Abstract
Background
Promoter methylation of the DNA repair gene, O-6-methylguanine-DNA methyltransferase (MGMT), is associated with improved treatment outcome for newly diagnosed glioblastoma (GBM) treated with standard chemoradiation. To determine the prognostic significance of MGMT protein expression as assessed by immunohistochemistry (IHC) and its relationship with methylation, we analyzed MGMT expression and promoter methylation with survival in a retrospective patient cohort.
Methods
We identified 418 patients with newly diagnosed GBM at University of California Los Angeles Kaiser Permanente Los Angeles, nearly all of whom received chemoradiation, and determined MGMT expression by IHC, and MGMT promoter methylation by methylation-specific PCR (MSP) and bisulfite sequencing (BiSEQ) of 24 neighboring CpG sites.
Results
With use of the median percentage of cells staining by IHC as the threshold, patients with <30% staining had progression-free survival (PFS) of 10.9 months and overall survival (OS) of 20.5 months, compared with PFS of 7.8 months (P < .0001) and OS of 16.7 months (P < .0001) among patients with ≥30% staining. Inter- and intrareader correlation of IHC staining was high. Promoter methylation status by MSP was correlated with IHC staining. However, low IHC staining was frequently observed in the absence of promoter methylation. Increased methylation density determined by BiSEQ correlated with both decreased IHC staining and increased survival, providing a practical semiquantitative alternative to MSP. On the basis of multivariate analysis validated by bootstrap analysis, patients with tandem promoter methylation and low expression demonstrated improved OS and PFS, compared with the other combinations.
Conclusions
Optimal assessment of MGMT status as a prognostic biomarker for patients with newly diagnosed GBM treated with chemoradiation requires determination of both promoter methylation and IHC protein expression.
Keywords: biomarker, glioblastoma, immunohisto-chemistry, methylation, MGMT
Glioblastoma multiforme (GBM) is the most common malignant primary brain tumor in adults. Current standard treatment for GBM includes cyto-reductive surgery followed by radiation (RT) and chemotherapy with temozolomide (TMZ).1 CpG methylation of the O-6-methylguanine-DNA methyltransferase (MGMT) gene promoter has emerged as the most robust predictor of TMZ treatment outcome for newly diagnosed GBM with prognostic implications.2–6 MGMT encodes for a DNA repair enzyme that provides resistance to alkylating chemotherapies, such as TMZ. Because MGMT transcription can be silenced by promoter methylation in tumor cells,7 it is widely assumed that MGMT promoter methylation in patient tumors causes decreased MGMT protein expression, thereby abrogating DNA repair activity necessary for TMZ resistance. However, the relationship between MGMT protein expression and promoter methylation in patient tumor samples has remained controversial.8–13 Furthermore, studies examining MGMT immunohistochemistry (IHC) and survival have not shown consistent correlation with outcome.4,5,9,10,13–19 The lack of definitive evidence has been attributed primarily to difficulty with MGMT IHC scoring; however, other possible reasons include small cohort size,4,5,10,13,18 nonuniform treatments,20,21 and various IHC scoring categories and cutoffs.4,9,10,21
Because of the importance of MGMT status for predicting treatment outcome, stratifying clinical trial patients, and guiding treatment decisions, the aims of the current single-arm retrospective study were to evaluate MGMT protein expression as a prognostic marker for patients with newly diagnosed GBM who were receiving standard of care RT/TMZ, to investigate the relationship between MGMT promoter methylation and protein expression, and to optimize the assessment of MGMT status. On the basis of a large cohort of 418 patients with newly diagnosed primary GBM, 410 of whom received chemoradiation with TMZ, we show that, although MGMT IHC is prognostic of patient survival, the combination of IHC and methylation testing provides optimized assessment of MGMT status.
Materials and Methods
Patients
Pretreatment formalin-fixed, paraffin-embedded tumor samples were available for 418 adult patients with newly diagnosed confirmed GBM, 410 of whom were treated with TMZ and RT. A total of 281 patients who received a diagnosis during 2000–2007 were retrospectively identified on the basis of an electronic database query of adult patients with primary GBM receiving upfront TMZ and treated at the University of California Los Angeles (UCLA) or Kaiser Permanente Los Angeles (KPLA). One hundred thirty-seven additional patients (who received a diagnosis during 2007–2010) whose samples were directed to the laboratory in an unselected manner were also included. The collection of human brain tumor samples was approved by the UCLA Institutional Review Board, and informed consent was obtained from all patients. Some MGMT MSP and IDH1 sequencing data have been previously reported.22–26 The range of follow-up was 2–137 months. The median follow-up period determined by method of Schemper and Smith27 was 70 months. We had missing overall survival data on 1 patient and missing progression data on 6 patients.
Complete methods can be found in the Supplementary materials. All multivariate analyses used the same additional variables (age, sex, Karnofsky performance status (KPS), extent of resection, bevacizumab treatment at any time, and IDH1R132 mutation status).
Progression was determined by central retrospective review of imaging and records.
Results
Patient Characteristics
As described in Methods, we derived a cohort of 418 patients with newly diagnosed primary GBM with available formalin-fixed, paraffin-embedded tumor samples from the initial surgery prior to any treatment. The clinical characteristics of the patients are listed in Table 1. The RT/TMZ treatment protocol varied slightly among the 410 total patients: 235 patients received concurrent daily RT/TMZ followed by TMZ (Stupp),1 127 patients received maintenance dose TMZ overlapping with RT (modified Stupp), and 48 patients received TMZ after RT (pre- Stupp). Variation in treatment protocol depended mostly on when patients were diagnosed and reflected evolving treatment patterns over time. In addition, some patients received additional upfront agents, such as isotretinoin or bevacizumab.26 As determined by Kaplan-Meier survival analysis (data not shown), the overall survival (OS) for the entire cohort was 18.2 months, consistent with recent studies,28 and the progression-free survival (PFS) was 9.0 months, possibly elevated because of inclusion of patients receiving upfront bevacizumab with RT/TMZ.26
Table 1.
