Abstract
Protists are ubiquitous members of soil microbial communities, but the structure of these communities, and the factors that influence their diversity, are poorly understood. We used barcoded pyrosequencing to survey comprehensively the diversity of soil protists from 40 sites across a broad geographic range that represent a variety of biome types, from tropical forests to deserts. In addition to taxa known to be dominant in soil, including Cercozoa and Ciliophora, we found high relative abundances of groups such as Apicomplexa and Dinophyceae that have not previously been recognized as being important components of soil microbial communities. Soil protistan communities were highly diverse, approaching the extreme diversity of their bacterial counterparts across the same sites. Like bacterial taxa, protistan taxa were not globally distributed, and the composition of these communities diverged considerably across large geographic distances. However, soil protistan and bacterial communities exhibit very different global-scale biogeographical patterns, with protistan communities strongly structured by climatic conditions that regulate annual soil moisture availability.
Keywords: biodiversity, biogeography, microbial ecology, soil protists
Introduction
Protists are unicellular eukaryotes (Adl et al., 2005) and critical components of microbial communities in both aquatic and terrestrial environments, where they are integral constituents of trophic chains and nutrient cycles (Luxton and Petersen, 1982; Sherr and Sherr, 2002; Cuvelier et al., 2010; Steele et al., 2011). As parasites and disease agents, they impact the health of humans, livestock and agricultural crops (Aurrecoechea et al., 2010). Despite their ecological and economic importance, and regular study since the time of van Leeuwenhoek, protistan diversity remains poorly characterized (Caron et al., 2009). The biogeographical patterns exhibited by protistan communities are also poorly understood, and have been the subject of recent debate (Foissner, 2006; Bass et al., 2007), with suggestions that the diversity of microbial eukaryotes may show limited variability across large spatial scales (Finlay, 2002).
Historically, surveys of protistan diversity have largely relied on morphological taxon identification; however, the limitations of such approaches are well known (Bass et al., 2007). Thus, more recent work has utilized DNA sequence-based methods to survey protists from a range of localities within oceans (López-García et al., 2001; Cuvelier, et al., 2010; Demir-Hilton et al., 2011; Steele et al., 2011) and freshwater lakes (Katz et al., 2005; Triadó-Margarit and Casamyor, 2012). Such studies continue to reveal a broad spectrum of previously unrecognized diversity within the group (López-García et al., 2001; Demir-Hilton et al., 2011; Kim et al., 2011), just as similar molecular approaches have revolutionized our understanding of bacterial diversity (Pace, 1997). However, previously published molecular studies have primarily focused on protistan diversity in aquatic systems. Molecular surveys of terrestrial protistan diversity are rare even though soils harbor large numbers of protists (typically 104–107 active individuals g−1) that are important components of biogeochemical cycles (Adl and Gupta, 2006; Howe et al., 2009). As a result, we have an inadequate understanding of the factors structuring soil protistan communities and their biogeographical patterns, nor do we know how protistan diversity patterns compare with those exhibited by soil bacterial communities, which have been relatively well-studied.
Here we used high-throughput pyrosequencing of the 18S ribosomal RNA (rRNA) gene to conduct a detailed and comprehensive survey of soil protistan diversity. We also examined the continental-scale biogeographical patterns exhibited by soil protists, just as broad-scale molecular surveys have been used previously to reveal the biogeographical patterns exhibited by soil prokaryotes (Lauber et al., 2009; Bates et al., 2011), terrestrial eukaryotic mesofauna (Wu et al., 2011) and marine protists (Katz et al., 2005; Cuvelier et al., 2010; Demir-Hilton et al., 2011). We collected samples across 40 broadly distributed sites that were selected to represent a wide range of soil and biome types. As each of the soils and sites were well-characterized and their bacterial communities surveyed in detail, we were also able to investigate the influence of edaphic and climatic factors on protistan community structure, and directly compare patterns of protistan diversity to those exhibited by soil bacteria.
Methods
Sampling, site characterization and isolation of soil DNA
The 40 soil samples collected for this study include sites across North and South America, as well as Antarctica. These represented a wide variety of biomes and soil types from diverse climates. Fierer and Jackson (2006) previously described the soil sampling protocol, outlined briefly as follows. During peak growing season for vascular plants (with the exception of the mainly plant-free Antarctic sites), samples were collected at undisturbed areas from the top 5 cm of mineral soil at 5–10 randomly selected locations within an area of ∼100 m2. Each collection consisted of ∼10 g of soil, sieved to remove roots and other debris. A single composite soil was then prepared by combining and thoroughly mixing all the randomly selected samples collected at a given locality. DNA was extracted from a 0.25 g subsample of the composite soil using the MoBio PowerSoil DNA extraction kit (MoBio Laboratories, Carlsbad, CA, USA) and following the manufacturer's instructions, with the exception of an additional incubation step at 65 °C for 10 min, followed by 2 min of bead beating to limit DNA shearing (Lauber et al., 2009). The aim of this study was to examine variability of microbial communities across a range of geographically and climatically diverse sites, rather than focusing specifically on spatial or temporal variability within individual sites; thus, single, composited soil samples were used to represent each site. To assess variation of soil protistan diversity at the local scale, some regions (for example, Mojave Desert) were represented by more than one sample site. These regional sampling sites were separated by distances of approximately 200–500 m and represented localities with distinct vegetation cover and unique edaphic qualities (see Supplementary Table S1).
