.Stupp R, Hegi ME, Mason WP, et al. Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial. Lancet Oncol. 2009;10(5):459–466. doi: 10.1016/S1470-2045(09)70025-7. [DOI] [PubMed] [Google Scholar]
- 3.Denny BJ, Wheelhouse RT, Stevens MF, Tsang LL, Slack JA. NMR and molecular modeling investigation of the mechanism of activation of the antitumor drug temozolomide and its interaction with DNA. Biochemistry. 1994;33(31):9045–9051. doi: 10.1021/bi00197a003. [DOI] [PubMed] [Google Scholar]
- 4.Stupp R, Hegi ME, Gilbert MR, Chakravarti A. Chemoradiotherapy in malignant glioma: standard of care and future directions. J Clin Oncol. 2007;25(26):4127–4136. doi: 10.1200/JCO.2007.11.8554. [DOI] [PubMed] [Google Scholar]
- 5.Johannessen TC, Bjerkvig R, Tysnes BB. DNA repair and cancer stem-like cells—potential partners in glioma drug resistance? Cancer Treat Rev. 2008;34(6):558–567. doi: 10.1016/j.ctrv.2008.03.125. [DOI] [PubMed] [Google Scholar]
- 6.Aas PA, Otterlei M, Falnes PO, et al. Human and bacterial oxidative demethylases repair alkylation damage in both RNA and DNA. Nature. 2003;421(6925):859–863. doi: 10.1038/nature01363. [DOI] [PubMed] [Google Scholar]
- 7.Gilljam KM, Feyzi E, Aas PA, et al. Identification of a novel, widespread, and functionally important PCNA-binding motif. J Cell Biol. 2009;186(5):645–654. doi: 10.1083/jcb.200903138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sundheim O, Talstad VA, Vagbo CB, Slupphaug G, Krokan HE. AlkB demethylases flip out in different ways. DNA Repair (Amst) 2008;7(11):1916–1923. doi: 10.1016/j.dnarep.2008.07.015. [DOI] [PubMed] [Google Scholar]
- 9.Ringvoll J, Nordstrand LM, Vagbo CB, et al. Repair deficient mice reveal mABH2 as the primary oxidative demethylase for repairing 1meA and 3meC lesions in DNA. EMBO J. 2006;25(10):2189–2198. doi: 10.1038/sj.emboj.7601109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Johannessen TC, Wang J, Skaftnesmo KO, et al. Highly infiltrative brain tumours show reduced chemosensitivity associated with a stem cell-like phenotype. Neuropathol Appl Neurobiol. 2009;35(4):380–393. doi: 10.1111/j.1365-2990.2008.01008.x. [DOI] [PubMed] [Google Scholar]
- 11.Tsujikawa K, Koike K, Kitae K, et al. Expression and sub-cellular localization of human ABH family molecules. J Cell Mol Med. 2007;11(5):1105–1116. doi: 10.1111/j.1582-4934.2007.00094.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Murat A, Migliavacca E, Gorlia T, et al. Stem cell-related “self-renewal” signature and high epidermal growth factor receptor expression associated with resistance to concomitant chemoradiotherapy in glioblastoma. J Clin Oncol. 2008;26(18):3015–3024. doi: 10.1200/JCO.2007.15.7164. [DOI] [PubMed] [Google Scholar]
- 13.He J, Reifenberger G, Liu L, Collins VP, James CD. Analysis of glioma cell lines for amplification and overexpression of MDM2. Genes Chromosomes Cancer. 1994;11(2):91–96. doi: 10.1002/gcc.2870110205. [DOI] [PubMed] [Google Scholar]
- 14.The Cancer Genome Atlas Research Network. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455(7216):1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vassilev LT, Vu BT, Graves B, et al. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science. 2004;303(5659):844–848. doi: 10.1126/science.1092472. [DOI] [PubMed] [Google Scholar]
