Abstract
Archaea are usually minor components of a microbial community and dominated by a large and diverse bacterial population. In contrast, the SM1 Euryarchaeon dominates a sulfidic aquifer by forming subsurface biofilms that contain a very minor bacterial fraction (5%). These unique biofilms are delivered in high biomass to the spring outflow that provides an outstanding window to the subsurface. Despite previous attempts to understand its natural role, the metabolic capacities of the SM1 Euryarchaeon remain mysterious to date. In this study, we focused on the minor bacterial fraction in order to obtain insights into the ecological function of the biofilm. We link phylogenetic diversity information with the spatial distribution of chemical and metabolic compounds by combining three different state-of-the-art methods: PhyloChip G3 DNA microarray technology, fluorescence in situ hybridization (FISH) and synchrotron radiation-based Fourier transform infrared (SR-FTIR) spectromicroscopy. The results of PhyloChip and FISH technologies provide evidence for selective enrichment of sulfate-reducing bacteria, which was confirmed by the detection of bacterial dissimilatory sulfite reductase subunit B (dsrB) genes via quantitative PCR and sequence-based analyses. We further established a differentiation of archaeal and bacterial cells by SR-FTIR based on typical lipid and carbohydrate signatures, which demonstrated a co-localization of organic sulfate, carbonated mineral and bacterial signatures in the biofilm. All these results strongly indicate an involvement of the SM1 euryarchaeal biofilm in the global cycles of sulfur and carbon and support the hypothesis that sulfidic springs are important habitats for Earth's energy cycles. Moreover, these investigations of a bacterial minority in an Archaea-dominated environment are a remarkable example of the great power of combining highly sensitive microarrays with label-free infrared imaging.
Keywords: Archaea, microbial ecology, PhyloChip, SR-FTIR, SRB, CTC
Introduction
Although the Archaea-scientific community is evolving fast, the lack of knowledge with respect to mesophilic and cold-loving archaea is still enormous. The recent cultivation success of thaumarchaeal representatives is revealing novel and fascinating information, as are alternative procedures that allow in situ studies of archaea in their natural environment or in microcosm experiments (Hatzenpichler et al., 2008; Dekas et al., 2009; Walker et al., 2010; Tourna et al., 2011). One major challenge in understanding the ecological role of archaea is that they are underrepresented in most natural systems, typically accounting for much less than 50% of the microbial cells present. Although some reports have revealed a predominance of (cren-) archaeal cells in marine water columns, reaching numbers of up to 90% Archaea versus Bacteria (Karner et al., 2001), the archaeal part is composed of a broad diversity in these settings (DeLong, 1998). Natural environments that are predominated by one single species of Archaea are rare; the most famous are the anaerobic methane-oxidizing (AMO) consortium (Orphan et al., 2001) and the ‘string-of-pearls community' (Rudolph et al., 2001). Both of these consortia seem to be based on syntrophy, in which both partners are mutually dependent on each other for nutrient exchange (Moissl-Eichinger and Huber, 2011). The AMO consortium has been the subject of numerous analyses and is currently fairly understood but the string-of-pearls community, and in particular the archaeal partner therein (SM1 Euryarchaeon), is still mysterious in many aspects.
The SM1 Euryarchaeon is found in sulfide-containing fresh and marine waters all over Europe (Rudolph et al., 2004), but only two sites (close to Regensburg, Bavaria, Germany) were studied extensively during the past 10 years: The Sippenauer Moor and the Muehlbacher Schwefelquelle (“Islinger Muehlbach” Henneberger et al., 2006). Both of these sites are characterized by a main, sulfidic spring, emanating into a streamlet where whitish mats of sulfide-oxidizing bacteria cover the submerged surfaces. These aquifers are very similar to sulfidic cave springs that are rich in sulfide, ammonia and sulfate (Engel et al., 2004) but poor in dissolved organic carbon, suggesting that the major microbial community of the biotopes are chemolithoautothrophs (Engel et al., 2003; Kodama and Watanabe, 2004). Although, sulfidic springs represent <10% of terrestrial fresh water springs (Palmer, 1991), they are believed to have an important role in global sulfur-cycling (Engel et al., 2003), as they can spawn huge amounts of microbial biomass mainly consisting of sulfur-oxidizing bacteria such as Thiothrix, Beggiatoa and Sulfuricuvum. These filamentous bacteria live as microbial mats or streamers in fluctuating gradients of sulfide and oxygen, and may also be responsible for the environmental success of the SM1 Euryarchaeon under oxygen-rich conditions (Moissl et al., 2002; Rudolph et al., 2004). Surrounding the archaeal colony, Thiothrix (Sippenauer Moor) and Sulfuricurvum (Muehlbacher Schwefelquelle) form the string-of-pearls community and possibly interact with the archaeon through an inter-species sulfur cycle (Moissl et al., 2002). In these communities, the bacterial partner and the SM1 Euryarchaeon are present in almost equal abundance, pointing at a ‘real' partnership and possibly at a symbiotic/syntrophic relation. The proposed sulfur cycle suggests the SM1 Euryarchaeon being an anaerobic sulfate reducer surrounded by sulfur-oxidizing bacteria. The latter metabolize products from sulfate reduction (H2S), and provide simultaneously the educts (sulfate) for the SM1 Euryarchaeon. In addition, the sulfur-oxidizing bacteria protect the SM1 Euryarchaeon from oxygen exposure by respiration (Moissl et al., 2002).
In contrast to other sulfidic springs that have been microbiologically studied, samples at the Muehlbacher Schwefelquelle can also be taken from ∼ 1 m below the water table, where the upwelling water is not yet mixed with atmospheric oxygen. By placing an in situ trapping system in this subsurface setting, slime-like biofilm structures consisting almost exclusively of SM1 euryarchaeal cells can be caught from the water stream, in stark contrast to the abovementioned string-of-pearls community (Henneberger et al., 2006). This second life-style of the SM1 Euryarchaeon differs also from other described microbial systems, in which archaea are involved in biofilm formation (Lapaglia and Hartzell, 1997; Tyson et al., 2004; Frols et al., 2008): first, the SM1 euryarchaeal biofilm represents the only known naturally occurring Archaea-dominated biofilm, revealing a purity of up to 95% based on microscopic counts (Henneberger et al., 2006). Second, the small archaeal cocci form porous colonies with defined distances between the single cells mediated by their unique cell surface appendages (Moissl et al., 2005; Henneberger et al., 2006). Third, bacteria in the biofilm are either randomly distributed or form dense microcolonies, and their varied morphological appearance hints at a broader genetic diversity. Lastly, no other archaea have been detected within the biofilm, using fluorescence in situ hybridization (FISH) or conventional cloning strategies, suggesting that the SM1 euryarchaeal biofilm is a natural ‘archaeal monospecies biofilm' (Henneberger et al., 2006). The Muehlbacher Schwefelquelle spring therefore represents an extraordinary window to an anoxic subsurface biotope of an unusual archaeon.