Each site was quantitatively categorized into one of four general environment types according to the climate moisture index (CMI) of Willmott and Feddema (1992). This index reflects the local deficiency or surplus of moisture from annual precipitation (P) in relation to the annual potential rate of evapotranspiration (PET; Thornthwaite, 1948), and is calculated as follows: CMI=(P/PET)−1 when P<PET or CMI=1−(PET/P) when P⩾PET. Values indicate sites with arid (−1 to <−0.5), semiarid (−0.5 to 0), semihumid (>0 to 0.5) or humid (>0.5 to 1) status. Climate data were obtained from the National Climatic Data Center (http://www.ncdc.noaa.gov/). Location and environmental characteristics for each site are provided in Supplementary Table S1.
Polymerase chain reaction amplification of 18S rRNA genes and bar-coded pyrosequencing
Genomic DNA extracted from the samples was prepared for pyrosequencing according to the protocol of Lauber et al. (2009), with the exception that eukaryotic-specific primers were used. This method includes targeted polymerase chain reaction amplification of ca. 600 bp of the 18S small subunit rRNA gene using the eukaryotic-specific primer set F515 (5′-GTGCCAGCMGCCGCGGTAA-3′) and R1119 (5′-GGTGCCCTTCCGTCA-3′), followed by triplicate polymerase chain reaction product pooling (per sample) to mitigate reaction-level polymerase chain reaction biases, and barcoded pyrosequencing. This primer set has been shown to amplify a portion of the 18S rRNA gene from a wide range of eukaryotic groups with few biases (Bates et al., 2012), and the read length is well suited for community analysis (Liu et al., 2007). Protocol and conditions follow exactly those outlined in Bates et al. (2012) for polymerase chain reaction, amplicon pooling, as well as barcoded pyrosequencing using recently developed FLX+ technology. Pyrosequencing was carried out at Roche Applied Science (Indianapolis, IN, USA).
Sequence processing and statistical analyses
Raw sequence data was processed, assessed for quality (filtered on the basis of quality score, sequence length and primer mismatch thresholds) and analyzed using the QIIME 1.4.0 software pipeline (Caporaso et al., 2010). Chimera checking and operational taxonomic unit (OTU) grouping were carried in QIIME using USEARCH (Edgar et al., 2011), as were taxonomic assignments of recovered eukaryotic OTUs (determined at ⩾97% similarity) using BLAST (Altschul et al., 1997) against the SILVA comprehensive ribosomal RNA database (http://www.arb-silva.de/) at a sequence similarity threshold of 0.97 and a maximum E-value of 10−10. After taxonomies had been assigned, a data set comprised of only protistan (excluding fungi and metazoans) taxa was culled from all high-quality sequence reads.
Taxonomy was determined for some protistan OTUs of uncertain affiliations after further BLAST searches in GenBank (http://www.ncbi.nlm.nih.gov/genbank/). Phylogenetic distance matrices were generated using the UniFrac method (Luzapone and Knight, 2005) against a tree generated in QIIME from a PyNAST alignment of the SILVA sequence set with a dynamic entropy and gap calculation filter applied (McDonald et al., 2012). QIIME was also used for Procrustes analysis (Caporaso et al., 2011), whereas principal coordinate analysis, Mantel tests, permutational multivariate analysis of variance and Bray–Curtis distance matrices generation were carried out using the PRIMER v.6 software package (PRIMER-E, Plymouth, WA, USA) (Clark and Gorley, 2006). Principal coordinate analysis used both unweighted (based on OTU presence or absence) and weighted (also considering OTU relative abundance) distance matrices (UniFrac and Bray–Curtis). All diversity and statistical analyses were performed with R statistical software (http://www.r-project.org/) with the aid of the packages Fields and Vegan. With the exception of the rarefaction curves (generated at a depth of 1000 sequences), analyses were carried out on the eukaryotic data set rarefied to 150 sequences to correct for unequal sampling efforts. A bacterial data set previously generated from the same soil samples (Bates et al., 2011), and used here to compare community structure between these two groups of soil microbes, was also rarefied at the same level of sampling effort (150 sequences). For these rarefied data sets, richness was estimated as the number of unique OTUs, whereas diversity values were calculated for the Shannon index using relative abundance as the number of sequences representing each OTU within a given sample.
Results and discussion
Diversity of soil protists
A total of 29 564 high-quality protistan sequences were recovered from the 40 unique soils examined, averaging 429 sequences per sample (ranging from 151 to 1228). A total of 1014 OTUs (determined at ⩾97% similarity) were identified across the sample set, including representatives from five supergroups and 13 major phyla: Amoebozoa (Acanthamoebidae, Eumycetozoa, Flabellinea, Tubulinea, Incertae sedis: Phalansterium), Archaeplastida (Chloroplastida), Excavata (Euglenozoa, Malawimonas, Jakobida), Opisthokonta (Choanomonada) and SAR (Alveolata, Cercozoa, Stramenopiles).