Patient Characteristics
| Characteristics | Current study |
|
|---|---|---|
| (n = 418) | ||
| No. of Patients | % | |
| Enrollment by site | ||
| UCLA | 317 | 76 |
| KPLA | 101 | 24 |
| Age, years | ||
| Median | 57.6 | |
| Range | 22.3–90.0 | |
| <50 | 123 | 29 |
| ≥50 | 295 | 71 |
| Sex | ||
| Male | 254 | 61 |
| Female | 164 | 39 |
| Karnofsky performance status | ||
| 100 | 58 | 14 |
| 90 | 199 | 48 |
| 80 | 103 | 25 |
| 70 | 26 | 6 |
| ≤60 | 30 | 7 |
| Recursive partitioning analysis by class | ||
| Class III/IV | 298 | 72 |
| Class V/VI | 117 | 28 |
| Extent of surgery | ||
| Biopsy | 38 | 9 |
| Sub Total resection | 200 | 49 |
| Gross Total resection | 175 | 42 |
| Upfront Treatment Protocol | ||
| Pre-Stupp | 48 | 11 |
| Pre-Stupp and isotretinoin | 32 | |
| Modified Stupp | 127 | 30 |
| Modified Stupp and isotretinoin | 51 | |
| Stupp | 235 | 56 |
| Stupp and bevacizumab | 80 | |
| Stupp and isotretinoin | 15 | |
| Other | 8 | 2 |
| Recurrent treatment | ||
| Progressed (n = 412) | 353 | 86 |
| Progressed with chemotherapy | 271 | |
| Progressed with bevacizumab | 161 | |
| Deaths | 356 | 85 |
MGMT IHC Is Prognostic of Survival
To determine whether MGMT IHC was predictive of survival in our cohort, we assessed MGMT expression levels by IHC in 355 patients. MGMT expression was determined by a trained neuropathologist (W.H.Y.) using a scoring system consistent with published recommendations based on percentage of tumor cells with positive immunostaining results, with particular attention placed on excluding staining from nontumor sources, such as endothelial cells and normal brain.11,17 The pathologists were blinded to the results of other MGMT assays or to clinical data. Consistent with other published studies,10,14–17 a histogram of the IHC scores shows that low MGMT expression is a common feature of untreated GBMs (Figure S1).
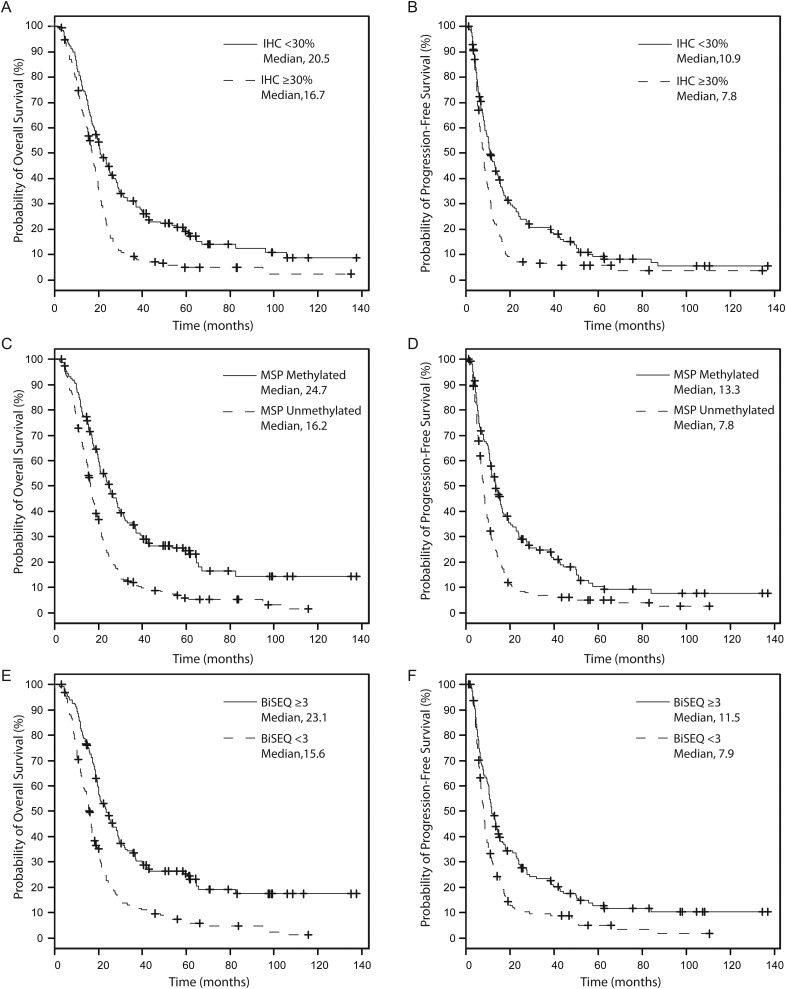
Using the median percentage of cells staining for MGMT by IHC as the staining threshold, we separated patients into a low expression group defined as <30% (183/355, 52% of patients) and a high expression group defined as ≥30% (172/355, 48% of patients). By Kaplan-Meier analysis, patients with low MGMT protein expression demonstrated a median OS of 20.5 months, whereas patients with high MGMT protein expression demonstrated a median OS of 16.7 months (log-rank, P < .0001; Fig. 1A). PFS was also higher among low-expressing patients than among high-expressing patients (10.9 months vs. 7.8 months; P < .0001; Fig. 1B). Multivariate analysis including other parameters, such as age, sex, KPS, extent of resection, bevacizumab treatment at any time, and IDH1R132 mutation status, showed MGMT IHC as a prognostic biomarker for both OS (hazard ratio [HR] = 1.63; P < .0001) and PFS (HR = 1.49; P = .001; Table S1). These results were validated by bootstrap analysis.29–31
Fig. 1.
Kaplan-Meier analysis of overall survival (OS, A, C, E) and progression-free survival (PFS, B, D, F) comparing (A and B) IHC <30% (solid) to IHC ≥30% (dashed), (C and D) MSP methylated (solid) to MSP unmethylated (dashed), and (E and F) BiSEQ ≥3 (hypermethylated, solid) to BiSEQ <3 (hypomethylated, dashed). A and B, IHC <30% showed improved outcomes compared to IHC ≥30% (OS, log rank P < .0001; PFS, log rank P < .0001). C and D, MSP methylated (solid) showed improved outcomes compared to MSP unmethylated (dashed) (OS, log rank P < .0001; PFS, log rank P < .0001). E and F, BiSEQ ≥3 (solid) showed improved outcomes compared to BiSEQ <3 (dashed) (OS, log rank P < .0001; PFS, log rank P = .0001).
In the intrarater assessment (W.H.Y.), the mean difference ± standard deviation (SD) between the 2 readings was 3.5 ± 15.6, and the median was 0 (interquartile range [IQR], −5 to 13). The estimated intraclass correlation coefficient was 0.86 (95% confidence interval [CI], 0.80–0.90). If the IHC value was dichotomized into 2 levels (low and high) with use of median as the cutoff point, 97 (83.6%) patients were classified into the same level by both readings, and 19 (16.4%) patients were classified into different levels. The agreement between the 2 readings was significantly high: the estimated κ was 0.67 (95% CI, 0.54–0.81; P < .0001). In the interrater assessment (W.H.Y. and N.K.), the mean difference ±SD between the 2 reviewers was 2.5 ± 18.9 (median, 3.0; IQR, −4.5 to 15). The estimated intraclass correlation coefficient was 0.80 (95% CI, 0.74–0.85). Eighty-three percent of patients were classified into the same level, and 17% of patients were classified into a different level by the 2 neuropathologists if the IHC value was dichotomized by median. The agreement between these 2 reviewers was significantly high; the estimated κ was 0.66 (95% CI, 0.51–0.80; P < .0001). Both assessments indicate that IHC can be reliably scored.
