Abstract
Although the influence of ozone (O3) on plants has been well studied in agroecosystems, little is known about the effect of elevated O3 (eO3) on soil microbial functional communities. Here, we used a comprehensive functional gene array (GeoChip 3.0) to investigate the functional composition, and structure of rhizosphere microbial communities of Yannong 19 (O3-sensitive) and Yangmai 16 (O3-relatively sensitive) wheat (Triticum aestivum L.) cultivars under eO3. Compared with ambient O3 (aO3), eO3 led to an increase in soil pH and total carbon (C) percentages in grain and straw of wheat plants, and reduced grain weight and soil dissolved organic carbon (DOC). Based on GeoChip hybridization signal intensities, although the overall functional structure of rhizosphere microbial communities did not significantly change by eO3 or cultivars, the results showed that the abundance of specific functional genes involved in C fixation and degradation, nitrogen (N) fixation, and sulfite reduction did significantly (P<0.05) alter in response to eO3 and/or wheat cultivars. Also, Yannong 19 appeared to harbor microbial functional communities in the rhizosphere more sensitive in response to eO3 than Yangmai 16. Additionally, canonical correspondence analysis suggested that the functional structure of microbial community involved in C cycling was largely shaped by soil and plant properties including pH, DOC, microbial biomass C, C/N ratio and grain weight. This study provides new insight into our understanding of the influence of eO3 and wheat cultivars on soil microbial communities.
Keywords: elevated ozone, functional gene, rhizosphere microbial community, wheat cultivar
Introduction
Ozone (O3) is an atmospheric gas that has been rising due to human activity and industrial development. For agriculture systems, O3 is considered as the most phytotoxic air pollutant potentially suppressing plant growth, productivity and yield (Schoene et al., 2004; Ainsworth, 2008; Booker et al., 2009), and more importantly, it may substantially impact below-ground functional processes, such as root growth and carbon (C) allocation (Morgan et al., 2003; Rämö et al., 2006; Feng and Kobayashi, 2009; Wittig et al., 2009; Betzelberger et al., 2010). It is expected that such effects will be much more rapid and have significant impacts on plant productivity, soil C and nitrogen (N) dynamics, and ecosystem functioning if anthropogenic activities leading to ozone formation continue unabated in the future (IPCC, 2007).
Wheat is the second largest food crop with an annual production of >650 million metric tons and harvested area of over 200 million hectares worldwide (Zhu et al., 2011). In the Yangtze River Delta region of China, O3 pollution resulted in about 10% yield loss of wheat in 1999 as predicted from results of local open-top chamber studies and monitoring data (Feng et al., 2003). Currently, the mean O3 concentrations (July–October) range from 38 to 46 ppb in the Yangtze River Delta of China (Wang et al., 2006), and it is predicted that the average O3 concentration will increase from 42 to 63 ppb by the end of this century, which could drive a further 10% decrease in yield for soybean, wheat and rice, and 20% for bean according to meta-analysis conducted by Feng and Kobayashi (2009). One of the strategies for reducing such negative O3 effects on wheat yield is to screen cultivars that are better adapted to eO3 conditions (Barnes et al., 1990; Pleijel et al., 2006). Recently, some O3-relatively sensitive wheat cultivars have been reported in China, which may avoid yield reduction in a high O3 environment (Cao et al., 2009; Zhu et al., 2011).
As soil systems depend on nutrient inputs from plants and nutrient cycling of soil microorganisms, a change in nutrient flux due to eO3 could also alter structural and functional aspects of soil microbial biodiversity (Andersen, 2003). In recent years, most studies on the effect of eO3 on rhizosphere microbial communities were largely based on open-top chambers, and the results mainly focused on the changes of microbial biomass and structure by phospholipid fatty acid analysis, denaturing gradient gel electrophoresis methods (Kasurinen et al., 2005; Chung et al., 2006; Kanerva et al., 2008), and/or single-strand conformation polymorphism method (Dohrmann and Tebbe, 2005). Those studies indicated that eO3 had little or significant effects on soil microbial communities with different plants/ecosystems. However, there are only few reports about effects of eO3 on soil microbial functional processes (Larson et al., 2002; Phillips et al., 2002; Zak et al., 2007; Chen et al., 2009). It is important to comprehensively examine the effect of eO3 on the functional composition, structure and metabolic potential of rhizosphere microbial communities.
Functional gene microarray-based technology (for example, GeoChip) has become a routine molecular tool to analyze the functional composition, structure and dynamics of microbial communities from a variety of ecosystems (He et al., 2012a, 2012c). For example, GeoChip 3.0 was used to examine how elevated CO2 affected soil microbial communities, and the results showed that the functional composition, structure and metabolic potential of soil microbial communities were shifted, which was significantly correlated with soil C and N contents and plant productivity (He et al., 2010b). Also, it was applied to examine the effects of global warming (temperature) on soil microbial communities (Zhou et al., 2012), and to profile arsenic-contaminated soil microbial communities (Xiong et al., 2010) and rhizosphere microbial communities of Candidatus Liberibacter asiaticus infected citrus trees (Trivedi et al., 2012). All results demonstrate that GeoChip is a robust and high-throughput tool to specifically, sensitively and quantitatively profile microbial communities and link their structure with environmental factors and ecosystem functioning.
In this study, we hypothesized that changes in plant and soil properties resulting from eO3 would alter the functional diversity, composition, structure and metabolic potential of rhizosphere microbial communities, and such effects would vary with wheat cultivars. To test those hypotheses, our objectives were (i) to examine the effect of eO3 on rhizosphere microbial functional genes of Yannong 19 (O3-sensitive) and Yangmai 16 (O3-relatively sensitive) wheat cultivars; (ii) to distinguish whether there were differential responses between those two wheat cultivars; and (iii) to understand the correlation between the functional structure of rhizosphere microbial communities and the soil and plant properties under eO3. The study was conducted on a free-air ozone enrichment (O3-FACE) experimental site located in a suburb of Jiangdu City, China. GeoChip 3.0 was used to determine the gene abundance from GeoChip hybridization signal intensities. Although the overall functional structure of rhizosphere microbial communities did not significantly change under eO3 or between cultivars, the results showed that eO3 did alter the structure of functional genes involved in C cycling and change the abundance of some key functional genes involved in C, N and sulfur (S) cycling, which appeared to be cultivar dependent. This study provides new insights into our understanding of the eO3 and wheat cultivars effect on rhizosphere microbial communities.
Materials and methods
Experimental site and sample collections
The experimental site is located in the suburb of Jiangdu City in Jiangsu province of China (32°35′ N, 119°42′ E), and the soil type is Shajiang Aquic Cambosols (Chinese Soil Taxonomy) (Li et al., 2009) with a sandy-loamy texture, with 15 g kg−1 total C, 1.59 g kg−1 total N, pH 6.8, and 25.1% clay (<0.001 mm) and bulk density 1.2 g cm−3 at 0–15 cm depth (Zhu et al., 2011). An experimental platform of O3-FACE was established in 2007 over a rice-wheat rotation system, with rice transplanted in mid-June and harvested in middle-to-late October and winter wheat sown in early November and harvested in late May or early June of the next year. This study was conducted during the wheat growing season of 2010 (for example, November 2009 to June 2010) after three growth seasons with O3 fumigation at three periods: 14 April to 22 May in 2007, 5 March to 26 May in 2008 and 1 March to 24 May in 2009, and their average concentrations of O3 were 42 ppb for aO3 and 53.4 ppb for eO3 (Zhu et al., 2011).