Using basic biochemical analyses, the water content of the SM1 Euryarchaeon biofilm has been determined to be extraordinary high (99.6% Amann T et al., unpublished data) and the composition of the extracellular polymeric substance has been shown to have a high ratio of protein versus carbohydrates (1.5:1). No nucleic acids, however, were found in the matrix surrounding the SM1 Euryarchaeon cells (Henneberger et al., 2006). The protein content is mainly owing to its extraordinary cell surface structures, called hami, which are highly-complex, filamentous attachment tools with a nano-sized grappling hook at their end (Moissl et al., 2005).
The biochemical analyses performed to date have been based on protocols that necessitate a complete extraction of chemical compounds from the biofilm and do not allow the assignment of organic and inorganic compounds to the different microbes in the biofilm (for example, to Bacteria or Archaea). Moreover, for the underrepresented bacteria in the biofilm neither their metabolic role nor their (possible metabolic) interaction with the archaea are defined or have been subject to deeper studies besides FISH (Henneberger et al., 2006). Hence, it is uncertain, whether bacterial key species coexist with the SM1 Euryarchaeon, or the detected bacterial diversity is randomly attached to the biofilm.
In order to understand the bacterial (and archaeal) diversity in the biofilm and a possible occurrence of certain key species therein, we have conducted highly sensitive PhyloChip analyses based on the 16S rRNA gene pool of the biofilm. In addition, we used synchrotron radiation-based Fourier transform infrared (SR-FTIR) spectromicroscopy to provide a nucleic-acid independent method to link the phylogenetic diversity information with the spatial distribution of the chemical composition and metabolic activity of the bacterial and archaeal cells within the biofilm. SR-FTIR is a non-invasive and label-free molecular imaging technique capable of micrometer spatial resolution (Holman et al., 2010). In this study, the capability of SR-FTIR to differentiate Bacteria from archaeal cells has been evaluated.
Material and methods
Sampling site and physical characteristics
Biofilm samples were collected from the cold (∼10.5 °C), sulfidic spring Muehlbacher Schwefelquelle. Its physical characteristics (pH and water composition) have already been described previously (Rudolph et al., 2004; Henneberger et al., 2006), and are found to be very constant over several years of measurement (including sulfate 16 mg l−1, thiosulfate 14 mg l−1, ammonia 0.33 mg l−1). Oxygen concentrations at different locations of the spring and the stream were re-measured using a highly sensitive oxygen dipping probe (PSt6) coupled with temperature measurement (Fibox 3, LCD trace; PreSens, Regensburg, Germany).
Sample collection
An in situ biofilm trapping system was used to catch biofilm pieces washed up from the deeper subsurface. The nets were incubated for 3 days as deep as possible in the spring bore. Sampling was performed as described earlier (Henneberger et al., 2006). Samples for FISH analysis were incubated in phosphate-buffered saline-containing paraformaldehyde (final concentration 3% (wt/vol)) for 1 h at room temperature (22 °C±2 °C); samples for PhyloChip G3 assays were frozen at −20 °C and samples for SR-FTIR spectromicroscopy necessitated air-drying of the biofilm on gold screens (G225G1, Plano GmBH, Wetzlar, Germany). In addition, 25 ml of spring water were collected as a field control for PhyloChip experiments. Anaerobic sampling for incubation experiments was performed as follows: a double-opened Schott flask was placed on a funnel letting almost all water of the spring pass through. The flask had several layers of polyethylene nets to filter the spring water and catch biofilm fragments. After an incubation of 4 days the bottle was closed with rubber stoppers under water (oxygen-free conditions). All samples were kept on ice during the transport from the site to the laboratory.
Metagenomic DNA extraction
A measure of 250 μl of each biofilm sample were used for individual extraction procedures. Spring water was concentrated via a Millipore amicon 50 kDa cutoff centrifugal filter (Millipore, Billerica, MA, USA), according to manufacturer's specifications before undergoing DNA extraction as described previously (Tillett and Neilan, 2000; Moissl-Eichinger, 2011). Concentrations of double-stranded DNA in the samples were determined using Qubit Quantitation Platform (Invitrogen, Carlsbad, CA, USA).
Quantitative PCR and cloning of dsrB genes
Quantitative PCR (qPCR) was carried out in triplicates with 1 μl of metagenomic DNA as described previously (Moissl-Eichinger, 2011), and the following primer sets were used. Archaeal 16S rRNA genes: 345aF-517uR (Lane, 1991; Burggraf et al., 1992; Moissl-Eichinger, 2011); bacterial 16S rRNA genes: 338bF-517uR (Lane, 1991); dissimilatory sulfite reductase subunit B (dsrB) genes: DSRp2060F (Geets et al., 2006) and DSR4R (Wagner et al., 1998). 16S rRNA gene standards were developed from PCR products of Methanococcus aeolicus (DSM 17 508) and Bacillus safensis (DSM 19 292).
dsrB gene standard was generated from an environmental biofilm sample. After PCR-amplification of dsrB genes with the abovementioned primers the amplicons were cloned into pCR2.1-Topo vector. Fifty-two clones were randomly picked and inserts were sequenced using M13F and M13R primers. Forty-eight clones revealed high quality and were vector-trimmed, clustalW aligned and grouped into operational taxonomic units (OTU) at a 0.01 hard cutoff (Schloss et al., 2009). One representative sequence of each OTU was submitted to GenBank (Acc. no. JX515394–7); a representative clone of the dominant OTU (JX515394) was used for generating a qPCR standard (PCR amplicon generated with M13 primers). The coverage of the library was calculating according to Good (1953).
16S rRNA gene amplification
The template concentration for PCR was set to 3 ng for biofilm samples but DNA isolated from spring water revealed no measurable concentrations (<0.05 ng) due to low biomass. Consequently, 1 μl of template was used for single PCR; the same settings were also applied for the extraction blank (see below). Bacterial 16S rRNA genes were amplified in a gradient PCR using primers B27f and 1492r as described elsewhere (Hazen et al., 2010), and 30 cycles were run. For amplification of archaeal 16S rRNA genes the degenerated primer pair 345af (5′-CGGGGYGCASCAGGCGCGAA-3′ (Burggraf et al., 1992)) and 1406ur (5′-ACGGGCGGTGTGTRCAA-3′ (Lane, 1991)) with an annealing temperature of 60 °C were chosen. Running only 25 PCR cycles and an evaluation of the primers via RDP II (Cole et al., 2009) in comparison to previous Archaea-directed primers (Hazen et al., 2010) promised an increase of the detectable archaeal biodiversity (coverage of these and previous primers evaluated via RDP II, Supplementary Table S1). PCR products were gel-purified prior to cloning or microarray analysis (QIAquick Gel Extraction Kit, Qiagen, Germany).
