Abstract
Background
Ongoing characterization of glioma has revealed that Akt signaling plays a crucial role in gliomagenesis. In mouse models, however, Akt alone was not sufficient to induce glioma.
Methods
We established transgenic zebrafish that overexpressed dominant-active (DA) human Akt1 or Rac1G12V (DARac1) at ptf1a domain and investigated transgenic phenotypes and mechanisms leading to gliomagenesis.
Results
Transgene expressions were spatiotemporally restricted without any developmental abnormality of embryos and persisted at cerebellum and medulla in adult zebrafish. DAAkt1 alone induced glioma (with visible bumps at the head), with incidences of 36.6% and 49% at 6 and 9 months, respectively. Histologically, gliomas showed various histologic grades, increased proliferation, and frequent invasion into the fourth ventricle. Preferential location of small tumors at periventricular area and coexpression of Her4 suggested that tumors originated from Ptf1a- and Her4-positive progenitor cells at ventricular zone. Gliomagenesis was principally mediated by activation of survival pathway through upregulation of survivin genes. Although DARac1 alone was incapable of gliomagenesis, when coexpressed with DAAkt1, gliomagenesis was accelerated, showing higher tumor incidences (62.0% and 73.3% at 6 and 9 months, respectively), advanced histologic grade, invasiveness, and shortened survival. DARac1 upregulated survivin2, cyclin D1, β-catenin, and snail1a but downregulated E-cadherin, indicating that DARac1 promotes gliomagenesis by enhancing proliferation, survival, and epithelial-to-mesenchymal transition. On pharmacologic tests, only Akt1/2 inhibitor effectively suppressed gliomagenesis, inhibited cellular proliferation, and induced apoptosis in established gliomas.
Conclusions
The zebrafish model reinforces the pivotal role of Akt signaling in gliomagenesis and suggests Rac1 as an important protein involved in progression.
Keywords: Akt1, epithelial-mesenchymal transition, glioma, Rac1, transgenic zebrafish
Primary brain tumors account for 1.4% of all cancers and 2.4% of all cancer-related deaths in the United States, and malignant gliomas account for ∼70% of malignant primary brain tumors.1 Glioma can be classified histologically from grade I to grade IV, according to World Health Organization (WHO) criteria, with glioblastoma (grade IV) accounting for 70% of malignant gliomas.2 Primary glioblastoma is predominant, with 10% being secondary glioblastoma formed by progression of pre-existing low-grade glioma.
Glioblastomas are formed by the sequential accumulation of genetic alterations, including the loss of the tumor-suppressor function, deregulation, and activation of growth factor signaling and activation of the survival pathway.1,3 Although molecular signatures vary between primary and secondary glioblastomas, p16, p53, retinoblastoma, and PTEN are among the genes most frequently lost in glioblastoma. Deregulation of growth factor signaling includes overexpression or activating mutations of platelet-derived growth factor (PDGF), PDGF receptor, epidermal growth factor receptor (EGFR), mouse double minute-2 (mdm2), CDK4, and PI3K.4,5 These alterations most often result in the activation of the Akt pathway.
Akt (also known as PKB), a serine-threonine protein kinase, is a key mediator of the phosphatidylinositol 3-kinase (PI3K)-Akt-mammalian target of rapamycin (mTOR) signal pathway, which is activated in up to 90% of all glioblastomas.6 The activation of Akt signaling can be initiated by PTEN inactivation, PI3K activation, or upregulation of growth factors.7,8 Studies of Akt have showed its involvement in such diverse physiological actions as nutrient metabolism, protein synthesis, cell survival, transcriptional regulation, the cell cycle, cell apoptosis, and proliferation. The anti-apoptotic signal is largely mediated by phosphorylation resulting in the inhibition of Bad, caspase-9, and forkhead transcriptional factor and the activation of I-κB kinase and Mdm2.9,10 Akt has also been identified as a positive regulator of the survivin gene,11 which is a member of the inhibitors of apoptosis (IAP) family expressed during embryonic development but not in terminally differentiated adult tissues.12–14 Subsequent studies have revealed that survivin is re-expressed in transformed cell lines and in a variety of human tumors and is essential for the anti-apoptotic function.15,16 mTOR plays a key role in mediating Akt-induced cell proliferation. Activated mTOR complex activates ribosomal protein S6 kinase (RS6K) and inactivates the eukaryotic translation initiation factor 4E binding protein 1 (4EBP1), which inhibits eukaryotic translation initiation factor 1 (Eif1a). Akt also inhibits GSK3β, which acts as a negative regulator of cell cycle progression through inhibition of cyclin D1.17,18
Rho GTPase family proteins mediate distinct cytoskeletal rearrangements in response to receptor stimulations and have been implicated in the establishment and maintenance of cadherin-based cell-cell adhesions.19,20 Rac is a member of this family and counteracts Rho activity. The reciprocal balance between Rho and Rac activity is a major determinant of cellular morphology and motility.21 Active Rac1 signaling is associated with acquiring the mesenchymal phenotype of cancer cells and with enhanced motility and invasion.22,23 In vitro studies have shown that suppression of Rac1 activity is associated with the immobilization and induction of apoptosis of glioma cells.24,25 In addition, Rac1 controls the nuclear localization of β-catenin by phosphorylation, which induces cell proliferation.26
Although evidence has been reported on the critical role of Akt signaling in gliomagenesis, the expression of Akt alone was insufficient to induce glioma in mouse models3,27; coactivation of Kras signaling was needed to induce glioblastoma. In the current study, we established transgenic zebrafish expressing dominant-active (DA) DAAkt1 or DARac1 at the ptf1a domain. We found that DAAkt1 alone induced glioma, and this process was accelerated by coexpression of DARac1; the relevant mechanism in which Akt1 and Rac1 are involved for glioma formation and progression was also investigated.
Materials and Methods
Transgenesis
We used Ptf1aGal4 fish previously established for targeted expression of transgenes under the regulation of the upstream activating sequence (UAS) by a binary expression system (Fig. 1A).28,29 Human DAAkt1 (1036 pcDNA3 Myr-HA-Akt1; myristoylated AKT1 targeted to the membrane independently of PtdIns-3,4,5-P3 and referred to as dominant-active human AKT1) was purchased from Addgene (Cambridge, MA), and human Rac1 cDNA was purchased from Open Biosystems (Huntsville, AL).
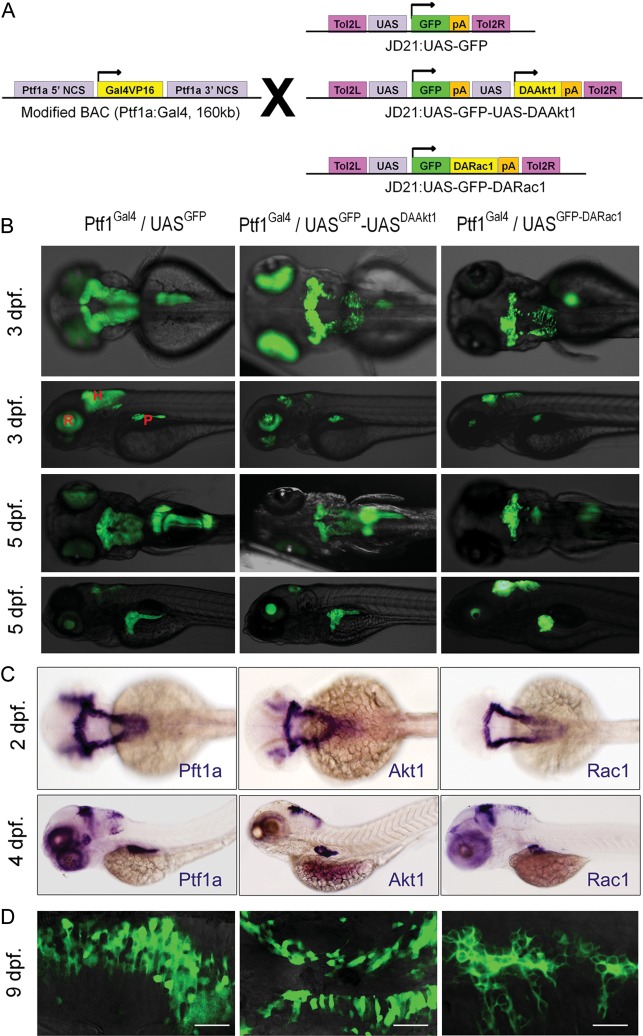
Fig. 1.
Targeted expression of transgenes in embryos. (A) Transgenesis strategy. Gal4-UAS system allows targeted expression of transgenes. (B) Inverted fluorescence images (upper column, dorsal views; lower column, lateral views). Transgenic GFP expression is spatiotemporally restricted to the Ptf1a domain. Exocrine pancreatic expression of DARac1 disturbs the posterior expansion of the exocrine pancreas. The morphology of the hindbrain is not altered by either DAAkt1 or DARac1 expression. (C) Whole-mount ISH at 2 dpf (dorsal views) and 4 dpf (lateral views). (D) Confocal images showing membrane localization of GFP fused with DARac1. Abbreviations: H, hindbrain; R, retina; P, exocrine pancreas. Bars, 20 μm.