- 16.Tovar C, Rosinski J, Filipovic Z, et al. Small-molecule MDM2 antagonists reveal aberrant p53 signaling in cancer: implications for therapy. Proc Natl Acad Sci USA. 2006;103(6):1888–1893. doi: 10.1073/pnas.0507493103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Weller M, Stupp R, Reifenberger G, et al. MGMT promoter methylation in malignant gliomas: ready for personalized medicine? Nat Rev Neurol. 2010;6(1):39–51. doi: 10.1038/nrneurol.2009.197. [DOI] [PubMed] [Google Scholar]
- 18.Hegi ME, Diserens AC, Gorlia T, et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. N Engl J Med. 2005;352(10):997–1003. doi: 10.1056/NEJMoa043331. [DOI] [PubMed] [Google Scholar]
- 19.Cetica V, Genitori L, Giunti L, et al. Pediatric brain tumors: mutations of two dioxygenases (hABH2 and hABH3) that directly repair alkylation damage. J Neurooncol. 2009;94(2):195–201. doi: 10.1007/s11060-009-9837-0. [DOI] [PubMed] [Google Scholar]
- 20.Gao W, Li L, Xu P, Fang J, Xiao S, Chen S. Frequent down-regulation of hABH2 in gastric cancer and its involvement in growth of cancer cells. J Gastroenterol Hepatol. 2011;26(3):577–584. doi: 10.1111/j.1440-1746.2010.06531.x. [DOI] [PubMed] [Google Scholar]
- 21.Yang CG, Yi C, Duguid EM, et al. Crystal structures of DNA/RNA repair enzymes AlkB and ABH2 bound to dsDNA. Nature. 2008;452(7190):961–965. doi: 10.1038/nature06889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Etcheverry A, Aubry M, de Tayrac M, et al. DNA methylation in glioblastoma: impact on gene expression and clinical outcome. BMC Genomics. 2010;11:701. doi: 10.1186/1471-2164-11-701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee SY, Luk SK, Chuang CP, Yip SP, To SS, Yung YM. TP53 regulates human AlkB homologue 2 expression in glioma resistance to Photofrin-mediated photodynamic therapy. Br J Cancer. 2010;103(3):362–369. doi: 10.1038/sj.bjc.6605797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Konishi N, Nakamura M, Ishida E, et al. High expression of a new marker PCA-1 in human prostate carcinoma. Clin Cancer Res. 2005;11(14):5090–5097. doi: 10.1158/1078-0432.CCR-05-0195. [DOI] [PubMed] [Google Scholar]
- 25.Tasaki M, Shimada K, Kimura H, Tsujikawa K, Konishi N. ALKBH3, a human AlkB homologue, contributes to cell survival in human non-small-cell lung cancer. Br J Cancer. 2011;104(4):700–706. doi: 10.1038/sj.bjc.6606012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Choi SY, Jang JH, Kim KR. Analysis of differentially expressed genes in human rectal carcinoma using suppression subtractive hybridization. Clin Exp Med. 2011;11(4):219–226. doi: 10.1007/s10238-010-0130-5. [DOI] [PubMed] [Google Scholar]
- 27.Shimada K, Nakamura M, Anai S, et al. A novel human AlkB homologue, ALKBH8, contributes to human bladder cancer progression. Cancer Res. 2009;69(7):3157–3164. doi: 10.1158/0008-5472.CAN-08-3530. [DOI] [PubMed] [Google Scholar]
- 28.Vousden KH, Ryan KM. p53 and metabolism. Nat Rev Cancer. 2009;9(10):691–700. doi: 10.1038/nrc2715. [DOI] [PubMed] [Google Scholar]
- 29.Lin T, Chao C, Saito S, et al. p53 induces differentiation of mouse embryonic stem cells by suppressing Nanog expression. Nat Cell Biol. 2005;7(2):165–171. doi: 10.1038/ncb1211. [DOI] [PubMed] [Google Scholar]
- 30.He L, He X, Lowe SW, Hannon GJ. MicroRNAs join the p53 network—another piece in the tumour-suppression puzzle. Nat Rev Cancer. 2007;7(11):819–822. doi: 10.1038/nrc2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang B, Xiao Z, Ko HL, Ren EC. The p53 response element and transcriptional repression. Cell Cycle. 2010;9(5):870–879. doi: 10.4161/cc.9.5.10825. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.