Archaeal clone library
The PCR products of one biofilm sample were used to generate an archaeal 16S rRNA gene clone library by using the TOPO TA cloning kit with TOP 10′ cells (Invitrogen). Colonies were manually picked and inserts were amplified using the abovementioned archaeal primer pair. For screening, restriction-fragment-length polymorphisms were performed using two restriction enzymes (HaeIII and HinfI, Promega, Madison, WI, USA (Vaneechoutte et al., 1992; Moissl-Eichinger, 2011)). Plasmids of clones with unique sequences were purified (Plasmid Mini DNA Purification Kit, Invitrogen) and bi-directionally sequenced using M13 primers (University of California, DNA Sequencing Facility, Berkeley, USA). After chimera check via Bellerophon (version 3 (http://greengenes.lbl.gov/)) and Pintail (Ashelford et al., 2005) sequences were compared with publicly available sequences and among each other using BLAST (http://blast.ncbi.nlm.nih.gov; Altschul et al., 1990).
16S rRNA gene microarray (PhyloChip G3) analysis
The PhyloChip G3 design, performance and analysis were already described (Hazen et al., 2010). Here, 500 ng of bacterial and 100 ng of archaeal 16S rRNA gene amplicons were used for PhyloChip analysis of biofilm samples. Hence, only 100 ng of bacterial PCR amplicons and 10 μl of archaeal amplicons (below detection limit) were hybridized on the chip for the background water sample. DNA extraction blanks yielded no quantifiable amounts and 14.5 μl of bacterial and 10.0 μl of archaeal post-PCR were used for PhyloChip assay. After combining amplicons, they were spiked with known amounts of non-16S rRNA genes (total 202 ng). Fluorescence intensities of these positive controls were used to normalize total array intensities among samples. Target fragmentation, biotin labeling, PhyloChip hybridization, scanning and staining, as well as background subtraction, noise calculation, detection and quantification criteria were performed as reported (Hazen et al., 2010).
PhyloChip data processing
Stage 1 and 2 analysis were performed as described elsewhere (Hazen et al., 2010) and thus, the threshold for identifying a bacterial OTU in a sample was set to a minimum of 18 perfect match probes. Quartiles of the ranked r-scores (response score to determine the potential of a probe pair responding to a target and not to the background) had to meet the following criteria: rQ1 ⩾0.70, rQ2 ⩾0.95, rQ3 ⩾0.98. In addition, subfamilies that had an rxQ3 value (cross-hybridization adjusted response score) of ⩾0.48, were considered as present but also requirement for the OTUs within this subfamily to be present.
For analysis of the archaeal OTUs the aforementioned parameters were adjusted to the smaller ∼1000 bp 16S rRNA gene amplicons. As shown in Supplementary Figure S1A, the number of probe pairs that could possibly be scored with these amplicons, varied among the archaeal OTUs on the chip. Hence, the criterion to call an archaeal OTU present was adjusted to a probe pair score of 14. Consequently, 92/639 of the archaeal OTUs present on the PhyloChip could not be included in the analysis (Supplementary Figure S1A); however, these OTUs were not restricted to one specific phylum and spread within the domain of the Archaea.
Subfamily based analysis was done by picking one representative sequence within an OTU per subfamily that was detected at least in 2/3 of the biofilm samples or in the background water. These OTUs were classified to family level using the Greengenes (DeSantis et al., 2006) database in combination with SILVA (Pruesse et al., 2007) and RDP II (Cole et al., 2009). Trees based on multiple sequence alignments were generated by retrieving 70 000 character alignments from SILVA database and the neighbor joining method (MEGA 4, (Tamura et al., 2007)). Trees with heatmaps were rendered in iTOL (Letunic and Bork, 2007).
Identification of significantly enriched OTUs in the biofilm
For identification of OTUs that were significantly enriched in the biofilm, three additional samples were included in the PhyloChip analysis that were taken from the Sippenauer Moor, where the SM1 Euryarchaeon can be cultivated in situ as a string-of-pearl community together with filamentous sulfur-oxidizing bacteria (Rudolph et al., 2001; Moissl et al., 2002). The samples from the Sippenauer Moor were collected as described previously (Moissl et al., 2003), and underwent the same molecular PhyloChip assay as described for the biofilm samples in this study. A two-tailed, homoscedastic Student's t-test was performed on abundance values of OTUs detected in at least one of the three biofilm or string-of-pearl-community samples. An adjusted P-value of 0.002 was chosen in order to avoid type one errors as 8114 different OTUs were included in the analysis. The resulting OTUs that met this requirement and had a higher average abundance in biofilm samples were then grouped in subfamilies by picking the OTU with the most drastic increase in abundance. Results of the string-of-pearls community analysis were used as a reference data set of an oxygen-exposed environment of the SM1 Euryarchaeon and were not included in this manuscript. Heatmaps of selected OTUs were generated in the R programming environment (http://www.r-project.org/).
Tracking the SM1 Euryarchaeon with PhyloChip technology
As the full 16S rRNA sequence of the SM1 Euryarchaeon is still not publicly available (Rudolph et al., 2001), this Euryarchaeon had not been included in the PhyloChip G3 design (Hazen et al., 2010). In order to track the abundance of the SM1 Euryarchaeon in samples analyzed with PhyloChip G3, the 1019 bp-long sequence of the dominant SM1 Euryarchaeon clone (IM-A1, JN861739) was bioinformatically broken up into 995 25-mers and compared with all probes present on the PhyloChip. Nine different probes were identified to perfectly match with the A1 clone sequence but only one of them revealed high specificity. By using RDP II probe match (Cole et al., 2009) the probe 5′-TGTGCAAGGAGCGGGGACATATTCA-3′ on the microarray (x=651, y=188; oligonucleotide sequences © 2011 Second Genome Inc.) was identified to match with only seven different archaeal sequences in the database, three of them belonging to SM1 euryarchaeal sequences. The relative hybridization intensities of this probe for biofilm, and water samples as well as positive and negative controls are given in Supplementary Figure S1B. As a matter of fact, the biofilm showed higher relative hybridization intensities (100 ng of archaeal PCR product) than the positive control (50 ng of purified PCR product of clone A1), reflecting the high abundance of the SM1 Euryarchaeon in the biofilm. The negative control and the extraction blank (see below) revealed very low intensities. Furthermore, the background water sample from the spring showed a very weak, relative hybridization intensity of 324 compared with an average value of 6913 retrieved from biofilm samples indicating a latent presence of the SM1 Euryarchaeon in the spring water (Supplementary Figure S1B).
Molecular analysis controls
Controls were included in each step mentioned above. For DNA extraction, a negative control performed with PCR grade water was used. The same control was then included in archaeal and bacterial 16S rRNA gene amplification and in PhyloChip analysis. Probes with positive response were masked in PhyloChip analysis of the actual samples in order to avoid false positives. In addition, a negative control of the PhyloChip analysis was performed that included nuclease- and nucleic-acid-free water as well as the spike genes only. However, no OTUs met the threshold requirement neither in the extraction blank nor in the negative control. As a positive control, 50 ng of SM1-Euryarcheon clone IM-A1 (JN861739) was run in microarray analysis in order to see probe responses to the SM1-Euryarchaeon amplicon in comparison to environmental samples, which detected also no other OTUs.
