Recent technological innovations allow us to measure gene expression, metabolic activity and the genotype of individual microbial cells retrieved from their natural environment. These technologies have been successful in delivering what they were primarily developed for - attributing microbial functions to identity (Kuypers and Jørgensen 2007; Wagner 2009). In addition, these measurements have also provided a glimpse at microbial individuality in the natural environment: we can compare the phenotype of individual cells, and investigate the basis of phenotypic differences between individuals. The surprising result of such investigations is that cells that belong to the same phylogenetic group, and reside in the same environment, sometimes show large phenotypic variability (Musat et al., 2008; Halm et al., 2009). What is the basis of these phenotypic differences? It is certainly possible that the environment might not be as homogeneous as we might think, or that genetic variation within a phylogenetic group causes different phenotypes. However, there is a third possibility that transcends the traditional concept of nature and nurture as determinants of the phenotype: that phenotypic diversity is produced independently of genetic for example, see environmental variation. It is this third possibility that is the focus of this commentary.
Experiments with genetic model systems in well-controlled laboratory settings revealed that genetically identical cells that live in the same environment often show marked variation in their phenotypic traits (Figure 1). One source of variation is random molecular processes during protein expression and cell division, and such variation is termed 'phenotypic noise' (Elowitz et al., 2002). In contrast to what the term might suggest, the degree of phenotypic noise is under genetic control, and thus an evolvable trait. Is phenotypic noise sometimes useful and thus promoted by natural selection? Genome-wide experiments with yeast and Escherichia coli indicate that most genes are under selection for lower levels of phenotypic noise (Lehner, 2008; Silander et al., 2012) but that certain type of traits and genes have higher levels of variation than expected. Experiments in the laboratory suggest that this phenotypic variation, and thus microbial individuality, can be beneficial and provide organisms with new functionality. The question is whether some of these potential benefits might matter in the natural environment, thus promoting the evolution of increased levels of phenotypic variation.
Figure 1.
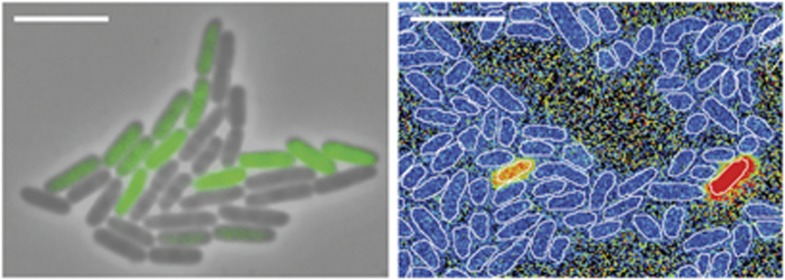
Phenotypic variation in gene expression (left) and metabolic activity (right) in clonal populations of bacteria. Left: a microcolony of Salmonella Typhimurium grown from a single cell; the green color corresponds to the expression of green fluorescent protein under the control of the flagellar promoter flgK (image: N Freed and M Ackermann). Right: cells from a clonal population of Klebsiella oxytoca, analyzed with nano-scale secondary ion mass spectrometry. Orange and red denote high levels of 15N in the biomass as a consequence of the fixation of experimentally provided 30N2 (image: F Schreiber, T Vagner, M Kuypers, M Ackermann). The white scale-bars correspond to about 5 μm.
A first potential adaptive function of phenotypic variation is that it might promote a genotype's survival in fluctuating environments. This phenomenon is known as ‘bet-hedging', and it is an alternative to signal transduction as a microbial survival strategy. Instead of adjusting their phenotypes in response to environmental cues that indicate changing conditions, some individuals attain alternative phenotypic states autonomously. If the environment changes suddenly, a small fraction of the cells within a clonal population might happen to express phenotypes that allows them to better survive the new conditions or resume growth more quickly. There are several convincing demonstrations of phenotypic variation that allows microbes to hedge their bets. A recent study in yeast (Levy et al., 2012) was even able to identify one specific protein that varies in abundance between genetically identical cells in benign conditions, and whose abundance predicts survival if temperature raises suddenly. Why would organisms use such a seemingly risky strategy instead of relying on environmental cues? It is conceivable that organisms might often not have enough time to adapt their phenotype after a shift in the environmental conditions, or that they might not have sufficient cellular resources to modify their phenotype. For example, microbes that live close to the metabolic limit might sometimes not be able to express catabolic pathways to access newly available substrates. Phenotypic variation produced before environmental shifts could thus allow genotypes to persist and proliferate under conditions where signal transduction and gene regulation fail.
There is a second potential adaptive function of phenotypic variation that has not received as much attention, but that might be equally relevant: genetically identical cells that express different phenotypes can interact with each other, and through these interactions gain new functionality. This concept is familiar from filamentous cyanobacteria, where genetically identical cells within a filament differentiate into two distinct phenotypes that specialize on different metabolic processes and complement each other. Similar examples are known from biofilms, where individual cells perform different functions. However, and importantly, differentiation and interaction does not require physical attachment between the cells. Many environments are spatially structured, so that the cells do not disperse much from where they emerge after division. Such structured environments can keep cells together as clonal groups, and thus promote interactions between different phenotypes within these groups. One type of environment that maintains microbes as nearly clonal groups is hosts that are infected by bacterial pathogens, and infection biology indeed offers several examples of interactions and the division of labor between phenotypes encoded by the same genotype (for example, see Ackermann et al., 2008). It is an open question whether such interactions between different phenotypes in clonal groups are also common in other natural environments.