With use of polymerase with a proofreading function (Invitrogen, Grand Island, NY), a green fluorescent protein (GFP) sequence including a polyA site was polymerase chain reaction (PCR) amplified from pEGFP1 vector (Clontech, Mountain View, CA) using F-GFP-Nco1/R-GFPpA-Xho1 primers. The PCR product was digested and inserted into the Nco1/Xho1 sites of JD21:UAS-GFP-Kras (a generous gift from Steven D. Leach) to generate JD21:UAS-GFPpA-Kras. DAAkt1 was PCR amplified using F-DAAkt1-Nco1/R-DAAkt1-Cla1 primers, digested, and inserted into Nco1/Cla1 sites of JD21:UAS-GFP-Kras to generate JD21:UAS-DAAkt1. Then, the UAS-DAAkt1 sequence was PCR amplified using F-UAS-Xho1/R-DAAkt1-Cla1, digested, and inserted into the Xho1/Cla1 sites of JD21:UAS-GFPpA-Kras to generate the final transgene construct, JD21:UAS-GFP-UAS-DAAkt1 (Fig. 1A). For targeted expression of DARac1G12V, Rac1 was PCR amplified using F-Rac1(G12V)-Nde1/R-Rac1-Cla1, digested, and inserted into the Nde1/Cla1 site of JD21:UAS-GFP-Kras to generate JD21:UAS-GFP-DARac1 (Fig. 1A). In this construct, the DARac1 is expressed as a fusion protein with GFP. The coexpressed GFP was intended to enable real-time observation. A control construct was generated by digesting JD21:UAS-GFPpA-Kras with Xhol1/Cla1, blunting, and then self-ligation. JD21:UAS-mCherrypA was generated by replacing GFP of JD21:UAS-GFPpA with mCherry. All constructs were sequence verified using the appropriate primers. Primers used for transgene constructs are listed in Supplementary Table S1.
Each injection mixture was made by reconstituting Tol2-transposase mRNA (20 ng/µL) and a transgene construct (20 ng/µL) in Danieu's buffer mixed with 0.03% phenol red. Single-cell stage Ptf1aGal4 embryos were transferred to a molded agarose dish, and 4 pL of the injection mixture was introduced by yolk injection with use of a MMPI-2 microinjector. Approximately 50% of the injected embryos survived. On day 2, fluorescence was monitored using an Olympus inverted fluorescence microscope, and F0 founder embryos showing green fluorescence at the Ptf1a domain were selected. For each transgene construct, ∼100 founder embryos were raised until adulthood and outcrossed to generate F1 transgenic zebrafish (Fig. 1). The use of Tol2-mediated transgenesis greatly enhanced the transgenic efficiency, such that 25%–50% of F0 zebrafish from each construct produced F1 offspring expressing transgenes. In each clutch of F1 embryos, ∼10% showed transgene expression. Among the F1 progenies, embryos showing faithful expression were selected and raised to produce F2 progenies. All transgenes were transmitted in normal Mendelian ratios. Through outcrossing with AB lines, all experiments were performed using zebrafish heterozygous for each transgene.
Animal Stocks and Embryo Care
All zebrafish were raised in a standardized aquarium system (Genomic-Design Co., Daejeon, Korea) according to standard protocols at 28°C on a 14:10-h light-dark cycle. The mitfaw2; roya9 (AB) zebrafish were purchased from Zebrafish International Resource Center (ZIRC, Eugene, OR). GFAPGFP,30 Her4RFP,31 and Olig2RFP32 transgenic fish were kind gifts from Hae-Chul Park. Embryos to be processed for whole-mount analyses were placed in E3 medium with 0.003% phenylthiourea at 24 h after fertilization to inhibit pigmentation. We strictly followed the Guidelines for the Welfare and Use of Animals in Cancer Research.33
Histology, Immunohistochemistry, In Situ Hybridization, and Immunofluorescence
Histologic evaluation was performed using 4-μm sections of 4% paraformaldehyde-fixed, paraffin-embedded tissues. Hematoxylin and eosin staining was performed according to standard protocols.34 Gene expression analyses were performed either by immunohistochemistry (IHC), if an antibody cross-reactive to zebrafish was available, or by in situ hybridization (ISH), as previously described.29,34 Primary antibodies were as follows: rabbit anti-Akt1 (1:500), mouse anti-Rac1 (1:500), mouse anti-proliferating cell nuclear antigen (PCNA) (1:2000), and rabbit anti-pancadherin (1:500) from Abcam (Cambridge, MA); rabbit anti-GFAP (1:200), rabbit anti-snail1a (1:500), rabbit anti-β-catenin1 (1:200), rabbit anti-caspase a (1:500), and rabbit anti-caspase b (1:500) from Anaspec (Fremont, CA, USA); and rabbit anti-phospho-histoneH3 (pHH3; 1:200), rabbit anti-phospho-mTOR (1:200), rabbit anti-phospho-4EBP1 (1:200), and rabbit anti-phospho-RS6K (1:200) from Cell Signaling (Danvers, MA). HRP-conjugated secondary antibodies were used and colored using diaminobenzidine solution. For labeling with 5-bromo-2′-deoxyuridine (BrdU), 2-month-old zebrafish were deprived of food for 24 h, bathed in a 1-L water tank containing 0.5% BrdU (Sigma-Aldrich, St. Louis, MO) for 2 h, rinsed, kept for 48 h, and processed for IHC, as previously described,35 with use of mouse anti-BrdU antibody (1:200; Dako, Gostrup, Denmark). Slides were counterstained with hematoxylin and mounted with Histomount (Zymed, San Francisco, CA).
Riboprobes were generated by PCR amplification of coding sequences from cDNA, TA cloning into pCRII vector (Invitrogen), and in vitro transcription. Primers used for TA cloning are listed in Supplementary Table S2. Hybridized embryos or sections were bound with an alkaline phosphatase-conjugated anti-digoxigenin antibody and colored using an NBT/BCIP solution. Sections were counterstained with neutral red and mounted with Histomount.
Immunofluorescence was performed using 8-μm cryosections of 4% paraformaldehyde-fixed tissues as previously described.36 Cryosections were incubated overnight in 10% goat serum with rabbit anti-S100 (1:400; Dako) antibody, washed with PBST, and then incubated overnight in 10% goat serum with a Cy3-conjugated secondary antibody (Jackson Labs, West Grove, PA).
Imaging
An Olympus MVX10 fluorescence microscope was used for whole-mount embryo imaging. Photographs from slide sections or live embryos were obtained using an Olympus BX51 microscope or a Carl Zeiss 700 confocal microscope.
Reverse-Transcription PCR
Real-time PCR was performed using dissected brain tissue from 3-month-old zebrafish. The coexpressed GFP enabled precise dissection of brain tissue. For each group, samples were collected from 3 or 4 zebrafish and used for total RNA extraction, followed by cDNA reverse transcription. Reverse-transcription (RT) PCR was performed as previously described29,34 with use of the 7300 Real Time PCR System (Applied Biosystems, Foster City, CA) with the QuantiTectTMSYBR Green PCR Kit (Qiagen, Valencia, CA). Samples were analyzed in triplicate, and all experiments were performed three times with use of separately prepared samples. Primer sequences for RT-RCR are shown in Supplementary Table S3.
Treatment with Akt Pathway Inhibitors
To counteract active Akt signaling in live zebrafish, rapamycin (mTOR inhibitor; IC50 50 pM; Sigma-Aldrich A8781), miltefosine (interferes with membrane localization of Akt; IC50 3–25 µM; Sigma-Aldrich M5571),37 and Akt1/2 inhibitor (kinase inhibitor; IC50 58 nM for Akt1, 210 nM for Akt2; Sigma-Aldrich A6730)38 were used. To determine treatment doses, 2-week-old embryos were treated with a serial escalation of each inhibitor for 7 days. We selected maximum tolerated doses (MTDs) that caused fatality rates of no more than 25%, which were 200 nM, 1 μM, and 500 nM for rapamycin, miltefosine, and Akt1/2 inhibitor, respectively. First, groups of 2-week-old Ptf1aGal4/UASGFP-UASDAAkt1 larvae (32 per group) were separately treated with the MTD of each inhibitor in 1-L water tanks up to 8 weeks and then processed for histology. Next, groups (3 per group) of 3-month-old Ptf1aGal4/UASGFP-UASDAAkt1 zebrafish having overt brain tumors were treated for 10 days and then processed for histologic evaluation. During the course of treatment, tank water was refreshed daily and inhibitors were newly added.
Statistical Analyses
Statistical analyses were performed using SPSS, version 11 (SPSS); P< 0.05 was considered to indicate a statistically significant difference.
Results
Transgene Expression in Embryos and Adult Zebrafish
Multiple independent transgenic lines were established: 7 for Ptf1aGal4/UASGFP-UASDAAkt1, 8 for Ptf1aGal4/UASGFP-DARac1, and 7 for Ptf1aGal4/UASGFP. Binary expression by a Gal4-UAS system allowed faithful expression of transgenes at the ptf1a domain (Fig. 1B and C). The expression levels, when estimated by GFP expression, were different among the F1 progenies, depending on their parental zebrafish. By selecting F1 embryos with faithful and robust expression of transgenes, we successfully established stable lines.