FISH and fluorescence microscopy
Whole-cell hybridizations were performed as mentioned elsewhere (Rudolph et al., 2001), using domain- and species-directed probes (Bacteria: EUB338 (Amann et al., 1990b)), Archaea: ARCHmix (Moissl et al., 2002; Henneberger et al., 2006), SM1 Euryarchaeon: SMARCH714 (Moissl et al., 2003). For the detection of bacteria involved in sulfate reduction, the sulfate-reducing bacteria (SRB) 385 probe (Amann et al., 1990a) and the Delta495a/b/c probe mix was applied (Loy et al., 2002). Probes were labeled with Rhodamine Green (RG), Cy3 and Texas Red. Specimens were afterwards analyzed using either confocal laser scanning microscopy (CLSM, LSM 510 Meta, Zeiss, Oberkochen, Germany; exc. 488 and 543/ em. LP 505 and LP 585; multi-track for RG and Cy3) or epifluorescence microscopy (Olympus BX-60, Hamburg, Germany). For controls, a fluorescent dye-labeled nonsense probe (NONEUB338) were applied to the samples, and separate bacterial controls were also included (Bacillus atrophaeus DSM7264, Escherichia coli K12 DSM30083). Theoretical coverage of FISH probes and representative sequences of PhyloChip OTUs (in silico FISH) was assessed using the ARB software package (Ludwig et al., 2004).
CTC-FISH to measure activity of specific microorganisms
Biofilm samples were sampled anaerobically as described above and handled in an anaerobic glove box (Coy, Grass Lake, MI, USA). Biofilms were supplemented with 100 μl spring water and 10 μl 50 mℳ CTC (5-cyano-2,3-ditoryl tetrazolium chloride; Stellmach, 1984; Stellmach and Severin, 1986; Yoshida and Hiraishi, 2004), which was prepared under anaerobic conditions (N2 gas phase). After an anaerobic incubation of 2 h at 11 °C in a waterbath, the biofilms were removed and underwent fixation, FISH in suspension (Delta495a/b/c mix, RG, performed similar to Wallner et al., 1993) and subsequent DAPI (4′,6-diamidino-2-phenylindole) staining.
SR-FTIR spectromicroscopy imaging and data analysis
SR-FTIR spectromicroscopy is a non-invasive and label-free chemical imaging technology that provides molecular information at micrometer spatial resolution (Carr et al., 1995; Dumas et al., 2009). SR-FTIR takes advantage of three technologies: (i) the well-known sensitivity of infrared spectroscopy to the bond vibration frequencies in a molecule for determining molecular functional groups, (ii) the convenience of a light microscope to locate areas for molecular and composition analysis, and (iii) the 100- to 1000-fold increase in signal-to-noise provided by a bright SR-based infrared light source. Using photons in the mid-infrared region (∼2.5 to ∼15.5 μm wavelength, or ∼4000 to ∼650 wavenumber in cm−1), SR-FTIR spectromicroscopy has been successfully used to characterize microbial activities in geological materials and in both hydrated and dried biofilms (Holman et al., 2009; Hazen et al., 2010; Holman et al., 2010), in spite of the limitation that some signals may be ambiguous.
Freshly harvested samples (four replicates) were gently air dried onto gold-coated copper disks. Although drying affects the three-dimensional structure of the biofilms, prior microscopy experiments with other biofilms suggest that the two-dimensional structure is largely unaffected. Therefore, the measured spatial distribution of Bacteria, Archaea and the biogeochemical features could represent their native two-dimensional distribution within the biofilm. All SR-FTIR spectromicroscopy measurements were performed in the transflectance mode at the infrared beamline of the Advanced Light Source (http://infrared.als.lbl.gov/), where mid-infrared photons emitted from the synchrotron are focused with a 0.65 numerical aperture objective in a Nicolet Nic-Plan infrared microscope. In transflectance mode, the beam is transmitted through the sample, reflected off the gold-coated copper surface and then transmitted through the sample a second time before striking the mercury cadmium telluride detector. Each spectrum is an average of eight scans at a spectral resolution of 4 cm−1. Background spectra were obtained on the cell-free area of the discs.
For each SR-FTIR imaging measurement, the 200 μm × 150 μm field-of-view for the biofilm was divided into equal-sized 2 μm × 2 μm pixels before raster scanning. The resulting data cube, which consists of position-associated FTIR spectra, was subjected to data preprocessing and processing calculations, including spectrum baseline removal, using both Thermo Scientific Omnic version 7.3 (Thermo Scientific, Madison, WI, USA) and Matlab (MathWorks, Nattick, MA, USA). The absorption spectra were then subjected to univariate and unsupervised multiple curve resolution (MCR) image analyses. The univariate approach, which integrates infrared absorbance of an individual peak of interest, relates the absorbance intensity to the relative concentration of a particular chemical component through the Beer−Lambert law. The unsupervised MCR approach, on the other hand, is based on the principal component analysis (PCA) of the entire fingerprint region (1800–700 cm−1) and of the C-H region (3100–2800 cm−1) instead of individual peaks. MCR analysis of SR-FTIR spectra was applied to reveal the distributions of Archaea, Bacteria and chemical variations in the biofilms, which were hidden in the univariate approach. In this study, the unsupervised MCR analysis was performed with non-negative constraints on both concentration and spectral values (Budevska et al., 2003).
Validation of SR-FTIR for differentiating Archaea and Bacteria in biofilms
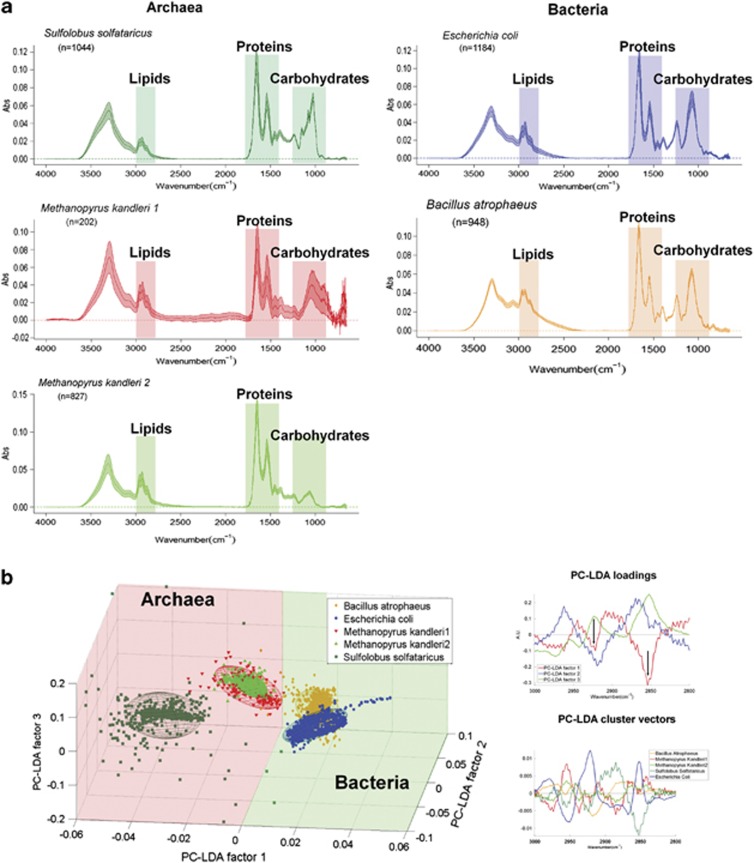
Our SR-FTIR approach assumed that Bacteria can be distinguished from Archaea by comparing spectral features of their lipids in the C-H region due to differences in cell envelope compositions. To confirm this, we performed validation experiments using the following four strains of archaea and bacteria: the archaeon Sulfolobus solfataricus DSMZ 1616T (grown at 80 °C in 0.25 × SME medium) with glycosylated surface layer protein on its surface, the archaeon Methanopyrus kandleri DSMZ 6324T (98 °C, in SME medium) with a pseudopeptidoglycan-containing cell wall covered by a proteinaceous layer, the Gram-negative bacterium E. coli K12 DSMZ 30 083T (37 °C, in LB medium) with a comparably less amount of peptidoglycan but large amount of lipopolysaccharides in its cell envelope, and the Gram-positive bacterium B. atrophaeus DSMZ 7264T (32 °C, in TSB (tryptic soy broth) medium) with a high amount of peptidoglycan in its cell wall. We first made SR-FTIR measurements on the four archaea and bacteria strains, and results were compared and summarized in Figure 1.
