SUMMARY
Studies of Parkinson’s disease (PD) have been greatly hindered by lack of access to affected human dopaminergic (DA) neurons. Here, we report generation of induced pluripotent stem cells that carry the p.G2019S mutation (G2019S-iPSCs) in the Leucine-Rich Repeat Kinase-2 (LRRK2) gene, the most common PD-related mutation. We demonstrate that these G2019S-iPSCs were able to differentiate into DA neurons and showed increased expression of key oxidative stress response genes and α-synuclein protein. Moreover, G2019S-mutant DA neurons were more sensitive to caspase-3 activation, caused by exposure to hydrogen peroxide, MG-132, and 6-hydroxydopamine, compared to unaffected DA neurons. These findings suggest that G2019S-iPSC-derived DA neurons exhibit early phenotypes linked to PD. Due to high penetrance of the LRRK2 mutation and its clinical resemblance to sporadic PD, these neurons may provide a valuable platform for identification of novel pharmacological agents and diagnostics for modeling and alleviation of a subset of disease phenotypes.
Keywords: Parkinson’s disease, p.G2019S, Leucine Rich Repeat Kinase 2 (LRRK2), induced pluripotent stem cells (iPSCs), human embryonic stem cells (hESCs), α-synuclein(SNCA), midbrain dopaminergic neurons (mDA), hydrogen peroxide, 6-hydroxydopamine, MG-132
INTRODUCTION
Parkinson’s disease (PD) is one of the most common neurodegenerative disorders of aging, afflicting 0.9% of the population 65 years and older and increasing to 5.1% by the age of 85 (de Rijk et al., 2000). Although it is not well understood, the pathogenesis of PD has been linked to both environmental and genetic factors (Dawson and Dawson, 2003). A common autosomal dominant missense mutation, G2019S in the Leucine-Rich Repeat Kinase 2 (LRRK2) gene, has previously been identified in 0.6 – 1.6% and 2 – 8% of sporadic and familial PD cases, respectively (Hernandez et al., 2005; Nichols et al., 2005; Paisan-Ruiz et al., 2005). The penetrance of the G2019S mutation is age-dependent, increasing from 17% at age 50 to 85% penetrance at age 70 (Kachergus et al., 2005). Clinical symptoms associated with the G2019S mutation are nearly indistinguishable from those of sporadic PD with pathologies affecting both motor and non-motor bodily functions. Histological studies of postmortem brain tissue from PD patients, including those with the p.G2019S LRRK2 mutation, showed cell loss in the substantia nigra and formation of Lewy bodies, protein aggregates containing α-synuclein (Baba et al., 1998; Ross et al., 2006). While therapeutic drugs may treat some symptoms of PD, curative therapies and methods to halt disease progression have yet to be developed. Moreover, although transgenic animal and cell models expressing known PD-associated genes have provided important insights into the disease (Betarbet et al., 2002; Heo et al., 2010; Ng et al., 2009; Paisan-Ruiz et al., 2004; Schüle et al., 2009), it has been difficult to demonstrate that implicated mechanisms are also present in neurons from an affected individual. iPSC-derived neurons from affected individuals hold promise of providing a missing link between current models and PD pathology.
Although derivation of iPSC lines from sporadic PD patients has been reported (Park et al., 2008; Soldner et al., 2009), it is not known whether PD iPSC–derived neurons exhibit PD phenotypes and whether sporadic PD lines will demonstrate overt phenotypes. Thus, proof of principle that iPSC lines may recapitulate key phenotypes of PD may be most appropriately revealed with monogenic mutations.
RESULTS
Generation and Characterization of PD-Derived iPSCs
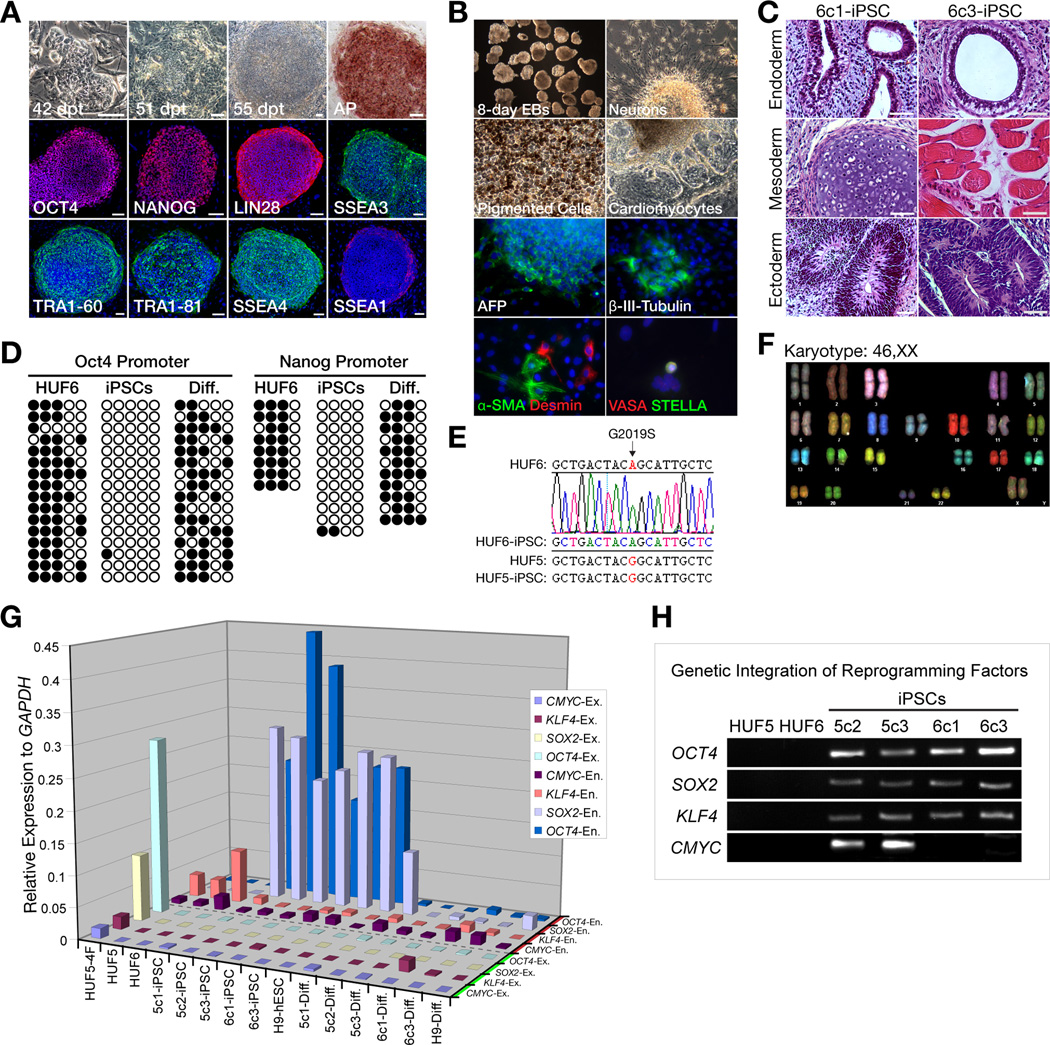
A 60-year-old female patient, with early onset (48 years), typical L-DOPA (L-3,4-dihydroxyphenylalanine)-responsive PD, and homozygous for the p.G2019S mutation in the LRRK2 gene, consented to a skin biopsy and iPSC derivation. After expansion of the primary dermal fibroblast line (termed HUF6), cells were transduced with retroviruses carrying three reprogramming factors, OCT4, SOX2, and KLF4, as previously described (Takahashi et al., 2007; Nakagawa et al., 2008). Approximately two months post-infection, three hESC-like colonies were picked for propagation under standard human embryonic stem cell (hESC) culture conditions. Two clones (6c1-iPSC and 6c3-iPSC, collectively referred to as G2019S-iPSCs) were then selected for detailed analysis. G2019S-iPSCs exhibited morphology similar to hESCs, were alkaline phosphatase positive and immunoreactive for OCT4, NANOG, LIN28, SSEA3, SSEA4, TRA1–60, TRA1–81, while being negative for SSEA1 (Figure 1A). iPSCs were successfully cultured in an undifferentiated state for more than 30 passages (8 months). G2019S-iPSCs were able to differentiate into cell types of all three germ layers both spontaneously in vitro and in vivo. (Figure 1B–C, Movie S1). Further analysis showed that undifferentiated G2019S-iPSCs were hypomethylated at the OCT4 and NANOG promoter regions compared to donor fibroblasts and differentiated iPSCs, retained the missense homozygous G2019S mutation and maintained a normal karyotype (Figure 1D–F). Gene expression profiling showed that undifferentiated and differentiated G2019S-iPSCs had a similar expression pattern as control iPSCs from a healthy adult human female previously derived with four factors including CMYC (termed HUF5-iPSC) and H9 hESCs (Thomson et al., 1998) (Figure S1). Importantly, RT-PCR analysis of all iPSC lines showed complete silencing of exogenous factors after reprogramming and exogenous factors remained silent after 35 days of in vitro directed neuronal differentiation (Figure 1G). This suggested that whether 3 or 4 reprogramming factors were used, exogenous genes were silenced. Genomic DNA analysis also confirmed that G2019S-iPSCs have integrated 3 reprogramming factors without CMYC (Figure 1H). Taken together, these findings demonstrate that both undifferentiated and differentiated G2019S-iPSCs are very similar to wild-type iPSCs and hESCs in cell morphology, pluripotency markers, differentiation potential, epigenetics and gene expression profile.
Figure 1. Generation and Characterization of Human iPSCs Carrying a p.G2019S LRRK2 Mutation.
(A) Progression of iPSC colony formation during reprogramming and subsequent expression of pluripotency markers: Alkaline phosphatase (AP), OCT4, NANOG, LIN28, SSEA3, TRA1–60, TRA1–81, SSEA4, and mostly negative for SSSEA1. Cell nuclei were counterstained with DAPI (blue). dpt = day post transduction; Scale bar = 100 µm.
(B) In vitro differentiation of G2019S-iPSCs to cells representative of each germ layer: α-fetoprotein (AFP, endoderm), β-III-tubulin (ectoderm), α-smooth muscle actin and desmin (mesoderm), and VASA and STELLA (germ cell precursors). Also, neurons, beating cardiomyocytes and pigmented cells are easily observed under a phase microscope.