Confirmation of Prognostic Value of MGMT Methylation-Specific PCR
To determine whether our cohort showed the expected survival stratification by methylation status,4–6,12 we performed standard 2-stage (nested) MSP6 and derived results for 402 (96%) of 418 patients. By Kaplan-Meier analysis, methylated patients demonstrated a median OS of 24.7 months, whereas unmethylated patients demonstrated a median OS of 16.2 months (log-rank, P < .0001; Fig. 1C). PFS was also higher among methylated than among unmethylated patients (13.3 months vs. 7.8 months; log-rank, P < .0001; Fig. 1D). Multivariate analysis including other parameters, such as age (continuous variable), sex, KPS, extent of resection, bevacizumab treatment at any time, and IDH1R132 mutation status, showed that MGMT promoter methylation status determined by MSP was a prognostic variable for both OS (HR = 0.47; P < .0001) and PFS (HR = 0.53; P < .0001; Table S2). These results were validated by bootstrap analysis.
MGMT Methylation Can Be Determined by BiSEQ Assessment of CpG Methylation Density
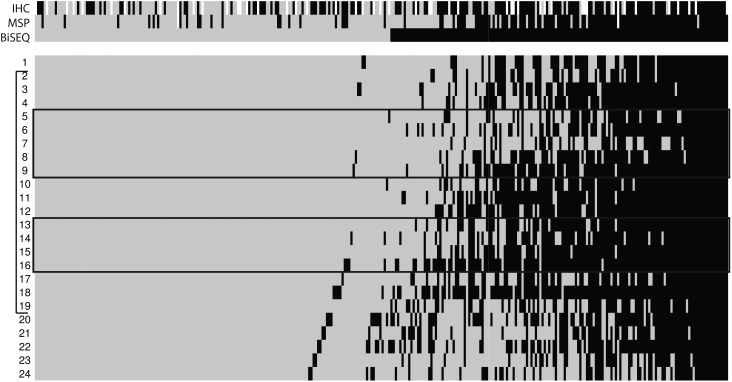
In contrast to MSP evaluation, BiSEQ using conventional sequencing instruments provides information for individual CpG sites within the entire sequenced region. To investigate whether BiSEQ could provide an alternative to MSP, we sequenced 24 CpG sites in the differentially methylated region 2 (DMR2) region32 of the MGMT promoter that contains the MSP region (Fig. 2). We obtained BiSEQ data on 312 patients and derived semiquantitative CpG methylation densities, ranging from 0 to 24 methylated CpG sites of the 24 total sites in this region (Fig. 2). We used the median number of methylated CpG sites as the threshold defining hypomethylated (<3 sites, 160/312, 51%) and hypermethylated (≥3 sites, 152/312, 49%) patients. By Kaplan-Meier analysis, we found that patients with ≥3 methylated CpG sites (hypermethylated) had OS of 23.1 months, whereas patients with <3 methylated CpG sites (hypomethylated) had OS of 15.6 months (log-rank, P < .0001; Fig. 1E). PFS was also higher among hypermethylated patients than among hypomethylated patients (11.5 months vs. 7.9 months; log-rank, P = .0001; Fig. 1F). Multivariate analysis using the same variables as previously described showed that BiSEQ dichotomized at 3 CpG sites was a prognostic variable for both OS (HR = 0.46; P < .0001) and PFS (HR = 0.64; P = .0006, Table S3). These results were validated by bootstrap analysis.
Fig. 2.
Heatmap diagram of BiSEQ results (n = 312). From left to right, samples are arranged in order of CpG methylation density from low to high. Black indicates methylation detected, gray indicates no methylation detected. CpG sites 1–24 are labeled on left. CpG sites interrogated by MSP are indicated in black boxes. DMR2 region indicated by a bracket (left). Results were derived from bisulfite sequencing of uncloned tumor DNA. Upper identification strip indicates IHC (gray: ≥30%, black: <30%, white: no data), MSP (black: methylated, gray: unmethylated, white: no data) and BiSEQ (black: ≥3, gray: <3).
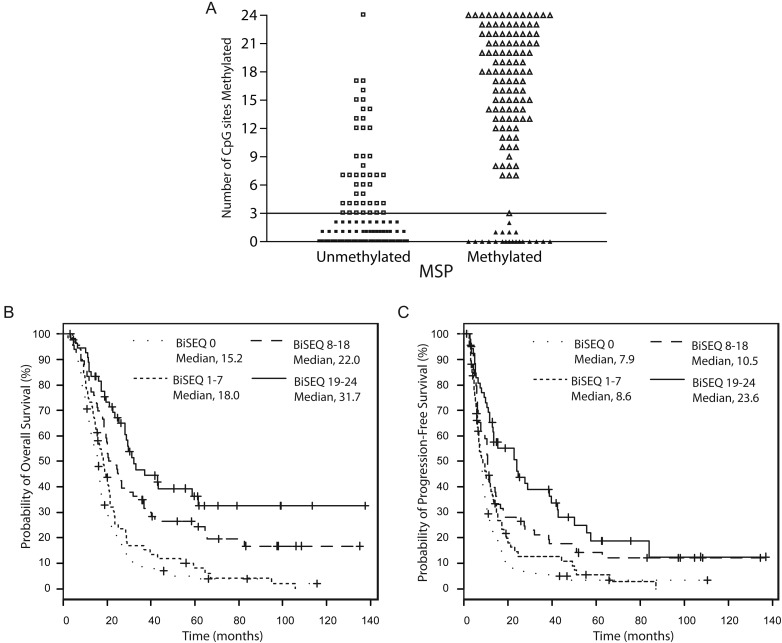
As shown in Fig. 3A, the majority of BiSEQ hypermethylated patients were also determined to be MSP methylated (open triangles), and the majority of BiSEQ hypomethylated patients were found to be MSP unmethylated (filled squares). Both the nondichotomized (point biserial correlation coefficient = 0.71, P < .0001) and dichotomized (κ = 0.58; 95% CI, 0.50–0.67; P < .0001) BiSEQ results were highly concordant with MSP status. There were 21 patients methylated on MSP but had <3 methylated CpG sites on BiSEQ (filled triangles). Survival (OS and PFS) among these 21 patients was similar to that among MSP unmethylated patients (data not shown), suggesting that these discordant results represent clinically silent methylation detectable by MSP but not BiSEQ. In addition, there were 43 patients who were MSP unmethylated who had evidence of ≥3 methylated CpG sites (open squares). The occurrence of these cases can be explained by the inability of MSP to detect methylation in regions outside the primer regions or incomplete methylation within the specific MSP primer location.2 Of interest, BiSEQ density (binned into 4 groups) was prognostic of patient survival, with increasing methylation density correlating with increased OS and PFS (trend test, P < .0001 for both; Fig. 3B and C). These results suggest that BiSEQ may provide added value, compared with MSP, by providing clinically relevant methylation density information.