The O3-FACE system has three O3-FACE rings and three similar ambient rings. Three O3-FACE replicate rings, each 14.5 m in diameter, were set randomly to continuously provide an eO3 concentration of 60 ppb from 0900 h to 1800 h during 3 March and 31 May 2010, while three ambient (40 ppb) replicate rings, each with the same size, were set randomly within the same area. All of the rings were far enough apart to prevent O3 from spilling over from one ring to another to avoid the influence of O3 from the O3-FACE rings on the ambient plots. In each O3-FACE or ambient ring, two winter wheat cultivars (Yannong 19 and Yangmai 16) were sown in November in each subplot of a ring. Previous studies showed different responses of Yannong 19 and Yangmai 16 to eO3, and Yannong 19 (Y19) was referred to as O3-sensitive wheat cultivar, and Yangmai 16 (Y16) as O3-relatively sensitive cultivar (Cao et al., 2009; Zhu et al., 2011). A total of 12 rhizosphere and 12 corresponding bulk samples were taken from 2 cultivars of 6 rings at the ripening stage on 10 June 2010. For GeoChip analysis, rhizosphere soil was taken by collecting soil that adhered to plant roots after the plant had been removed from soil and shaken, and each sample was a composite from the roots of five randomly selected plants and then stored at −20 °C until DNA extraction. For soil property analysis, soil cores (2.5-cm diameter at the depth of 0–15 cm) were taken from the corresponding site of plants from which the rhizosphere soil had been collected, and each soil sample was pooled from five soil cores and stored at −20 or 4 °C until further analyses.
Analysis of plant and soil properties
The total carbon (TC) and nitrogen (TN) of plant or soil were determined by a TruSpec CN Elemental Analyzer (Leco Corporation, St. Joseph, MI, USA). Dissolved organic carbon (DOC) was determined by a Multi N/C 3100 analyzer (Analytik Jena AG, Jena, Germany). Soil pH was determined with a glass electrode in 1:2.5 (soil:water) solution (w/v). Fifteen winter wheat plants from each subplot were harvested, and partitioned into grain and litter. Litter and grain samples were dried at 65 °C until a constant weight was obtained, and then weighed for 1000 kernel weight (grain weight) and individual plant weight (plant weight).
Nucleic acid extraction, purification, amplification and labeling
Community DNA of 12 rhizosphere soil samples was extracted by the freeze-grinding method (Zhou et al., 1996). Extracted DNA was stored at −80 °C until it was used. The quality of purified DNA was assessed by an ND-1000 spectrophotometer (NanoDrop Technologies Inc., Wilmington, DE, USA) and the concentration of DNA was measured using a Quant-It PicoGreen kit (Invitrogen, Carlsbad, CA, USA) using a FLUOstar Optima (BMG Labtech, Jena, Germany). An aliquot of 100 ng DNA from each sample was amplified in triplicate using the TempliPhi kit (Amersham Biosciences, Piscataway, NJ, USA) in a modified buffer containing single strand binding protein (200 ng μl−1) and spermidine (0.04 mℳ) to increase the sensitivity of amplification at 30 °C for 3 h (Wu et al., 2006). Amplified DNA (∼3.0 μg) was mixed with 20 ml 2.5 × random primers (Invitrogen), heated to 99 °C for 5 min, and immediately placed on ice, then fluorescently labeled in a reaction solution containing 50 μℳ dATP, dCTP, dGTP, 20 μM dTTP (USB Corporation, Cleveland, OH, USA), 1 mℳ Cy5 dUTP (Amersham Pharmacia Biotech, Piscataway, NJ, USA),and 40 U of Klenow fragment (Invitrogen), incubating at 37 °C for 3 h. The labeled products were purified with a QIAquick PCR purification kit (Qiagen, Valencia, CA, USA), and then dried down in a SpeedVac (Thermo Fisher Scientific Inc., Milford, MA, USA) for 45 min at 45 °C.
Microarray hybridizations, scanning and array data processing
The fluorescently labeled DNA was suspended in the hybridization mix (50% formamide, 3 × SSC, 0.3% SDS, 0.7 mg ml−1 herring sperm DNA), and 0.86 mM DTT incubated at 95 °C for 5 min, and then maintained at 60 °C until hybridization. Samples were hybridized with GeoChip 3.0 (He et al., 2010a) on a MAUI Hybridization System (BioMicro Systems, Salt Lake City, UT, USA) at 42 °C for 12 h. Microarrays were scanned on a Pro ScanArray Microarray Scanner (Perkin-Elmer, Boston, MA, USA), and signal intensities of each spot were measured with ImaGene 6.0 (Biodiscovery, El Segundo, CA, USA). Empty and poor spots were removed before the signal intensities were normalized by the mean signal across the slide, and spots with signal-to-noise ratio (SNR=(signal mean−background mean)/background standard deviation)>2.0 were used as the cutoff for positive spots for further analyses (He and Zhou, 2008). A gene was considered as positive if it was detected in at least 2 of 12 samples.
Statistical analyses
All data were analyzed through a general linear model for split-plot design to determine the effects of O3 concentration (ambient vs. elevated), different wheat cultivars (Y19 vs. Y16) and their interactions. Diversity indices were calculated as previously described (He et al., 2010b). Permutational multivariate analysis of variance (adonis) was based on Euclidean distance matrices to partition differences among different treatments using permutations (999 times), and the significance test was based on pseudo-F ratio (Oksanen et al., 2010). Mantel and partial Mantel analyses were used to link the functional structure of microbial communities with plant and soil variables (He et al., 2010b). All statistical analyses were performed by the Vegan package in R (Dixon, 2003). Detrended correspondence analysis (DCA) and canonical correspondence analysis (CCA) were performed using CANOCO for Windows version 4.5 (Biometris—Plant Research International, Wageningen, The Netherlands).
Results
Effects of eO3 on soil and plant properties
To understand whether eO3 affects soil and wheat properties, 5 soil variables and 10 plant variables were measured (Supplementary Table S1). ANOVA results showed that soil pH and the TC in both grain (TC-grain) and straw (TC-straw) were significantly (P<0.05) increased under eO3, while DOC and grain weight were significantly (P<0.05) decreased under eO3. Also, the decreases in DOC and grain weight were significantly (P<0.05) different between Yannong 19 and Yangmai 16 cultivars with an average of 15.4% decrease for the Yangmai 16 cultivar and an average of 17.4% decrease for the Yannong 19 cultivar (Figure 1). However, microbial biomass C (MBC), TC, TN, C/N ratio (CNR), total N in grain (TN-grain) and straw (TN-straw), grain and straw CNRs (CNR-grain, CNR-straw), panicle length, plant weight and root-straw weight did not significantly change at eO3 (Supplementary Table S1). In addition, no significant effects were observed for the interaction of O3 and cultivar (Supplementary Table S1; Figure 1). Such changes in soil and plant properties may affect the functional composition and structure of rhizosphere microbial communities, which was further examined by GeoChip 3.0 in this study.
Figure 1.
Significantly changed soil and plant properties under eO3 and with Yannong 19 and Yangmai 16 wheat cultivars. C-Y19: Yannong 19 (O3-sensitive) cultivar under control conditions; C-Y16: Yangmai 16 (O3-relatively sensitive) cultivar under control conditions; O-Y19: Yannong 19 cultivar under elevated O3 conditions; O-Y16: Yangmai 16 cultivar under elevated O3 conditions. P-values shown in the figure are based on split-plot ANOVA (O: O3; C: cultivar; O × C: O3 × cultivar).