(C) In vivo (teratoma) differentiation of G2019S-iPSCs (clones 6c1 and 6c3) with evidence of all three germ layers: gut-like epithelium (endoderm), cartilage and muscle (mesoderm) and neural-like tissues (ectoderm). Scale bar = 100 µm.
(D) Methylation analysis of OCT4 and NANOG promoters. Open and closed circles represent unmethylated and methylated CpG islands, respectively.
(E) Genomic DNA sequencing showing presence of homozygous p.G2019S mutation in dermal skin fibroblasts (HUF6) and G2019S-derived iPSCs, but not in normal HUF5 skin fibroblasts or iPSCs.
(F) Spectral karyotyping (SKY) analysis of G2019S-iPSCs (clone 6c3-iPSCs).
(G) Gene expression of endogenous (En.) and exogenous (Ex.) reprogramming factors in iPSCs and differentiated 35-day neurons. HUF5-4F is a positive control, RNA was collected 3 days after virus transduction; HUF5 and HUF6 are non-transduced human dermal skin fibroblast.
(H) PCR analysis of the iPSCs. 6c1- and 6c3-iPSCs (G2019S) have three integrated reprogramming factors without CMYC and that 5c2- and 5c3-iPSCs have four factors with cMYC.
See also Figure S1 and Movie S1 for more characterization of iPSCs.
Directed Differentiation of iPSCs into Midbrain Dopaminergic Neurons
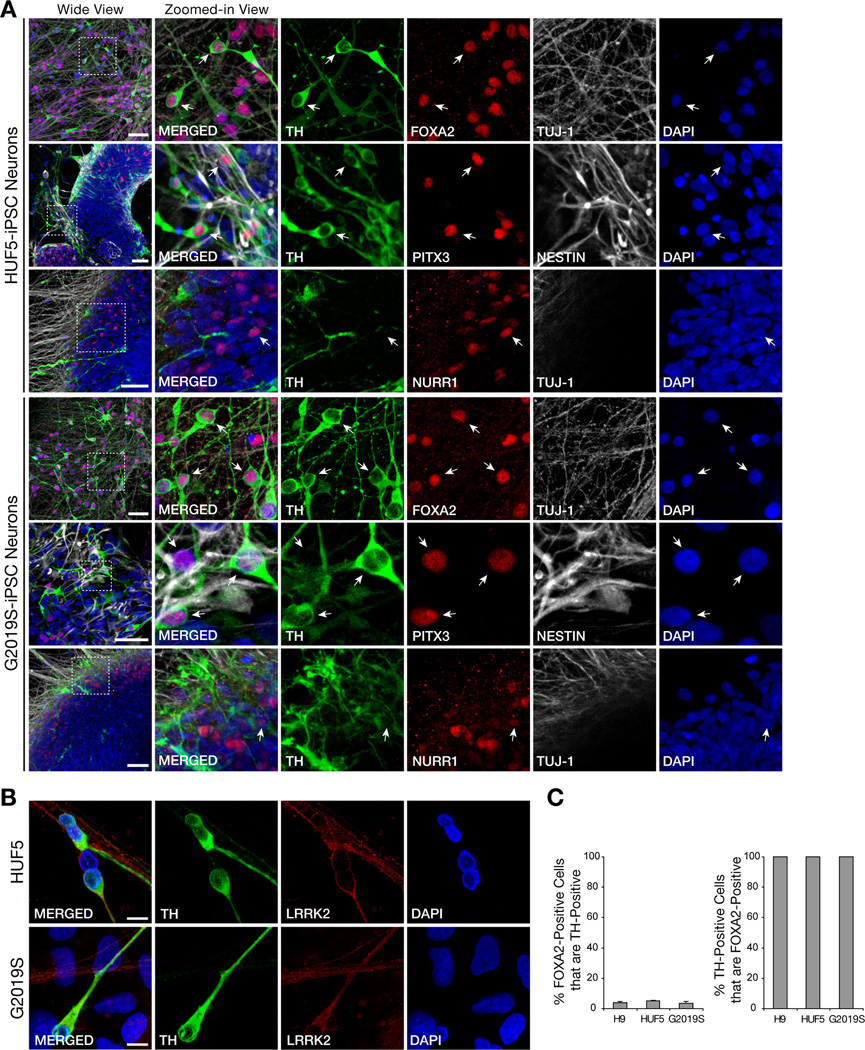
Since PD is associated with degeneration of midbrain dopaminergic (mDA) neurons, we sought to determine if both normal and PD iPSC lines, HUF5-iPSCs and G2019S-iPSCs, respectively, could differentiate into mDA neurons in vitro. We followed an established protocol, with modifications (Figure S2A), that used a combination of small molecules and midbrain-selective patterning factors, FGF8b (fibroblast growth factor 8 beta) and SHH (sonic hedgehog), to direct human iPSC differentiation to DA neurons (Chambers et al., 2009). During the course of differentiation, iPSC lines generated numerous neural progenitor cells (immuno-positive for PAX6 and nestin) and, subsequently, neurons (immuno-positive for doublecortin and β-III-tubulin (data not shown)). A significant number of these neurons also expressed tyrosine hydroxylase (TH; rate-limiting enzyme in synthesis of dopamine) (Figure 2A). Other markers consistent with a midbrain phenotype were also present including: forkhead box A2 (FOXA2), orphan nuclear receptor (NURR1), paired-like homeodomain transcription factor 3 (PITX3) and LRRK2 (Figure 2A–B). Orthodenticle homeobox 2 (OTX2) and calbindin (CALB) were also expressed in a portion of TH-positive neurons (Liang et al., 1996; Vernay et al., 2005) (Figure S2). Notably, OTX2 expression in TH-positive cells appeared to be most-consistent with a ventral tegmental area (A10, VTA) phenotype as described in mouse brain (Di Salvio et al., 2010). Whereas, FOXA2, NURR1, PITX3 and LRRK2 may play an important role in development of midbrain neurons, specifically substantia nigra pars compacta (A9, SNpc) subtype (Han et al., 2008; Ichinose et al., 1999; Kittappa et al., 2007; Nunes et al., 2003).
Figure 2. Directed Differentiation of iPSCs into Midbrain Dopaminergic (mDA) Neurons.
(A) Expression of midbrain dopaminergic neurons markers, tyrosine hydroxylase (TH), FOXA2, PITX3, and NURR1, in iPSC-derived 30- to 35-day neurons. Note the co-localization of FOXA, PITX3 and NURR1 with TH, characteristic of mDA neurons (arrows). NESTIN and TUJ1 positive immunoreactivity also show the presence of neurons. Scale bar = 50 µm.
(B) Expression of LRRK2 proteins in dopaminergic neurons. Scale bar = 10 µm.
(C) Quantification of the yield of mDA, based on TH and FOXA2 colocalization, in 35-day neurons derived from H9, HUF5, and G2019S lines.
See also Figure S2 for more characterization of iPSC-derived neurons.
Quantification of mDA neurons was based on average cell counts from 3 different clones of HUF5-iPSC, two clones of G2019S-iPSC and an hESC control (H9) line. We observed that 97–98% of neurons after 35 days of differentiation and enrichment were positive for FOXA2, consistent with the FGF8/SHH patterning environment. We also observed that a significant, though minority fraction, of these cells (3.6% – 5%) also differentiated into TH-expressing neurons within the course of differentiation (Figure 2C). Colocalization of multiple midbrain-specific markers with TH suggested that many iPSC-derived neurons may be of an mDA phenotype, the cell type lost in PD neurodegeneration.
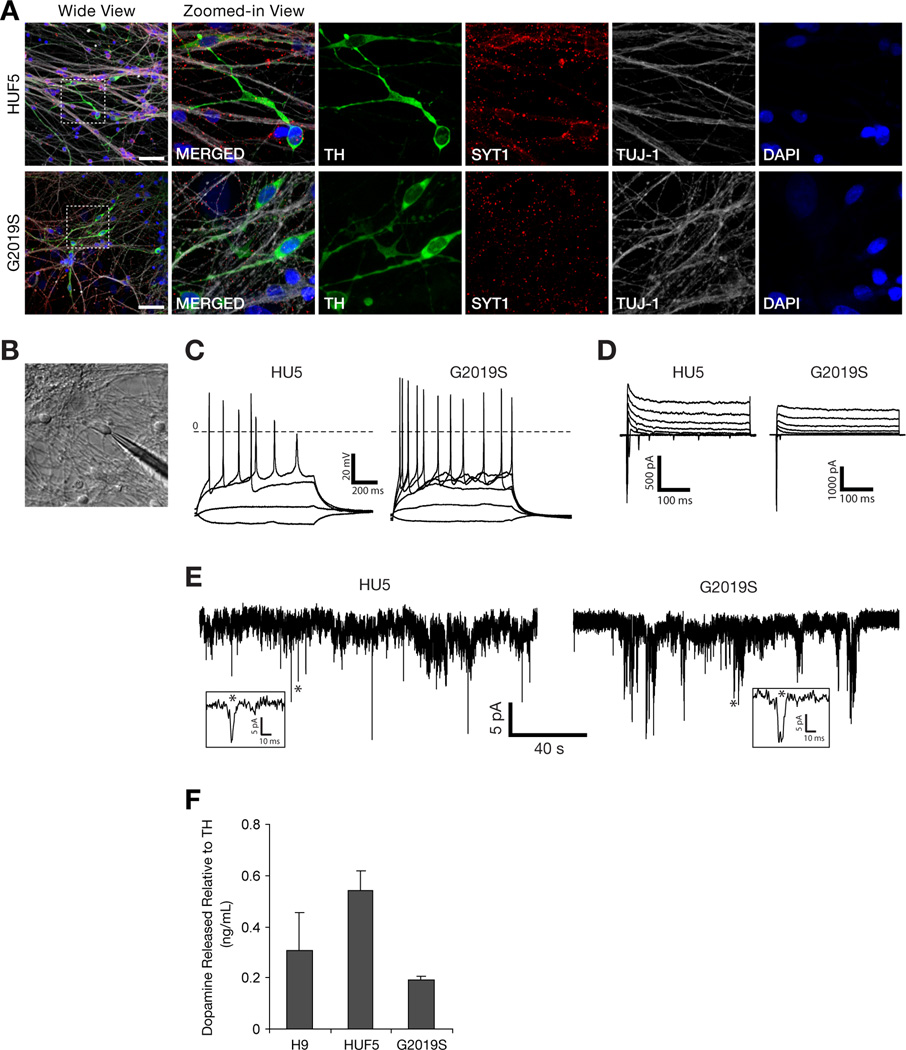
We also observed properties that suggested that TH-positive neurons derived from HUF5- and G2019S-iPSCs were mature. First, the neurons expressed synaptotagmin-1 (SYT1), a protein localized to synaptic vesicles (Figure 3A). Second, electrophysiological recordings of these neurons, 30–35 days after differentiation, demonstrated that they could fire action potentials (Figure 3B–D). Approximately 60% of recorded cells fired action potentials in response to depolarizing current injections (Table S1) and showed spontaneous synaptic activities (Figure 3E). Third, these DA neurons were able to synthesize and release dopamine in response to stimulation with high potassium (Figure 3F). Collectively, these results suggest that iPSC-derived neurons produced by this protocol are sufficiently mature to be functional and form spontaneously active neural networks in culture.