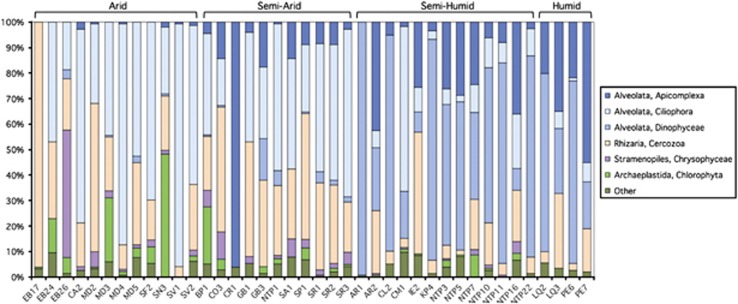
The rarified sequence data set suggests that soils from our sites were overwhelmingly dominated by taxa in the SAR supergroup (Figure 1 and Table 1), with the Alveolata and Cercozoa representing 66.5% and 22.5%, respectively, of all of the protistan sequences recovered. The green algae (Archaeplastida, Chloroplastida, Chlorophyta) were also highly abundant in a few soils from arid areas, as were the golden algae (SAR, Stramenopiles, Chrysophyceae; Figure 1). Our finding that Cercozoa (diverse flagellates and amoebae within Rhizaria) and Ciliophora (one of three major groups within Alveolata, along with dinoflagellates and Apicomplexa) are common members of eukaryotic soil microbial communities is congruent with what we know from previously published direct observation studies of soil protistan communities (Adl and Gupta, 2006; Chao et al., 2006). The only comparable study using high-throughput pyrosequencing to examine terrestrial protistan diversity (Urich et al., 2011, who examined a single site in Germany) also found alveolates and Rhizaria to be the dominant protists in soil. However, as detailed below, we also found a number of higher-level protistan taxa that have rarely been reported from soil.
Figure 1.
Relative abundance of soil protistan taxa (y axis as group percentage of the total number of 18S rRNA gene sequences per sample, after rarefaction to correct for uneven sampling effort) grouped by general CMI class. Identities of the sites sampled are given on the x axis (see Supplementary Table S1 for specific details on site and soil characteristics).
Table 1. Dominant protistan taxa of soils (data have been rarefied to correct for uneven sampling effort).
| ID | Taxon string | Frequency (% of samples) | Abundance (% of total) |
|---|---|---|---|
| Eukaryotes | |||
| OTU00001 | Alveolata, Ciliophora, Intramacronucleata, Spirotrichea | 77.5 | 9.07 |
| OTU00003 | Alveolata, Apicomplexa, Coccidia, Eucoccidiorida | 70.0 | 4.57 |
| OTU00033 | Rhizaria, Cercozoa, Cercomonadida, Heteromitidae | 67.5 | 1.43 |
| OTU00014 | Rhizaria, Cercozoa, Thecofilosea, Chlamydophryidae | 65.0 | 3.80 |
| OTU00729 | Alveolata, Ciliophora, Intramacronucleata, Spirotrichea | 65.0 | 2.65 |
| OTU04795 | Alveolata, Apicomplexa, Coccidia, Eucoccidiorida | 57.5 | 2.32 |
| OTU00346 | Alveolata, Apicomplexa, Coccidia, Eucoccidiorida | 55.0 | 1.40 |
| OTU00017 | Alveolata, Ciliophora, Intramacronucleata, Spirotrichea | 52.5 | 1.00 |
| OTU00074 | Alveolata, Ciliophora, Intramacronucleata, Colpodea | 50.0 | 0.65 |
| OTU00046 | Stramenopiles, Chrysophyceae, Chromulinales, Chromulinaceae | 47.5 | 1.95 |
| OTU00085 | Alveolata, Ciliophora, Intramacronucleata, Colpodea | 47.5 | 1.02 |
| OTU00077 | Rhizaria, Cercozoa, Cercomonadida, Heteromitidae | 47.5 | 0.67 |
| OTU03988 | Rhizaria, Cercozoa, Cercomonadida, Heteromitidae | 45.0 | 0.60 |
| OTU00103 | Rhizaria, Cercozoa, Cercomonadida, Heteromitidae | 40.0 | 0.80 |
| OTU04137 | Alveolata, Ciliophora, Intramacronucleata, Spirotrichea | 40.0 | 0.60 |
Abbreviation: OTU, operational taxonomic unit.
Relative abundance (percentage of all 18S rRNA gene sequences) and frequency (percentage occurrence of a given OTU within all soils sampled).
The rarefaction curves (Supplementary Figure S1) and the high numbers of rare taxa that were present in these soils (70% of rarified OTUs were represented by <5 sequences) underscore the high diversity of these protistan communities, even when a more conservative approach to OTU binning was used (that is, 97% as opposed to 99% similarity). Bacteria are among the most species-rich organisms in the terrestrial environment (Torsvik et al., 2002; Fierer and Lennon, 2011), yet we found diversity levels of protists to be within the same order of magnitude as bacteria when directly comparing these communities across the same soil samples at equivalent survey depths (Supplementary Figure S1). The full extent of diversity within either of these groups, however, remains largely unknown as the rarefaction curves failed to asymptote. Protistan and bacterial communities were also similar in their structure, exhibiting comparable trends for both rank frequency and abundance across the soils sampled (Supplementary Figure S2).