These two adaptive explanations–bet-hedging and interactions–are not mutually exclusive, but they are clearly different. They differ with respect to the conditions under which they can manifest, and with respect to how the benefits arise. Bet-hedging is a strategy to decrease variation in growth and survival in the face of environmental fluctuations, and it thus requires such fluctuations to be beneficial; constant environments would simply select for genotypes that grow and survive best under these conditions. In contrast, interactions between different phenotypes in clonal populations do not require changing environments, but they require that the growth and survival of individual cells be affected by the presence of other individuals and by the phenotypes they express. In order to test whether the phenotypic variation that we observe is potentially adaptive, and to find out why it is adaptive, we thus need to follow individual cells over time, analyze the phenotypes they express and ask how their growth and survival depends on their phenotype, on the environment and of the phenotypes of the cells around them.
Is microbial individuality and the concept of beneficial phenotypic variation in clonal groups an important aspect of the biology of microbes in their natural environment? One might argue that microorganisms in their natural environment are always exposed to diverse signals and diffusion gradients to which they will respond, and that phenotypic variation promoted by intracellular processes is thus not important. However, as discussed above, phenotypic variation can provide advantages that are not accessible if microbes strictly follow environmental cues. To address the potential importance of microbial individuality in nature, we need to analyze phenotypic variation between individuals in their natural environment, and investigate the functional consequences of this variation. While such experiments are undoubtedly challenging, technological innovations make single-cell measurements in environmental systems increasingly feasible, even for microbes that are not genetic model systems (Brehm-Stecher and Johnson 2004; Wagner 2009; Lencastre Fernandes et al., 2011). Ideally, these experiments would be driven by existing (for example, Kussell and Leibler 2005; Ackermann et al., 2008) and newly developed theory, and address clear concepts and hypotheses about how phenotypic variation could allow the persistence of types in fluctuating environments and promote interactions within clonal populations. It will be fascinating to find out whether microbial populations in natural environments gain benefits from phenotypic variation among individuals–that is, whether microbial individuality can give rise to collective functionality.
Acknowledgments
We thank the members of the Molecular Microbial Ecology group at ETH and Eawag, Marcel Kuypers, Michael Wagner and Matthias Zimmermann for valuable discussions, and Dave Johnson, Frank Schreiber and an anonymous reviewer for comments on the paper. Our work on this topic is funded by SNF, ETH Zurich, Eawag, and the Marie Curie program of the European Commission
The authors declare no conflict of interest.
References
- Ackermann M, Stecher B, Freed NE, Songhet P, Hardt WD, Doebeli M. Self-destructive cooperation mediated by phenotypic noise. Nature. 2008;454:987–990. doi: 10.1038/nature07067. [DOI] [PubMed] [Google Scholar]
- Brehm-Stecher BF, Johnson EA. Single-cell microbiology: tools, technologies, and applications. Microbiol Mol Biol Rev. 2004;68:538–559. doi: 10.1128/MMBR.68.3.538-559.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elowitz MB, Levine AJ, Siggia ED, Swain PS. Stochastic gene expression in a single cell. Science. 2002;297:1183–1186. doi: 10.1126/science.1070919. [DOI] [PubMed] [Google Scholar]
- Halm H, Musat N, Lam P, Langlois R, Musat F, Peduzzi S, et al. Co-occurrence of denitrification and nitrogen fixation in a meromictic lake, Lake Cadagno (Switzerland) Environ Microbiol. 2009;11:1945–1958. doi: 10.1111/j.1462-2920.2009.01917.x. [DOI] [PubMed] [Google Scholar]
- Kussell E, Leibler S. Phenotypic diversity, population growth, and information in fluctuating environments. Science. 2005;309:2075–2078. doi: 10.1126/science.1114383. [DOI] [PubMed] [Google Scholar]
- Kuypers MMM, Jørgensen BB. The future of single cell environmental microbiology. Environ Microbiol. 2007;9:6–7. doi: 10.1111/j.1462-2920.2006.01222_5.x. [DOI] [PubMed] [Google Scholar]
- Lehner B. Selection to minimise noise in living systems and its implications for the evolution of gene expression. Mol Syst Biol. 2008;4:170. doi: 10.1038/msb.2008.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lencastre Fernandes R, Nierychlo M, Lundin L, Pedersen AE, Puentes Tellez P, Dutta A, et al. Experimental methods and modeling techniques for description of cell population heterogeneity. Biotechnol Adv. 2011;29:575–599. doi: 10.1016/j.biotechadv.2011.03.007. [DOI] [PubMed] [Google Scholar]
- Levy SF, Ziv N, Siegal ML. Bet Hedging in Yeast by heterogeneous, age-correlated expression of a stress protectant. PLoS Biol. 2012;10:e1001325. doi: 10.1371/journal.pbio.1001325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musat N, Halm H, Winterholler B, Hoppe P, Peduzzi S, Hillion F, et al. A single-cell view on the ecophysiology of anaerobic phototrophic bacteria. Proc Natl Acad Sci. 2008;105:17861–17866. doi: 10.1073/pnas.0809329105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silander OK, Nikolic N, Zaslaver A, Bren A, Kikoin I, Alon U, et al. A genome-wide analysis of promoter-mediated phenotypic noise in Escherichia coli. Plos Genet. 2012;8:e1002443. doi: 10.1371/journal.pgen.1002443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner M. Single-cell ecophysiology of microbes as revealed by Raman microspectroscopy or secondary ion mass spectrometry imaging. Annu Rev Microbiol. 2009;63:411–429. doi: 10.1146/annurev.micro.091208.073233. [DOI] [PubMed] [Google Scholar]