Figure 1.
SR-FTIR validation experiments: comparison of reference archaea and bacteria. (a) Comparison of SR-FTIR spectra of reference archaea and bacteria in the 4000–650 cm−1 region reflecting individual membrane lipids and cell envelope compositional characteristics (see Materials and methods section). All spectra are mean±standard deviation (colored area). Archaea: S. solfataricus (glycosilated surface layer); M. kandleri (pseudopeptidoglycan and proteinaceous sheath; please note, M. kandleri exhibits two types of spectra, depending on the observed accumulation of extracellular material (type 1: with extracellular material, type 2: without extracellular material)). Bacteria: E. coli (Gram-negative cell wall), B. atrophaeus (Gram-positive cell wall). The numbers of reference spectra per species measured are given (n). (b) PC-LDA of the same spectra in the CH vibration region (3000–2800 cm−1). Left: three-dimensional PC-LDA score plots reveal an excellent separation of archaea and bacteria along the first PC-LDA factor; each ellipse covers an area of 95% confidence level. The three components explain 92.7% of the variance. Right: The first PC-LDA loading spectrum has two distinct peaks at 2920 cm−1 and 2850 cm−1 (see arrows), which are associated with CH2 bond stretching. The corresponding cluster vector spectra reveal more specific membrane lipids composition and organization variations among the reference strains.
Baseline corrected and vector-normalized spectra in the C−H region between 3000 and 2800 cm−1 were then subjected to the multivariate PCA and then linear discriminant analysis (LDA) using MathLab (7.0). PCA and LDA were used to generate new variables (factors) that were linear combinations (that is, weighted sum) of the original variables (wavenumbers). PCA was first applied to the spectra to reduce the hundreds of absorbance intensities at different wavenumbers to just a few factors that could capture more than 95% of the variance. We typically selected seven components based on the 95% percentage of variance explained and on the spectral features of the loading plot. LDA was then applied to maximize the ‘inter-class' variance over the ‘intra-class' variance of the factors. We visualized the multivariate analysis results in the form of score plots (Figure 1b, left panel) and cluster vector plots (Figure 1b, right panels). In this study, score plots were three-dimensional plots where the first three PC-LDA components were the x-, y- and z-axes; the nearness between classes (clusters) indicates the similarity, whereas the distance between classes implies dissimilarity.
Detailed analyses (Figure 1b, left and right panels) revealed that, in spite of the significant variations in the cell envelope (including cell wall) compositions, Bacteria can be distinguished from Archaea solely by comparing spectral features of their lipids in the C−H regions (3100–2800 cm−1). As expected, bacterial membrane lipids consist of fatty acids with long alkylic (−CH2−) chains which have only one to two terminal methyl (CH3−) groups. In contrast, archaeal membrane lipids generally consist of branched and saturated hydrocarbon isoprene, and therefore relatively less CH2− and more CH3− groups (Mancuso et al., 1986). Our earlier study showed that the SM1 Euryarchaeon possesses a typical CH3-rich lipid (archaeol, Rudolph, 2003). In this context, the ratio of CH2 to CH3 could be used to detect Bacteria in an Archaea-dominated biofilm (Supplementary Figures S2A and S2B).
To confirm this observation further, we made measurements and performed spatial correlation analysis on FISH and MCR SR-FTIR images (Supplementary Figures S3A and S3B) using the image processing software ImageJ (http://www.macbiophotonics.ca/imagej/) and Manders approach of interpretation (Manders et al., 1993). Field-collected biofilm samples were labeled with Archaea-directed probes (ARCHmix; Moissl et al., 2002; Henneberger et al., 2006). The samples' fluorescence and the corresponding MCR SR-FTIR images were acquired by means of a Nicolet Continuum XL infrared microscope equipped with a fluorescence attachment and a WG fluorescence cube. Although the MCR recovered image and the fluorescence image have different ‘brightness', ‘intensity' and effective spatial resolution (microns for infrared and hundreds of nanometers for fluorescence microscopy), Pearson's coefficient was 0.504, Manders' overlap coefficient 0.798, the co-localization coefficient M1 was 0.988 and M2 was 1.000 (Manders et al., 1993). This demonstrated that the two images were quite similar (Manders et al., 1993).
Results
Using an in situ trapping system (Henneberger et al., 2006), fragments of the SM1-Euryarchaeon biofilm from the subsurface were collected for an in-depth characterization of the unique subsurface biotope that can be accessed through the Muehlbacher Schwefelquelle.
Oxygen concentration in the spring area revisited
Using a highly sensitive oxygen probe, a chemocline could be detected with decreasing oxygen concentrations towards the spring (Supplementary Figure S4). No oxygen could be measured in the subsurface water before mixing with the atmosphere in the streamlet, indicating a complete oxygen-free environment in the subsurface. These on-site measurements are in contrast to previous investigations that reported low amounts of oxygen in the spring water (Rudolph et al., 2004; Henneberger et al., 2006).