Figure 3. Differentiation of iPSCs into Functional DA Neurons.
(A) Expression of synaptotagmin-1 (SYT1), a synaptic vesicle protein, in DA neurons generated from HUF5- and G2019S-iPSCs. Scale bar = 50 µm.
(B) Exemplar of electrophysiological recordings from cells with typical neuronal morphology, residing remotely from the cell clusters.
(C) Current clamp recordings of voltage deflections induced by injections of current of different amplitude (ΔIinj = 5 pA).
(D) Voltage clamp recordings of the net currents at various membrane potentials (−80 – +20, ΔV = 10 mV, Vhold = −90 mV).
(E) Spontaneous synaptic activity of the neurons measured at −70 mV.
(F) HPLC measurements of total level of dopamine release from iPSC-derived neurons relative to TH protein expression. Data represent mean ± SEM (n = 2).
See also Table S1 for electrophysiological recordings of neurons.
Characterization of iPSC-Derived Neurons
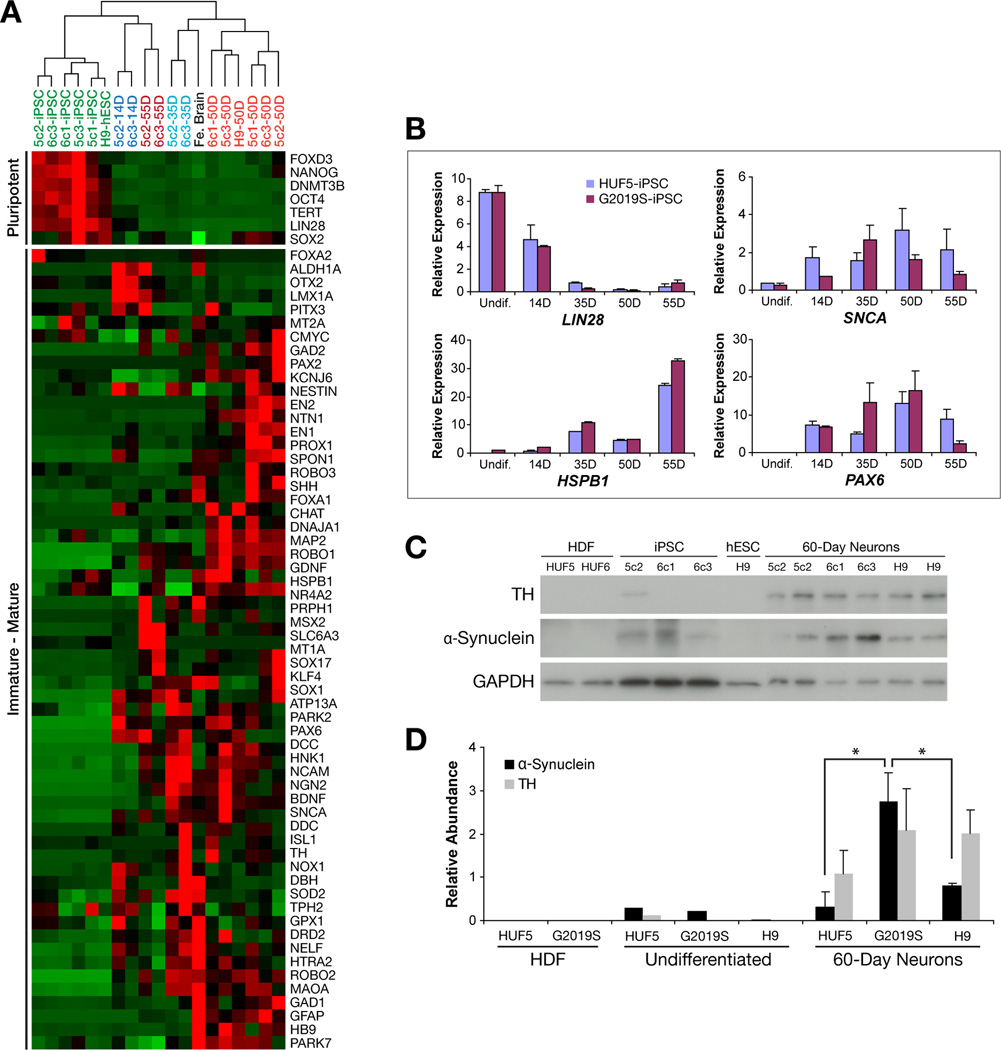
We next sought to characterize iPSC-derived neurons in more detail. Thus, we directed iPSC differentiation into mDA neurons, picked neuronal aggregates of approximately 100 cells (both manually and after enzymatic treatment) and assayed for gene expression at various time points (undifferentiated, 14, 35, 50 and 55 days of differentiation) via microfluidic qPCR. Cluster analysis of gene expression showed that samples correlated primarily by stage of neuronal differentiation and demonstrated expected expression patterns (Figure 4A). Undifferentiated iPSC lines and H9 hESCs strongly expressed pluripotency genes, clustered together and had closest relatedness to 14-day neurons, while 35-, 50- and 55-day neurons, expressed neuronal-specific genes, and grouped together on separate clusters along with a whole human fetal brain sample. This suggested that the neuronal differentiation procedure follows expected developmental pathways, provided credence to using small-scale samples in RT-PCR analysis and showed that these iPSC-derived neurons gene expression patterns bear similarity to that of developing human fetal brain. Detailed analysis of representative genes, such as LIN28, SNCA, HSPB1 and PAX6, from a group of neurons and from single neurons verified cluster analysis (Figure 4B–E, Figure S3A–F). Note, however, that a subset of markers presumed to be midbrain-specific, primarily FOXA2 and PITX3 were not consistently expressed during some stages of differentiation. This observation could be due to analysis of subsets of neurons with unique midbrain phenotypes or alternative subtypes. We observed that other mature midbrain markers (other than FOXA2) such as Engrailed-1 and -2 were often highly expressed.
Figure 4. Characterization of Human iPSC-Derived Neurons.
(A) Gene expression analysis of HUF5- and G2019S-iPSC-derived neurons at various stages of in vitro differentiation. Cluster analysis of iPSCs and various neuronal differentiation stages (14–55 days). Fe. Brain = whole human fetal brain; Red = High expression; Green = Low; Black = Zero.
(B) Quantitative RT-PCR analysis showing average expression of LIN28, α-synuclein (SNCA), heat shock 27kD protein 1 (HSPB1), and paired box gene 6 (PAX6) in normal HUF5-iPSC- and G2019S-iPSC-derived neurons. Gene expression normalized to GAPDH, CENTB3, EEF1α and CNNTB1 (based on a geometric mean) by qBasePlus. Data represent mean ± SEM (n = 3 to 7 biological replicates per clone).
(C) Western analysis of human dermal fibroblasts (HDF), iPSCs, hESCs, and 60-day neurons derived from iPSCs and hESCs.
(D) Graphical representation of western blot performed in Figure 4F. Relative protein abundance was normalized to GAPDH. Error bars represent standard deviation from two independent neuronal differentiation. * p < 0.05.
See also Figure S3 for RT-PCR analysis of iPSC-derived neurons.
To further characterize neurons generated, we manually isolated 35-day single neurons and assayed for gene expression using the same techniques mentioned above. 35-day HUF5-iPSC- and G2019S-iPSC-derived single neurons self-clustered together and correlated with other bulk neuronal samples, including human fetal brain (data not shown). Using direct gene-to-gene comparison between G2019S- and HUF5-derived neurons, the average expression of single neurons showed similar relative gene expression pattern as neurons prepared in bulk (Figure S3D–E). We also noticed that there was a cell-to-cell variability in gene expression, consistent with the fact that the neurons represent multiple stages of differentiation and include both TH-positive and TH-negative neuronal subtypes. In summary, data from small aggregates and from single neurons demonstrated gene expression patterns indicative of successful neural lineage determination and differentiation into mature neurons though variation in gene expression was observed.
Emergence of disease-related phenotypes in iPSC-Derived Neurons
Direct comparison between G2019S- and HUF5-iPSC-derived neurons showed differential gene expression at various stages of differentiation, some of which indicated differential steady-state oxidative stress. Genes involved in oxidative stress pathways such as HSPB1, NOX1 and MAOB were expressed higher (3.7-fold for NOX1 and MAOB; 1.4-fold for HSPB1) at day 35 in G2019S-iPSC-derived neurons than HUF5-iPSC-derived neurons (Figure S3C). We noted that expression levels of some other key oxidative stress response genes were also slightly higher in G2019S-iPSC-derived neurons than HUF5-iPSC-derived neurons at day 35, but not at day 50 and 55.
Accumulation of α-synuclein protein is also implicated in LRRK2 gain of function mutations. α-Synuclein is a major component of Lewy bodies, protein aggregates that form in neurodegenerative disorders such as Lewy body disease, Alzheimer’s disease and PD (Bayer et al., 1999; Giasson et al., 2006; Hashimoto and Masliah, 1999; Spillantini et al., 1998). Previous studies demonstrated that over expression of LRRK2 G2019S protein can lead to abnormal accumulation of α-synuclein, either as a direct consequence of phosphorylation of SNCA protein by LRRK2 or indirectly through alterations in kinase signaling leading to increased SNCA gene expression or protein stabilization (Cookson, 2009).