Overall review of soil microbial functional structure under eO3
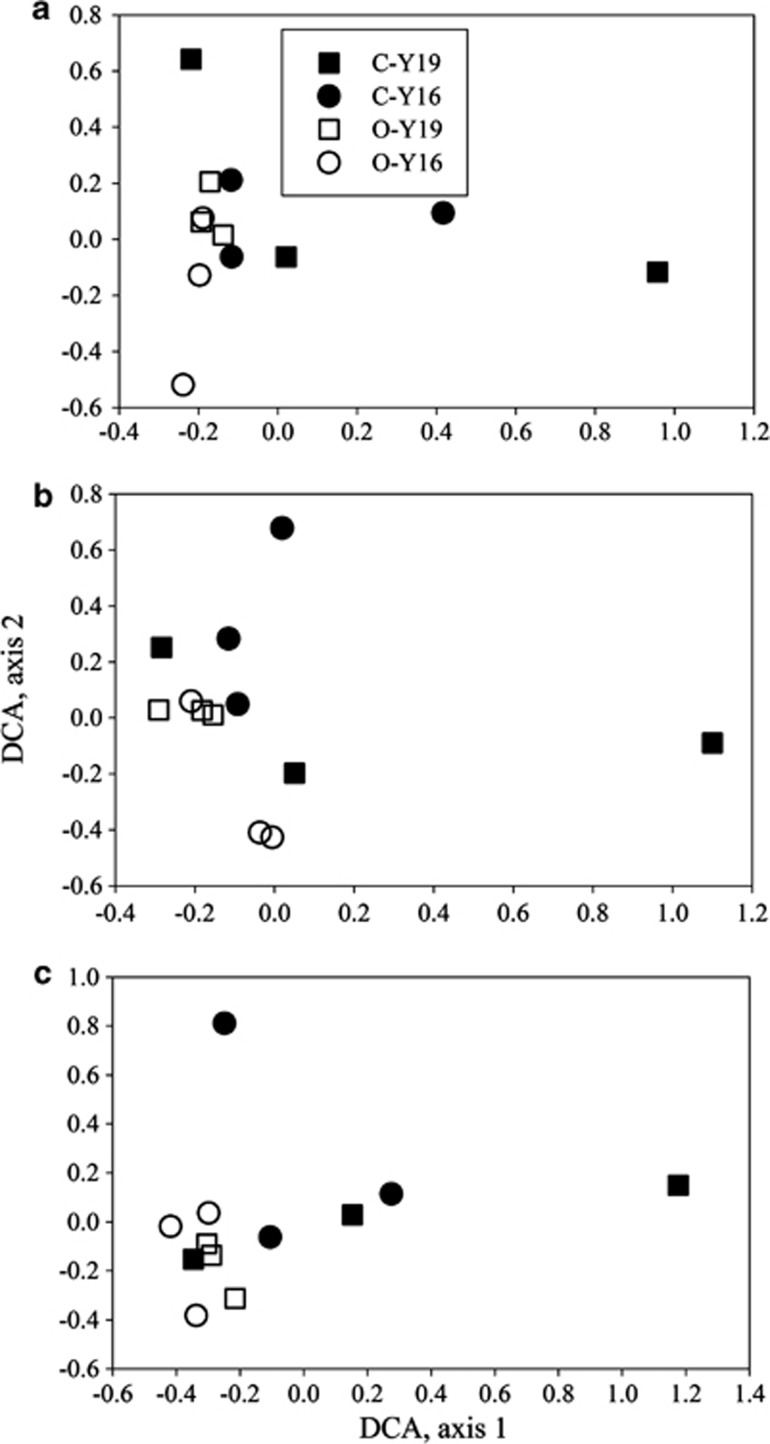
A total of 3691 genes were detected by GeoChip 3.0 in at least 2 out of 12 samples, including 244 genes derived from fungi, and 131 from archaea. The number of genes detected for individual samples ranged from 1485 to 2858, but no significant differences in the number of detected genes were observed between aO3 and eO3 samples or between both cultivars. Similarly, the Shannon index (H′) and the Simpson's reciprocal index (1/D) were not significantly different between aO3 and eO3 or between both cultivars. However, the Simpson's evenness was higher at aO3 for both cultivars than at eO3, at which this index was also higher in the Yangmai 16 cultivar than in the Yannong 19 cultivar (Supplementary Table S2). Adonis analysis of abundances of all detected genes showed that O3, cultivar and their combination had little effect upon the overall functional structure of rhizosphere microbial community (Table 1). Also, DCA of all detected genes, or subsets of fungal or archaeal genes indicated that aO3 and eO3 samples or Yannong 19 and Yangmai 16 samples could not be well separated, though eO3 samples appeared to be more closely clustered together than aO3 samples (Figure 2). In addition, the ratios of fungi to bacteria, and archaea to bacteria were significantly decreased and increased, respectively, at eO3 for the Yannong 19 cultivar only, while no significant changes were seen for the Yangmai 16 cultivar (Supplementary Figure S1). Therefore, all these results indicated that the overall functional diversity and structure of rhizosphere microbial communities appeared not to be significantly altered by eO3.
Table 1. Adonis analysis of the effect of eO3 and cultivars on the functional structure of rhizosphere microbial communities based on the signal intensity of all detected genes by GeoChip 3.0.
| F-value | P-value | R2 | |
|---|---|---|---|
| O3 | 0.7905 | 0.495 | 0.0805 |
| Cultivar | 0.5213 | 0.889 | 0.0531 |
| O3:cultivar | 0.5047 | 0.904 | 0.0514 |
Abbreviation: eO3, elevated ozone.
Figure 2.
DCA of all detected genes (a), and subsets of genes derived from archaea (b) and fungi (c) based on hybridization signal intensities detected by GeoChip 3.0. For C-Y19, C-Y16, O-Y19 and O-Y16, please see Figure 1 for details.
Relationships between functional structure and environmental variables
To explore possible linkages between the functional structure of rhizosphere microbial communities and environmental factors, including pH, MBC, DOC, TC, TN, CNR, TC-grain, TN-grain, CNR-grain, TC-straw, TN-straw, CNR-straw, panicle length, grain weight, plant weight and straw-root weight (Supplementary Table S1), we analyzed GeoChip data and those environmental variables by Mantel or partial Mantel tests and CCA. First, Mantel analysis of all environmental factors and the signal intensity of all detected genes showed significant (P<0.05) correlations between 11 individual functional genes (for example, aceA, aceB, nirK, ppk and dsrA) and those soil variables, but none of those genes were significantly correlated with the plant variables (Table 2). Second, CCA was used to further explore what environmental factors largely shaped the functional structure of rhizosphere microbial communities. Although no significant correlations were found between the signal intensity of all detected functional genes and environmental variables selected, the detected C cycling genes were found to have a significant (P<0.05) relationship with the selected environmental variables, including soil pH, MBC, DOC, CNR, grain weight and O3 concentration (mean concentrations for each plot during the wheat growth season) (Figure 3). Based on this model, a total of 60.7% of the total variance could be explained by the first two constrained axes, with the first axis explaining 48.4% and the second axis for 12.3%. Both the first canonical axis (F=4.692, P=0.022) and the sum of all canonical axes (F=2.694, P=0.010) were significant by the Monte Carlo test. The structure of C cycling genes was quite different between aO3 and eO3, which was well separated by the first axis, and positively correlated with DOC and negatively correlated with O3 concentration, soil pH and MBC. The second axis was positively correlated with soil C/N and negatively with grain weight. The first axis indicated that DOC decreased, and pH and MBC increased following an exposure to eO3 for both wheat cultivars, while MBC was higher in the Yangmai 16 cultivar than in the Yannong 19 cultivar. The second axis indicated grain weight decreased and soil C/N increased under eO3 compared with aO3 for both cultivars (Figure 3). The results indicated that the functional diversity and structure of microbial communities involved in C cycling was closely correlated with soil and plant properties, and largely shaped by O3 concentration, soil pH, DOC, MBC and CNR as well as by grain weight.
Table 2. Significantly correlated functional genes involved in C, N, S and P cycling with soil and/or plant properties analyzed by partial Mantel analysis.
| Gene or enzyme | Functional process | Soil with plant as control | Plant with soil as control | ||
|---|---|---|---|---|---|
| |
|
r-value |
P-value |
r-value |
P-value |
| aceA | C degradation | 0.357 | 0.025 | 0.017 | 0.446 |
| aceB | 0.326 | 0.044 | −0.017 | 0.536 | |
| CODH | 0.316 | 0.039 | 0.123 | 0.209 | |
| Phenol oxidase | 0.352 | 0.037 | 0.121 | 0.240 | |
| pcc | C fixation | 0.326 | 0.031 | 0.010 | 0.462 |
| nasA | Assimilatory N reduction | 0.362 | 0.022 | 0.075 | 0.306 |
| nirB | 0.355 | 0.040 | 0.069 | 0.308 | |
| nirK | Denitrification | 0.366 | 0.020 | 0.052 | 0.348 |
| norB | 0.434 | 0.017 | 0.278 | 0.089 | |
| ppk | P utilization | 0.421 | 0.026 | 0.118 | 0.236 |
| dsrA | Sulfite reduction | 0.405 | 0.010 | 0.254 | 0.056 |
Abbreviations: C, carbon; CODH, carbon monoxide dehydrogenase; N, nitrogen; P, phosphorus.