Distribution patterns for soil protists
The biogeographical patterns exhibited by protistan communities have been the subject of vigorous debate (Green and Bohannan, 2006; Caron et al., 2009). It has been hypothesized that organisms with small body sizes, such as soil protists, should have the capacity for continuous large-scale dispersal, such that nearly all-possible taxa will be found within any sample (Finlay, 2002). Models suggest that microbial taxa with body sizes in the 10 μm diameter range, such as many protists, are capable of global dispersal (Wilkinson et al., 2012); however, the viability of protistan organisms or their encysting structures after long-distance areal travel is not well understood (Foissner, 2006). Our results do not support the Finlay hypothesis of cosmopolitan distribution patterns as no protistan taxa were found across all the sampled soils (Table 1); in the rarified data set, only one of the 672 OTUs was found in more than 75% of our soil samples and the majority of phylotypes (84%) were restricted to five or fewer individual soil samples. Although deeper sequencing may identify more cosmopolitan taxa, our results highlight that numerous protistan taxa exhibit some degree of endemism, as hypothesized by Foissner (2006) and demonstrated previously for terrestrial ciliates (Chao et al., 2006).
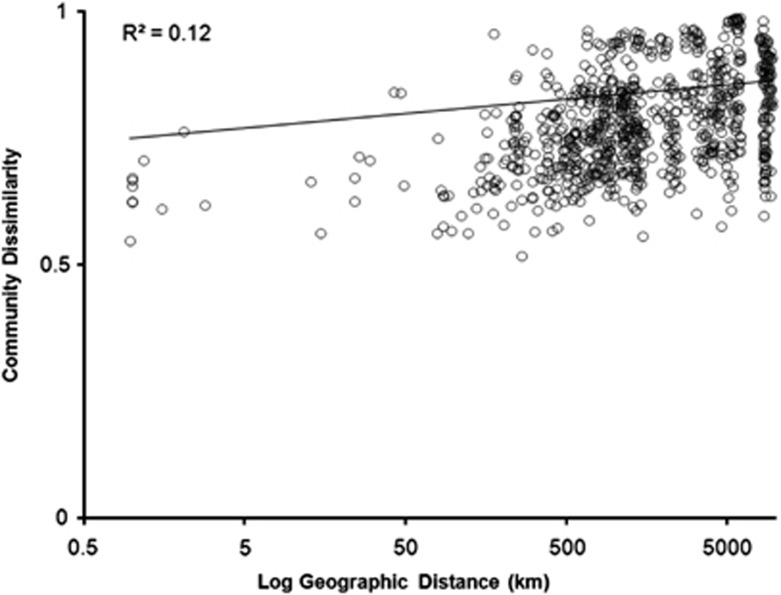
Distance–decay relationships for microbial community similarity have been demonstrated previously for soil fungi (Green et al., 2004), and the soil protistan communities examined here were likewise found to be less similar, generally, with increasing geographic distance (Figure 2). Distance, however, was not a very strong predictor of community dissimilarity (linear regression R2=0.12, P<0.001), which varied considerably within most levels of geographic separation. This result, along with those of other recent sequence-based studies characterizing the diversity of eukaryotic soil macro- and mesofauna (Robeson et al., 2011; Wu et al., 2011), suggests that terrestrial protists, and soil eukaryotes in general, are much more diverse than previously recognized and that they are not cosmopolitan in their distributions as had previously been hypothesized.
Figure 2.
Protistan community dissimilarity (unweighted UniFrac) with increasing geographic distance (best-fitted linear regression line). UniFrac values range from 1 (completely different phylogenetic assemblages) to 0 (no differences observed between communities).
Factors influencing soil protistan community structure
It is well known that, at the continental scale, alpha diversity levels of macroscopic organisms frequently correlate with energy, water availability and latitude (Hawkins et al., 2003; Davies et al., 2008). Diversity of soil bacteria, on the other hand, is strongly correlated with soil pH levels (Lauber et al., 2009; Griffiths et al., 2011). In contrast, protistan richness (Supplementary Figure S3) and diversity (Supplementary Figure S4; Shannon index) were only marginally influenced by pH or any other of the measured environmental variables (Supplementary Table S2). Alpha diversity of soil protists, therefore, does not appear to be driven by environmental factors known to strongly affect the diversity of many plant and animal taxa or soil bacteria. Furthermore, levels of protistan and bacterial diversity were only moderately correlated across samples (Supplementary Figure S5; Pearson's R=0.46, P=0.003 for richness and R=0.38, P=0.015 for Shannon index), indicating that distinct factors may influence diversity levels within these individual microbial groups.
Although alpha diversity levels were not strongly correlated with environmental factors and community structure was highly heterogeneous across large spatial scales, we did observe predictable beta diversity patterns for both soil protistan and bacterial communities. As has been reported in a number of other studies (Fierer and Jackson, 2006; Lauber et al., 2009; Griffiths et al., 2011), we also found bacterial beta diversity patterns in soil were strongly related to pH (Mantel global R=0.6–0.7, P<0.01, Bray–Curtis or UniFrac for both weighted and unweighted matrices). Shifts in community composition for soil protists, on the other hand, were most strongly correlated with CMI values (Mantel global R=0.5–0.6, P<0.01, all matrices), which is an index of annual soil moisture availability. Soil protistan and bacterial community structure also differed significantly in ordination space (Supplementary Figure. S6; Procrustes M2=0.52, P>0.05), again suggesting that the factors structuring these microbial communities are fundamentally distinct.