In DAAkt1- and DARac1-expressing embryos, hindbrain morphologies did not differ from those of control embryos. Exocrine pancreas in DARac1-expressing embryos, however, revealed morphological changes: the posterior growth of the exocrine pancreas was retarded, revealing a doughnut-shaped exocrine pancreas circumscribing the principal islet even at 5 days after fertilization. This phenotype might be attributable to the DARac1-induced cytoskeletal derangement, but detailed pancreatic phenotypes were beyond the scope of this study and were not further explored.
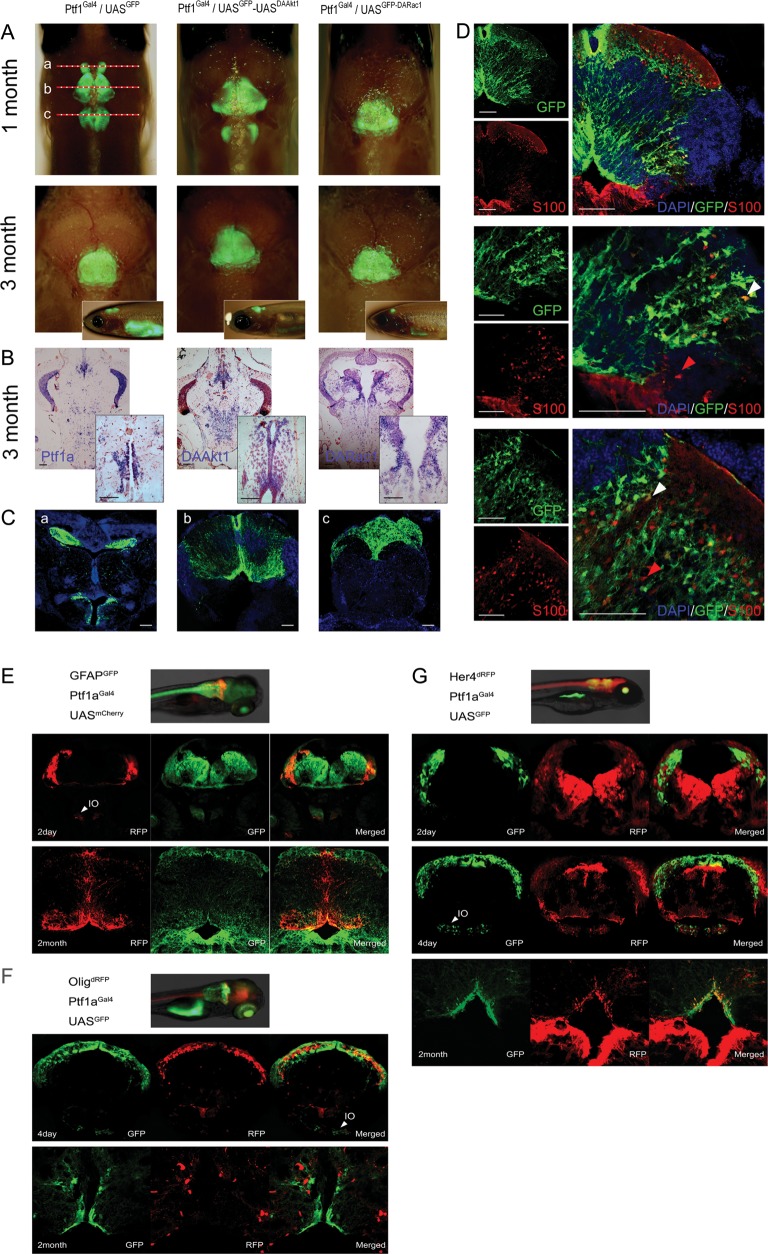
The precise expression domain of ptf1a has not yet been characterized. During embryonic development, Ptf1a expression was noted in cells at the dorsum of the cerebellum and inferior olive nucleus (Figs 1 and 2E). Although hindbrain expression of ptf1a has been reported to gradually decrease from 72 h after fertilization in zebrafish,39 we observed persistent expression at the cerebellum and medulla based on the observation of GFP expression (Fig. 2). Transgene expression was especially robust in cells along the midline of the cerebellum and at the intermediate layer and ventricular zone (dorsal lining of fourth ventricle), which has been assumed to be a niche for progenitor cells. Immunofluorescence revealed that a small proportion of ptf1a-positive cells expressed an astrocyte marker, S100 (Fig. 2D).
Fig. 2.
Transgene expression in juvenile and adult zebrafish. (A) Merged bright and fluorescence images showing GFP expression at the cerebellum and medulla. Depigmented phenotypes were obtained by successive crossing with the mitfaw2;roya9 line. Although 1-month-old zebrafish showed sustained GFP expression at the cerebellum and medulla, transgene expression was more localized at the cerebellum at 3 months. The top is anterior. Inlets are lateral views. (B) ISH for Ptf1a and transgenes. Transgene expression is stronger at the intermediate layer and the ventricular zone. Inlets are enlarged views of the ventricular area. (C) Coronal images of 1-month-old Ptf1aGal4/UASGFP zebrafish of (A). (b) GFP expression is robust along the midline and at the dorsal lining of the fourth ventricle. (D) Immunofluorescence for S100 at the similar plane level of (Cb). S100-positive cells are concentrated at the ventral lining of the fourth ventricle and the intermediate layer and scattered in the granular layer. Strong Ptf1a expression (GFP) persists in cells at intermediate layer, ventricular zone, and along the midline. Images reveal cerebellar cells positive for either GFP or S100 (red arrowheads) alone or for both (white arrowheads). (E–G) Coexpression analyses. During embryonic development, Ptf1a expression is noted in cells at the dorsum of cerebellum and inferior olive nucleus (IO). (E) GFAPGFP/Ptf1aGal4/UASmCherry zebrafish. Most cells expressing Ptf1a (RFP) also express GFAP (GFP). (F) Olig2dRFP/Ptf1aGal4/UASGFP zebrafish. Cerebellar cells do not coexpress Olig2 and Ptf1a both in embryo and adult. (G) Her4dRFP/Ptf1aGal4/UASGFP zebrafish. During embryonic development, ptf1a-positive cells (GFP) are at just underneath Her4-expressing (RFP) cells and rarely coexpress Her4. In adults, Ptf1a-positive cerebellar cells at ventricular and supraventricular areas occasionally coexpress both Ptf1a and Her4. Bars, 50 μm.
For further clarification, we performed coexpression analyses by crossing them with transgenic lines expressing biomarkers at the GFAP, Her4, or Olig2 domain. Most Ptf1a-positive cerebellar cells also expressed GFAP from embryo to adulthood (Fig. 2E). Olig2 expression, however, was not noted in Ptf1a-positive cells (Fig. 2F). Although Ptf1a-positive cells rarely coexpressed Her4 in embryos, cells at the supraventricular and ventricular zone occasionally coexpressed both ptf1a and Her4 in adult cerebellum (Fig. 2G). Ventricular Her4-positive cells have been assumed to be neuronal progenitors that proliferate after injury and produce neural cells in the adult zebrafish brain.40,41 The aforementioned findings suggest that ventricular ptf1a-positive cells encompass progenitor cells that can produce neuronal and glial cells.
DAAkt1 Induces Glioma of Various Histologic Grades
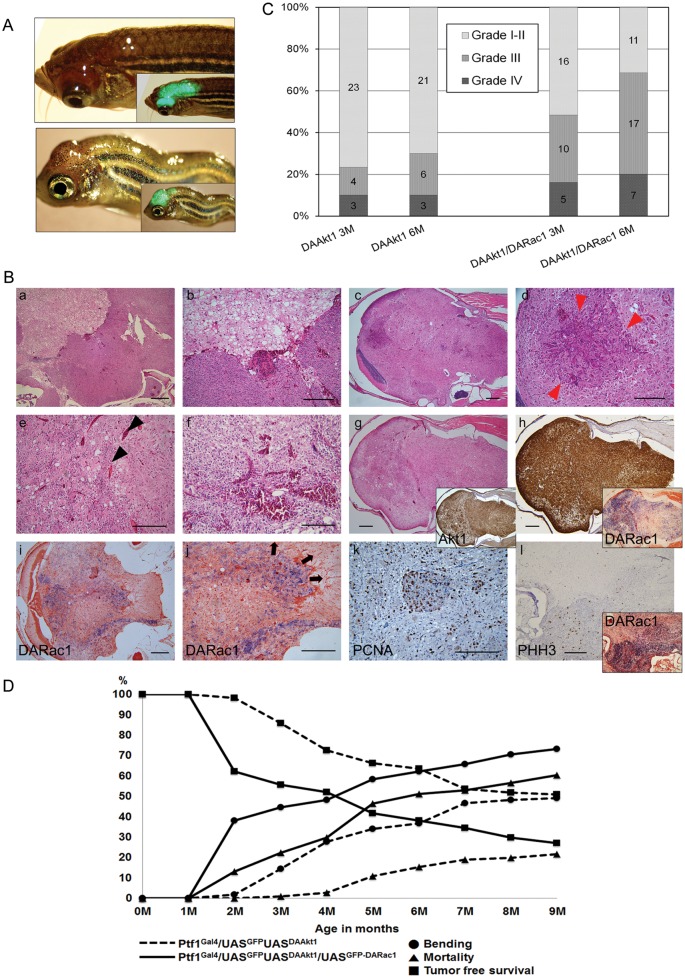
At 2 months of age, some of the DAAkt1-expressing zebrafish started to show morphologic changes in their body configuration. Their body became bent and then gradually tortuous. Approximately 1 month later, fish started to show visible bumps on the head, which had strong GFP expression (Fig. 3A and B). The bending of the body seemed to be attributable to motor disturbance caused by a tumor formed in the cerebellum. Because body bending always preceded the appearance of a visible tumor, we considered this phenotype to be a surrogate marker. On the basis of this surrogate marker, tumor incidence rates were 36.6% and 49% at 6 and 9 months, respectively.