Fig. 3.
Correlation of BiSEQ determined methylation density with MSP and survival. (A) Scatterplot (open squares (MSP unmethylated, BiSEQ ≥3; n = 43), filled squares (MSP unmethylated, BiSEQ < 3; n = 138), open triangles (MSP methylated, BiSEQ ≥3; n = 108), and filled triangles (MSP methylated, BiSEQ < 3; n = 21) showing correlation between BiSEQ and MSP (non-dichotomized BiSEQ, point biserial correlation coefficient = 0.71, P < .0001, and dichotomized BiSEQ, Kappa = 0.58, P < .0001)). Horizontal line indicates 3 CpG sites. Kaplan-Meier analysis of (B) OS and (C) PFS showing correlation of increasing CpG methylation density (0 CpG sites methylated = dotted line, 1–7 CpG sites methylated = small dashed line, 8–18 CpG sites methylated = large dashed line, 19–24 CpG sites methylated = solid line) and survival (OS, trend test P < .0001; PFS, P < .0001).
MGMT Promoter Methylation Correlates with Reduced Expression
To determine the relationship between MGMT expression level (IHC) and MGMT methylation status as determined by either MSP or BiSEQ, we plotted expression levels for MSP methylated and MSP unmethylated patients (Fig. 4A) and BiSEQ hypermethylated and hypomethylated patients (Figure S2A). We found that MGMT MSP was correlated with nondichotomized MGMT IHC scores (point biserial correlation coefficient = −0.41; P < .0001) and with IHC dichotomized at 30% (κ = −0.39; 95% CI, −0.29 to −0.48; P < .0001). Seventy-eight percent of MSP methylated patients had reduced MGMT expression (100/129), whereas 37% of unmethylated patients (79/215) also had reduced MGMT expression (Fig. 4A; χ2 test, P < .0001). When BiSEQ data were binned into 4 groups of increasing CpG density, we observed that increasing number of methylated CpG sites was also correlated with decreasing IHC score in our samples (Fig. 4B; Spearman correlation coefficient = −0.48; P < .0001). These observations are consistent with prior studies that showed that increasing MGMT promoter methylation density in DMR2 is correlated with decreasing MGMT protein expression in vitro.32,33 The 29 MSP methylated patient samples that demonstrated high MGMT expression could result from low-level methylation or persistent expression that escapes methylation silencing (Fig. 4A). Overall, these results clearly demonstrate the relationship between promoter methylation and reduced expression in patient samples and confirm the importance of this CpG island region in down-regulating protein expression. However, our data also suggest that reduced expression can occur via other mechanisms, such as methylation outside the DMR2 region32,34 or non–methylation-dependent transcriptional silencing.35–37
Fig. 4.
Correlation of methylation and IHC. (A) Scatterplot (open squares (MSP unmethylated, IHC ≥30%; n = 136), filled squares (MSP unmethylated, IHC <30%; n = 79), open triangles (MSP methylated, IHC ≥30%; n = 29), and filled triangles (MSP methylated, IHC <30%; n = 100) showing correlation between MSP and IHC (non-dichotomized IHC, point biserial correlation coefficient = −0.41 P < .0001, and dichotomized IHC, Kappa = −0.39, P < .0001)). (B) Scatterplot showing correlation between CpG density and decreased protein expression (Spearman correlation coefficient = −0.48, P < .0001). Open squares (0 CpG sites methylated), triangles (1–7 CpG sites methylated), circles (8–18 CpG sites methylated), and diamonds (19–24 CpG sites methylated) indicate IHC ≥30%. Filled squares (0 CpG sites methylated), triangles (1–7 CpG sites methylated), circles (8–18 CpG sites methylated), and diamonds (19–24 CpG sites methylated) indicate IHC < 30%. Kaplan-Meier analysis of (C) OS and (D) PFS showing survival curves for groups with different MSP/IHC combinations (MSP methylated, IHC <30% = solid line; MSP methylated, IHC ≥30% = alternating dashed and dotted line; MSP unmethylated, IHC <30 = large dashed line; MSP unmethylated, IHC ≥30% = dotted line). Patients with the combination of MSP methylated and IHC <30% demonstrated improved OS and PFS compared to all other groups (P < .0001 for both). No significant difference in OS or PFS was observed among the other three groups.
MGMT Methylation and Low Expression by IHC in Combination Are Additively Prognostic of Outcome
By Kaplan-Meier analyses, combined testing of MSP and IHC enabled identification of a long-term survival group when methylation and low expression were observed in tandem (Fig. 4C; OS: P < .0001, compared with other 3 combinations; PFS: P < .0001; Fig. 4D). Further inspection of the Kaplan-Meier curves shows that patients with high protein expression have poor outcomes despite the presence of methylation and that patients with low expression without methylation also have poor outcomes. These results are summarized in Table 2.
Table 2.
Combined MGMT Analysis with IHC and Methylation
| IHC | Methylation | Percentage of patients | Overall Survival Months (Median, 95% CI) | Progression-Free Survival Months (Median, 95% CI) | Note |
|---|---|---|---|---|---|
| <30% | MSP Methylated | 29% | 27.8 (21.0, 36.2)* | 15.2 (11.5, 22.4)* | Best survival group |
| ≥30% | MSP Methylated | 8% | 18.5 (11.7, 22.9)^ | 9.6 (4.4, 11.6)^ | Methylated alone inadequate for good outcome |
| <30% | MSP Unmethylated | 23% | 16.4 (13.6, 20.0)^ | 8.2 (6.6, 9.9)^ | Low expression alone inadequate for good outcome |
| ≥30% | MSP Unmethylated | 40% | 16.3 (14.5, 18.2)^ | 7.8 (6.5, 8.7)^ | Unmethylated or high expression either together or alone adequate for poor outcome |
| <30% | BiSEQ ≥3 sites methylated | 38% | 26.3 (20.5, 29.2)* | 13.3 (10.4, 17.6)* | Best survival group |
| ≥30% | BiSEQ ≥3 sites methylated | 11% | 19.3 (13.6, 22.4)^ | 9.6 (6.3, 11.2)^ | Low methylation alone inadequate for good outcome |
| <30% | BiSEQ <3 sites methylated | 20% | 15.3 (12.3, 20.0)^ | 8.3 (6.6, 10.9)^ | Low expression alone inadequate for good outcome |
| ≥30% | BiSEQ <3 sites methylated | 31% | 16.3 (11.9, 18.3)^ | 7.7 (5.9, 8.1)^ | Low methylation or high expression either together or alone adequate for poor outcome |
Abbreviations: IHC, immunohistochemistry; MSP, methylation specific PCR; BiSEQ, bisulfite sequencing.