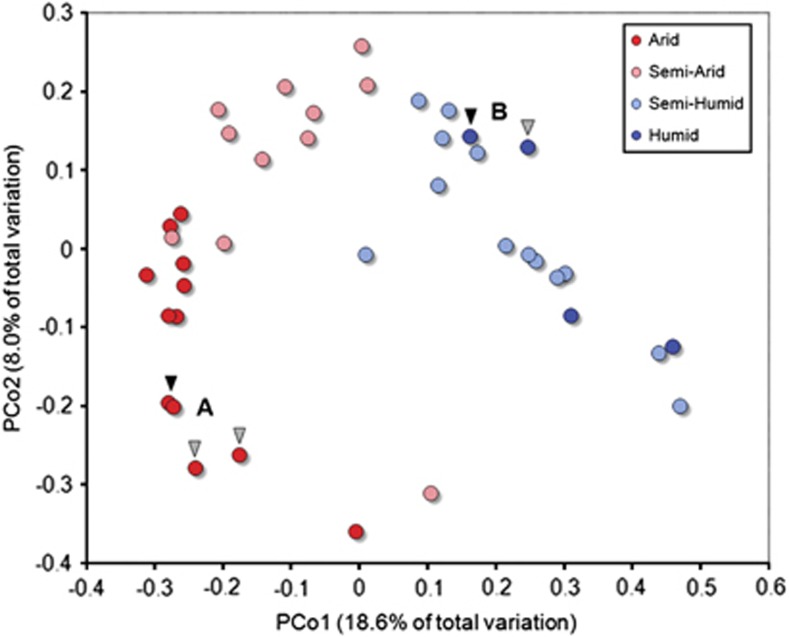
The relationship between protistan community structure and the general CMI class (arid, semiarid, semihumid and humid) was significant for both taxonomic- and phylogenetic-based distance measures (Bray–Curtis or UniFrac, respectively, for both weighted and unweighted matrices; each permutational multivariate analysis of variance at P<0.05). This trend was most evident when using the unweighted UniFrac distance metric (permutational multivariate analysis of variance, pseudo-F=3.64, P=0.001) as shown in Figure 3. As a result of this correlation between annual soil moisture availability and community similarity, assemblages of soil protists from very distant localities could be highly similar. For example, the extremely arid Antarctica and Mojave Desert (western United States) sites shared relatively similar protistan communities (Figure 3a), as did humid sites from Peru and Puerto Rico (Figure 3b).
Figure 3.
Principal coordinates analysis (PCoA) plot on the unweighted UniFrac distance matrix generated from rarefied taxon abundances and depicting patterns of beta diversity for protistan communities of soil. Points that are closer together on the ordination have communities that are more similar. permutational multivariate analysis of variance indicated that differences between communities mapped according to site CMI classes were highly significant (P<0.001). Sites where community composition was highly similar despite large geographic separation are highlighted with arrows indicating (A) Antarctic Dry Valleys (black; EB24 and EB26) and Mojave Desert (gray; MD4 and MD5) of California, USA and (B) Peru (black; PE7) and Puerto Rico (gray; LQ3).
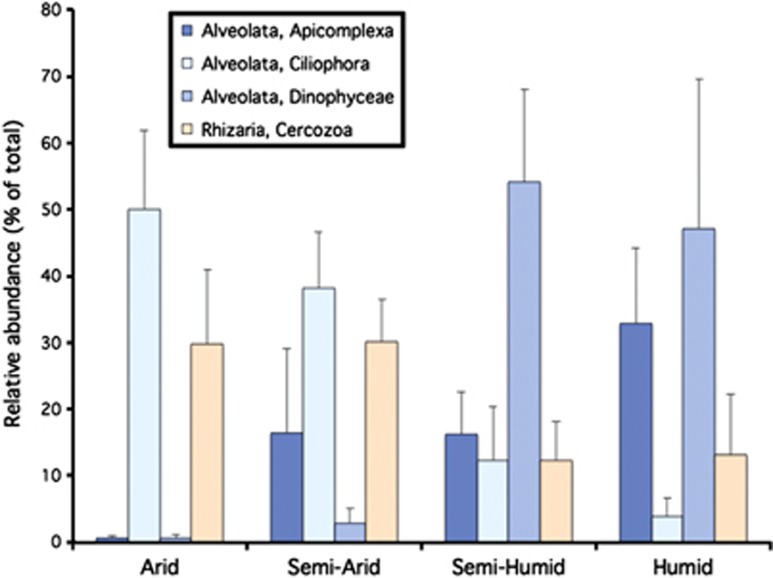
Within each general CMI class, the relative abundance of certain higher-level (for example, phylum or class) protistan taxa varied considerably (Figure 4). Dinophyceae clearly dominated more arid soils, whereas the Ciliophora and Apicomplexa were relatively more abundant in soils of humid sites. The Cercozoa were widespread, but tended to be more abundant in arid soils. The prevalence of Cercozoans in soil was expected, as these protists are known to be abundant and broadly dispersed in terrestrial environments. The structure of soil cercozoan communities has been shown to vary between tropical and temperate sites (Bass et al., 2007; Howe et al., 2009), and climate has also been suggested as an important factor in shaping the structure of other protistan goups, namely protosteloid amoebae communities (Aguilar and Lado, 2012). Likewise, the diversity of particular ciliate taxa are known to shift according to the biome (primarily arid versus mesic types) in which they are found (Bamforth, 1995; Chao et al., 2006), whereas some ciliates form cysts that allow these organisms to survive in arid areas even under drought conditions (Adl and Gupta, 2006). Our findings are therefore congruent with what is known about the ecology of some soil protist groups, and highlight the importance of higher-level taxa in shaping the biogeographical patterns that we observed.