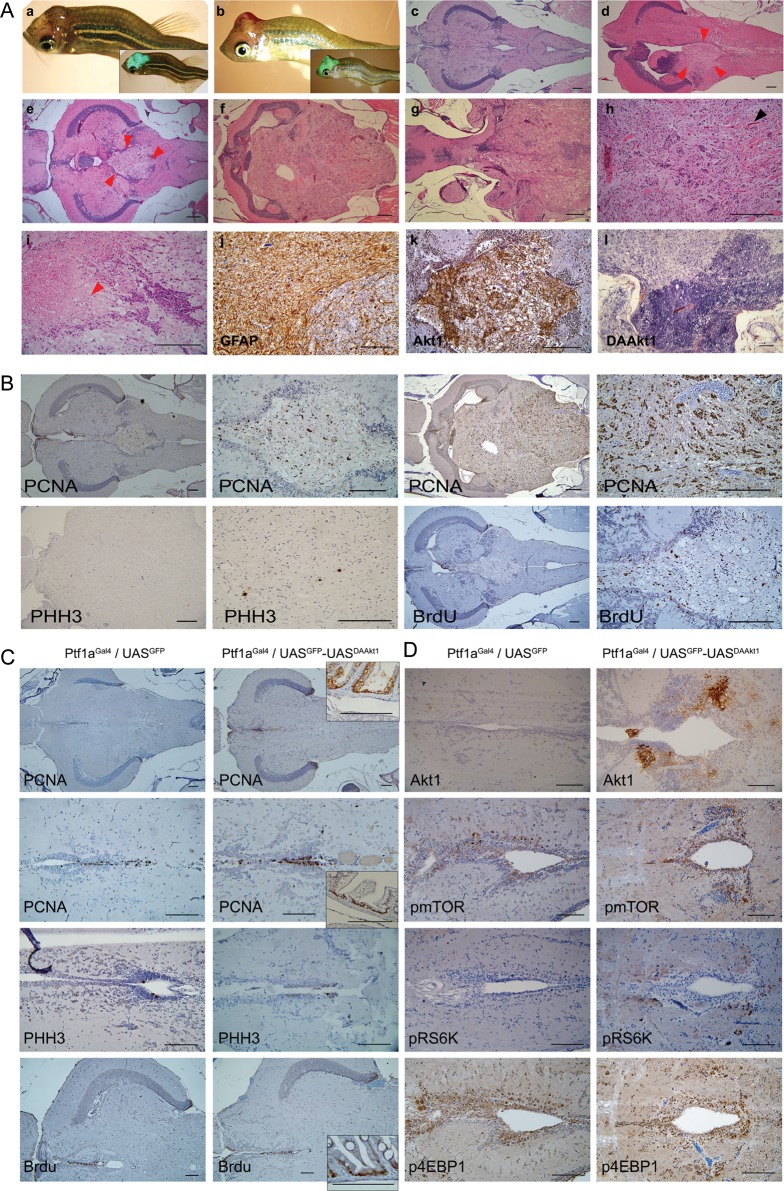
Fig. 3.
DAAkt1-induced gliomas. (A) Gross and histologic findings. (a,b) Glioma-bearing 3-month-old Ptf1aGal4/UASGFP-UASDAAkt1 zebrafish with bent body and visible bumps at the head, showing strong GFP expression on merged bright and fluorescence images (inlets). (c) Hematoxylin and eosin stains of cerebellum in control zebrafish. (d–i) Hematoxylin and eosin stains of gliomas. (d and e) Small tumors (boundaries by red arrowheads) at periventricular area showing invasion into 4th ventricle. (f) A large glioma replacing almost the whole cerebellum. (g) A large glioma showing invasion into the midbrain. (h) A high-grade glioma showing increased vascularity (black arrowhead). (i) A high-grade glioma showing necrosis (red arrowhead). (j) IHC for GFAP showing robust expression. (k) IHC for Akt1 on a small tumor invading 4th ventricle showing expression at tumor. (l) ISH for DAAkt1 showing stronger expression at the hypercellular area. (B) Increased proliferation in DAAkt1-induced gliomas. IHC indicates highly frequent positivity to PCNA, which is even higher in high-grade tumor. Tumors occasionally show positive staining to PHH3, a mitotic marker. BrdU labeling also reveals frequently positive cells. (C) Proliferation analyses in the pre-neoplastic cerebellum of 2-month-old zebrafish. Reactivity at the intestinal cells is used as an internal positive control (Inlets). Cells at the ventricular lining and the intermediate layer frequently reveal immunoreactivity to proliferative markers. The non–tumor-bearing cerebellum of DAAkt1-expressing zebrafish is not associated with increased proliferation. Average numbers of positive cells counted at the periventricular are 4.5 ± 2.2 and 4.7 ± 2.6 for PHH3 and 18.7 ± 6.8 and 19.9 ± 6.9 for BrdU in Ptf1aGal4/UASGFP and Ptf1aGal4/UASGFP-UASDAAkt1 zebrafish, respectively, which are not significantly different. Bars, 50 μm. (D) Activation of Akt downstream components in nonneoplastic DAAkt1-expressing cerebellum. IHC analyses reveal slightly increased expression of phospho-mTOR, -RS6K, and -4EBP1 in cells at the ventricular and periventricular zone.
In histologic evaluation, the zebrafish with bent bodies invariably had tumors at the cerebellum (Fig. 3A). The tumors were gliomas immunoreactive to GFAP, with a robust expression of Akt1 and histologic grades varying from I to IV according to the WHO classification. Larger tumors tended to have advanced histologic grades. High-grade tumors invariably showed hypercellularity, increased vascularity, and frequent mitosis, with occasional necrosis in ∼10% of tumors. Necrosis was infrequently observed because tumor-bearing zebrafish died of tumor before the tumor grew large enough for the development of necrosis. Advanced tumors frequently revealed various histologic grades in a tumor, showing areas with hypercellularity and a more robust expression of DAAkt1.
Gliomas Originate from Ptf1a- and Her4-Positive Progenitor Cells at the Ventricular Zone
Small tumors were invariably located at the vicinity of the fourth ventricle, showing disruption of the ventricular lining and a space-occupying mass in the ventricle (Fig. 3A). As a tumor grew, it gradually replaced the whole cerebellum and invaded the midbrain. For lineage evaluation, DAAkt1-expressing zebrafish were outcrossed to express RFP in the Her4 or Olig2 domain (Supplementary Fig. 2). Most of the DAAkt1-positive glioma cells also expressed Her4 but not Olig2. Taking into consideration that Her4-positive cells are neuronal progenitors in zebrafish brain, the aforementioned findings suggest that the tumor originated from Ptf1a- and Her4-expressing progenitor cells at the ventricular zone, which is known as a putative stem cell niche in the brain.42
DAAkt1 Expression Induced Akt Signaling Without Enhanced Proliferation
In the cerebellum of the control and DAAkt1-expressing zebrafish, proliferating cells were principally located at the ventricular zone and intermediate layer, where transgene expression was also robust (Figs 2B and 3C). DAAkt1-induced glioma cells showed a striking increase in cellular proliferation (Fig. 3B). However, non–tumor-bearing cerebellum from DAAkt1-expressing zebrafish did not show increased proliferation, compared with control (Fig. 3C). To see Akt signaling activation, we evaluated phosphorylated downstream components by IHC in preneoplastic cerebellum. In contrast to the GFP expression shown on confocal imaging and the DAAkt1 expression shown by ISH, activation of downstream components was noted in a small subset of cells at the ventricular and periventricular zone, showing an increased number of cells positive for pmTOR and pRS6K but not for p4EBP1 (Fig. 3D). However, an established glioma revealed strong expression of those active downstream proteins of Akt signaling (Fig. 6B). This finding suggests that DAAkt1-induced gliomagenesis occurs in the subset of cells in which active Akt signaling has been induced by DAAkt1 expression in this model.