Bold values are significant, with P<0.05.
Figure 3.
CCA of GeoChip hybridization signal intensities of detected functional genes involved in C cycling and environmental variables, including O3, DOC, MBC, soil pH, soil C/N and grain weight. For C-Y19, C-Y16, O-Y19 and O-Y16, please see Figure 1 for details.
Responses of functional genes involved in C, N, S and phosphorus (P) cycling under eO3
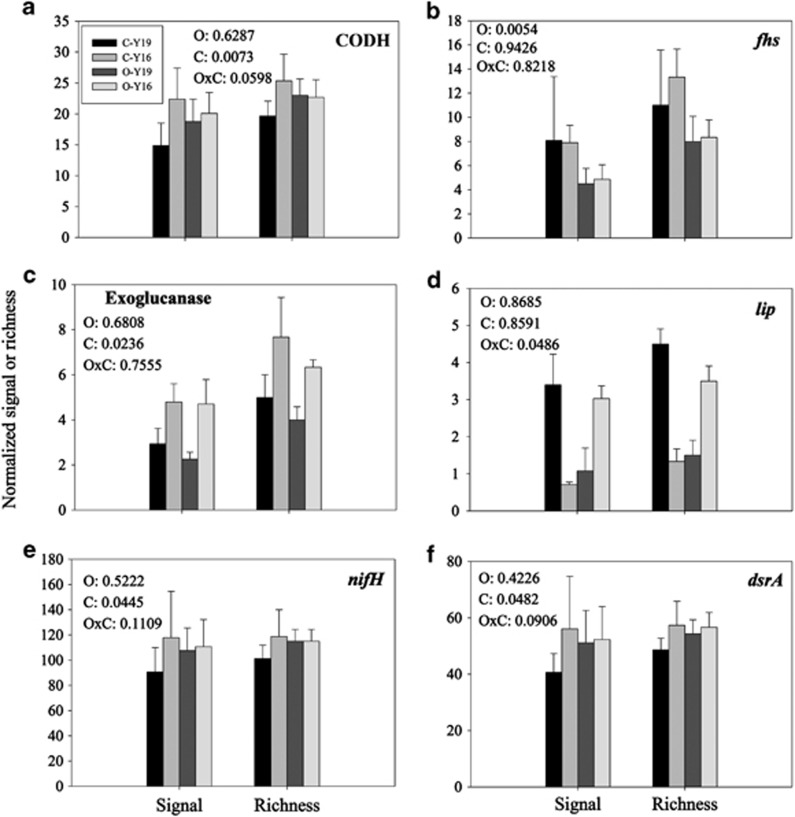
Although eO3 did not have significant effects on the overall microbial functional structure, ANOVA of all detected genes showed significant changes at the gene family level under eO3. The abundances of 17 gene families were significantly (P<0.05) affected by eO3 (fhs and corC), by cultivars (limEH, aclB, chrA, copA, nifH, pimF, pobA, dsrA and carbon monoxide dehydrogenase (CODH), exoglucanase, aliphatic nitrilase and small multidrug resistance genes), or by their interaction (lip, alkH and chnB) (Supplementary Table S3). With a focus on functional genes involved in C, N, P and S cycling, some general trends of microbial community responses to eO3 and cultivars were described in detail below.
C cycling genes
The abundances of four C cycling genes were significantly (P<0.05) changed by eO3 (fhs), cultivars (CODH and exoglucanase genes), or eO3 and cultivar (lip). The abundances of fhs were lower at eO3 than at aO3 for both cultivars (Figure 4b); the abundances of CODH and exoglucanase genes were higher in Yangmai 16 than in Yannong 19 (Figures 4a and c); under eO3, a significant decrease for Yannong 19 and a significant increase for Yangmai 16 were observed for lip (Figure 4d). Also, the abundance of C cycling genes generally decreased at eO3, but the abundances of a few genes increased, including mcrA, and mannanase and xylanase genes for both cultivars, amyX, nplT and lip for Yangmai 16, and pcc, aceA/B, bacterial ara, and CODH and phenol oxidase genes for Yannong 19 (Table 3). In addition, for the rhizosphere microbial communities, the abundances of C cycling genes were generally higher in Yangmai 16 than in Yannong 19 under both aO3 and eO3 conditions (Table 3). The results generally indicated that the C cycling process of rhizosphere microbial communities from both Yannong 19 and Yangmai 16 cultivars might be inhibited by eO3, but functional genes of microbial communities from Yangmai 16 maintained relatively high abundances, indicating that rhizosphere microbial communities of Yangmai 16 (O3-relatively sensitive) may be also more tolerant under eO3 conditions.
Figure 4.
Representatives of key functional genes significantly affected by eO3 and/or wheat cultivars via ANOVA. (a) CODH; (b) fhs encoding formyltetrahydrofolate synthetase; (c) Exoglucanase; (d) lip; (e) nifH; (f) dsrA. Details of C-Y19, C-Y16, O-Y19 and O-Y16 are described in Figure 1. P-values shown in the figure are based on split-plot ANOVA (O: O3; C: cultivar; O × C: O3 × cultivar).
Table 3. Effects of elevated O3 and wheat cultivars on abundances of key functional genes involved in C cycling analyzed by ANOVA. Only genes with >3 probes detected were shown.
| Gene or enzyme | Functional process | O3 (%)a | Cultivar (%)b | P-valuec | ||||
|---|---|---|---|---|---|---|---|---|
| |
|
Y19 |
Y16 |
aO3 |
eO3 |
O3 |
Cultivar |
O3:cultivar |
| CODH | C fixation | 25.98 | −10.24 | 50.25 | 7.05 | 0.6287 | 0.0073 | 0.0598 |
| pcc | 10.54 | −15.88 | 36.83 | 4.12 | 0.6759 | 0.1223 | 0.2280 | |
| Rubisco | −5.21 | −16.74 | 21.70 | 6.90 | 0.4621 | 0.4169 | 0.6630 | |
| fhs | Acetogenesis | −44.37 | −38.62 | −2.25 | 7.85 | 0.0054 | 0.9426 | 0.8218 |
| aceA | Glyoxylate cycle | 22.50 | −13.07 | 43.92 | 2.13 | 0.9152 | 0.1802 | 0.2340 |
| aceB | 5.90 | −8.45 | 24.39 | 7.52 | 0.8213 | 0.1135 | 0.4216 | |
| amyA | Starch degradation | −6.81 | −11.87 | 14.80 | 8.57 | 0.3071 | 0.2547 | 0.7335 |
| amyX | −8.24 | 124.84 | −58.02 | 2.87 | 0.2292 | 0.7732 | 0.7298 | |
| Glucoamylase | −33.45 | −6.77 | −12.92 | 21.99 | 0.4231 | 0.9721 | 0.5746 | |
| nplT | −44.24 | 27.04 | −37.11 | 43.30 | 0.6793 | 0.8439 | 0.3539 | |
| pulA | −16.23 | −0.91 | 0.32 | 18.67 | 0.6362 | 0.6594 | 0.6724 | |
| Cellobiase | Hemi-cellulose degradation | −17.60 | −7.36 | 4.09 | 17.03 | 0.6117 | 0.7158 | 0.8416 |
| Mannanase | 29.91 | 112.68 | 62.12 | 165.41 | 0.1579 | 0.0675 | 0.3079 | |
| ara-bacteria | 10.35 | −10.73 | 12.28 | −9.17 | 0.9730 | 0.9656 | 0.6549 | |
| ara-fungi | −3.67 | −9.40 | 3.64 | −2.53 | 0.7180 | 0.9742 | 0.8700 | |
| xylA | −48.45 | −21.45 | −22.94 | 17.41 | 0.0567 | 0.6811 | 0.3479 | |
| Xylanase | 20.18 | 6.12 | 26.61 | 11.80 | 0.5302 | 0.3593 | 0.7800 | |
| Endoglucanase | Cellulose degradation | −22.09 | −13.52 | 8.29 | 20.21 | 0.4212 | 0.5986 | 0.8704 |
| Exoglucanase | −23.22 | −1.97 | 63.17 | 108.31 | 0.6806 | 0.0236 | 0.7555 | |
| vanA | Aromatics degradation | 12.84 | −19.23 | 29.16 | −7.54 | 0.7606 | 0.5999 | 0.3388 |
| NAG* | Chitin degradation | −24.27 | −16.72 | −6.83 | 2.46 | 0.2560 | 0.8875 | 0.8043 |
| Endochitinase | −17.45 | −7.24 | 3.47 | 16.27 | 0.2640 | 0.4491 | 0.6556 | |
| glx | Lignin degradation | −38.68 | −34.61 | 32.03 | 40.78 | 0.1337 | 0.3092 | 0.9001 |
| lip | −68.33 | 327.27 | −79.14 | 181.42 | 0.8685 | 0.8591 | 0.0486 | |
| mnp | −86.30 | −57.76 | −11.15 | 173.85 | 0.0539 | 0.8953 | 0.2185 | |
| Phenol oxidase | 16.21 | −11.16 | 23.71 | −5.43 | 0.9536 | 0.6727 | 0.4663 | |
| mcrA | Methanogenesis | 19.73 | 34.46 | 29.98 | 45.97 | 0.4396 | 0.3089 | 0.7638 |
| pmoA | CH4 oxidation | −23.20 | −9.22 | 28.92 | 52.38 | 0.4147 | 0.1085 | 0.7924 |
Abbreviations: aO3, ambient ozone; C, carbon; eO3, elevated ozone; CODH, carbon monoxide dehydrogenase; NAG*, N-acetyl-glucosaminidase.