*P < .001 vs. all other groups. ^P> 0.1 among these 3 groups.
To confirm this, we performed multivariate analysis by including the 4 (2 × 2 table) combinations of methylation and IHC into the Cox regression model adjusting for the same clinical variables and setting the combination of methylation and low expression (<30) as the reference. We found that combined methylation by MSP and low expression by IHC prognosticated improved outcome for OS and PFS, compared with the other combinations (Table 3). Using bootstrap analysis to provide unbiased estimates of HR and P values, we found similar results.
Table 3.
Cox Proportional Hazard Analysis Including MSP and IHC Combinations with Bootstrap Validation
| Overall Survival | Overall Survival (Bootstrap) | Progression Free Survival | Progression Free Survival (Bootstrap) | ||
|---|---|---|---|---|---|
| Factor | Estimate HR (95% CI), P-value | Estimate HR (95% CI), P-value | Estimate HR (95% CI), P-value | Estimate HR (95% CI), P-value | |
| MGMT MSP and IHC | 1. MSP = M, IHC < 30 (n = 100) | 1.92 (1.18, 3.14), P = .0087 | 1.92 (1.18, 3.14), P = .0087 | 1.92 (1.18, 3.14), P = .0087 | 1.92 (1.18, 3.14), P = .0087 |
| 2. MSP = M, IHC≥30 (n = 29) | 1.92 (1.18, 3.14), P = .0087 | 1.91 (1.16, 3.16), P = .0114 | 1.78 (1.10, 2.88), P = .0186 | 1.82 (1.13, 2.93), P = .0132 | |
| 3. MSP = U, IHC < 30 (n = 79) | 2.20 (1.54, 3.12), P < .0001 | 2.14 (1.47, 3.12), P < .0001 | 1.98 (1.41, 2.79), P < .0001 | 2.02 (1.41, 2.90), P = .0001 | |
| 4. MSP = U, IHC≥30 (n = 136) | 2.62 (1.91, 3.60), P < .0001 | 2.57 (1.84, 3.60), P < .0001 | 2.15 (1.59, 2.92), P < .0001 | 2.23 (1.60, 3.10), P < .0001 | |
| Gender | Male vs. Female | 1.48 (1.15, 1.91), P = .0021 | 1.47 (1.10, 1.97), P = .0091 | 1.30 (1.02, 1.67), P = .0371 | 1.28 (0.98, 1.68), P = .0691 |
| Age | 1.02 (1.01, 1.04), P < .0001 | 1.02 (1.01, 1.04), P < .0001 | 1.02 (1.01, 1.03), P = .0039 | 1.02 (1.01, 1.03), P = .0046 | |
| KPS | 100 | 1.48 (1.15, 1.91), P = .0021 | 1.48 (1.15, 1.91), P = .0021 | 1.48 (1.15, 1.91), P = .0021 | 1.48 (1.15, 1.91), P = .0021 |
| 90 | 1.21 (0.83, 1.75), P = 0.3246 | 1.24 (0.87, 1.78), P = .2310 | 1.05 (0.73, 1.52), P = .7908 | 1.04 (0.74, 1.46), P = .8306 | |
| 80 | 1.24 (0.82, 1.88), P = .2989 | 1.27 (0.85, 1.88), P = .2384 | 0.91 (0.60, 1.38), P = .6595 | 0.90 (0.63, 1.28), P = .5544 | |
| 70 or lower | 1.91 (1.19, 3.05), P = .0070 | 2.03 (1.08, 3.83), P = .0279 | 1.68 (1.05, 2.68), P = .0309 | 1.72 (1.04, 2.84), P = .0336 | |
| Resection | GTR vs. STR and Bx | 0.68 (0.53, 0.88), P = .0028 | 0.66 (0.50, 0.87), P = .0028 | 0.65 (0.51, 0.84), P = .0008 | 0.64 (0.49, 0.83), P = .0006 |
| Bevacizumab | Yes vs. No | 0.78 (0.60, 0.99), P = .0444 | 0.75 (0.57, 0.99), P = .0418 | 1.07 (0.84, 1.37), P = .5778 | 1.08 (0.84, 1.38), P = .5464 |
| IDH1 | WT vs. MUT | 0.52 (0.26, 1.01), P = .0528 | 0.50 (0.25, 0.97), P = .0416 | 0.59 (0.32, 1.08), P = .0867 | 0.59 (0.31, 1.11), P = .1015 |
Abbreviations: Bx, biopsy; GTR, gross-total resection; IHC, immunohistochemistry; KPS, Karnofsky performance status; MGMT, O6-methylguanine DNA methyltransferase; MSP, methylation specific PCR; M, methylated; MUT, mutant; STR, sub-total resection; U, unmethylated; WT, wildtype.
These data indicate that both MSP and IHC were individually inferior in prognosticating survival for a significant portion of patients, compared with when they were used in combination, and suggest that low IHC expression is only prognostic of improved outcome when associated with methylation (Table 3).
Discussion
Although CpG island promoter methylation represents an established mechanism of transcriptional silencing and resultant reduced protein expression, the prognostic significance of MGMT protein expression as assessed by IHC and its relationship with methylation remain unclear in patients with newly diagnosed GBM treated with standard radiation and TMZ.3,4,8–11,15,17,19,32,34 On the basis of concurrent analyses of MGMT IHC and promoter methylation by MSP and BiSEQ in a 418-patient cohort, our results demonstrate that optimal assessment of MGMT status as a prognostic biomarker for newly diagnosed GBM treated with RT and TMZ should take into consideration both protein expression and methylation status. Using both evaluations, we observed substantially improved outcomes for patients with GBM that demonstrated simultaneous methylation and low expression. In addition, we showed that methylation was correlated with reduced protein expression, although low expression occurred frequently in the absence of methylation (79/215, 37% of unmethylated patients; Table 3). In this 2 institution study, it is important to note that most but not all cases were subjected to central review of pathology (85% of cases) and central review of progression (88% of cases).