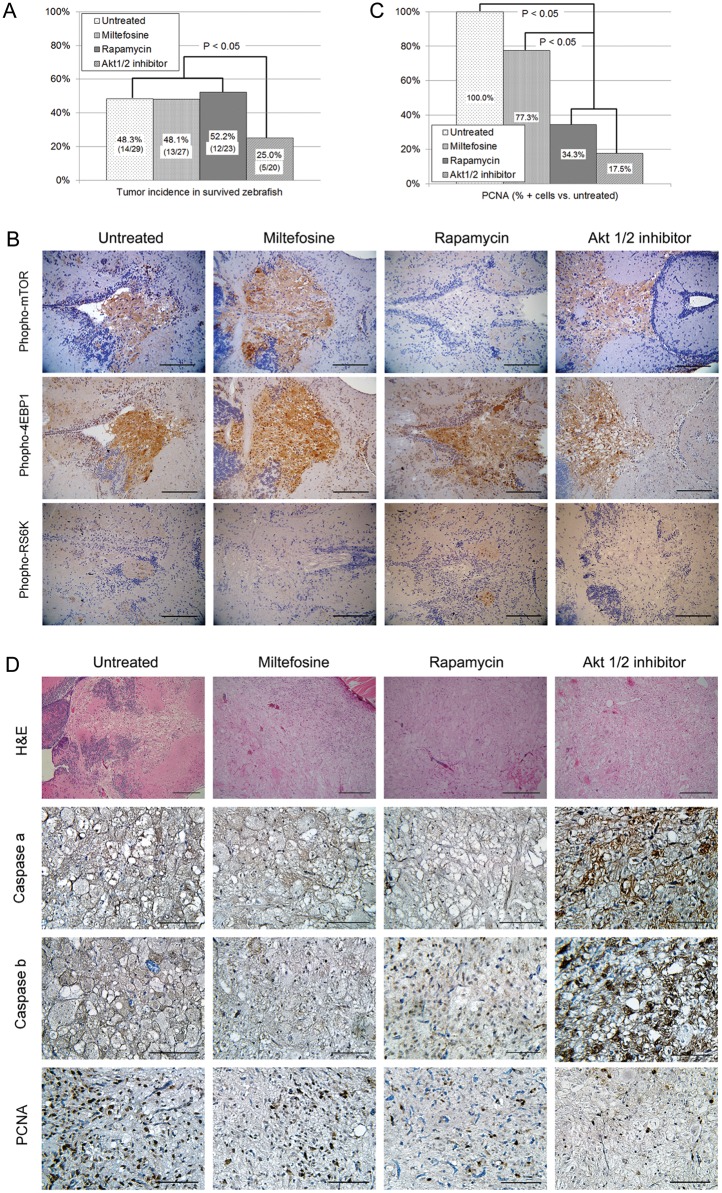
Fig. 6.
Treatment with Akt pathway inhibitors in groups of ptf1aGal4/UASGFP-UASDAAkt1 larvae. (A and B) Prolonged treatment from 2 weeks to 2 months. (A) Tumor incidence is significantly lowered by Akt1/2 inhibitor but not by miltefosine or rapamycin (χ2 test). (B) Expression of Akt1 downstream component. Untreated glioma cells express phospho-mTOR, -4EBP1, and RS6K, suggesting activation of Akt1 signaling. Although rapamycin treatment decreased phospho-mTOR expression, miltefosine and Akt1/2 inhibitor decreased phospho-RS6K expression. (C, D) Treatment in 3-month-old zebrafish with glioma. (C) Counted PCNA-positive cells from 10 high-power fields are significantly lowered by treatment with rapamycin or Akt1/2 inhibitor (ANOVA test). (D) IHC analyses indicated increased cells positive for active caspases in gliomas treated with Akt1/2 inhibitor and decreased cells positive for PCNA in gliomas treated with Akt1/2 inhibitor or rapamycin. Bars, 50 μm.
Coexpression of DARac1 Accelerates DAAkt1-Induced Tumorigenesis
Rac has been implicated in the migration and invasion of glioma cells.23,26 In the current model, DARac1 alone did not induce brain tumors during the 18-month follow-up. When DARac1-expressing zebrafish were crossed with DAAkt1-expressing fish to produce coexpression, gliomagenesis was greatly accelerated. On the basis of the surrogate marker, incidence rates were 44.4% at 3 months, 62.0% at 6 months, and 73.3% at 9 months (Fig. 4D). The coexpression of DARac1 not only enhanced tumor incidence but also increased histologic grade and invasiveness. The proportions of high-grade tumor (grade III or IV) were 48.4% and 67% at 3 and 6 months, respectively (Fig. 4C). Frequently, individual high-grade tumors showed heterogeneous histologic grades, manifested as intratumoral cell nests with increased cellularity (Fig. 4B); the intratumoral cell nests further revealed an increase in reactivity to proliferative markers and more robust expression of transgene DARac1 (Fig. 4C). Tumor cells at the invasion front showed more robust expression of DARac1 (Fig. 4C), suggesting enhanced invasiveness by coexpression of DARac1. This accelerated gliomagenesis and invasiveness resulted in earlier and higher mortality by DARac1 coexpression (Fig. 4D).
Fig. 4.
DARac1 accelerates DAAkt1-induced glioma. (A) Two-month-old zebrafish showing bumps at the head. Inlets are merged bright field and fluorescence images. (B) Histologic findings of gliomas in Ptf1aGal4/UASGFP-UASDAAkt1/UASGFP-DARac1 zebrafish. (a-g) Hematoxylin and eosin staining. Tumors frequently reveal heterogeneous grade of glioma showing a hypercellular nest within the tumor (red arrowheads) and increased vascularity (black arrowheads). (g and h) IHC and ISH for transgenes in a high-grade glioma. Inlets of (g) and (h) are IHC for Akt1 and ISH for DARac1, respectively. (i and j) ISH for DARac1. Cells at the invasion front show stronger expression of DARac1. Arrows indicate the direction of invasion. (k and l) Proliferation analyses reveal increased positivity to PCNA and PHH3, especially at the hypercellular areas, which accompanies a more robust expression of DARac1 on ISH (inlet). (C) Histologic grade of gliomas. Coexpression of DARac1 increased not only the tumor incidence but also the histologic grades of gliomas (D) Tumor incidence and mortality. 112 zebrafish from each Ptf1aGal4/UASGFP-UASDAAkt1 and Ptf1aGal4/UASGFP-UASDAAkt1/UASGFP-DARac1 line were followed. Tumor incidence was estimated by body bending, which preceded the appearance of an obvious bump in the head. The incidence rates of glioma in Ptf1aGal4/UASGFP-UASDAAkt1 and Ptf1aGal4/UASGFP-UASDAAkt1/UASGFP-DARac1 zebrafish are 14.3% and 44.4% at 3 months, 36.6% and 62.0% at 6 months, and 49.1% and 73.2% at 9 months, respectively. The mortality rates are also increased by the coexpression of DARac1: 0.9% vs. 22.2% at 3 months, 15.2% vs. 50.9% at 6 months, and 21.4% vs. 60.2% at 9 months. The tumor-free survival rates are much lower in the Ptf1aGal4/UASGFP-UASDAAkt1/UASGFP-DARac1 line. Bars, 50 μm.
Differentially Expressed Genes
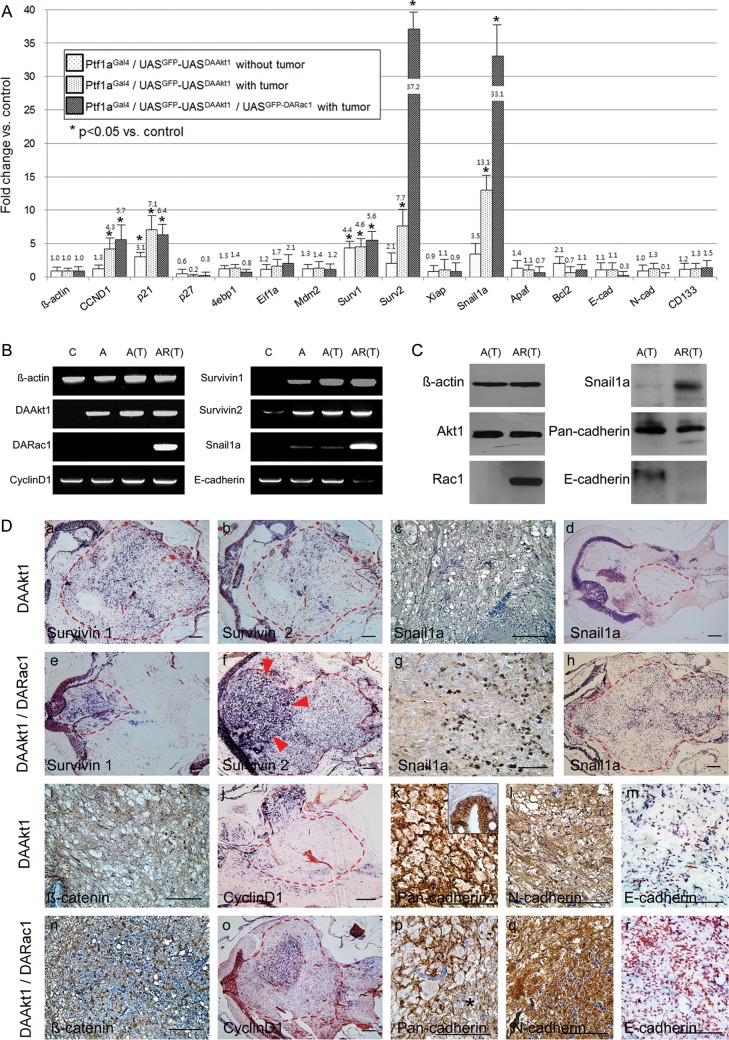
We selected a list of genes that might be modulated by the aberrant expression of DAAkt1 and DARac1. Among the genes evaluated, survivin1 and p21 were upregulated by DAAkt1 expression in non–tumor-bearing zebrafish. DAAkt1-induced glioma resulted in significant upregulation of p21, cyclin D1, survivin1, survivin2, and snail1a, compared with the control brain tissue (Fig. 5A and B). Coexpression of DARac1 was associated with striking upregulation of survivin2 and snail1a, modest upregulation of cyclin D1 and survivin1, and downregulation of E-cadherin. Western blot analyses confirmed the RT-PCR findings (Fig. 5C). This finding suggested that activation of survivin genes played an important role in the current model and that snail1a upregulation was involved in DARac1-induced acceleration of gliomagenesis.