Figure 4.
Relative abundance of soil protistan taxa (y axis as group percentage of the total number of 18S rRNA gene sequences per sample, after rarefaction to correct for uneven sampling effort) averaged across CMI classes.
We also made an unexpected discovery that soils harbored relatively large numbers of protistan taxa long considered to be absent, or at least rarely encountered, in soil. For example, Apicomplexa taxa have been known to inhabit soils during encysting stages (Ruiz et al., 1973), yet they have not previously been recognized as being abundant in soil. We found Apicomplexa (81% of these sequences represented the Eucoccidiorida) to be abundant in soils from more mesic sites (Figure 4); however, the possibility that some taxa recovered in this study may only represent resting stages from non-indigenous organisms accumulated over time cannot be explicitly ruled out by our methods. Eucoccidiorida are known as common animal parasites such as Toxoplasma that can also encyst like ciliates. Their high abundance in more humid soils could be attributed to the fact that oocyst viability is likely dependent on adequate soil moisture (Ruiz et al., 1973). Trends in Toxoplasma-related diseases follow similar patterns, with incidences being more frequent in areas with higher annual rainfall (Gómez-Marin et al., 2011). Soil invertebrates are also known to harbor commensal or parasitic apicomplexans (Olsen, 1986); however, the numbers of these taxa associated with a host versus freely distributed throughout the soils sampled here cannot be conclusively determined by our methods. The Dinophyceae (99% of the sequences represented the Heterocapsaceae) were also found to be highly abundant in humid soils (Figure 4). Members of this class are generally thought to be restricted to aquatic habitats; however, we found that they can also be abundant in soils. Taken together, these results and those reported in other recent molecular surveys (Lejzerowicz et al., 2010; Coolen et al., 2011) suggest that protistan taxa long considered to be restricted to aquatic environments may also be quite common in terrestrial environments.
Conclusions
The results of this study, perhaps the most comprehensive assessment of terrestrial protists conducted to date, suggests that, in addition to commonly known soil groups (for example, Cercozoa and Ciliophora), previously unrecognized taxa (for example, Apicomplexa and Dinophyceae) are also important members of soil protistan communities. Future studies using RNA-based approaches will be useful in confirming the presence of these previously unrecognized taxa as being biologically active soil community members. As soil studies conducted in recent decades have stressed the incredible diversity of soil prokaryotic communities (Curtis et al., 2002), we found that bacteria are not unique in this respect, as their diversity is similar in magnitude to that of soil protists. Protistan taxa were generally not cosmopolitan across the soils sampled here, and like their bacterial counterparts, soil protists exhibit predictable biogeographical patterns. Whereas pH has frequently been identified as the dominant factor driving global patterns of bacterial biogeography (Fierer and Jackson, 2006; Lauber et al., 2009; Griffiths et al., 2011), protistan biogeography is best predicted by climatic conditions that regulate annual moisture availability in soils at comparable scales of inquiry. Despite debate regarding the existence of biogeographic patterns for microorganisms (Green and Bohannan, 2006; Martiny et al., 2006), our survey adds to the growing body of literature suggesting that soil microbes, even those with the potential for sustained global dispersal, have distinct biogeographies.
Acknowledgments
Members of the Fierer group helped in the laboratory and with the analyses. Chinnappa Kodira of Roche Applied Science supervised the pyrosequencing. This research was supported in part by grants to NF and RK from the National Institutes of Health, the National Science Foundation, the US Department of Agriculture and the Howard Hughes Medical Institute.
Footnotes
Supplementary Information accompanies the paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Adl M, Simpson AGB, Farmer MA, Andersen RA, Anderson OR, Barta JR, et al. The new higher level classification of eukaryotes with emphasis on the taxonomy of protists. J Eukaryot Microbiol. 2005;52:399–451. doi: 10.1111/j.1550-7408.2005.00053.x. [DOI] [PubMed] [Google Scholar]
- Adl SM, Gupta VVSR. Protists in soil ecology and forest nutrient cycling. Can J Forest Res. 2006;36:1805–1817. [Google Scholar]
- Aguilar M, Lado C. Ecological niche models reveal the importance of climate variability for the biogeography of protosteloid amoebae. ISME J. 2012;6:1506–1514. doi: 10.1038/ismej.2012.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Madden TJ, Schaffer AA, Zhang J, Zhang Z, Miller W, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aurrecoechea C, Brestelli J, Brunk BP, Fischer S, Gajria B, Gao X, et al. EuPathDB: a portal to eukaryotic pathogen databases. Nucleic Acids Res. 2010;38 ((Suppl 1)::D415–D419. doi: 10.1093/nar/gkp941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bamforth SS.1995Interpreting soil ciliate biodiversityIn: Collins HP, Robertson GP, Klug MJ (eds.)The Significance and Regulation of Soil Biodiversity Kluwer Academic Publishers: Dordrecht; 179–184. [Google Scholar]