Percentage change by eO3 was calculated using the following formula: (eO3−aO3) × 100/aO3, where aO3 and eO3 were the average signal intensities of genes detected by GeoChip 3.0 at aO3 or eO3, respectively.
Percentage change by cultivar is calculated using the following formula: (cultY16−cultY19) × 100/cultY19 under aO3 or eO3 conditions.
P-values based on ANOVA using the model: Y∼O3+cultivar+O3: cultivar.
Bold values are significant, with P<0.05.
N cycling genes
The abundances of most N cycling genes generally decreased or remained unchanged for microbial communities of Yangmai 16 but increased for Yannong 19 at eO3; however, the abundances of N cycling genes were generally higher in Yangmai 16 than those in Yannong 19 (Supplementary Table S4). For example, nifH was detected as the only significantly (P<0.05) changed gene in the N cycle in this study. Compared with aO3, the percentage changes were −6.1% and 18.6% for Yangmai 16 and Yannong 19, respectively, at eO3, while those percentage changes of Yangmai 16 over Yannong 19 were 2.9% and 30.0% at eO3 and aO3, respectively, indicating a higher abundance for Yangmai 16 than for Yannong 19 at both aO3 and eO3 conditions (Supplementary Table S4; Figure 4e). These results indicated that eO3 might affect N cycling processes (for example, N fixation) of microbial communities, and that the rhizosphere microbial community of Yannong 19 (O3-sensitive) cultivar may be also more sensitive under eO3 than that from the Yangmai 16 (O3- relatively sensitive) cultivar.
P and S cycling genes
The changes in abundance of most P and S cycling genes were similar to those for N cycling genes with a general increase for Yannong 19, and a decrease for Yangmai 16 at eO3. dsrA was detected as the only significantly (P<0.05) changed gene in S cycling and no significantly changed genes were detected for P utilization in this study. Compared with aO3, the percentage changes of dsrA were −6.7% and 25.9% for Yangmai 16 and Yannong 19, respectively, at eO3, while the percentages of changes for Yangmai 16 over Yannong 19 were 2.3% and 38.0% at eO3 and aO3, respectively (Supplementary Table S5; Figure 4f). Similarly to N cycling genes, these results indicated that eO3 might modify S and P cycling with Yannong 19 (O3-sensitive) cultivar microbial communities being more sensitive under eO3.
gyrB
GeoChip 3.0 also targets gyrB (He et al., 2010a), a phylogenetic marker to examine the phylogenetic diversity, composition and structure of microbial communities. A total of 195 gyrB probes showed positive signals, but the number or signal intensity of detected genes was not significantly different between aO3 and eO3, or between two cultivars (Supplementary Figure S2A), and nor did DCA of all detected gyrB genes show a clear separation by aO3 and eO3, or two cultivars (Supplementary Figure S2B). The results indicated that the phylogenetic diversity and structure of the rhizosphere microbial communities was not significantly impacted by eO3 or by wheat cultivars, which was generally consistent with our analyses of functional genes above.
Discussion
Understanding the response of soil microbial communities to eO3 is essential for establishing sustainable agroecosystems in an eO3 environment. In this study, we examined the functional composition and structure of rhizosphere microbial communities of Yannong 19 and Yangmai 16 wheat cultivars under eO3. Our results indicated that the overall microbial functional diversity or structure was not significantly affected by eO3, but the structure of functional genes involved in C cycling altered, and significantly changed functional genes involved in C, N and S cycling were identified. Also, the abundance of functional genes generally decreased, indicating an inhibitory effect of eO3 on soil microbial communities. In addition, the Yannong 19 (O3-sensitive) cultivar appears to harbor rhizosphere microbial communities more sensitive under eO3 than the Yangmai 16 cultivar.
One of our hypotheses is that eO3 significantly affects the overall functional diversity and structure of rhizosphere microbial communities via the changes in plant properties and soil microenvironments (Biswas et al., 2008; Feng et al., 2008; Feng and Kobayashi, 2009; Zhu et al., 2011). Previous studies showed that eO3 decreased plant aboveground and root biomass and grain yield, altered litter and soil chemistry, and adjusted antioxidant capacity (Morgan et al., 2003; Liu et al., 2005; Rämö et al., 2006; Betzelberger et al., 2010), especially in wheat (Biswas et al., 2008; Feng et al., 2008; Feng and Kobayashi, 2009; Zhu et al., 2011). However, only few studies have been focused on the effect of eO3 on microbial community functions. For example, a significant discrimination of sole-carbon source utilization patterns was observed for the wheat rhizosphere soil but not for the bulk soil, and the functional diversity of the rhizosphere microbial communities was reduced under eO3 (Chen et al., 2009). Also, a few studies at the Aspen FACE site (Larson et al., 2002; Phillips et al., 2002; Chung et al., 2006; Holmes et al., 2006; Zak et al., 2007) and with open-top chambers (Kasurinen et al., 2005) or mesocosms (Kanerva et al., 2008) indicated that eO3 altered the functional diversity, structure and/or metabolic potential of soil microbial communities. In this study, our results did not fully support the above hypothesis that eO3 would alter the overall functional diversity and structure of rhizosphere microbial communities. There are a few possibilities. First, the difference of concentrations between aO3 (40 ppb) and eO3 (60 ppb) was not large enough to cause significant changes in the overall soil microbial community diversity and structure. However, it should also be noted that although this increase in O3 is relatively small, those differences identified in this study are highly relevant to the near future as well as for current years when O3 concentration is relatively high. Second, the eO3 exposure time may be still too short for soil microbial communities to respond to eO3. Third, since DOC is largely composed of low molecular weight compounds (for example, organic acids, amino acids and sugars) with low pH, a decrease in DOC could lead to an increase in pH, but the overall effect of both factors may result no significant changes observed. Fourth, plant biomass or root biomass was not significantly changed between aO3 and eO3, or Yannong 19 and Yangmai 16 cultivars, indicating that soil C inputs may remain unchanged. In addition, only three replicates were used in this study so that the statistical power may not be enough to distinguish subtle changes.