We observed that α-synuclein protein was not detected by Western blot in HUF5 and G2019S (HUF6) dermal fibroblasts, was only detectable at very low levels in undifferentiated HUF5-iPSCs, G2019S-iPSCs and H9-hESCs, and was expressed at much higher levels in mature neurons (Figure 4C–D). In comparison to HUF5-iPSC- and H9-hESC-derived neurons, G2019S-iPSC-derived 60-day neurons showed a significant increase (3- to 8-fold) in monomeric α-synuclein protein levels (p < 0.05, n = 2 independent differentiations with 2 replicate wells per line in each experiment). Results also indicated that, in these samples, there was no significant difference in TH expression (Figure 4D), suggesting that higher α-synuclein expression was not due to the presence of more TH-positive cells but instead correlated with genotype. Based on this analysis, G2019S-iPSCs appeared to present at least a subset of native PD-related phenotypes of increased indicators of oxidative stress and α-synuclein accumulation even after reprogramming to pluripotency and differentiation into dopaminergic neurons in vitro.
G2019S-iPSC-Derived DA Neurons are Differentially Susceptible to Peroxide-Induced Cell Death
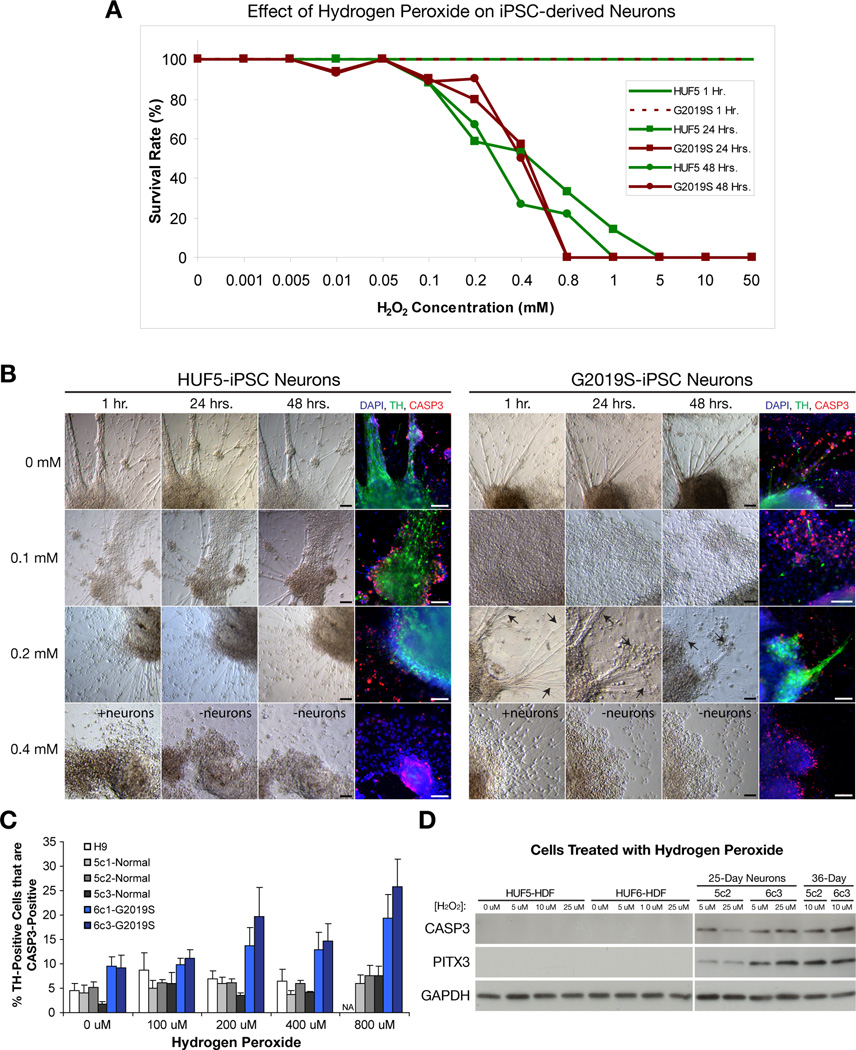
Next, we compared susceptibility of H9-, HUF5-iPSC- and G2019S-iPSC-derived neurons to hydrogen peroxide (H2O2). iPSC-derived neuronal clusters were transferred to 48-well plates (5 clusters per well). Following at least two days of recovery, 35-day neurons were treated with various concentrations of H2O2 and visually evaluated after 24 or 48 hours in order to establish a survival curve (Figure 5A). We found that, at 0 – 0.05 mM, most neurons survived after 24 – 48 hours of treatment (n = 257 wells, 3 independent experiments). In contrast, at 0.1 – 1 mM, neurons progressively degenerated after 24 – 48 hours (n = 230 wells); and at 5 – 50 mM, there were no surviving neurons at 24 hours (n = 36 wells). At 48 hrs, cells were fixed and stained for type III beta tubulin (Tuj1, a generic marker for neurons), TH and activated (or cleaved) form of caspase-3 (CASP3), an apoptotic marker. Confocal imaging was used to score the fraction of TH-positive or TH-negative neurons that were co-labeled for CASP3. Apoptotic index was determined for 3 independently derived clones of HUF5-iPSC (5c1, 5c2 and 5c3), 2 independently derived clones of G2019S-iPSC (6c1 and 6c3) and H9-derived DA neurons (Figure 5B–C, Table S2). In some clones, experiments were independently repeated to validate internal reproducibility. Based on 2- way analysis of variance (2-way ANOVA), the apparent increase in cell death was not significant between all lines at different H2O2 treatments. However, when ignoring treatment dosage, we observed a significant effect of cell type (F(5,67)=8.53, p<0.000003, n=12–24 per cell line). Duncan’s multiple range post-hoc analysis revealed that independent replicates of G2019S-iPSC-derived DA neuron cultures had significantly more cell death than both wild-type HUF5-iPSC- and H9-derived DA neuron cultures. To determine if the effect of H2O2 was specific to DA neurons, we also performed the same analysis on TH-negative (TUJ1+) neurons. We observed that TH-negative neurons were less susceptible to peroxide-induced cell death at all concentrations tested, and there was no significant difference between unaffected and diseased cell lines (Figure S4A–B). These findings suggested that G2019S-iPSC-derived DA neurons may be more susceptible to oxidative stress and CASP3 activation than normal H9- and HUF5-iPSC-derived DA neurons.
Figure 5. Effect of Hydrogen Peroxide on hESC- and iPSC-Derived DA Neurons.
(A) Effect of H2O2 treatment on 35-day neurons after 24 and 48 hours. Survival rate from 1 to 48 hours was measured by observation under the phase-constrast microscope and immunostaining for Tyrosine Hydroxylase (TH) after 48 hours. At least 7 – 21 wells from 3 independent experiments at each time point and concentration within 0.01 – 1 mM range.
(B) Hydrogen Peroxide (H2O2) causes neuronal degeneration in vitro. Note presence of spontaneous and induced cell death, identified by activated (or cleaved) form of caspase-3 (CASP3) immunofluorescence, at all concentrations. Arrows indicate the regression of neuronal axons after 0.1 mM H2O2 treatment. Scale bar = 100 µm.
(C) Quantification of cells double positive for TH and CASP3 in different clones of normal (wild-type) H9 and HUF5-iPSC versus disease G2019S-iPSC-derived DA neurons in the treatment of various of H2O2 concentrations. Data represent mean ± SEM (n = 3 to 6 wells per clone, two independent experiments).
(D) Western analysis comparing the levels of cell death (CASP3) between normal HUF5-iPSC-derived neurons (5c2) and PD G2019S-iPSC-derived neurons (6c3) at various H2O2 treatments and stages of neuronal differentiation.
See also Figure S4 and Table S2 for RT-PCR analysis and cell counts
We also analyzed gene expression profiles of iPSC-derived neurons via qPCR as in previous experiments, but this time, following treatment of neurons with a sub-lethal dose (25 µM) of H2O2 for 48 hours. Expression of some of the oxidative stress response genes, such as CASP9 and HSPA1A, was increased in G2019S-iPSC-derived neurons relative to unaffected H9- and HUF5-iPSC-derived neurons (Figure S4C). In particular, HSPB1 (Heat Shock 27kDa Protein 1) expression was significantly higher in G2019S-iPSC-derived neurons than normal HUF5-iPSC-derived neurons (p = 0.009, n = 4); on the other hand, expression of the neuronal genes, LRRK2 and Engrailed 2 (EN2), was lower in G2019S-iPSC-derived neurons than HUF5- and H9-derived DA neurons (p = 0.04 and 0.07, respectively, n = 4). Expression of SNCA (α-synuclein) in G2019S-iPSC-derived neurons was also higher than in H9-derived neurons (p = 0.07, 2-fold, n = 4) and slightly higher (no significance, n = 4) than HUF5-iPSC-derived neurons.
We also sought to quantitate and verify potential effects of H2O2 on differentiated neurons by Western blotting. Data indicated that in all treatments and stages of neuronal differentiation, relative abundance of activated CASP3 expression in G2019S-iPSC-derived neurons was higher than in HUF5-iPSC-derived neurons (Figure 5D). Differences were most notable (2 – 3-fold) at 10 µM (36-day) and 25 µM (25-day) H2O2 treatments. These findings suggest that mature iPSC-derived neurons, carrying a p.G2019S mutation in LRRK2, were more vulnerable to peroxide-induced oxidative stress than normal HUF5-iPSC- and H9-hESC-derived neurons.