Dominance of the SM1 Euryarchaeon in the subsurface biofilm
FISH with Archaea-directed and SM1 Euryarchaeon-specific probes confirmed previous results showing the archaeal dominance within the biofilm. The predominance of the SM1 Euryarchaeon was additionally confirmed by using domain-specific qPCR. The ratio of archaeal and bacterial 16S rRNA gene copy numbers was 97:3 (Table 1), which is similar to previously reported ratio of Archaea:Bacteria being 95:5 (Henneberger et al., 2006). A newly constructed clone library of archaeal 16S rRNA gene sequences generated from biofilm samples resulted in four different restriction-fragment-length polymorphism patterns after analyzing 48 clones. The dominant clone sequence (88% of all clones, IM-A1, JN861739) and two others (2%, IM-C8, JN861741; 2% IM-4–1, JN861742) showed high similarity to publicly available 16S rRNA gene sequences of the SM1 Euryarchaeon and among each other (>99%). One clone sequence (8%, IM-C4, JN861740) was closely related to the environmental clone sequence SMK5 (Rudolph et al., 2004), which was retrieved from Sippenauer Moor string-of-pearls community in 2005 (99% similarity) but shows a genetic distance of 20% to the SM1 Euryarchaeon sequence.
Table 1. Quantification of archaeal/bacterial 16S rRNA and dsrB gene sequences in 1 ng metagenomic DNA from SM1 Euryarchaeon biofilm samples taken at two different sampling times.
| Biofilm replicate |
Number of gene copies |
||
|---|---|---|---|
| Archaeal 16S rRNA | Bacterial 16S rRNA | dsrB | |
| 1 | 2.26E+06 | 6.88E+04 | 5.68E+03 |
| 2 | 3.33E+06 | 8.40E+04 | 5.46E+03 |
| 3 | 3.09E+06 | 7.49E+04 | 4.22E+03 |
| Mean | 2.89E+06 | 7.59E+04 | 5.12E+03 |
Abbreviation: dsrB, dissimilatory sulfite reductase subunit B.
The currently most sensitive method available (PhyloChip G3 16S rRNA gene microarray technology, detection limit 2 pℳ of 16S rRNA PCR product; (Hazen et al., 2010)) was used for characterizing the archaeal and bacterial composition in the biofilm and the spring water itself based on 16S rRNA gene analysis. Besides a comprehensive detection of Bacteria, the setup of the PhyloChip G3 technology was geared towards the identification of (also underrepresented) archaeal signatures. To avoid primer mismatches of typical, Archaea-directed primers binding to the front region of the 16S rRNA gene (Rudolph et al., 2001), a differrent primer set for amplification of (SM1 eury-) archaeal 16S rRNA genes was used and an adjusted bioinformatical approach for the shorter PCR amplicons was necessary. Although the SM1 Euryarchaeon was originally not included in PhyloChip G3 design (Hazen et al., 2010), we developed a method to track its abundance in our samples based on the hybridization intensity of a specific probe on the microarray. With the aid of PhyloChip technology, the SM1 Euryarchaeon was detected highly enriched in the biofilm samples (∼2130% increase in abundance) compared with the spring water.
Microbial diversity in spring water and biofilm
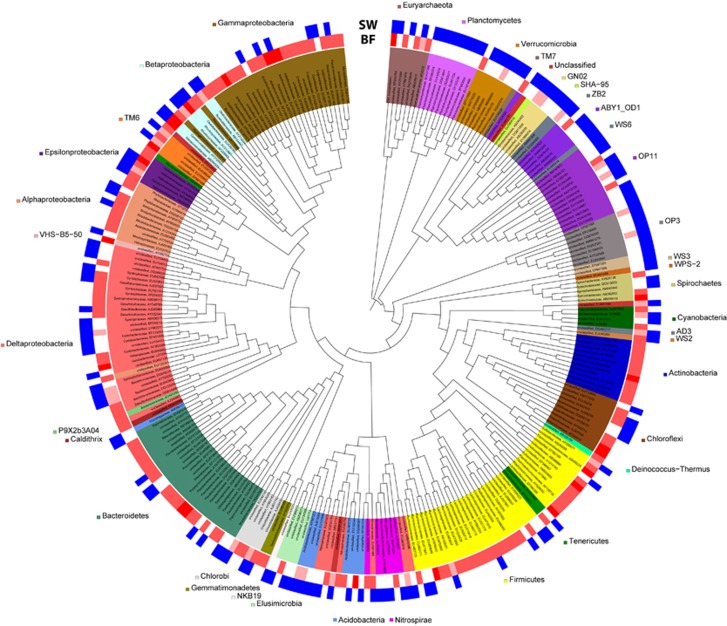
In the spring water and biofilm microbiome, in total 4444 OTUs in 869 different subfamilies were detected by PhyloChip analyses with 10 OTUs (in 10 subfamilies) belonging to the archaeal domain in addition to the SM1 Euryarchaeon. The overall distribution of the microbial taxa on subfamily level ranged from 14% for Firmicutes to 0.1%, for example, for Aquificales. Only 36% of the subfamilies detected in the water were also present in at least one of the three biofilm replicates (Figure 2).
Figure 2.
Difference in microbial richness between the spring water and the biofilm: presence and absence of subfamilies in spring water (SW) and biofilm samples (BF). Color intensities (red) of the biofilm samples reflect the number of times the subfamily was called present in one of the three replicates. As the SW sample was not replicated, heatmap reflects presence (blue label) or absence only. The Neighbor Joining tree was constructed with one representative OTU per subfamily (branch length is ignored). Leaf IDs give the classification on family level and the accession number of the representative OTU. Only those subfamilies that occurred in the water sample or in at least 2/3 of the biofilm replicates are shown.
The diversity of the spring water was dominated by Deltaproteobacteria (14%); however, signatures of Methanomicrobia and Thermosplasmata of the archaeal domain were also retrieved (distribution of spring water diversity in Supplementary Figure S5A). The diversity of Firmicutes, Gammaproteobacteria and Bacteroidetes increased in the biofilm, whereas members of the OP11-group and the Planctomycetes were less diverse than in the spring water. Considering the diversity of subfamilies that occurred in at least 2/3 of the biofilm samples, Firmicutes were again the most diverse taxon, followed by Gammaproteobacteria and Bacteroidetes (Supplementary Figure S5B).
Core microbiome of the biofilm
Biofilm subfamilies detectable in 2/3 replicates were then analyzed in depth at the OTU level: If the coefficient of variation of the abundance values of a single OTU was <10% among biofilm replicates, the OTU was assumed to be non-fluctuating, and thus a constant member (potential key species) of the biofilm. Abundance values and a detailed description of these constant OTUs (263, including the SM1 Euryarchaeon) are presented in Supplementary Figure S6. This community is considered to represent the core microbiome of the biofilm.
As OTUs such as Thiothrix clone sipK4 (AJ307941) and the Sulfuricurvum clone IMB1 (AJ307940) were found to fluctuate, they were not considered as representatives of the core microbiome. However, both of these OTUs have been identified as key species in the string-of-pearls communities at various sampling sites (Rudolph et al., 2001; Moissl et al., 2002). Eleven other string-of-pearls community related OTUs, which had been reported but not as key species, were also identified and found to be mostly fluctuating (Supplementary Table S2).