Although the overall microbial diversity and structure did not significantly change in response to eO3, we did observe eO3-induced and cultivar-specific changes for key functional genes involved in C, N and S cycling. For example, the abundance of fhs signficantly decreased at eO3. Formyltetrahydrofolate synthetase encoded by fhs catalyzes a key step in the reductive acetogenesis pathway and produces acetate from C1 precursors (for example, CO2). Acetogenesis is of great important to the global C cycle, producing ∼10% of acetate annually in anaerobic habitats (Leaphart and Lovell, 2001). Actate is considered as an important substrate for many micoorganisms in soil, and a decrease of fhs abundance indicates a decrease of acetate production at eO3, which may result in an increase in pH. Interestingly, the abundance of lip signifcantly decreased in Yannong 19, and signifcantly increased in Yangmai 16 at eO3. Lignin peroxidase encoded by lip uses two substrates, 1,2-bis(3,4-dimethoxyphenyl)propane-1,3-diol and H2O2, and produces 3,4-dimethoxybenzaldehyde, 1-(3,4-dimethoxyphenyl)ethane-1,2-diol, and H2O, where H2O2 is mainly responsible for plant adjustements in response to eO3 (Heath, 2008). A significant increase of the lip abundance at eO3 may be related to a reduction of H2O2 concentration in the rhizosphere, and faciliate detoxication of H2O2 in the rhizosphere and/or in plants. Also, more genes showed cultivar-specific changes at aO3 or/and eO3. For example, the abundances of genes encoding CODH, exoglucanase, NifH and DsrA were signifcantly higher in Yangmai 16 than those in Yannong 19 although they were not significantly different between aO3 and eO3. These results are generally consistent with previous studies upon enzyme activities of 1,4-β-glycosidase, 1,4-β-N-acetylglucosaminidase and other C degradation enzymes under eO3 (Larson et al., 2002; Chung et al., 2006). Furthermore, the changes of functional gene abundances showed two clear patterns: (i) most C cycling genes decreased under eO3 with a generally higher abundance for Yangmai 16 under both eO3 and eO3 and (ii) most N, S and P cycling genes increased for Yannong 19 and decreased for Yangmai 16 under eO3 although a general higher abundance remained for Yangmai 16. Therefore, our results indicated that the abundances of some key genes signifcantly changed at eO3 and/or between two cultivars, which may modify ecosystem functional processes, strongly supporting one of our hypotheses that eO3 would affect the composition and metablic potential of rhizosphere microial commuities. In addition, this study identifies some important functional genes invovled in the reductive acetogenesis pathway (for example, fhs), lignin degradation (for example, lip) and N fixation (for example, nifH) in response to eO3; thus, our future studies may focus on the diversity and quantification of those key genes by quantitative PCR and/or high-throughput sequencing approaches.
The microbial community diversity, structure and functional activity are also shaped by environmental factors, such as soil and plant properties. Previous studies indicated that soil pH and C inputs significantly affected soil microbial community diversity and structure (Fierer and Jackson, 2006; Aciego Pietri and Brookes, 2009). In this study, an increase in soil pH and a decrease in DOC were observed. Mantel analyses of all detected genes also showed that the abundances of many genes (for example, aceA, aceB, pcc, nasA, nirK, ppk and dsrA) involved in C, N, P and S cycling were significantly correlated with soil variables. Further CCA indicated that the structure of functional genes involved in C cycling could be largely shaped by soil pH, DOC, MBC, CNR and grain weight. Therefore, this study indicated that the microbial community structure might be shaped by soil environmental factors and plant properties.
Root exudates have an important role in plant–microbe interactions and shape the diversity, structure and function of soil, especially rhizosphere microbial communities (Bais et al., 2006; Dennis et al., 2010; Doornbos et al., 2012). A previous study showed a degree of specificity in the interaction of wheat cultivars and fluorescent Pseudomonas species via analysis of phlD genes (Mazzola et al., 2004). Also, a recent study with Arabidopsis thaliana suggested that root exudation differences could influence rhizosphere bacterial communities (Micallef et al., 2009). Similarly, another study with different plants (wheat, maize, rape and barrel clover) demonstrated that plant species root exudates significantly shaped the rhizosphere bacterial community structures (Haichar et al., 2008). In addition, artificial root exudate solutions were added to soil micrososms, and the results indicated that organic acids might have an important role in shaping soil bacterial communities (Shi et al., 2011). However, it is unclear if root exudates shape the rhizosphere micorbial communities from Yannong 19 and Yangmai 16 cultivars, and further investigations are needed by a comprehesive profiling of root exudates and linking them with microbial functional structure and activity.
The rhizosphere microbial communities of Yannong 19 may be more sensitive than those from Yangmai 16 in response to eO3. First, based on the number of significantly changed genes and the abundance changes of all detected genes, more shifts were found by cultivars than by O3, or by their interaction. Second, many genes detected had higher abundances at eO3 than at aO3 for Yannong 19 rhizosphere microbial communities, while the abundances of detected genes in Yangmai 16 rhizosphere microbial communities remained largely unchanged. Third, CCA suggests that the signal intensity of detected genes involved in C cycling had a positive correlation with MBC for Yangmai 16 rhizosphere samples, but a negative correlation for Yannong 19 rhizosphere samples, indicating that Yangmai 16 may be more favorable for soil microbial growth than Yannong 19. Therefore, our results suggested that the rhizosphere microbial communities from Yannong 19 may be also more sensitive in response to eO3 than those from Yangmai 16. However, further investigations are necessary to understand the mechanism using different approaches, such as quantitative PCR or high-throughput sequencing analysis of 16S rRNA and/or key functional genes (for example, fhs, nifH and lip) identified in this study. It should be noted that two crops were grown at the FACE site per year with wheat being planted after rice, and rice was also exposed to eO3 in the same ring/plot as wheat. Although rice is also sensitive to ozone, it is expected that rice has little effect on wheat rhizosphere microbial communities during the wheat growth season. Previous studies also suggest that the effect of O3 on soil microbial communities largely came from wheat in the current growth season, especially after anthesis (Slaughter et al., 1989; Pleijel et al., 1998; Soja et al., 2000).
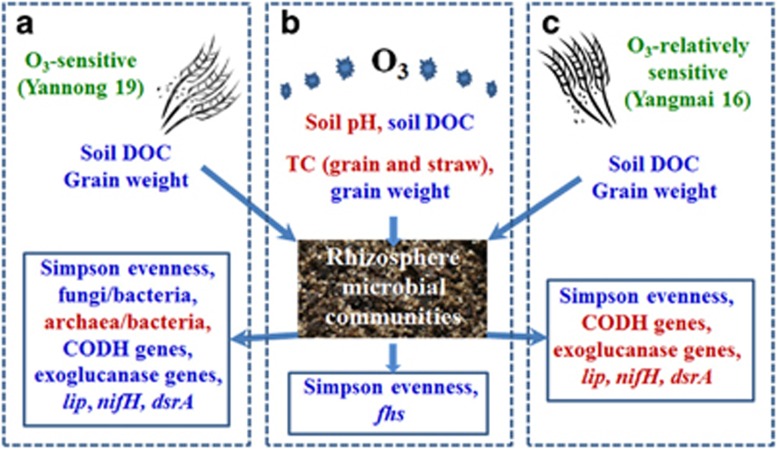
In summary, this study indicates that continuously increasing O3 may be a threat for sustainable production of wheat agroecosystems in the future (Figure 5). First, eO3 significantly caused an increase in soil pH, total C concentration of straw and grain, and a decrease in soil DOC and grain weight. Second, although the overall rhizosphere microbial functional diversity and structure was not significantly affected by eO3 or cultivars, the Simpson evenness and fungi/bacteria decreased, and many key microbial processes, especially C cycling showed a trend of inhibition under eO3. Third, Yannong 19 appeared to harbor microbial communities more sensitive in response to eO3 than Yangmai 16 (Figure 5). This study provides new insights into our understanding of the influence of eO3 and wheat cultivars on soil microbial communities.