Aside from challenges in scoring IHC staining,9,11,17 possible reasons for the lack of agreement between previous studies attempting to correlate IHC with survival include small cohort size,4,5,10,13,18 various IHC scoring categories and cutoffs,4,9,10,21 small tissue sample size with tissue arrays,9 multiple glioma subtypes,15 and nonuniform treatment protocols.5,9,10,13 In contrast, our study investigated a large number of newly diagnosed GBM treated with TMZ and RT. Furthermore, we scored IHC staining as a percentage of positive tumor cells, whereas many other studies divided IHC staining into categories.5,9,10,18 Studies suggest that it is critical to ensure that only staining from tumor cells are included with specific consideration to exclude staining from normal brain tissue, endothelium, or macrophages.11,17 From the median percentage of cells staining for MGMT by IHC, we used 30% staining as the threshold separating percentage IHC staining into 2 groups. This threshold is similar to the 10%–20% thresholds used in several smaller studies showing positive correlation between MGMT IHC staining and outcome in malignant gliomas.14–17,19 In addition, stratification of our IHC data using other thresholds of 10%–50% also yielded significant prognostic value, as evidenced by Kaplan-Meier analysis (data not shown). Our results indicate that a single threshold delineating high and low expression is sufficient, compared with stratification of immunostaining into multiple levels. As demonstrated in several smaller published studies,14–17,19,38 we believe that IHC scoring applying a single threshold is amenable in different histopathology laboratories. Our analyses of interreader and intrareader variability indicate that ∼17% of slides could be classified differently by separate reviewers or after re-review by the same reviewer, indicating that the reproducibility and reliability of IHC scoring may be difficult for a small number of patients.
On the basis of our results demonstrating that IHC and methylation additively prognosticated outcome, we recommend that evaluation of MGMT methylation and IHC be incorporated into standard molecular testing for newly diagnosed GBM. Because of the relative ease and low expense, many diagnostic histopathology laboratories have the ability to establish and offer MGMT IHC evaluation on resected tumor tissue. The most significant benefit in tandem evaluation of IHC and methylation is to identify patients with both low protein expression and promoter methylation, because our results show that outcome for this group is most favorably separated from the 3 other possible combinations (Tables 2 and 3).
In terms of how IHC can enhance methylation testing by MSP, our results show that IHC status can stratify outcome in the methylated group. In other words, 22% of methylated patients (29/129) would be predicted to have poor outcomes on the basis of having high protein expression. This methylated with high IHC subgroup is relatively small, making it less clear whether this delineates a biologically relevant set of tumors showing MGMT expression that can escape methylation-dependent downregulation or whether this is attributable to detection of clinically silent levels of methylation. Despite this uncertainty, the addition of IHC to MSP will optimize prognosis for methylated patients with high IHC staining whom we observed to have poor outcome despite the presence of methylation.
In situations in which MSP testing is unavailable, our results indicate that IHC has clinical use for identifying patients with high expression, because OS among these patients is independent of MGMT methylation. High-expressing (≥30%) patients had poor outcome regardless of methylation status, whereas low-expressing patients with associated promoter methylation (56% of low IHC) have clinically significant improved outcomes, indicating that patients with low IHC staining should undergo methylation testing. A possible explanation for the poor outcomes of low-expressing tumors without associated methylation could be that these tumors have inducible MGMT protein expression.39–44 Alternatively, MGMT promoter methylation may be associated with additional survival/treatment benefits unrelated to MGMT expression.
Another reason for the perceived lack of utility of IHC in prognosticating treatment outcome for GBM has been the apparent lack of correlation with promoter methylation.8–11,13 In our cohort, we found that promoter methylation by MSP is correlated with protein expression, as evidenced by low expression in the majority of patients with methylation. This observation is consistent with several previous studies that showed correlation between MGMT promoter methylation and protein expression.4,18,32 This is also consistent with in vitro studies in which methylation of the MGMT promoter caused protein downregulation.45 However, our data and those from other studies10,13 show that nearly half (44%) of the patients with low IHC staining do not have promoter methylation. It is possible that the presence of this population obscured the correlation between methylation and IHC in other studies,8–11,13 although factors, such as mixed tumor subtypes in the study cohort8,11 and nonuniform treatment protocols,9,10,13 are also possible limitations.
The correlation between methylation and protein expression is further supported by data from BiSEQ, which we performed as an alternative evaluation of methylation to address the major limitation of MSP, because it is nonquantitative and has limited interrogation of promoter CpG sites. BiSEQ is straightforward and can be performed on conventional DNA sequencing instruments without any specialized procedures beyond bisulfite conversion, which is also necessary for MSP. The 24 CpG sites evaluated coincide with 1 of the 2 differentially methylated regions in the MGMT promoter, DMR2, found to be particularly important for MGMT downregulation.32 By showing that expression level correlates with CpG methylation density in this region, we confirm the importance of DMR2 methylation for transcriptional regulation of MGMT. Consistent with the study by Dunn et al.,3 we also found that methylation density was correlated with survival. Overall, BiSEQ results were highly correlated with MSP, and it provides an attractive semiquantitative alternative to MSP. We did not determine the contribution of CpG sites outside this region or of individual CpG sites towards expression and prognosis, such as previously described.46
On the basis of our results, we recommend that both IHC and methylation determination be obtained and considered in combination for prognostication of GBM treatment outcome. In institutions where only IHC is available, high expression alone is prognostic of poor outcome. However, low expression is less informative, because these patients can have significantly different outcomes depending on methylation status. Our results highlight the possibility that low expression can occur for likely non–methylation-dependent reasons. Further investigation is necessary to understand why low-expressing patients with associated methylation outperform those without methylation and whether our results are applicable to low-grade and anaplastic gliomas. Our findings need to be validated in an independent dataset, preferably in the context of controlled clinical trials.
Supplementary Material
Funding
This work was supported by National Institutes of Health (NCI-K08CA124479 to A. L.), Merck/Schering Plough, Henry E. Singleton Brain Tumor Program, Art of the Brain, Ben and Catherine Ivy Foundation, and American Brain Tumor Association Medical Student Summer Fellowship Supported by the Eric Perry Memorial 5 k Run/Walk (to S. L.).
Supplementary Material
Acknowledgments
We thank SiliconMed for providing data aggregation and search support and the UCLA Brain Tumor Translational Resource for logistical and technical support. This work was previously presented at the ASCO Annual Meeting 2011 as an oral presentation.
Conflict of interest statement. None declared.