Fig. 5.
Differential genes involved in gliomagenesis. (A) Real-time RT-PCR using dissected glioma and control cerebellum shows changes in transcripts. Survivin1 and p21 are upregulated in pre-neoplastic cerebellum by DAAkt1 expression. DAAkt1-induced glioma shows upregulation of cyclin D1, p21, Survivin1, 2, and snail1a. Coexpression of DARac1 reveals further upregulation of survivin2 and snail1a, and downregulation of E-cadherin. *P< 0.05. Mann-Whitney U test was used for statistical differences. Electrophoretic (B) and Western blot (C) images confirm quantitative RT-PCR findings. C, control; A, Ptf1aGal4/UASGFP-UASDAAkt1 without tumor; A(T), Ptf1aGal4/UASGFP-UASDAAkt1 with tumor; Ptf1aGal4/UASGFP-UASDAAkt1/UASGFP-DARac1 with tumor. (D) Expression analyses of differential genes by IHC (c, g, i, k, l, n, p, and q) or ISH (a, b, d, e, f, h, j, m, o, and r). Dotted red lines indicate tumor boundaries. Gliomas coexpressing DAAkt1 and DARac1 show stronger expression of survivin2, snail1a, β-catenin, and cyclin D1, especially at the hypercellular area (red arrowheads). Expression of pan-cadherin and E-cadherin is downregulated in glioma coexpressing DARac1, especially at the higher-grade area (* in p). Bars, 50 μm.
Next, expression patterns were evaluated at the histologic level (Fig. 5D). Gliomas coexpressing DAAkt1 and DARac1 showed higher expressions of β-catenin, cyclin D1, and survivin2, especially at intratumoral hypercellular nests, compared with tumors expressing DAAkt1 alone. Snail1a expression was also increased by coexpression of DARac1, especially at the invasion front and hypercellular area. Although the expressions of pan-cadherin and E-cadherin were downregulated, N-cadherin expression was increased by coexpression of DARac1. These findings suggest that DARac1 further enhanced cell proliferation and survival, along with cell migration and invasion, through epithelial-mesenchymal transition (EMT) by snail1a induction and through altering cell-to-cell adhesion.
Akt1/2 Inhibitor Suppressed Glioma Formation
Each group of 14-day-old DAAkt1-expressing larvae was treated with Akt pathway inhibitors up to 2 months of age. Prolonged treatment increased mortality rates, which were 9.4% (3/32), 15.6% (5/32), 28.1% (9/32), and 37.5% (12/32) for control, miltefosine, rapamycin, and Akt1/2 inhibitor, respectively. Akt1/2 inhibitor was effective in suppressing glioma formation (Fig. 6A), showing a 23.3% point decrease of tumors. Miltefosine or rapamycin treatment, however, was not preventive for gliomagenesis. Next, we treated 3-month-old adult zebrafish with established tumors with these agents for 10 days. Although none of these 3 agents resulted in tumor shrinkage, Akt1/2 inhibitor increased the number of tumor cells positive for active caspases (Fig. 6C). Proliferation analysis showed that both Akt1/2 inhibitor and rapamycin significantly reduced the number of tumor cells reactive to PCNA (Fig. 6B and D). The ineffectiveness of miltefosine probably resulted from an insufficient dose because of dose-limiting toxicity (MTD. 1 µM). Rapamycin effectively suppressed mTOR activation (Fig. 6B) and decreased PCNA-positive cells (Fig. 6C and D) but did not prevent gliomagenesis. These findings suggest that enhanced survival but not proliferation mediated by mTOR played a key role in Akt1-induced gliomagenesis in this model, which was more likely because of the preferential deregulation of survival genes found on RT-PCR.
Discussion
To our knowledge, this is the first study reporting that the targeted expression of DAAkt1 at the ptf1a domain alone initiates gliomagenesis in the zebrafish cerebellum. The tumor incidence was high, with half of DAAkt1-expressing zebrafish exhibiting glioma at 9 months of age. The discrepancy between zebrafish and a mammalian model may be attributable to the inherent susceptibility of zebrafish to Akt1-induced gliomagenesis or the different transgenic strategies used in the current model, which allowed faithful and robust expression by the GAL4-UAS system. DAAkt1 transgenic zebrafish made it possible to study single gene-mediated tumorigenesis, which allows a more straightforward approach to elucidating the pivotal role of Akt signaling and its molecular events in glioma formation. Unfortunately, antibodies reactive to zebrafish antigens are rarely available, and thus, it was impractical to investigate all Akt signaling targets; among the long lists of humanized antibodies against Akt downstream components, anti-pmTOR, anti-pRS6K, and anti-p4EBP1 were active only for IHC but not for Western blot.
Akt has multiple targets and is involved in mediating various biological responses, including cell proliferation, cytoskeleton, and survival, and transcription, protein synthesis, and nutrient metabolism, all of which are implicated in the process of tumorigenesis.8,10,43,44 Recent studies have suggested that PI3K-Akt signaling plays a crucial role in glioma formation and progression.43 PTEN inactivation frequently occurs in glioma, either by deletion, mutation, or promoter methylation, resulting in downstream activation of Akt signaling.7,45,46 Amplification of PDGFR or EGFR frequently occurs in glioma and also leads to Akt activation.
In the current model, we aberrantly expressed DAAkt1 at the Ptf1a domain in zebrafish. Ptf1a expression is induced in the hindbrain, spinal cord, retinal, and the exocrine pancreas during embryonic development in zebrafish. However, the precise cellular compartment of ptf1a expression in the brain has not yet been defined. Our current study revealed a persistent and strong expression of ptf1a in the cells at the ventricular zone and intermediate layer. Most of the ptf1a-positive cells were also positive for GFAP, and some were positive for Her4. A recent study suggested that neuronal cells can be derived from ventricular ptf1a-expressing progenitor cells.47 GFAP-positive radial glia cells can lead to both neurons and oligodendrocystes.32 In addition, Her4-positive ventricular progenitor cells proliferate and generate neuronal cells.40 In the current model, DAAkt1-induced gliomas were positive for GFAP and Her4, and tumors were invariably located at the periventricular area, where transgene expression was robust. Growing tumors gradually invaded both the ventricle and the granular layer of the cerebellum. Although the cell origin of glioma is still enigmatic, if the ventricular area is assumed to be a stem cell niche,42 the current study suggests that gliomas arise from a subset of progenitor cells expressing Ptf1a, Her4, and GFAP at the ventricular zone.
The predominant tumor type in human cerebellum is, in fact, medulloblastoma. The medulloblastoma differs from glioma in genetics and cell origin:48 the hedgehog and Wnt pathways play key roles in medulloblastoma, whereas the Akt pathway plays a key role in gliomagenesis. The finding of only gliomagenesis in the current model suggests that the genetic events occurred in a specific cell type, with Akt activation being the major factor determining tumor type. Although current models develop gliomas, which are infrequent in cerebellum, we believe these models also recapitulate gliomas occurring in cerebrum at molecular and genetic levels and are suitable for exploring mechanisms involved in gliomagenesis by activated Akt pathway.
Current data suggest that the survival pathway played a major role in the current model of AKT-induced gliomagenesis for several reasons. First, RT-PCR showed frank upregulation of survivin genes, which are members of IAP family, whereas Eif1a was not upregulated. Survivin genes regulate cell division and cell death,44 protect cells against apoptosis,49 are highly expressed in malignant glioma, and are also associated with a poor prognosis.50 Activation of the Akt pathway leads to survivin expression,51 and inhibition of Akt induces downregulation of survivin.11 Akt-induced proliferation relies on mTOR protein, whose activation leads to transcriptional activation of Eif1a, which revealed subtle change on RT-PCR. Second, when cellular proliferation was evaluated before tumor development, DAAkt1 did not induce widespread activation of downstream pathways or increased proliferation in ptf1a-positive ventricular cells; only established tumors did so. Third, although rapamycin treatment in established tumors reduced the number of PCNA-positive tumor cells and inhibited mTOR activation, gliomagenesis was not inhibited when larvae were treated for long periods. Therefore, it is more plausible that the survival pathway and not the proliferation pathway played a major role in the current glioma models.