Figure 5.
Summary of significant effects of eO3 on plant and soil properties, and key microbial functional genes involved in C, N, S and P cycling (b), and cultivar-specific responses in Yannong 19 (a), and Yangmai 16 (c). In three rectangle boxes, the red and blue text is for significantly increased and decreased functional gene abundances, respectively. Soil, plant properties or functional genes without significant changes between aO3 and eO3, or between two cultivars are not shown.
Acknowledgments
This research was supported by the Knowledge Innovation Project of the Chinese Academy of Sciences (KZCX2-YW-QN403 and KZCX2-EW-414), the International S&T Cooperation Program of China (Grant No. 2009DFA31110), and the Global Environment Research Fund by the Ministry of the Environment, Japan (Grant No. C-062), and by Oklahoma Applied Research Support (OARS), Oklahoma Center for the Advancement of Science and Technology (OCAST) through the Projects AR062-034 and AR11-035. The GeoChip development and associated computational pipelines used in this study were partially supported by the United States Department of Energy, Biological Systems Research on the Role of Microbial Communities in Carbon Cycling Program (DE-SC0004601).
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Aciego Pietri JC, Brookes PC. Substrate inputs and pH as factors controlling microbial biomass, activity and community structure in an arable soil. Soil Biol Biochem. 2009;41:1396–1405. [Google Scholar]
- Ainsworth EA. Rice production in a changing climate: a meta-analysis of responses to elevated carbon dioxide and elevated ozone concentration. Glob Change Biol. 2008;14:1642–1650. [Google Scholar]
- Andersen CP. Source–sink balance and carbon allocation below ground in plants exposed to ozone. New Phytol. 2003;157:213–228. doi: 10.1046/j.1469-8137.2003.00674.x. [DOI] [PubMed] [Google Scholar]
- Bais HP, Weir TL, Perry LG, Gilroy S, Vivanco JM. The role of root exudates in rhizoshpere interactions with plants and other organisms. Annu Rev Plant Biol. 2006;57:233–266. doi: 10.1146/annurev.arplant.57.032905.105159. [DOI] [PubMed] [Google Scholar]
- Barnes JD, Velissariou D, Davison AW, Holevas CD. Comparative ozone sensitivity of old and modern Greek cultivars of spring wheat. New Phytol. 1990;116:707–714. [Google Scholar]
- Betzelberger AM, Gillespie KM, McGrath JM, Koester RP, Nelson RL, Ainsworth EA. Effects of chronic elevated ozone concentration on antioxidant capacity, photosynthesis and seed yield of 10 soybean cultivars. Plant Cell Environ. 2010;33:1569–1581. doi: 10.1111/j.1365-3040.2010.02165.x. [DOI] [PubMed] [Google Scholar]
- Biswas DK, Xu H, Li YG, Sun JZ, Wang XZ, Han XG, et al. Genotypic differences in leaf biochemical, physiological and growth responses to ozone in 20 winter wheat cultivars released over the past 60 years. Glob Change Biol. 2008;14:46–59. [Google Scholar]
- Booker F, Muntifering R, McGrath M, Burkey K, Decoteau D, Fiscus E, et al. The ozone component of global change: potential effects on agricultural and horticultural plant yield, product quality and interactions with invasive species. J Integr Plant Biol. 2009;51:337–351. doi: 10.1111/j.1744-7909.2008.00805.x. [DOI] [PubMed] [Google Scholar]
- Cao J-L, Wang L, Zeng Q, Liang J, Tang H-Y, Xie Z-B, et al. Characteristics of photosynthesis in wheat cultivars with different sensitivities to ozone under O3-free air concentration enrichment conditions. Acta Agron Sin. 2009;35:1500–1507. [Google Scholar]
- Chen Z, Wang X, Feng Z, Xiao Q, Duan X. Impact of elevated O3 on soil microbial community function under wheat crop. Water Air Soil Pollut. 2009;198:189–198. [Google Scholar]
- Chung H, Zak D, Lilleskov E. Fungal Community Composition and Metabolism Under Elevated CO2 and O3. Springer Verlag GMBH: Germany; 2006. pp. 143–154. [DOI] [PubMed] [Google Scholar]
- Dennis PG, Miller AJ, Hirsch PR. Are root exudates more important than other sources of rhizodeposits in structuring rhizosphere bacterial communities. FEMS Microbiol Ecol. 2010;72:313–327. doi: 10.1111/j.1574-6941.2010.00860.x. [DOI] [PubMed] [Google Scholar]
- Dixon P. VEGAN, a package of R functions for community ecology. J Veg Sci. 2003;14:927–930. [Google Scholar]
- Dohrmann AB, Tebbe CC. Effect of elevated tropospheric ozone on the structure of bacterial communities inhabiting the rhizosphere of herbaceous plants native to Germany. Appl Environ Microbiol. 2005;71:7750–7758. doi: 10.1128/AEM.71.12.7750-7758.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doornbos RF, van Loon LC, Bakker PAH. Impact of root exudates and plant defense signaling on bacterial communities in the rhizosphere. A review. Agron Sustain Dev. 2012;32:227–243. [Google Scholar]
- Feng Z, Kobayashi K. Assessing the impacts of current and future concentrations of surface ozone on crop yield with meta-analysis. Atmos Environ. 2009;43:1510–1519. [Google Scholar]
- Feng Z, Kobayashi K, Ainsworth EA. Impact of elevated ozone concentration on growth, physiology, and yield of wheat (Triticum aestivum L.): a meta-analysis. Glob Change Biol. 2008;14:2696–2708. [Google Scholar]
- Feng Z-W, Jin M-H, Zhang F-Z, Huang Y-Z. Effects of ground-level ozone (O3) pollution on the yields of rice and winter wheat in the Yangtze River delta. J Environ Sci. 2003;15:360–362. [PubMed] [Google Scholar]
- Fierer N, Jackson RB. The diversity and biogeography of soil bacterial communities. Proc Natl Acad Sci USA. 2006;103:626–631. doi: 10.1073/pnas.0507535103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haichar FZ, Marol C, Berge O, Rangel-Castro JI, Prosser JI, Balesdent J, et al. Plant host habitat and root exudates shape soil bacterial community structure. ISME J. 2008;2:1221–1230. doi: 10.1038/ismej.2008.80. [DOI] [PubMed] [Google Scholar]
- He Z, Deng Y, Van Nostrand JD, Tu Q, Xu M, Hemme CL, et al. GeoChip 3.0 as a high-throughput tool for analyzing microbial community composition, structure and functional activity. ISME J. 2010a;4:1167–1179. doi: 10.1038/ismej.2010.46. [DOI] [PubMed] [Google Scholar]
- He Z, Deng Y, Zhou J. Development of functional gene microarrays for microbial community analysis. Curr Opin Biotechnol. 2012a;23:49–55. doi: 10.1016/j.copbio.2011.11.001. [DOI] [PubMed] [Google Scholar]
- He Z, Piceno Y, Deng Y, Xu M, Lu Z, DeSantis T, et al. The phylogenetic composition and structure of soil microbial communities shifts in response to elevated carbon dioxide. ISME J. 2012b;6:259–272. doi: 10.1038/ismej.2011.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Z, Van Nostrand JD, Zhou J. Applications of functional gene microarrays for profiling microbial communities. Curr Opin Biotechnol. 2012c;23:460–466. doi: 10.1016/j.copbio.2011.12.021. [DOI] [PubMed] [Google Scholar]
- He Z, Xu M, Deng Y, Kang S, Kellogg L, Wu L, et al. Metagenomic analysis reveals a marked divergence in the structure of belowground microbial communities at elevated CO2. Ecol Lett. 2010b;13:564–575. doi: 10.1111/j.1461-0248.2010.01453.x. [DOI] [PubMed] [Google Scholar]