References
- 1.Stupp R, Mason WP, van den Bent MJ, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352(10):987–996. doi: 10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- 2.Weller M, Stupp R, Reifenberger G, et al. MGMT promoter methylation in malignant gliomas: ready for personalized medicine? Nat Rev Neurol. 2009;6(1):39–51. doi: 10.1038/nrneurol.2009.197. [DOI] [PubMed] [Google Scholar]
- 3.Dunn J, Baborie A, Alam F, et al. Extent of MGMT promoter methylation correlates with outcome in glioblastomas given temozolomide and radiotherapy. Br J Cancer. 2009;101(1):124–131. doi: 10.1038/sj.bjc.6605127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Karayan-Tapon L, Quillien V, Guilhot J, et al. Prognostic value of O6-methylguanine-DNA methyltransferase status in glioblastoma patients, assessed by five different methods. J Neurooncol. 2010;97(3):311–322. doi: 10.1007/s11060-009-0031-1. [DOI] [PubMed] [Google Scholar]
- 5.Cao VT, Jung TY, Jung S, et al. The correlation and prognostic significance of MGMT promoter methylation and MGMT protein in glioblastomas. Neurosurgery. 2009;65(5):866–875. doi: 10.1227/01.NEU.0000357325.90347.A1. discussion 875. [DOI] [PubMed] [Google Scholar]
- 6.Hegi ME, Diserens AC, Gorlia T, et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. N Engl J Med. 2005;352(10):997–1003. doi: 10.1056/NEJMoa043331. [DOI] [PubMed] [Google Scholar]
- 7.Kulis M, Esteller M. DNA methylation and cancer. Adv Genet. 2010;70:27–56. doi: 10.1016/B978-0-12-380866-0.60002-2. [DOI] [PubMed] [Google Scholar]
- 8.Brell M, Ibanez J, Tortosa A. O6-Methylguanine-DNA methyltransferase protein expression by immunohistochemistry in brain and non-brain systemic tumours: systematic review and meta-analysis of correlation with methylation-specific polymerase chain reaction. BMC Cancer. 2011;11(1):35. doi: 10.1186/1471-2407-11-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Preusser M, Charles Janzer R, Felsberg J, et al. Anti-O6-methylguanine-methyltransferase (MGMT) immunohistochemistry in glioblastoma multiforme: observer variability and lack of association with patient survival impede its use as clinical biomarker. Brain Pathol. 2008;18(4):520–532. doi: 10.1111/j.1750-3639.2008.00153.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rodriguez FJ, Thibodeau SN, Jenkins RB, et al. MGMT immunohistochemical expression and promoter methylation in human glioblastoma. Appl Immunohistochem Mol Morphol. 2008;16(1):59–65. doi: 10.1097/PAI.0b013e31802fac2f. [DOI] [PubMed] [Google Scholar]
- 11.Sasai K, Nodagashira M, Nishihara H, et al. Careful exclusion of non-neoplastic brain components is required for an appropriate evaluation of O6-methylguanine-DNA methyltransferase status in glioma: relationship between immunohistochemistry and methylation analysis. Am J Surg Pathol. 2008;32(8):1220–1227. doi: 10.1097/PAS.0b013e318164c3f0. [DOI] [PubMed] [Google Scholar]
- 12.Weller M, Felsberg J, Hartmann C, et al. Molecular predictors of progression-free and overall survival in patients with newly diagnosed glioblastoma: a prospective translational study of the German Glioma Network. J Clin Oncol. 2009;27(34):5743–5750. doi: 10.1200/JCO.2009.23.0805. [DOI] [PubMed] [Google Scholar]
- 13.Yachi K, Watanabe T, Ohta T, et al. Relevance of MSP assay for the detection of MGMT promoter hypermethylation in glioblastomas. Int J Oncol. 2008;33(3):469–475. [PubMed] [Google Scholar]
- 14.Capper D, Mittelbronn M, Meyermann R, et al. Pitfalls in the assessment of MGMT expression and in its correlation with survival in diffuse astrocytomas: proposal of a feasible immunohistochemical approach. Acta Neuropathol. 2008;115(2):249–259. doi: 10.1007/s00401-007-0310-x. [DOI] [PubMed] [Google Scholar]
- 15.Friedman HS, McLendon RE, Kerby T, et al. DNA mismatch repair and O6-alkylguanine-DNA alkyltransferase analysis and response to Temodal in newly diagnosed malignant glioma. J Clin Oncol. 1998;16(12):3851–3857. doi: 10.1200/JCO.1998.16.12.3851. [DOI] [PubMed] [Google Scholar]
- 16.Nakagawa T, Ido K, Sakuma T, et al. Prognostic significance of the immunohistochemical expression of O6-methylguanine-DNA methyltransferase, P-glycoprotein, and multidrug resistance protein-1 in glioblastomas. Neuropathology. 2009;29(4):379–388. doi: 10.1111/j.1440-1789.2008.00983.x. [DOI] [PubMed] [Google Scholar]
- 17.Nakasu S, Fukami T, Baba K, et al. Immunohistochemical study for O6-methylguanine-DNA methyltransferase in the non-neoplastic and neoplastic components of gliomas. J Neurooncol. 2004;70(3):333–340. doi: 10.1007/s11060-004-9170-6. [DOI] [PubMed] [Google Scholar]
- 18.Sonoda Y, Yokosawa M, Saito R, et al. O(6)-Methylguanine DNA methyltransferase determined by promoter hypermethylation and immunohistochemical expression is correlated with progression-free survival in patients with glioblastoma. Int J Clin Oncol. 2010;15(4):352–358. doi: 10.1007/s10147-010-0065-6. [DOI] [PubMed] [Google Scholar]
- 19.Watanabe R, Nakasu Y, Tashiro H, et al. O(6)-Methylguanine DNA methyltransferase expression in tumor cells predicts outcome of radiotherapy plus concomitant and adjuvant temozolomide therapy in patients with primary glioblastoma. Brain Tumor Pathol. 2011;28(2):127–135. doi: 10.1007/s10014-011-0022-8. [DOI] [PubMed] [Google Scholar]
- 20.Kang SH, Park KJ, Kim CY, et al. O(6)-methylguanine DNA methyltransferase status determined by promoter methylation and immunohistochemistry in gliosarcoma and their clinical implications. J Neurooncol. 2010;101(3):477–486. doi: 10.1007/s11060-010-0267-9. [DOI] [PubMed] [Google Scholar]
- 21.Felsberg J, Thon N, Eigenbrod S, et al. Promoter methylation and expression of MGMT and the DNA mismatch repair genes MLH1, MSH2, MSH6 and PMS2 in paired primary and recurrent glioblastomas. International Journal of Cancer. 2011;129(3):659–670. doi: 10.1002/ijc.26083. [DOI] [PubMed] [Google Scholar]