Significantly enriched OTUs in the biofilm compared with the string-of-pearls community
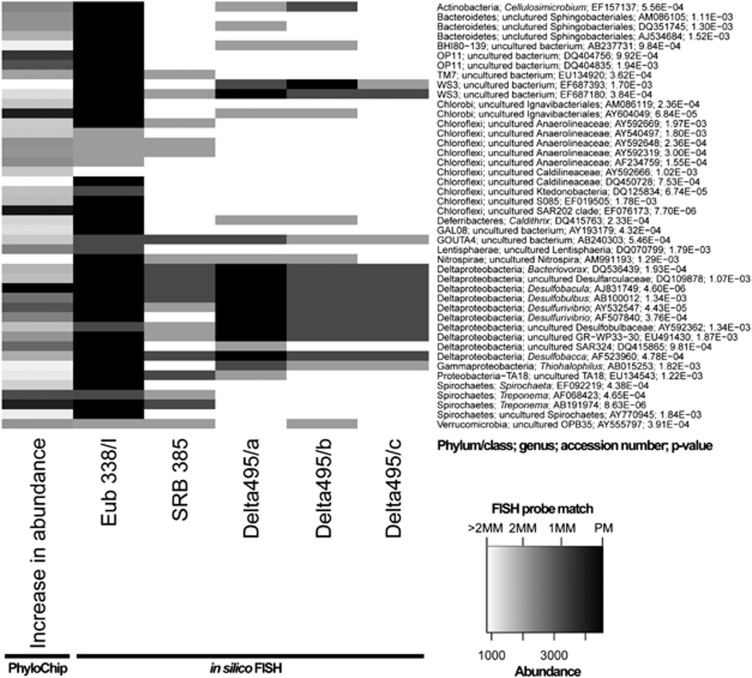
PhyloChip G3 analyses revealed 2139 OTUs that increased in abundance in the biofilm samples compared with the reference sample set (string-of-pearls community, data not shown). Eighty-three OTUs met the requirement of being highly significantly enriched (adjusted P-value 0.002), which were grouped into 44 subfamilies and are displayed in Figure 3. The OTU with the greatest increase in abundance (accession number of representative sequence AJ831749; increase in abundance: 4559) was also the OTU with the most significant P-value of 4.60E-06. The representative sequence grouped this OTU in the Deltaproteobacteria, genus Desulfobacula. However, also OTUs of other phyla and genera were detected as significantly enriched, among those many Chloroflexi and Spirochetes.
Figure 3.
Significantly enriched OTUs (one representative of each subfamily) in the SM1 Euryarchaeon biofilm and in silico FISH-probe match. Heatmap of OTUs that increased highly significantly (P<0.002) in biofilm compared with string-of-pearls community samples and were called present in at least one of the samples (first column). Probes used for FISH experiments in this study were in silico matched to representative sequences of the enriched OTUs using the ARB software package. The theoretical coverage of the FISH probes is displayed in columns 2−5; the decreasing heatmap intensity reflects the number of mismatches of each probe per OTU (MM=mismatch, PM=perfect match).
Detection of SRB via FISH and correlation with PhyloChip data
The presence and amount of (potentially) SRB in the biofilm was further confirmed by FISH with two different (sets of) probes targeting bacterial sulfate reducers: bacterial sulfate-reducer probe SRB385 and Delta495 probe mix. Each approach was backed up by probes directed for Bacteria (Eub 338/I, Texas Red) or Archaea (ArchMix, RG), and DAPI staining or combinations thereof. The specificity of FISH experiments was confirmed by using appropriate controls and a nonsense probe (NONEUB338), which showed no signal when applied to biofilm samples.
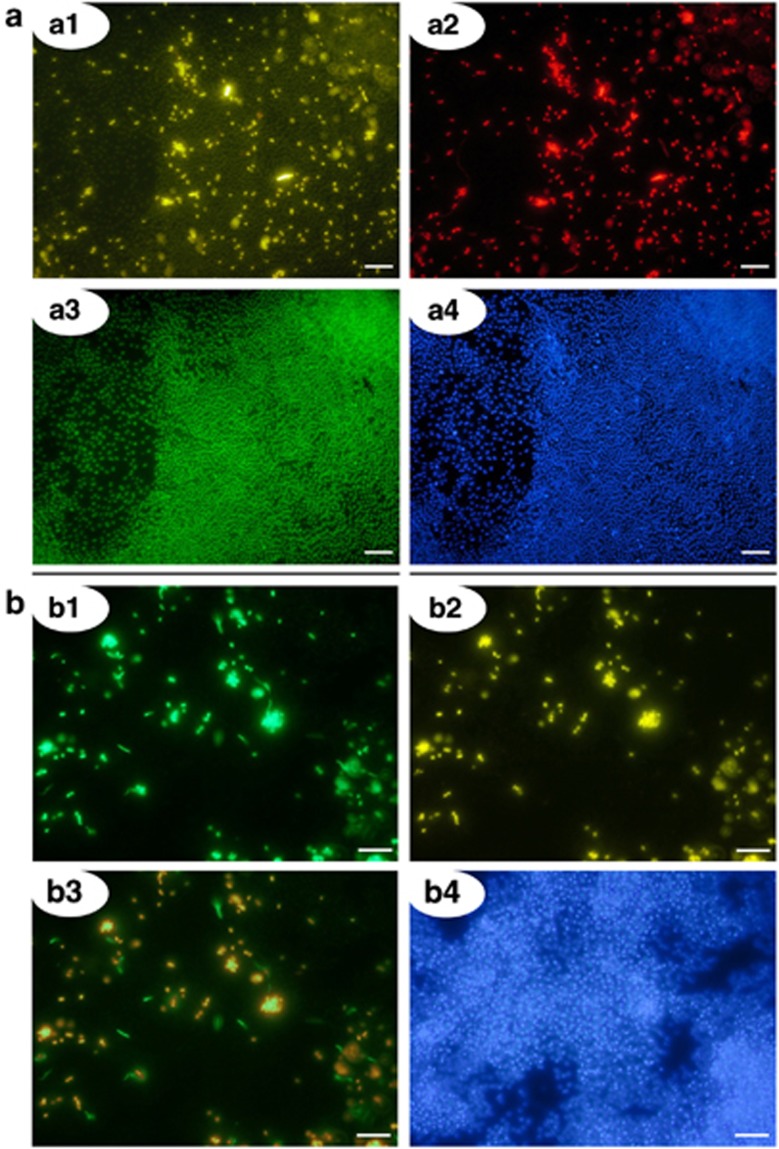
The morphology of the bacteria in the biofilm was diverse, ranging from single cocci to aggregates, filaments, oval-, rod- and helix-shaped. The percentage of bacteria was estimated at 5%, confirming results from qPCR and previous studies (Henneberger et al., 2006). Interestingly, 85.4% (±4.7% s.d.; 15 biofilm samples analyzed) of cells stained with the bacterial probe also exhibited signals for the SRB385 probe (Figure 4a). This percentage of SRB was confirmed by the usage of the Delta495 probe mix, which revealed an amount of 89.2% (±0.9% standard deviation; four biofilm samples analyzed) SRB (Figure 4b).
Figure 4.