The acceleration of gliomagenesis by coexpression of DARac1 was quite striking. A series of in vitro and in vivo studies suggested that activation of Rac promotes the migratory and metastatic abilities of tumor cells by enhancing EMT23,52 and also leads to an enhanced nuclear localization of β-catenin, resulting in increased cellular proliferation.53 Present data showed that coexpression of DARac1 not only enhanced tumor incidence but also aggravated histologic grade and invasiveness of glioma. On histologic evaluation, gliomas coexpressing DAAkt1 and DARac1 frequently revealed a mixture of low-grade and high-grade portions. The high-grade area of glioma showed an increase in proliferative markers accompanied by stronger expression of DARac1 and downregulation of E-cadherin. DARac1 induced further increases in cyclin D1 and β-catenin expression and striking increases in survivin2 and snail1a. Although Akt alone induces snail1 expression45,54 and tumor progression through EMT,55–57 the above findings indicate that Rac1 acts as an important player in glioma progression by enhancing EMT and promoting the survival and proliferation of glioma cells.
The current models can be a useful platform for the screening and evaluation of small molecular drugs targeting the Akt pathway. The zebrafish model provides a unique opportunity for high throughput in vivo screening of anticancer candidate drugs, which is not feasible in a mouse model. The feasibility of acquiring a toxicity profile in a physiologic context is an additional benefit of using the zebrafish as a model for drug screening. These inherent advantages will allow cost-effective and high-throughput in vivo study to search for effective small molecules that target the Akt pathway. The current model reflects the Akt1-induced enhancement of survival signaling and suggests that survivin is a promising target for the treatment of glioma.
In summary, this study identifies novel functions of Akt1 and Rac1 in the initiation and progression of gliomagenesis in zebrafish. Overexpression of DAAkt1 is sufficient to induce gliomagenesis through activation of anti-apoptosis signals. Furthermore, we identified Rac1 as an important molecule involved in the progression of glioma, which is mediated by enhanced EMT and a boost in the activation of survival and proliferation signaling. These transgenic models will be a valuable platform for exploring the mechanisms of gliomagenesis and may allow screening of potential targeted agents for the treatment of glioma.
Funding
This study was supported by Mid-Career Researcher Program (7-2011-0043) and Basic Science Research Program (7-2012-0531) through the National Research Foundation of Korea funded by the Ministry of Education, Science and Technology, and the Institutional Grant of Yonsei University College of Medicine (6-2012-0007).
Conflict of interest statement. None declared.
Supplementary material
References
- 1.Louis DN. Molecular pathology of malignant gliomas. Ann Rev Pathol. 2006;1:97–117. doi: 10.1146/annurev.pathol.1.110304.100043. [DOI] [PubMed] [Google Scholar]
- 2.Rousseau A, Mokhtari K, Duyckaerts C. The 2007 WHO classification of tumors of the central nervous system - what has changed? Curr Opin Neurol. 2008;21:720–727. doi: 10.1097/WCO.0b013e328312c3a7. [DOI] [PubMed] [Google Scholar]
- 3.Gladson CL, Prayson RA, Liu WM. The pathobiology of glioma tumors. Ann Rev Pathol. 2010;5:33–50. doi: 10.1146/annurev-pathol-121808-102109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rossig L, Jadidi AS, Urbich C, Badorff C, Zeiher AM, Dimmeler S. Akt-dependent phosphorylation of p21(Cip1) regulates PCNA binding and proliferation of endothelial cells. Mol Cell Biol. 2001;21:5644–5657. doi: 10.1128/MCB.21.16.5644-5657.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mayo LD, Donner DB. A phosphatidylinositol 3-kinase/Akt pathway promotes translocation of Mdm2 from the cytoplasm to the nucleus. Proc Natl Acad Sci USA. 2001;98:11598–11603. doi: 10.1073/pnas.181181198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cancer Genome Atlas Research Network. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dahia PL, Aguiar RC, Alberta J, et al. PTEN is inversely correlated with the cell survival factor Akt/PKB and is inactivated via multiple mechanisms in haematological malignancies. Hum Mol Genet. 1999;8:185–193. doi: 10.1093/hmg/8.2.185. [DOI] [PubMed] [Google Scholar]
- 8.Bos JL. A target for phosphoinositide 3-kinase: Akt/PKB. Trends Biochem Sci. 1995;20:441–442. doi: 10.1016/s0968-0004(00)89097-0. [DOI] [PubMed] [Google Scholar]
- 9.Daly C, Wong V, Burova E, et al. Angiopoietin-1 modulates endothelial cell function and gene expression via the transcription factor FKHR (FOXO1) Genes Dev. 2004;18:1060–1071. doi: 10.1101/gad.1189704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dan HC, Jiang K, Coppola D, et al. Phosphatidylinositol-3-OH kinase/AKT and survivin pathways as critical targets for geranylgeranyltransferase I inhibitor-induced apoptosis. Oncogene. 2004;23:706–715. doi: 10.1038/sj.onc.1207171. [DOI] [PubMed] [Google Scholar]
- 11.Hideshima T, Catley L, Raje N, et al. Inhibition of Akt induces significant downregulation of survivin and cytotoxicity in human multiple myeloma cells. Br J Haematol. 2007;138:783–791. doi: 10.1111/j.1365-2141.2007.06714.x. [DOI] [PubMed] [Google Scholar]
- 12.Papapetropoulos A, Fulton D, Mahboubi K, et al. Angiopoietin-1 inhibits endothelial cell apoptosis via the Akt/survivin pathway. J Biol Chem. 2000;275:9102–9105. doi: 10.1074/jbc.275.13.9102. [DOI] [PubMed] [Google Scholar]
- 13.Adida C, Crotty PL, McGrath J, Berrebi D, Diebold J, Altieri DC. Developmentally regulated expression of the novel cancer anti-apoptosis gene survivin in human and mouse differentiation. Am J Pathol. 1998;152:43–49. [PMC free article] [PubMed] [Google Scholar]
- 14.Ambrosini G, Adida C, Altieri DC. A novel anti-apoptosis gene, survivin, expressed in cancer and lymphoma. Nat Med. 1997;3:917–921. doi: 10.1038/nm0897-917. [DOI] [PubMed] [Google Scholar]
- 15.Li F, Ambrosini G, Chu EY, et al. Control of apoptosis and mitotic spindle checkpoint by survivin. Nature. 1998;396:580–584. doi: 10.1038/25141. [DOI] [PubMed] [Google Scholar]
- 16.Tamm I, Wang Y, Sausville E, et al. IAP-family protein survivin inhibits caspase activity and apoptosis induced by Fas (CD95), Bax, caspases, and anticancer drugs. Cancer Res. 1998;58:5315–5320. [PubMed] [Google Scholar]
- 17.Karra L, Shushan A, Ben-Meir A, et al. Changes related to phosphatidylinositol 3-kinase/Akt signaling in leiomyomas: possible involvement of glycogen synthase kinase 3α and cyclin D2 in the pathophysiology. Fertil Steril. 2010;93:2646–2651. doi: 10.1016/j.fertnstert.2009.03.100. [DOI] [PubMed] [Google Scholar]
- 18.Gotoh J, Obata M, Yoshie M, Kasai S, Ogawa K. Cyclin D1 over-expression correlates with β-catenin activation, but not with H-ras mutations, and phosphorylation of Akt, GSK3β and ERK1/2 in mouse hepatic carcinogenesis. Carcinogenesis. 2003;24:435–442. doi: 10.1093/carcin/24.3.435. [DOI] [PubMed] [Google Scholar]
- 19.Hall A. Rho GTPases and the actin cytoskeleton. Science. 1998;279:509–514. doi: 10.1126/science.279.5350.509. [DOI] [PubMed] [Google Scholar]
- 20.Braga VM, Machesky LM, Hall A, Hotchin NA. The small GTPases Rho and Rac are required for the establishment of cadherin-dependent cell-cell contacts. J Cell Biol. 1997;137:1421–1431. doi: 10.1083/jcb.137.6.1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sander EE, ten Klooster JP, van Delft S, van der Kammen RA, Collard JG. Rac downregulates Rho activity: reciprocal balance between both GTPases determines cellular morphology and migratory behavior. J Cell Biol. 1999;147:1009–1022. doi: 10.1083/jcb.147.5.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yamazaki D, Kurisu S, Takenawa T. Involvement of Rac and Rho signaling in cancer cell motility in 3D substrates. Oncogene. 2009;28:1570–1583. doi: 10.1038/onc.2009.2. [DOI] [PubMed] [Google Scholar]