- 22.Lai A, Tran A, Nghiemphu PL, et al. Phase II study of bevacizumab plus temozolomide during and after radiation therapy for patients with newly diagnosed glioblastoma multiforme. J Clin Oncol. 2011;29(2):142–148. doi: 10.1200/JCO.2010.30.2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pope WB, Lai A, Mehta R, et al. Apparent Diffusion Coefficent Histogram Analysis Stratifies Progression-Free Survival in Newly Diagnosed Bevacizumab-Treated Glioblastoma. AJNR Am J Neuroradiol. 2011;32(5):882–889. doi: 10.3174/ajnr.A2385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Carrillo JA, Lai A, Nghiemphu PL, et al. Relationship between Tumor Enhancement, Edema, IDH1 Mutational Status, MGMT Promoter Methylation, and Survival in Glioblastoma. AJNR Am J Neuroradiol. 2012;33(7):1349–1355. doi: 10.3174/ajnr.A2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ellingson BM, Cloughesy TF, Pope WB, et al. Anatomic localization of O6-methylguanine DNA methyltransferase (MGMT) promoter methylated and unmethylated tumors: a radiographic study in 358 de novo human glioblastomas. Neuroimage. 2012;59(2):908–916. doi: 10.1016/j.neuroimage.2011.09.076. [DOI] [PubMed] [Google Scholar]
- 26.Lai A, Kharbanda S, Pope WB, et al. Evidence for sequenced molecular evolution of IDH1 mutant glioblastoma from a distinct cell of origin. J Clin Oncol. 2011;29(34):4482–4490. doi: 10.1200/JCO.2010.33.8715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schemper M, Smith TL. A note on quantifying follow-up in studies of failure time. Controlled clinical trials. 1996;17(4):343–346. doi: 10.1016/0197-2456(96)00075-x. [DOI] [PubMed] [Google Scholar]
- 28.Grossman SA, Ye X, Piantadosi S, et al. Survival of patients with newly diagnosed glioblastoma treated with radiation and temozolomide in research studies in the United States. Clin Cancer Res. 2010;16(8):2443–2449. doi: 10.1158/1078-0432.CCR-09-3106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Altman DG, Andersen PK. Bootstrap investigation of the stability of a cox regression model. Statistics in medicine. 1989;8(7):771–783. doi: 10.1002/sim.4780080702. [DOI] [PubMed] [Google Scholar]
- 30.Sauerbrei W, Schumacher M. A bootstrap resampling procedure for model building: Application to the cox regression model. Statistics in medicine. 1992;11(16):2093–2109. doi: 10.1002/sim.4780111607. [DOI] [PubMed] [Google Scholar]
- 31.Verweij PJM, Van Houwelingen HC. Cross-validation in survival analysis. Statistics in medicine. 1993;12(24):2305–2314. doi: 10.1002/sim.4780122407. [DOI] [PubMed] [Google Scholar]
- 32.Malley DS, Hamoudi RA, Kocialkowski S, et al. A distinct region of the MGMT CpG island critical for transcriptional regulation is preferentially methylated in glioblastoma cells and xenografts. Acta Neuropathol. 2011;121(5):651–661. doi: 10.1007/s00401-011-0803-5. [DOI] [PubMed] [Google Scholar]
- 33.Nakagawachi T, Soejima H, Urano T, et al. Silencing effect of CpG island hypermethylation and histone modifications on O6-methylguanine-DNA methyltransferase (MGMT) gene expression in human cancer. Oncogene. 2003;22(55):8835–8844. doi: 10.1038/sj.onc.1207183. [DOI] [PubMed] [Google Scholar]
- 34.Shah N, Lin B, Sibenaller Z, et al. Comprehensive analysis of MGMT promoter methylation: correlation with MGMT expression and clinical response in GBM. PLoS One. 2011;6(1):e16146. doi: 10.1371/journal.pone.0016146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ma LC, Kuo CC, Liu JF, et al. Transcriptional repression of O6-methylguanine DNA methyltransferase gene rendering cells hypersensitive to N,N'-bis(2-chloroethyl)-N-nitrosurea in camptothecin-resistant cells. Mol Pharmacol. 2008;74(2):517–526. doi: 10.1124/mol.107.043620. [DOI] [PubMed] [Google Scholar]
- 36.Bocangel D, Sengupta S, Mitra S, et al. p53-Mediated down-regulation of the human DNA repair gene O6-methylguanine-DNA methyltransferase (MGMT) via interaction with Sp1 transcription factor. Anticancer Res. 2009;29(10):3741–3750. [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang W, Zhang J, Hoadley K, et al. miR-181d: a predictive glioblastoma biomarker that downregulates MGMT expression. Neuro-Oncology. 2012;0:0–0. doi: 10.1093/neuonc/nos089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chinot OL, Barrie M, Fuentes S, et al. Correlation between O6-methylguanine-DNA methyltransferase and survival in inoperable newly diagnosed glioblastoma patients treated with neoadjuvant temozolomide. J Clin Oncol. 2007;25(12):1470–1475. doi: 10.1200/JCO.2006.07.4807. [DOI] [PubMed] [Google Scholar]
- 39.Grombacher T, Mitra S, Kaina B. Induction of the alkyltransferase (MGMT) gene by DNA damaging agents and the glucocorticoid dexamethasone and comparison with the response of base excision repair genes. Carcinogenesis. 1996;17(11):2329–2336. doi: 10.1093/carcin/17.11.2329. [DOI] [PubMed] [Google Scholar]
- 40.Ueda S, Mineta T, Nakahara Y, et al. Induction of the DNA repair gene O6-methylguanine-DNA methyltransferase by dexamethasone in glioblastomas. J Neurosurg. 2004;101(4):659–663. doi: 10.3171/jns.2004.101.4.0659. [DOI] [PubMed] [Google Scholar]
- 41.Zhang J, Stevens MF, Laughton CA, et al. Acquired resistance to temozolomide in glioma cell lines: molecular mechanisms and potential translational applications. Oncology. 2010;78(2):103–114. doi: 10.1159/000306139. [DOI] [PubMed] [Google Scholar]
- 42.Fritz G, Tano K, Mitra S, et al. Inducibility of the DNA repair gene encoding O6-methylguanine-DNA methyltransferase in mammalian cells by DNA-damaging treatments. Mol Cell Biol. 1991;11(9):4660–4668. doi: 10.1128/mcb.11.9.4660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Metellus P, Coulibaly B, Nanni I, et al. Prognostic impact of O6-methylguanine-DNA methyltransferase silencing in patients with recurrent glioblastoma multiforme who undergo surgery and carmustine wafer implantation: a prospective patient cohort. Cancer. 2009;115(20):4783–4794. doi: 10.1002/cncr.24546. [DOI] [PubMed] [Google Scholar]
- 44.Wiewrodt D, Nagel G, Dreimuller N, et al. MGMT in primary and recurrent human glioblastomas after radiation and chemotherapy and comparison with p53 status and clinical outcome. Int J Cancer. 2008;122(6):1391–1399. doi: 10.1002/ijc.23219. [DOI] [PubMed] [Google Scholar]
- 45.Harris LC, Remack JS, Brent TP. Identification of a 59 bp enhancer located at the first exon/intron boundary of the human O6-methylguanine DNA methyltransferase gene. Nucleic Acids Res. 1994;22(22):4614–4619. doi: 10.1093/nar/22.22.4614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Everhard S, Tost J, El Abdalaoui H, et al. Identification of regions correlating MGMT promoter methylation and gene expression in glioblastomas. Neuro Oncol. 2009;11(4):348–356. doi: 10.1215/15228517-2009-001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.