- Bass D, Richards TA, Matthai L, Marsh V, Cavalier-Smith T. DNA evidence for global dispersal and probable endemicity of protozoa. BMC Evol Biol. 2007;7:162. doi: 10.1186/1471-2148-7-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates ST, Berg-Lyons D, Caporaso JG, Walters WA, Knight R, Fierer N. Examining the global distribution of dominant archaeal populations in soil. ISME J. 2011;5:908–917. doi: 10.1038/ismej.2010.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates ST, Berg-Lyons D, Lauber CL, Walters WA, Knight R, Fierer N. A preliminary survey of lichen associated eukaryotes using pyrosequencing. Lichenologist. 2012;44:137–146. [Google Scholar]
- Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, et al. QIIME allows integration and analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Lozupone CA, Turnbaugh PJ, et al. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc Natl Acad Sci USA. 2011;108:4516–4522. doi: 10.1073/pnas.1000080107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caron D, Worden AZ, Countway PD, Demir E, Heidelberg KB. Protists are microbes too: a perspective. ISME J. 2009;3:4–12. doi: 10.1038/ismej.2008.101. [DOI] [PubMed] [Google Scholar]
- Chao A, Li PC, Agatha S, Foissner W. A statistical approach to estimate soil ciliate diversity and distribution based on data from five continents. Oikos. 2006;114:479–493. [Google Scholar]
- Clark KR, Gorley RN. PRIMER v.6: User Manual/Tutorial. PRIMER-E: Plymouth, WA; 2006. [Google Scholar]
- Coolen MJL, van de Giessen J, Zhu EY, Wuchter C. Bioavailability of soil organic matter and microbial community dynamics upon permafrost thaw. Environ Microbiol. 2011;13:2299–2314. doi: 10.1111/j.1462-2920.2011.02489.x. [DOI] [PubMed] [Google Scholar]
- Curtis TP, Sloan WT, Scannell JC. Estimating prokaryotic diversity and its limits. Proc Natl Acad Sci USA. 2002;99:10494–10499. doi: 10.1073/pnas.142680199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuvelier ML, Allen AE, Moniera A, McCrow JP, Messié M, Tringe SG, et al. Targeted metagenomics and ecology of globally important uncultured eukaryotic phytoplankton. Proc Natl Acad Sci USA. 2010;107:14679–14684. doi: 10.1073/pnas.1001665107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies JT, Fritz SA, Grenyer R, Orme CDL, Bielby J, Bininda-Emonds ORP, et al. Phylogenetic trees and the future of mammalian biodiversity. Proc Natl Acad Sci USA. 2008;105:11556–11563. doi: 10.1073/pnas.0801917105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demir-Hilton E, Sudek S, Cuvelier ML, Gentemann CL, Zehr JP, Worden AZ. Global distribution patterns of distinct clades of the photosynthetic picoeukaryote Ostreococcus. ISME J. 2011;5:1095–1107. doi: 10.1038/ismej.2010.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics. 2011;27:2194–2200. doi: 10.1093/bioinformatics/btr381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fierer N, Jackson R. The diversity and biogeography of soil bacterial communities. Proc Natl Acad Sci USA. 2006;103:626–631. doi: 10.1073/pnas.0507535103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fierer N, Lennon J. The generation and maintenance of diversity in microbial communities. Am J Bot. 2011;98:439–448. doi: 10.3732/ajb.1000498. [DOI] [PubMed] [Google Scholar]
- Finlay BJ. Global dispersal of free-living microbial eukaryote species. Science. 2002;296:1061–1063. doi: 10.1126/science.1070710. [DOI] [PubMed] [Google Scholar]
- Foissner W. Biogeography and dispersal of micro-organisms: a review emphasizing protists. Acta Protozool. 2006;45:111–136. [Google Scholar]
- Gómez-Marin JE, de-la-Torre A, Angel-Muller E, Rubio J, Arenas J, Osorio E, et al. First Colombian multicentric newborn screening for congenital toxoplasmosis. PLoS Negl Trop Dis. 2011;5:e1195. doi: 10.1371/journal.pntd.0001195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green JL, Holmes AJ, Westoby M, Oliver I, Briscoe D, Dangerfield M, et al. Spatial scaling of microbial eukaryote diversity. Nature. 2004;432:747–750. doi: 10.1038/nature03034. [DOI] [PubMed] [Google Scholar]
- Green J, Bohannan JM. Spatial scaling of microbial biodiversity. Trends Ecol Evol. 2006;21:501–507. doi: 10.1016/j.tree.2006.06.012. [DOI] [PubMed] [Google Scholar]
- Griffiths RI, Thomson BC, James P, Bell T, Bailey M, Whiteley AS. The bacterial biogeography of British soils. Environ Microbiol. 2011;13:1642–1654. doi: 10.1111/j.1462-2920.2011.02480.x. [DOI] [PubMed] [Google Scholar]
- Hawkins BA, Field R, Cornell HV, Currie DJ, Guégan J-F, Kaufman DM, et al. Energy, water, and broad-scale geographic patterns of species richness. Ecology. 2003;84:3105–3117. [Google Scholar]