Effect of 6-Hydroxydopamine on DA Neurons
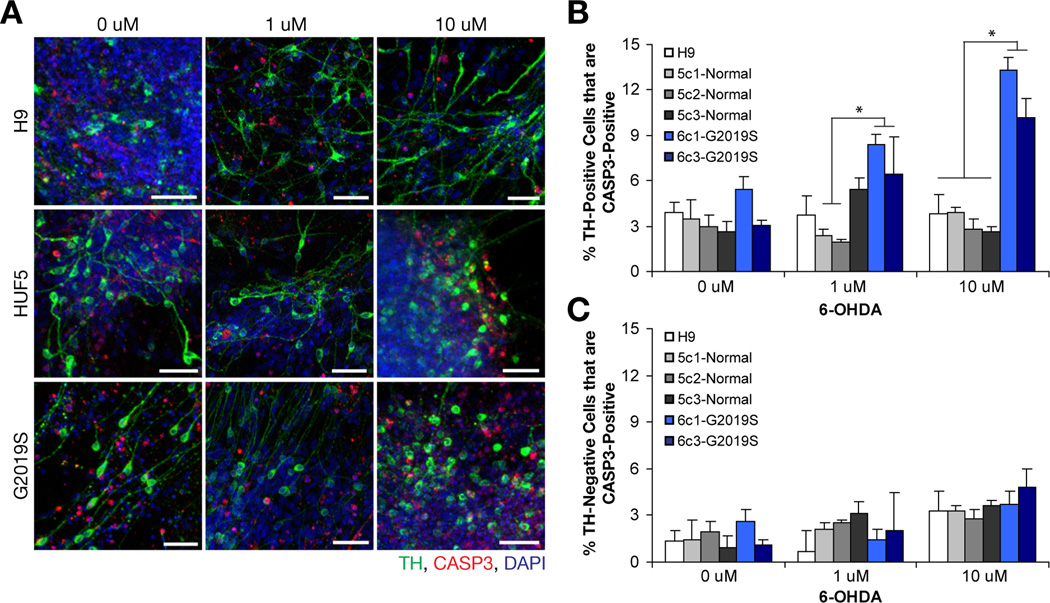
Mitochondrial defects are also implicated in PD. 6-Hydoxydopmine (6-OHDA) can directly lead to destruction of DA neurons by formation of free radicals and/or by inhibition of mitochondrial complexes I and IV (Berretta et al., 2005; Ding et al., 2004; Sauer and Oertel, 1994). To test and to compare effects of 6-OHDA on H9, HUF5- and G2019S-iPSC derived DA neurons, we treated 35-day neurons with various concentrations of 6-OHDA and assayed for cell death using methods described above (Figure 6A–B, Table S3). Analysis based on 2-way ANOVA at 1 µM treatment showed that clones with the G2019S genotype displayed increased (3–4 fold) cell death relative to control clones from HUF5 (p < 0.05, n = 3 wells per clone). At 10 µM treatment, all clones from G2019S showed significantly higher (3–5 fold) cell death than all clones from HUF5-iPSC- and H9-derived DA neurons (p < 0.05, n = 4 well per clone). In contrast, in the absence of treatment, there was no difference between all lines tested (n = 3 per clone). However, we could not rule out the possibility that there might be differential expression of dopamine transporter in the DA neurons between these lines. We also compared levels of cell death in TH-negative cell populations and observed only a slight increase in percentage of TH-negative neurons that co-labeled with CASP3 with increasing concentrations of 6-OHDA (Figure 6C); however, there was no detectable differences between disease and wild-type lines (n = 3–4 well per clone or per line at each concentration). These findings suggest that G2019S-iPSC-derived DA neurons were more sensitive to 6-OHDA than both HUF5-iPSC- and H9-derived DA neurons.
Figure 6. Effect of 6-Hydroxydopamine on DA Neurons.
(A) Representations of neurodegeneration in H9-, HUF5- and G2019S-iPSC derived neurons when treated with various concentrations of 6-OHDA. Scale bar = 50 µm.
(B) Quantification of cells double positive for TH and CASP3. Data represent mean ± SEM (n = 3 to 4 wells per clone).
(C) Quantification of TH-negative (TUJ+) neurons that are also CASP3-positive. Data represent mean ± SEM (n = 3 to 4 wells per clone).
See also Figure S5 and Table S3 for RT-PCR analysis and cell counts.
RT-PCR analysis of neurons after 10 µM of 6-OHDA treatment showed differential gene expression patterns between lines (Figure S5). Results showed that ALDH1A1, HMOX1, MSX2, NEFL, SNCG, LRRK2 and EN2 expression levels were lower in G2019S line than H9 and HUF5 lines. However, significant differences (p < 0.05, n = 3 per clone) were not uniform across all clones of the same genotype, suggesting that there was clonal variability between lines.
Effect of MG-132 on DA Neurons
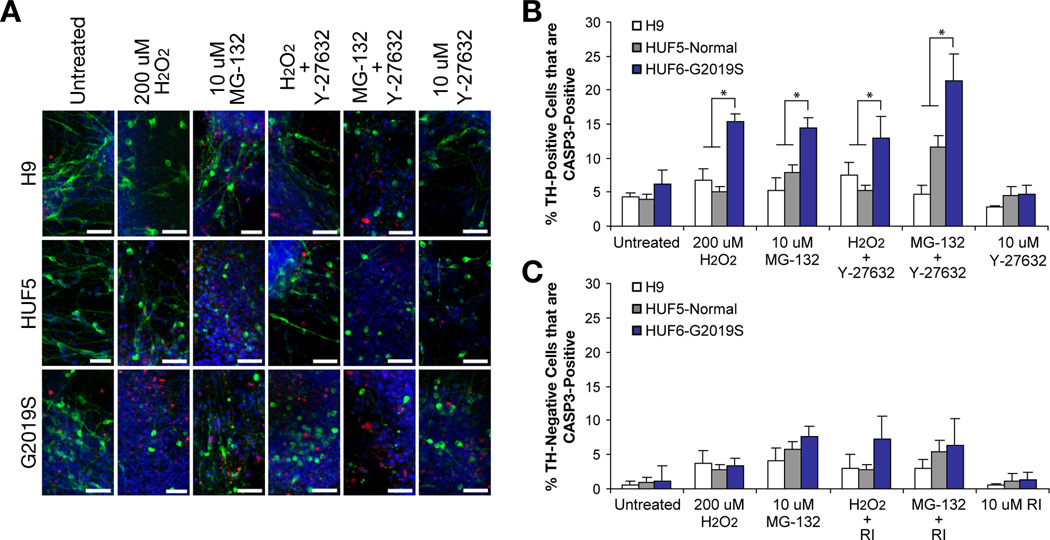
Finally, we tested for sensitivity to impaired protein degradation using the proteasome inhibitor MG-132, which has been shown to cause DA neuron degeneration both in vitro and in an animal model (Sun et al., 2006), on iPSC-derived neurons (Figure 7A–B, Table S4). G2019S-iPSC-derived DA neurons treated with 10 µM of MG-132 showed a higher number of TH-positive cells that are also CASP3-positive than unaffected H9 and HUF5 (p < 0.05, n = 2–3 wells per clone). When we compared MG-132 toxicity directly with 200 uM H2O2 treatment, the effect was similar. In addition, TH-negative cells showed less caspase-3 activation than TH-positive cells in the presence of MG-132 (Figure 7C). Taken together, G2019S-iPSC-derived DA neurons demonstrated increased susceptibility to H2O2, 6-OHDA and MG-132, relative to DA neurons derived from pluripotent cell lines carrying wild-type LRRK2.
Figure 7. Effect of ROCK Inhibitor, Y-27632, on Patient-derived DA Neurons.
(A) Representations of neurodegeneration in H9-, HUF5- and G2019S-iPSC derived neurons when treated with neurotoxins, H2O2 and MG-132, and ROCK inhibitor Y-27632. Scale bar = 50 µm.
(B) Quantification of cells double positive for TH and CASP3. Data represent mean ± SEM (n = 2–3 wells per iPSC clone; average of 3 clones of HUF5 and two clones of G2019S).
(C) Quantification of TH-negative (TUJ+) neurons also positive for CASP3 after treatments as described above. Data represent mean ± SEM (n = 2 to 3 wells per clone).
See also Table S4 for cell counts.
Effect of ROCK Inhibitor, Y-27632, on Patient-derived DA Neurons
West and colleagues have shown that the G2019S mutation increases LRRK2 kinase activity and that expression of a LRRK2 G2019S transgene increases susceptibility to peroxide-induced cell death (Cookson, 2009; West et al., 2005). They also showed that mutations which abolish kinase activity of the LRRK2 transgene also abolished the increase in peroxide-induced cell death. Subsequent work from Nichols and colleagues further showed that several ROCK inhibitors are able to effectively reduce kinase activity of a recombinant G2019S LRRK2 protein in cell-free systems (Nichols et al., 2009). Among these were ROCK inhibitor, Y-27632, which displayed selective inhibition of mutant LRRK2 over wild type LRRK2 using in vitro assays. The prediction from this work was that inhibition of LRRK2 kinase activity might attenuate the LRRK2 G2091S-mediated increase in neuron vulnerability.
To test this prediction, 35-day neurons were exposed to H2O2 or MG-132 for 48 hours in the presence or absence of Y-27632. Cells were then fixed and stained for Tuj1, TH and CASP3 (Figure 7A–B, Table S4). Differential sensitivity of TH-positive neurons from the LRRK2 G2019S genetic background was then observed following H2O2 or MG-132; however, we noted that Y-27632 had no measurable effect following exposure to peroxide and instead appeared to exacerbate cell death following treatment with MG-132. This data suggested that inhibition of kinase activity with ROCK inhibitor Y-27632 does not protect patient-derived DA neurons from H2O2- or MG-132-mediated caspase-3 activation.
DISCUSSION
Parkinson’s disease is a devastating disease that affects both motor and non-motor functions. A defining feature of PD is progressive loss of mDA neurons, yet the underlying etiology of most PD is completely unknown. Studies from animal and cellular models have shown that certain mutations in the LRRK2 gene locus can increase its kinase activity, which contributes to neurotoxicity, possibly via oxidative stress pathways (Heo et al., 2010; MacLeod et al., 2006; Smith et al., 2006; West et al., 2005). Moreover, cells expressing mutant LRRK2 have been shown to be more vulnerable to peroxide-induced cell death than LRRK2 wild-type cells (Cookson, 2009; West et al., 2005). Although the function of LRRK2 is currently not resolved, reduced expression of LRRK2 mRNA limits neural progenitor cell differentiation potential towards dopaminergic neurons and may enhance cell death (Milosevic et al., 2009). These findings suggest that LRRK2 plays a pivotal role in neuronal survival and disruption in this gene can lead to PD (Lin et al., 2009).