- 23.Symons M, Segall JE. Rac and Rho driving tumor invasion: who's at the wheel? Genome Biol. 2009;10:213. doi: 10.1186/gb-2009-10-3-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Goldberg L, Kloog Y. A Ras inhibitor tilts the balance between Rac and Rho and blocks phosphatidylinositol 3-kinase-dependent glioblastoma cell migration. Cancer Res. 2006;66:11709–11717. doi: 10.1158/0008-5472.CAN-06-1878. [DOI] [PubMed] [Google Scholar]
- 25.Senger DL, Tudan C, Guiot MC, et al. Suppression of Rac activity induces apoptosis of human glioma cells but not normal human astrocytes. Cancer Res. 2002;62:2131–2140. [PubMed] [Google Scholar]
- 26.Wu X, Tu X, Joeng KS, Hilton MJ, Williams DA, Long F. Rac1 activation controls nuclear localization of β-catenin during canonical Wnt signaling. Cell. 2008;133:340–353. doi: 10.1016/j.cell.2008.01.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Holland EC, Celestino J, Dai C, Schaefer L, Sawaya RE, Fuller GN. Combined activation of Ras and Akt in neural progenitors induces glioblastoma formation in mice. Nat Genet. 2000;25:55–57. doi: 10.1038/75596. [DOI] [PubMed] [Google Scholar]
- 28.Pisharath H, Parsons MJ. Nitroreductase-mediated cell ablation in transgenic zebrafish embryos. Methods Mol Biol. 2009;546:133–143. doi: 10.1007/978-1-60327-977-2_9. [DOI] [PubMed] [Google Scholar]
- 29.Jung IH, Jung DE, Park YN, Song SY, Park SW. Aberrant hedgehog ligands induce progressive pancreatic fibrosis by paracrine activation of myofibroblasts and ductular cells in transgenic zebrafish. PloS One. 2011;6:1–15. doi: 10.1371/journal.pone.0027941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bernardos RL, Raymond PA. GFAP transgenic zebrafish. Gene Expr Patterns. 2006;6:1007–1013. doi: 10.1016/j.modgep.2006.04.006. [DOI] [PubMed] [Google Scholar]
- 31.Yeo SY, Kim M, Kim HS, Huh TL, Chitnis AB. Fluorescent protein expression driven by her4 regulatory elements reveals the spatiotemporal pattern of Notch signaling in the nervous system of zebrafish embryos. Dev Biol. 2007;301:555–567. doi: 10.1016/j.ydbio.2006.10.020. [DOI] [PubMed] [Google Scholar]
- 32.Kim H, Shin J, Kim S, Poling J, Park HC, Appel B. Notch-regulated oligodendrocyte specification from radial glia in the spinal cord of zebrafish embryos. Dev Dyn. 2008;237:2081–2089. doi: 10.1002/dvdy.21620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Workman P, Aboagye EO, Balkwill F, et al. Guidelines for the welfare and use of animals in cancer research. Br J Cancer. 2010;102:1555–1577. doi: 10.1038/sj.bjc.6605642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Park SW, Davison JM, Rhee J, Hruban RH, Maitra A, Leach SD. Oncogenic KRAS induces progenitor cell expansion and malignant transformation in zebrafish exocrine pancreas. Gastroenterology. 2008;134:2080–2090. doi: 10.1053/j.gastro.2008.02.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Janezic G, Widni EE, Haxhija EQ, Stradner M, Frohlich E, Weinberg AM. Proliferation analysis of the growth plate after diaphyseal midshaft fracture by 5′-bromo-2′-deoxy-uridine. Virchows Arch. 2010;457:77–85. doi: 10.1007/s00428-010-0932-6. [DOI] [PubMed] [Google Scholar]
- 36.Davison JM, Woo Park S, Rhee JM, Leach SD. Characterization of Kras-mediated pancreatic tumorigenesis in zebrafish. Methods Enzymol. 2008;438:391–417. doi: 10.1016/S0076-6879(07)38027-0. [DOI] [PubMed] [Google Scholar]
- 37.Beckers T, Voegeli R, Hilgard P. Molecular and cellular effects of hexadecylphosphocholine (Miltefosine) in human myeloid leukaemic cell lines. Eur J Cancer. 1994;30A:2143–2150. doi: 10.1016/0959-8049(94)00438-b. [DOI] [PubMed] [Google Scholar]
- 38.Larson B, Banks P, Zegzouti H, Goueli SA. A simple and robust automated kinase profiling platform using luminescent ADP accumulation technology. Assay Drug Dev Techn. 2009;7:573–584. doi: 10.1089/adt.2009.0216. [DOI] [PubMed] [Google Scholar]
- 39.Lin JW, Biankin AV, Horb ME, et al. Differential requirement for ptf1a in endocrine and exocrine lineages of developing zebrafish pancreas. Dev Biol. 2004;270:474–486. doi: 10.1016/j.ydbio.2004.02.023. [DOI] [PubMed] [Google Scholar]
- 40.Kroehne V, Freudenreich D, Hans S, Kaslin J, Brand M. Regeneration of the adult zebrafish brain from neurogenic radial glia-type progenitors. Development. 2011;138:4831–4841. doi: 10.1242/dev.072587. [DOI] [PubMed] [Google Scholar]
- 41.Jung SH, Kim HS, Ryu JH, et al. Her4-positive population in the tectum opticum is proliferating neural precursors in the adult zebrafish brain. Mol Cells. 2012;33:627–632. doi: 10.1007/s10059-012-0091-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kaslin J, Ganz J, Geffarth M, Grandel H, Hans S, Brand M. Stem cells in the adult zebrafish cerebellum: initiation and maintenance of a novel stem cell niche. J Neurosci. 2009;29:6142–6153. doi: 10.1523/JNEUROSCI.0072-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Meadows KN, Iyer S, Stevens MV, et al. Akt promotes endocardial-mesenchyme transition. J Angiogenes Res. 2009;1:2. doi: 10.1186/2040-2384-1-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Altieri DC. Survivin, cancer networks and pathway-directed drug discovery. Nat Rev Cancer. 2008;8:61–70. doi: 10.1038/nrc2293. [DOI] [PubMed] [Google Scholar]
- 45.Ramnanan CJ, Groom AG, Storey KB. Akt and its downstream targets play key roles in mediating dormancy in land snails. Comp Biochem Physiol B Biochem Mol Biol. 2007;148:245–255. doi: 10.1016/j.cbpb.2007.06.002. [DOI] [PubMed] [Google Scholar]
- 46.Dutcher JP. Mammalian target of rapamycin (mTOR) inhibitors. Curr Oncol Rep. 2004;6:111–115. doi: 10.1007/s11912-004-0022-5. [DOI] [PubMed] [Google Scholar]
- 47.Hoshino M, Nakamura S, Mori K, et al. Ptf1a, a bHLH transcriptional gene defines GABAergic neuronal fates in cerebellum. Neuron. 2005;47:201–213. doi: 10.1016/j.neuron.2005.06.007. [DOI] [PubMed] [Google Scholar]
- 48.Onvani S, Etame AB, Smith CA, Rutka JT. Genetics of medulloblastoma; clues for novel therapies. Expert Rev Neurother. 2010;10:811–823. doi: 10.1586/ern.10.31. [DOI] [PubMed] [Google Scholar]
- 49.Liu T, Biddle D, Hanks AN, et al. Activation of dual apoptotic pathways in human melanocytes and protection by survivin. J Invest Dermatol. 2006;126:2247–2256. doi: 10.1038/sj.jid.5700381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wagenknecht B, Glaser T, Naumann U, et al. Expression and biological activity of X-linked inhibitor of apoptosis (XIAP) in human malignant glioma. Cell Death Diff. 1999;6:370–376. doi: 10.1038/sj.cdd.4400503. [DOI] [PubMed] [Google Scholar]
- 51.Zhao P, Meng Q, Liu LZ, You YP, Liu N, Jiang BH. Regulation of survivin by PI3K/Akt/p70S6K1 pathway. Biochem Biophy Res Comm. 2010;395:219–224. doi: 10.1016/j.bbrc.2010.03.165. [DOI] [PubMed] [Google Scholar]
- 52.Zondag GC, Evers EE, ten Klooster JP, Janssen L, van der Kammen RA, Collard JG. Oncogenic Ras downregulates Rac activity, which leads to increased Rho activity and epithelial-mesenchymal transition. J Cell Biol. 2000;149:775–782. doi: 10.1083/jcb.149.4.775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Phelps RA, Chidester S, Dehghanizadeh S, et al. A two-step model for colon adenoma initiation and progression caused by APC loss. Cell. 2009;137:623–634. doi: 10.1016/j.cell.2009.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Julien S, Puig I, Caretti E, et al. Activation of NF-κB by Akt upregulates Snail expression and induces epithelium mesenchyme transition. Oncogene. 2007;26:7445–7456. doi: 10.1038/sj.onc.1210546. [DOI] [PubMed] [Google Scholar]
- 55.Gos M, Miloszewska J, Przybyszewska M. Epithelial-mesenchymal transition in cancer progression. Postepy Biochem. 2009;55:121–128. [PubMed] [Google Scholar]
- 56.Micalizzi DS, Farabaugh SM, Ford HL. Epithelial-mesenchymal transition in cancer: parallels between normal development and tumor progression. J Mammary Gland Biol Neoplasia. 2010;15:117–134. doi: 10.1007/s10911-010-9178-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ansieau S, Caron de Fromentel C, Bastid J, Morel AP, Puisieux A. Role of the epithelial-mesenchymal transition during tumor progression. Bull Cancer. 2010;97:7–15. doi: 10.1684/bdc.2009.1025. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.