- Howe AT, Bass D, Vickerman K, Chao EE, Cavalier-Smith T. Phylogeny, taxonomy, and astounding genetic diversity of Glissomonadida ord. nov., the dominant gliding zooflagellates in soil (Protozoa: Cercozoa) Protist. 2009;160:159–189. doi: 10.1016/j.protis.2008.11.007. [DOI] [PubMed] [Google Scholar]
- Katz LA, McManus GB, Snoeyenbos-West OLO, Pirog K, Griffin A, Foissner W. Reframing the microbial ‘Everything is Everywhere' debate: evidence for high gene flow and diversity in ciliate morphospecies. Aquat Microb Ecol. 2005;41:55–65. [Google Scholar]
- Kim E, Harrison JW, Sudek S, Jones MDM, Wilcox HM, Richards TA, et al. Newly identified and diverse plastid-bearing branch on the eukaryotic tree of life. Proc Natl Acad Sci USA. 2011;108:1496–1500. doi: 10.1073/pnas.1013337108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauber CL, Knight R, Hamady M, Fierer N. Soil pH as a predictor of soil bacterial community structure at the continental scale: a pyrosequencing-based assessment. Appl Environ Microbiol. 2009;75:5111–5120. doi: 10.1128/AEM.00335-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lejzerowicz F, Pawlowski J, Fraissinet-Tachet L, Marmeisse R. Molecular evidence for widespread occurrence of Foraminifera in soils. Environ Microbiol. 2010;12:2518–2526. doi: 10.1111/j.1462-2920.2010.02225.x. [DOI] [PubMed] [Google Scholar]
- Liu Z, Lozupone C, Hamady M, Bushman FD, Knight R. Short pyrosequencing reads suffice for accurate microbial community analysis. Nucleic Acids Res. 2007;35:e120. doi: 10.1093/nar/gkm541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-García P, Rodríguez-Valera F, Pedrós-Alió C, Moreira D. Unexpected diversity of small eukaryotes in deep-sea Antarctic plankton. Nature. 2001;409:603–607. doi: 10.1038/35054537. [DOI] [PubMed] [Google Scholar]
- Luxton M, Petersen H. A comparative analysis of soil fauna populations and their role in decomposition processes. Oikos. 1982;39:288–388. [Google Scholar]
- Luzapone C, Knight R. UniFrac: a new phylogenetic method for comparing communities. Appl Environ Microbiol. 2005;71:8228–8235. doi: 10.1128/AEM.71.12.8228-8235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martiny JBH, Bohannan BJM, Brown JH, Colwell RK, Fuhrman JA, Green JL, et al. Microbial biogeography: putting microorganisms on the map. Nat Rev Microbiol. 2006;4:102–112. doi: 10.1038/nrmicro1341. [DOI] [PubMed] [Google Scholar]
- McDonald D, Price M, Goodrich J, Nawrocki E, Desantis T, Probst A, et al. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J. 2012;6:610–618. doi: 10.1038/ismej.2011.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen OW. Animal Parasites: Their Life Cycles and Ecology. Courier Dover: New York; 1986. [Google Scholar]
- Pace N. A molecular view of microbial diversity and the biosphere. Science. 1997;276:734–739. doi: 10.1126/science.276.5313.734. [DOI] [PubMed] [Google Scholar]
- Robeson MS, King AJ, Freeman KR, Birky CW, Martin AP, Schmidt SK. Soil rotifer communities are extremely diverse globally but spatially autocorrelated locally. Proc Natl Acad Sci USA. 2011;108:4406–4410. doi: 10.1073/pnas.1012678108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz A, Frenkel JK, Cerdas L. Isolation of Toxoplasma from soil. J Parasitol. 1973;59:204–206. [PubMed] [Google Scholar]
- Sherr EB, Sherr BF. Significance of predation by protists in aquatic microbial food webs. Antonie Van Leeuwenhoek. 2002;81:293–308. doi: 10.1023/a:1020591307260. [DOI] [PubMed] [Google Scholar]
- Steele JA, Countway PD, Xia L, Vigil PD, Beman JM, Kim DY, et al. Marine bacterial, archaeal and protistan association networks reveal ecological linkages. ISME J. 2011;5:1414–1425. doi: 10.1038/ismej.2011.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornthwaite CW. An approach toward a rational classification of climate. Geogr Rev. 1948;38:55–94. [Google Scholar]
- Torsvik V, Ovreas L, Thingstad TF. Prokaryotic diversity: magnitude, dynamics, and controlling factors. Science. 2002;296:1064–1066. doi: 10.1126/science.1071698. [DOI] [PubMed] [Google Scholar]
- Triadó-Margarit X, Casamyor EO. Genetic diversity of planktonic eukaryotes in high mountain lakes (Central Pyrenees, Spain) Environ Microbiol. 2012;14:2445–2456. doi: 10.1111/j.1462-2920.2012.02797.x. [DOI] [PubMed] [Google Scholar]
- Urich T, Lanzén A, Qi J, Huson DH, Schleper C, Schuster SC. Simultaneous assessment of soil microbial community structure and function through analysis of the meta-transcriptome. PLoS One. 2011;3:e2527. doi: 10.1371/journal.pone.0002527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson DM, Koumoutsaris S, Mitchell EAD, Bey I. Modelling the effect of size on the aerial dispersal of microorganisms. J Biogeogr. 2012;39:89–97. [Google Scholar]
- Willmott CJ, Feddema JJ. A more rational climatic moisture index. Prof Geogr. 1992;44:84–87. [Google Scholar]
- Wu T, Ayres E, Bardgett RD, Wall DH, Garey JR. Molecular study of worldwide distribution and diversity of soil animals. Proc Natl Acad Sci USA. 2011;108:17720–17725. doi: 10.1073/pnas.1103824108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.