Here, we observed a subset of cardinal PD features in DA neurons derived from a patient with symptomatic PD possessing the LRRK2 p.G2019S mutation. Although, this study was limited to a single PD-iPSC line, with multiple subclones and controls, methods and results described should facilitate direct comparisons between lines carrying different LRRK2 mutations or carrying different mutations that predispose to PD. Most importantly, cardinal features that we observed included differential accumulation of α-synuclein protein, upregulation of key oxidative stress response genes, and vulnerability of PD neurons to neurotoxins, including H2O2, MG-132 and 6-OHDA, relative to unaffected neurons. Although differences are modest, given the more limited duration of experiments in vitro (relative to the time course of disease in humans), the distinction between TH-positive and –negative neurons and the increased susceptibility of diseased DA neurons from the p.G2019S LRRK2 patient suggest that central characteristics of PD are retained and detected in this model system. We believe that the differential effect would be more dramatic between the normal control and the diseased neurons if we could allow neurons to mature longer in culture. However, it is difficult to maintain DA neurons in culture, based on our modified protocol, for an extended period of time (50+ days) as cells tend to detach and die spontaneously.
West and others (West et al., 2005) have shown that LRRK2 kinase activity is elevated by the G2019S mutation, leading many to speculate that either increased kinase activity or altered kinase target spectrum may underlie LRRK2-related PD (Cookson, 2009). Nichols et al. (Nichols et al., 2009) further identified ROCK inhibitors, including Y-27632, that were able to effectively reduce kinase activity of recombinant LRRK2 G2019S. Contrary to our expectations, Y-27632, had no protective effect on patient-derived G2019S LRRK2 DA neurons. This could be due to different systems and methods used in our studies. We used hESC- and iPSC-derived DA neurons; whereas Nichols et al. used a cell-free system.
Based on our findings, we propose that G2019S-iPSC derived neurons are more vulnerable to perturbation, caused by oxidative stress, mitochondrial impairment or defects in protein degradation. This vulnerability was selective for TH-positive neurons and accompanied by elevated α-synuclein content and a steady-state elevation in expression of several stress response genes, including activation of CASP3. In spite of the potential usefulness of using iPSC-derived neurons described here, however, there are a number of limitations that must be addressed. First, it is currently not possible to obtain a large number of DA neurons that are pure of contamination of other cell types. This means that results must be interpreted by a combination of experimental means to validate each data point, thus making high-throughput approaches difficult. Second, there is limited knowledge at this time of the endogenous programs and markers of human DA neurons, especially those with specific regionalization. Third, there is limited knowledge of the impacts of specific genetic and environmental parameters individually or in combination that trigger the onset of neurodegeneration. In spite of these limitations, the human iPSC system may provide a novel system that can be used to address specific hypotheses especially regarding the onset of PD in those with genetic determinants. Subsequently, results may be extended from genetically-defined PD to a further understanding of the ontogeny and molecular mechanisms underlying sporadic PD and possibly other neurological disorders that may share a similar etiology.
EXPERIMENTAL PROCEDURES
Isolation of Primary Human Skin Fibroblasts and Generation of iPSCs
Methods for isolation and culture of primary adult human fibroblasts, stem cell culturing, retroviral production and infection were as described (Byrne et al., 2009). HUF5 cells were obtained from a 46-year-old healthy female, and HUF6 (G2019S) cells were obtained from a 60-year-old female PD patient. Procedures for generation of iPSCs were adapted from Yamanaka and colleagues (Takahashi et al., 2007). Briefly, one day before transduction, fibroblasts were seeded at 8 × 104 cells per well of a 6-well plate. Equal volumes (0.5 ml) of each of the four or three (without CMYC) concentrated viral supernatant, supplemented with 8 µg/ml polyprene, were mixed to obtain a final concentration of 10X and added to growing fibroblasts. Next day, viral supernatants were washed with PBS and replaced with fresh MEF medium. The same infection process was repeated a day later. 5 days post-transduction, cells were detached with trypsin, resuspended in MEF medium, counted, and seeded at 5 to 10 × 104 cells, onto 10-cm dishes pre-plated with irradiated MEFs. After overnight incubation, MEF medium was replaced with hESC medium, and thereafter, medium was changed every other day or every day, as required. hESC-like colonies appeared among background colonies around 14–20 days (for 4 factors) or 50–55 days (for 3 factors) post-transduction. Colonies were manually picked and transferred to 12- or 24-well plates pre-plated with CF1 feeders for expansion. HUF5-iPSCs were derived with 4 factors and G2019S-iPSCs were derived with 3 factors without CMYC. For enzymatic passaging, iPSCs were incubated with 200 U/ml collagenase IV (Invitrogen) at 37°C for 3 – 5 min, washed with PBS and then culture medium, and scraped off the dish with a cell scraper. Colonies were further dissociated by pipetting, centrifuged, split at a 1:5 ratio and transferred to a new dish pre-coated with irradiated embryonic mouse feeders (CF1, Charles River). For manual passaging, individual colonies were simultaneously cut and scraped off from the plate using a customized hockey-style (half-loop) glass pipette tip and then transferred to a new dish.
Alkaline Phosphatase Staining and Immunocytochemistry
AP staining was done using the Vector® Red Alkaline Phosphatase Substrate Kit I (Vector Laboratories, CA), according to the manufacturer’s protocol. Standard immunostaining techniques were applied for surface, cytoplasmic and nuclear protein localization as described (Byrne et al., 2009). Primary antibodies and dilutions were as follows: OCT4 (1:100), nestin (1:100), NURR1 (1:100), and FOXA2 (1:50) were obtained from Santa Cruz Biotechnology; SOX2 (1:200), SSEA1 (1:200), SSEA4 (1:200), TRA1–60 (1:200), TRA1–81 (1:200), STELLA (1:200), and LRRK2 (1:200) were obtained from Millipore; NANOG (1:100), β-III-tubulin (TUJ1, 1:200), α-smooth muscle actin (1:200), PITX3 (1:200), OTX2 (1:200), CALB (1:200), sheep tyrosine hydroxylase (1:400), and vimentin (1:200) were obtained from Abcam; desmin (1:200) from Lab Vision; rabbit tyrosine hydroxylase (1:400) from Pel-freez; α-fetoprotein (1:200), VASA (1:200) from R&D Systems; cleaved caspase-3 (1:400) from Cell Signaling; synaptotagmin-1 (1:50) was obtained from Dr. Thomas Sudhof, Stanford University.
Western Blotting Analysis
Proteins were extracted from the cells by RIPA buffer (Sigma)/Protease inhibitor (Roche) mixture, measured using the BCA Protein Assay Kit (Thermo Scientific), separated by running in 15% SDS-PAGE gel and transferred to a PVDF membrane (Millipore). Signals were detected by ECL Plus Western Blotting Detection Kit (GE Healthcare) and analyzed by NIH ImageJ. Primary antibodies and their dilutions were used as follows: α-synuclein (1:1000, Thermo Scientific), cleaved caspase-3 (1:1000, Cell Signaling), GAPDH (1:10,000, Abcam), TH and PITX3 (1:1000, Abcam). We note that previous studies have suggested a close correlation between TUNEL staining and activated caspase-3 (Duan et al., 2003).
Neuronal and Spontaneous Differentiation
Directed differentiation of iPSCs into midbrain dopaminergic neurons was previously described (Chambers et al., 2009) with some adjustments. Briefly, stem cells were plated onto 6-well plates pre-coated with 2.5% Matrigel (BD Biosciences) and allowed to reach 80% confluent in standard hESC medium. Prior to neural induction, differentiated cells were manually cleared. For the first five days, cells were treated with SB431542 (10 uM; Tocris Bioscience) and Noggin (100 ng/mL) in serum placement medium (standard hESC medium without FGF2); for the next four days, cells were treated with human recombinant Sonic hedgehog (200 ng/mL) in DMEM:F12 (Invitrogen) and N2 supplement (Invitrogen); for the next three days, cells were switched to BDNF (20 ng/mL), ascorbic acid (200 uM, Sigma-Alderich), sonic hedgehog (200 ng/mL), and FGF8b (100 ng/mL) in DMEM:F12/N2; thereafter, cells were treated with DMEM:F12/N2 with BDNF, ascorbic acid, GDNF (10 ng/mL), TGF-b (1 ng/mL), and cAMP (500 uM, Sigma-Alderich). Note, we did not separate stem cells into single cells and avoided using Rho kinase (ROCK) inhibitor during the process of differentiation. All growth factors were purchased from R&D Biosystems. Initially, neurons were passed onto fresh plates after 15 days of induction and passed again when the culture expanded. The purity of neurons was improved with each round of collagenase treatment and passaging. Large neuronal clumps (or neurospheres) were manually dissected into smaller pieces with the aid of collagenase treatment.
For spontaneous embryoid body (EB) differentiation, iPSCs were seeded into ultra low attachment hydrogel (Corning) plates containing DMEM:F12 supplemented with 20% FBS (HyClone), 2 mM L-glutamine, 0.1 mM non-essential amino acids, and 0.1 mM 2-mercaptoethanol differentiation medium. After 8 days growing in the suspension, the EB’s were transferred to gelatin-coated dishes containing the same medium to allows the cells to expand for an additional 10–25 days.
Neurotoxicity Assays and ROCK Inhibitor Assays
Starting at day 15–30 days post neuronal differentiation, neurons were purified several rounds by enzymatic (Collagenase IV treatment) and manual (hand dissected) passaging. When the majority of the cells were of neuronal phenotype, neurons were manually and randomly picked and transferred to 48-well culture plates pre-coated with matrigel. Five neuronal clumps (neurospheres) were transferred into each well and maintained with DMEM:F12/N2 medium containing BDNF, GDNF, cAMP, TGFb and Ascorbic Acid. Prior to transfer, large neurospheres were delicately dissected to smaller pieces. One day prior to neurotoxin treatment, before day 35 post differentiation, medium with growth factor was replaced with DMEM:F12/N2 medium alone. Neurospheres that barely adhered to the well substrate were washed out at this step. For H2O2 assay, on day 35, fresh solution (30%, Acros) was diluted with DMEM:F12/N2 medium alone and added to each neuronal well to reach the desired final concentration. Without any disturbance, 24–48 hours later, cells were fixed with 4% PFA and analyzed using immunohistochemistry. For 6-hydroxydopamine assay, fresh agent (Invitrogen) was reconstituted in Phosphate Buffered Saline (Gibco) with 1 mM Ascorbic Acid and then diluted to the desired final concentration with DMEM:F12/N2 medium. 6-OHDA was freshly prepared and the entire procedure was done in the dark. 6-OHDA was added to the neurons at day 35 post differentiation. Treated cells were fixed following 48 hours of reincubation. MG-132 (Calbiochem) was dissolved in ethanol and added to 35-day-old neurons to the final concentration of 10 µM. After 48 hours, cells were fixed in a similar manner as described above. ROCK inhibitor, Y-27632, (Calbiochem) was diluted in water and used at a final concentration of 10 µM. For rescue attempts, Y-27632 was mixed together with neurotoxins and then added to 35-day-old neurons. After 48 hours, cells were fixed as above.