- He Z, Zhou J. Empirical evaluation of a new method for calculating signal-to-noise ratio for microarray data analysis. Appl Environ Microbiol. 2008;74:2957–2966. doi: 10.1128/AEM.02536-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heath RL. Modification of the biochemical pathways of plants induced by ozone: what are the varied routes to change. Environ Pollut. 2008;155:453–463. doi: 10.1016/j.envpol.2008.03.010. [DOI] [PubMed] [Google Scholar]
- Holmes WE, Zak DR, Pregitzer KS, King JS. Elevated CO2 and O3 alter soil nitrogen transformations beneath trembling aspen, paper birch, and sugar maple. Ecosystems. 2006;9:1354–1363. [Google Scholar]
- IPCC . Intergovernmental Panel on Climate Change. Cambridge University Press: Cambridge; 2007. [Google Scholar]
- Kanerva T, Palojärvi A, Rämö K, Manninen S. Changes in soil microbial community structure under elevated tropospheric O3 and CO2. Soil Biol Biochem. 2008;40:2502–2510. [Google Scholar]
- Kasurinen A, Keinänen MM, Kaipainen S, Nilsson L-O, Vapaavuori E, Kontro MH, et al. Below-ground responses of silver birch trees exposed to elevated CO2 and O3 levels during three growing seasons. Glob Change Biol. 2005;11:1167–1179. [Google Scholar]
- Larson J, Zak D, Sinsabaugh R. Extracellular enzyme activity beneath temperate trees growing under elevated carbon dioxide and ozone. Soil Sci Soc Am J. 2002;66:1848–1856. [Google Scholar]
- Leaphart AB, Lovell CR. Recovery and analysis of formyltetrahydrofolate synthetase gene sequences from natural populations of acetogenic bacteria. Appl Environ Microbiol. 2001;67:1392–1395. doi: 10.1128/AEM.67.3.1392-1395.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Xu C, Liang W, Zhong S, Zheng X, Zhu J. Residue incorporation and N fertilization affect the response of soil nematodes to the elevated CO2 in a Chinese wheat field. Soil Biol Biochem. 2009;41:1497–1503. [Google Scholar]
- Liu L, King JS, Giardina CP. Effects of elevated concentrations of atmospheric CO2 and tropospheric O3 on leaf litter production and chemistry in trembling aspen and paper birch communities. Tree Physiol. 2005;25:1511–1522. doi: 10.1093/treephys/25.12.1511. [DOI] [PubMed] [Google Scholar]
- Mazzola M, Funnell DL, Raaijmakers JM. Wheat cultivar-specific selection of 2,4-diacetylphloroglucinol-producing fluorescent Pseudomonas species from resident soil populations. Microb Ecol. 2004;48:338–348. doi: 10.1007/s00248-003-1067-y. [DOI] [PubMed] [Google Scholar]
- Micallef SA, Shiaris MP, Colón-Carmona A. Influence of Arabidopsis thaliana accessions on rhizobacterial communities and natural variation in root exudates. J Exp Bot. 2009;60:1729–1742. doi: 10.1093/jxb/erp053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan PB, Ainsworth EA, Long SP. How does elevated ozone impact soybean? A meta-analysis of photosynthesis, growth and yield. Plant Cell Environ. 2003;26:1317–1328. [Google Scholar]
- Oksanen J, Blanchet FG, Kindt R, Legendre P, O'Hara RB, Simpson GL, et al. 2010. vegan: Community Ecology Package.
- Phillips RL, Zak DR, Holmes WE, White DC. Microbial community composition and function beneath temperate trees exposed to elevated atmospheric carbon dioxide and ozone. Oecologia. 2002;131:236–244. doi: 10.1007/s00442-002-0868-x. [DOI] [PubMed] [Google Scholar]
- Pleijel H, Danielsson H, Gelang J, Sild E, Selldén G. Growth stage dependence of the grain yield response to ozone in spring wheat (Triticum aestivum L.) Agric Ecosyst Environ. 1998;70:61–68. [Google Scholar]
- Pleijel H, Eriksen AB, Danielsson H, Bondesson N, Selldén G. Differential ozone sensitivity in an old and a modern Swedish wheat cultivar - grain yield and quality, leaf chlorophyll and stomatal conductance. Environ Exp Bot. 2006;56:63–71. [Google Scholar]
- Rämö K, Kanerva T, Nikula S, Ojanperä K, Manninen S. Influences of elevated ozone and carbon dioxide in growth responses of lowland hay meadow mesocosms. Environ Pollut. 2006;144:101–111. doi: 10.1016/j.envpol.2006.01.009. [DOI] [PubMed] [Google Scholar]
- Slaughter LH, Mulchi CL, Lee EH, Tuthill K. Chronic ozone stress effects on yield and grain quality of soft red winter wheat. Crop Sci. 1989;29:1251–1255. [Google Scholar]
- Soja G, Barnes JD, Posch M, Vandermeiren K, Pleijel H, Mills G. Phenological weighting of ozone exposures in the calculation of critical levels for wheat, bean and plantain. Environ Pollut. 2000;109:517–524. doi: 10.1016/s0269-7491(00)00055-5. [DOI] [PubMed] [Google Scholar]
- Schoene K, Franz JT, Masuch G. The effect of ozone on pollen development in Lolium perenne L. Environ Pollut. 2004;131:347–354. doi: 10.1016/j.envpol.2004.03.014. [DOI] [PubMed] [Google Scholar]
- Shi S, Richardson AE, O'Callaghan M, DeAngelis KM, Jones EE, Stewart A, et al. Effects of selected root exudate components on soil bacterial communities. FEMS Microbiol Ecol. 2011;77:600–610. doi: 10.1111/j.1574-6941.2011.01150.x. [DOI] [PubMed] [Google Scholar]
- Trivedi P, He Z, Van Nostrand JD, Albrigo G, Zhou J, Wang N. Huanglongbing alters the structure and functional diversity of microbial communities associated with citrus rhizosphere. ISME J. 2012;6:363–383. doi: 10.1038/ismej.2011.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Zhou L, Tang X. Ozone concentrations in rural regions of the Yangtze Delta in China. J Atmos Chem. 2006;54:255–265. [Google Scholar]
- Wittig VE, Ainsworth EA, Naidu SL, Karnosky DF, Long SP. Quantifying the impact of current and future tropospheric ozone on tree biomass, growth, physiology and biochemistry: a quantitative meta-analysis. Glob Change Biol. 2009;15:396–424. [Google Scholar]
- Wu L, Liu X, Schadt CW, Zhou J. Microarray-based analysis of subnanogram quantities of microbial community DNAs by using whole-community genome amplification. Appl Environ Microbiol. 2006;72:4931–4941. doi: 10.1128/AEM.02738-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong J, Wu L, Tu S, Van Nostrand JD, He Z, Zhou J, et al. Microbial communities and functional genes associated with soil arsenic contamination and the rhizosphere of the arsenic-hyperaccumulating plant Pteris vittata L. Appl Environ Microbiol. 2010;76:7277–7284. doi: 10.1128/AEM.00500-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zak DR, Holmes WE, Pregitzer KS. Atmospheric CO2 and O3 alter the flow of 15N in developing forest ecosystems. Ecology. 2007;88:2630–2639. doi: 10.1890/06-1819.1. [DOI] [PubMed] [Google Scholar]
- Zhou J, Bruns MA, Tiedje JM. DNA recovery from soils of diverse composition. Appl Environ Microbiol. 1996;62:316–322. doi: 10.1128/aem.62.2.316-322.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Xue K, Xie J, Deng Y, Wu L, Cheng X, et al. Microbial mediation of carbon-cycle feedbacks to climate warming. Nature Clim Change. 2012;2:106–110. [Google Scholar]
- Zhu X, Feng Z, Sun T, Liu X, Tang H, Zhu J, et al. Effects of elevated ozone concentration on yield of four Chinese cultivars of winter wheat under fully open-air field conditions. Glob Change Biol. 2011;17:2697–2706. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.