Cell Counting
After neurons were fixed in 48-well plates, they were immunostained for TH, TUJ1, CASP3 as described above. Cell nuclei were also counterstained with DAPI. Data were collected and analyzed by a confocal microscope (Zeiss) with a 10X objective and at 1.5 optical zoom. Fields of neurons (TH-positive) were systematically scanned (top to bottom and left to right) and captured. TUJ1 and CASP3 signals were not used as criteria for scanning and capturing. Subsequently, raw images were merged with ImageJ and counted. TH-positive-cell channel in a particular field were all counted first, then CASP3-positive channel was turned on and counted together with TH-positive-cell channel. At least two entire random fields and up to 100 cells were counted per well. In some cases, especially at higher toxin concentrations, the entire well was counted due to massive cell death.
Bisulfite Sequencing
Bisulfite sequencing and analysis of OCT4 and NANOG promoter regions was performed as previously described (Kossack et al., 2009).
RNA Extraction and RT-PCR
Total RNA was purified using RNeasy Mini Kit (Qiagen) according to the manufacturer’s instructions. One microgram of RNA was used in reverse transcription reaction (20 µl total) with Superscript III (Invitrogen) and random hexamers. 1 µl of the cDNA products were treated with 10 µl of RT-PreAmp master mix consisting of 5 µl Cells Direct 2X reaction mix (Invitrogen), 2.5 µl 0.2X PPP mix (48-gene set, Taqman probes), 1 µl Platinum Taq (Invitrogen), and 1.5 µl TE Buffer (Qiagen). Reactions were pre-amplified with the following settings: 1 cycle at 50°C, 15 minutes; 1 cycle at 70°C, 2 minutes; and 14 cycles at 95°C, 15 seconds and at 60°C, 4 minutes. Reactions were diluted with TE buffer to 20 µl. 2.25 µl of cDNA were used in downstream RT-PCR analysis via the Biomark Fluidigm system (Fluidigm). Ct values for each sample and gene were normalized in relative to GAPDH, RPLPO, CNNTB1, CENTB3 and EEF1a by qBasePlus (Biogazelle).
Measurements of Dopamine Release
After 60 days of neuronal differentiation, media was removed and 1 mL of N2 supplemented with 56 mM KCl was added per well (6 well dish) and incubated at 37°C for 15 minutes. This was collected and immediately frozen in liquid nitrogen and stored at −80°C until assay. Subsequently, protein lysates of each well were made in ice cold 25 mM Tris supplemented with a Complete Mini protease inhibitor cocktail tablet (Roche). Lysates were sonicated and cleared by maximum speed centrifugation in a tabletop microcentrifuge. Soluble protein concentrations were measured by a standard Bradford assay. KCl samples were thawed, stabilized at a final concentration of 0.4N perchloric acid, and centrifuged at 15,000 rpm at 4°C for 12 minutes to clear debris. Supernatant was collected and dopamine assayed by HPLC with electrochemical detection (Coularray detector, ESA, Chelmsford, MA) using a reverse phase C18 column (Perkin Elmer Instruments, Shelton, CT). The mobile phase consisted of a mixture of 90 mM sodium acetate, 35 mM citric acid, 130 uM ethylene-diamine-tetra-acetic-acid (EDTA), 230 uM 1-octanesulfonic acid and 10% (v/v) methanol, with a flow rate of 1mL/min. DA concentration was quantified by comparison of AUC to known standard dilutions. Well to well variation was adjusted by protein determinations of cleared protein lysates.
Statistical Analysis
Student’s two-tailed t-test was used to determine statistical significance for data generated from qRT-PCR. For Western blot and cell counting, statistic analysis was performed using a one-way and two-way ANOVA, respectively, each with Duncan’s multiple range post-hoc tests. For H2O2 toxicity assay, the experiment was repeated twice with multiple biological replicates. Differences were considered significant at p < 0.05.
Supplementary Material
Highlights.
Generation of mature midbrain dopaminergic (DA) neurons from a patient with PD and LRRK2 mutation
PD-derived neurons show accumulation of α-synuclein and increase oxidative stress
PD-derived DA neurons are more susceptible to CASP3 activation than normal neurons
PD-derived DA neurons are sensitive to H2O2, 6-OHDA, and MG-132 treatments
ACKNOWLEDGEMENTS
We thank members of the Reijo Pera Lab for their helpful suggestions and feedback and Matha Isla and Ian Irwin for HPLC analysis of dopamine release studies. This research was supported by funds from CIRM (RL1-00670-1 and CL-00518-1, RRP, TG2-01159, AS), the Blume Foundation (TDP, BS), Kinetics Foundation (TDP), Parkinson’s Alliance (JWL, BS), Stanford Institute for Stem Cell Biology and Regenerative Medicine (RRP), and a generous donor (TDP, RRP).
REFERENCES
- Baba M, Nakajo S, Tu PH, Tomita T, Nakaya K, Lee VM, Trojanowski JQ, Iwatsubo T. Aggregation of alpha-synuclein in Lewy bodies of sporadic Parkinson's disease and dementia with Lewy bodies. Am. J. Pathol. 1998;152:879–884. [PMC free article] [PubMed] [Google Scholar]
- Bayer TA, Jakala P, Hartmann T, Havas L, McLean C, Culvenor JG, Li QX, Masters CL, Falkai P, Beyreuther K. Alpha-synuclein accumulates in Lewy bodies in Parkinson's disease and dementia with Lewy bodies but not in Alzheimer's disease beta-amyloid plaque cores. Neurosci. Lett. 1999;266:213–216. doi: 10.1016/s0304-3940(99)00311-0. [DOI] [PubMed] [Google Scholar]
- Berretta N, Freestone PS, Guatteo E, de Castro D, Geracitano R, Bernardi G, Mercuri NB, Lipski J. Acute effects of 6-hydroxydopamine on dopaminergic neurons of the rat substantia nigra pars compacta in vitro. Neurotoxicol. 2005;26:869–881. doi: 10.1016/j.neuro.2005.01.014. [DOI] [PubMed] [Google Scholar]
- Betarbet R, Sherer TB, Greenamyre JT. Animal models of Parkinson's disease. Bioessays. 2002;24:308–318. doi: 10.1002/bies.10067. [DOI] [PubMed] [Google Scholar]
- Byrne JA, Nguyen HN, Reijo Pera RA. Enhanced generation of induced pluripotent stem cells from a subpopulation of human fibroblasts. PLoS One. 2009;4:e7118. doi: 10.1371/journal.pone.0007118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers SM, Fasano CA, Papapetrou EP, Tomishima M, Sadelain M, Studer L. Highly efficient neural conversion of human ES and iPS cells by dual inhibition of SMAD signaling. Nat. Biotechnol. 2009;27:275–280. doi: 10.1038/nbt.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chartier-Harlin MC, Kachergus J, Roumier C, Mouroux V, Douay X, Lincoln S, Levecque C, Larvor L, Andrieux J, Hulihan M, et al. Alpha-synuclein locus duplication as a cause of familial Parkinson's disease. Lancet. 2004;364:1167–1169. doi: 10.1016/S0140-6736(04)17103-1. [DOI] [PubMed] [Google Scholar]
- Cookson MR. Alpha-synuclein and neuronal cell death. Mol. Neurodegener. 2009;4:9. doi: 10.1186/1750-1326-4-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson TM, Dawson VL. Molecular pathways of neurodegeneration in Parkinson's disease. Science. 2003;302:819–822. doi: 10.1126/science.1087753. [DOI] [PubMed] [Google Scholar]
- de Rijk MC, Launer LJ, Berger K, Breteler MM, Dartigues JF, Baldereschi M, Fratiglioni L, Lobo A, Martinez-Lage J, Trenkwalder C, et al. Prevalence of Parkinson's disease in Europe: A collaborative study of population-based cohorts Neurologic diseases in the elderly research group. Neurol. 2000;54:S21–S23. [PubMed] [Google Scholar]
- Di Salvio M, Di Giovannantonio LG, Omodei D, Acampora D, Simeone A. Otx2 expression is restricted to dopaminergic neurons of the ventral tegmental area in the adult brain. Int. J. Dev. Biol. 2010;54:939–945. doi: 10.1387/ijdb.092974ms. [DOI] [PubMed] [Google Scholar]
- Ding YM, Jaumotte JD, Signore AP, Zigmond MJ. Effects of 6-hydroxydopamine on primary cultures of substantia nigra: Specific damage to dopamine neurons and the impact of glial cell line-derived neurotrophic factor. J. Neurochem. 2004;89:776–787. doi: 10.1111/j.1471-4159.2004.02415.x. [DOI] [PubMed] [Google Scholar]
- Duan WR, Garner DS, Williams SD, Funckes-Shippy CL, Spath IS, Blomme EA. Comparison of immunohistochemistry for activated caspase-3 and cleaved cytokeratin 18 with the TUNEL method for quantification of apoptosis in histological sections of PC-3 subcutaneous xenografts. J. Pathol. 2003;199:221–228. doi: 10.1002/path.1289. [DOI] [PubMed] [Google Scholar]
- Giasson BI, Covy JP, Bonini NM, Hurtig HI, Farrer MJ, Trojanowski JQ, Van Deerlin VM. Biochemical and pathological characterization of Lrrk2. Ann. Neurol. 2006;59:315–322. doi: 10.1002/ana.20791. [DOI] [PubMed] [Google Scholar]
- Han BS, Iacovitti L, Katano T, Hattori N, Seol W, Kim KS. Expression of the Lrrk2 gene in the midbrain dopaminergic neurons of the substantia nigra. Neurosci. Lett. 2008;442:190–194. doi: 10.1016/j.neulet.2008.06.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto M, Masliah E. Alpha-synuclein in Lewy body disease and Alzheimer's disease. Brain Pathol. 1999;9:707–720. doi: 10.1111/j.1750-3639.1999.tb00552.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heo HY, Park JM, Kim CH, Han BS, Kim KS, Seol W. Lrrk2 enhances oxidative stress-induced neurotoxicity via its kinase activity. Exp. Cell. Res. 2010;316:649–656. doi: 10.1016/j.yexcr.2009.09.014. [DOI] [PubMed] [Google Scholar]
- Hernandez D, Paisan Ruiz C, Crawley A, Malkani R, Werner J, Gwinn-Hardy K, Dickson D, Wavrant Devrieze F, Hardy J, Singleton A. The dardarin G2019S mutation is a common cause of Parkinson's disease but not other neurodegenerative diseases. Neurosci. Lett. 2005;389:137–139. doi: 10.1016/j.neulet.2005.07.044. [DOI] [PubMed] [Google Scholar]
- Ichinose H, Ohye T, Suzuki T, Sumi-Ichinose C, Nomura T, Hagino Y, Nagatsu T. Molecular cloning of the human NURR1 gene: Characterization of the human gene and cDNAs. Gene. 1999;230:233–239. doi: 10.1016/s0378-1119(99)00065-7. [DOI] [PubMed] [Google Scholar]
- Kachergus J, Mata IF, Hulihan M, Taylor JP, Lincoln S, Aasly J, Gibson JM, Ross OA, Lynch T, Wiley J, et al. Identification of a novel LRRK2 mutation linked to autosomal dominant parkinsonism: Evidence of a common founder across European populations. Am. J. Hum. Genet. 2005;76:672–680. doi: 10.1086/429256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kittappa R, Chang WW, Awatramani RB, McKay RD. The foxa2 gene controls the birth and spontaneous degeneration of dopamine neurons in old age. PLoS Biol. 2007;5:e325. doi: 10.1371/journal.pbio.0050325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kossack N, Meneses J, Shefi S, Nguyen HN, Chavez S, Nicholas C, Gromoll J, Turek PJ, Reijo-Pera RA. Isolation and characterization of pluripotent human spermatogonial stem cell-derived cells. Stem Cells. 2009;27:138–149. doi: 10.1634/stemcells.2008-0439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang CL, Sinton CM, Sonsalla PK, German DC. Midbrain dopaminergic neurons in the mouse that contain calbindin-d28k exhibit reduced vulnerability to mptp-induced neurodegeneration. Neurodegen. 1996;5:313–318. doi: 10.1006/neur.1996.0042. [DOI] [PubMed] [Google Scholar]
- Lin X, Parisiadou L, Gu XL, Wang L, Shim H, Sun L, Xie C, Long CX, Yang WJ, Ding J, et al. Leucine-rich repeat kinase 2 regulates the progression of neuropathology induced by Parkinson's-disease-related mutant alpha-synuclein. Neuron. 2009;64:807–827. doi: 10.1016/j.neuron.2009.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liou AK, Leak RK, Li L, Zigmond MJ. Wild-type Lrrk2 but not its mutant attenuates stress-induced cell death via erk pathway. Neurobiol. Dis. 2008;32:116–124. doi: 10.1016/j.nbd.2008.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLeod D, Dowman J, Hammond R, Leete T, Inoue K, Abeliovich A. The familial parkinsonism gene LRRK2 regulates neurite process morphology. Neuron. 2006;52:587–593. doi: 10.1016/j.neuron.2006.10.008. [DOI] [PubMed] [Google Scholar]
- Milosevic J, Schwarz SC, Ogunlade V, Meyer AK, Storch A, Schwarz J. Emerging role of LRRK2 in human neural progenitor cell cycle progression, survival and differentiation. Mol. Neurodegener. 2009;4:25. doi: 10.1186/1750-1326-4-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa M, Koyanagi M, Tanabe K, Takahashi K, Ichisaka T, Aoi T, Okita K, Mochiduki Y, Takizawa N, Yamanaka S. Generation of induced pluripotent stem cells without myc from mouse and human fibroblasts. Nat. Biotechnol. 2008;26:101–106. doi: 10.1038/nbt1374. [DOI] [PubMed] [Google Scholar]
- Ng CH, Mok SZ, Koh C, Ouyang X, Fivaz ML, Tan EK, Dawson VL, Dawson TM, Yu F, Lim KL. Parkin protects against lrrk2 G2019S mutant-induced dopaminergic neurodegeneration in Drosophila. J. Neurosci. 2009;29:11257–11262. doi: 10.1523/JNEUROSCI.2375-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols RJ, Dzamko N, Hutti JE, Cantley LC, Deak M, Moran J, Bamborough P, Reith AD, Alessi DR. Substrate specificity and inhibitors of Lrrk2, a protein kinase mutated in Parkinson's disease. Biochem. J. 2009;424:47–60. doi: 10.1042/BJ20091035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols WC, Pankratz N, Hernandez D, Paisan-Ruiz C, Jain S, Halter CA, Michaels VE, Reed T, Rudolph A, Shults CW, et al. Genetic screening for a single common LRRK2 mutation in familial Parkinson's disease. Lancet. 2005;365:410–412. doi: 10.1016/S0140-6736(05)17828-3. [DOI] [PubMed] [Google Scholar]
- Nunes I, Tovmasian LT, Silva RM, Burke RE, Goff SP. Pitx3 is required for development of substantia nigra dopaminergic neurons. Proc. Natl. Acad. Sci. 2003;100:4245–4250. doi: 10.1073/pnas.0230529100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paisan-Ruiz C, Jain S, Evans EW, Gilks WP, Simon J, van der Brug M, Lopez de Munain A, Aparicio S, Gil AM, Khan N, et al. Cloning of the gene containing mutations that cause PARK8-linked Parkinson's disease. Neuron. 2004;44:595–600. doi: 10.1016/j.neuron.2004.10.023. [DOI] [PubMed] [Google Scholar]
- Paisan-Ruiz C, Lang AE, Kawarai T, Sato C, Salehi-Rad S, Fisman GK, Al-Khairallah T, St George-Hyslop P, Singleton A, Rogaeva E. LRRK2 gene in Parkinson disease: Mutation analysis and case control association study. Neurol. 2005;65:696–700. doi: 10.1212/01.wnl.0000167552.79769.b3. [DOI] [PubMed] [Google Scholar]
- Park IH, Arora N, Huo H, Maherali N, Ahfeldt T, Shimamura A, Lensch MW, Cowan C, Hochedlinger K, Daley GQ. Disease-specific induced pluripotent stem cells. Cell. 2008;134:877–886. doi: 10.1016/j.cell.2008.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polymeropoulos MH, Lavedan C, Leroy E, Ide SE, Dehejia A, Dutra A, Pike B, Root H, Rubenstein J, Boyer R, et al. Mutation in the alpha-synuclein gene identified in families with Parkinson's disease. Science. 1997;276:2045–2047. doi: 10.1126/science.276.5321.2045. [DOI] [PubMed] [Google Scholar]
- Ross OA, Toft M, Whittle AJ, Johnson JL, Papapetropoulos S, Mash DC, Litvan I, Gordon MF, Wszolek ZK, Farrer MJ, et al. LRRK2 and Lewy body disease. Ann. Neurol. 2006;59:388–393. doi: 10.1002/ana.20731. [DOI] [PubMed] [Google Scholar]
- Sauer H, Oertel WH. Progressive degeneration of nigrostriatal dopamine neurons following intrastriatal terminal lesions with 6-hydroxydopamine: A combined retrograde tracing and immunocytochemical study in the rat. Neurosci. 1994;59:401–415. doi: 10.1016/0306-4522(94)90605-x. [DOI] [PubMed] [Google Scholar]
- Schüle B, Reijo Pera RA, Langston J. Can cellular models revolutionize drug discovery in parkinson's disease? Biochim. Biophys. Acta. 2009;1792:1043–1051. doi: 10.1016/j.bbadis.2009.08.014. [DOI] [PubMed] [Google Scholar]
- Singleton AB, Farrer M, Johnson J, Singleton A, Hague S, Kachergus J, Hulihan M, Peuralinna T, Dutra A, Nussbaum R, et al. Alpha-synuclein locus triplication causes Parkinson's disease. Science. 2003;302:841. doi: 10.1126/science.1090278. [DOI] [PubMed] [Google Scholar]
- Smith WW, Pei Z, Jiang H, Dawson VL, Dawson TM, Ross CA. Kinase activity of mutant LRRK2 mediates neuronal toxicity. Nat. Neurosci. 2006;9:1231–1233. doi: 10.1038/nn1776. [DOI] [PubMed] [Google Scholar]
- Soldner F, Hockemeyer D, Beard C, Gao Q, Bell GW, Cook EG, Hargus G, Blak A, Cooper O, Mitalipova M, et al. Parkinson's disease patient-derived induced pluripotent stem cells free of viral reprogramming factors. Cell. 2009;136:964–977. doi: 10.1016/j.cell.2009.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spillantini MG, Crowther RA, Jakes R, Hasegawa M, Goedert M. Alpha-synuclein in filamentous inclusions of Lewy bodies from Parkinson's disease and dementia with Lewy bodies. Proc. Natl. Acad. Sci. 1998;95:6469–6473. doi: 10.1073/pnas.95.11.6469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun F, Anantharam V, Zhang D, Latchoumycandane C, Kanthasamy A, Kanthasamy AG. Proteasome inhibitor mg-132 induces dopaminergic degeneration in cell culture and animal models. Neurotoxicol. 2006;27:807–815. doi: 10.1016/j.neuro.2006.06.006. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- Thomson JA, Itskovitz-Eldor J, Shapiro SS, Waknitz MA, Swiergiel JJ, Marshall VS, Jones JM. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- Vernay B, Koch M, Vaccarino F, Briscoe J, Simeone A, Kageyama R, Ang SL. Otx2 regulates subtype specification and neurogenesis in the midbrain. J. Neurosci. 2005;25:4856–4867. doi: 10.1523/JNEUROSCI.5158-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West AB, Moore DJ, Biskup S, Bugayenko A, Smith WW, Ross CA, Dawson VL, Dawson TM. Parkinson's disease-associated mutations in Leucine-rich repeat kinase 2 augment kinase activity. Proc. Natl. Acad. Sci. 2005;102:16842–16847. doi: 10.1073/pnas.0507360102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.