(a) Ternary FISH analysis of the subsurface biofilm with SRB-, Bacteria and Archaea-specific probes. Analysis reveals a dominance of archaeal cocci (SM1 Euryarchaeon) and of SRB-385-stained bacteria in the bacterial minority: ∼85% of the detected Bacteria revealed a signal with the sulfate-reducer specific probe. Scale bars=10 μm. a1: biofilm, FISH-stained with probe SRB385 CY3 (targeting SRB, yellow). a2: same detail, stained with probe EUB 338/I Texas Red (targeting Bacteria, red). a3: same detail, stained with probe mixture ArchMix RG (targeting Archaea, green). a4: same detail, reference-stained with DAPI (blue). (b) biofilm sample FISH-stained with SRB-directed Delta495 probe mix. The overwhelming majority of bacteria in the biofilm showed signals with the Delta495 probe mix (89.2%). Scale bars=10 μm. b1: probe 338/I RG (targeting Bacteria, green). b2: same detail, stained with probe mix Delta495 (targeting SRB, yellow). b3: overlay of details 1 and 2. b4: same detail, reference-stained with DAPI (blue).
In order to correlate FISH data with enriched OTUs detected by PhyloChip analysis, the theoretical coverage of the Bacteria- and sulfate-reducer directed FISH probes was analyzed in silico (Figure 3). All probes (SRB385, Delta495 probe mix and EUB 338/I) showed theoretical coverage of the target group (SRB), and therefore confirmed our FISH results. Interestingly, 29.4% of all bacterial cells that did not stain with the Delta495 probe mix exhibited a typical Spirochaeta-like morphology. This genus was also found to be highly enriched in the biofilm, but whose 16S rRNA reveals >2 mismatches for the Delta495 probe mix (Figure 3).
Based on the high percentage of the SRB385 probe and the Delta495 probe mix stained bacteria, the PhyloChip and in silico FISH analysis it can be concluded that the major part of bacteria in the biofilm can be affiliated to members of the Deltaproteobacteria, most likely to one specific, enriched OTU (genus Desulfobacula AJ831749). Cultivated members within the genus Desulfobacula were described as oval-shaped; bacteria with this morphology were positively stained with SRB and Delta495 probes and formed aggregates in the biofilm (Figure 4b).
Detection of dsrB genes in biofilm samples
In order to further prove the presence of SRB and their metabolic capability, qPCR with dsrB-directed primers was performed. We were able to specifically detect the presence of genes encoding dsrB and to quantify their amount (Table 1). The abundance of detectable dsrB genes in biofilm samples allowed the conclusion that these signatures were derived from bacteria and not from the dominant SM1 Euryarchaeon (three-log difference in archaeal 16S rRNA and dsrB gene abundance). Moreover, the one-log difference of bacterial 16S rRNA and dsrB genes can be attributed to the fact that ribosomal genes can have up to 15 copies per genome (Klappenbach et al., 2001; Lee et al., 2009), whereas dsrB genes generally appear once (Heidelberg et al., 2004).
A clone library generated from the dsrB amplicons showed four different OTUs of dsrB genes belonging to the Deltaproteobacteria cluster, whereas one OTU was dominant (Accession no. JX515394: 33 clones; Accession no. JX515395: 11 clones; Accession no. JX515396: 3 clones, Accession no. JX515397: 1 clone). The coverage of the library was determined as 98%.
Metabolic activity of SRB in the biofilm
A combination of CTC staining and FISH analysis showed an overlap of signals from CTC and SRB-directed FISH probes (Delta495 probe mix) in biofilm samples that were incubated in spring water anaerobically (Supplementary Figure S7). The formation of CTC-formazan precipitates can be attributed to biological redox reactions, for example, respiratory electron transport, and thus provide evidence for the metabolic activity (Stellmach, 1984; Stellmach and Severin, 1986; Yoshida and Hiraishi, 2004).
SR-FTIR measurements of Bacteria, Archaea, and metabolic intermediates distributions in biofilms
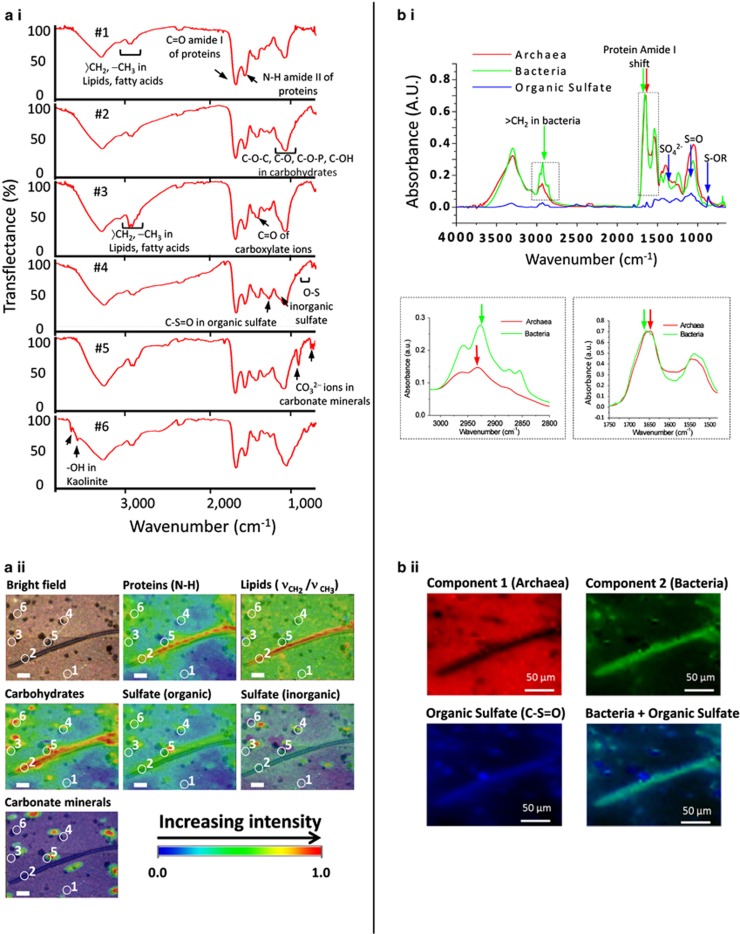
The high brightness of SR-FTIR spectromicroscopy enabled us to identify the presence of Bacteria, Archaea and a number of metabolic intermediates at a spatial resolution between 2 and 10 μm. Biofilm samples were first examined with crossed polarized microscopy and fluorescence microscopy because the biofilm fraction often exhibits visually interesting biogeochemical structures (Supplementary Figure S8). Then the spatial distribution of molecular composition and possible metabolites in the biofilm were analyzed by SR-FTIR spectromicroscopy. Figure 5a.i shows examples of a range of typical SR-FTIR spectra collected on the biofilm samples (Figure 5a.ii, white circles in bright field). A striking feature was that these spectra, although obtained at locations merely several tens of micrometer apart (see circles in Figure 5a.ii), contained distinctly different signatures known to be associated with organic and inorganic markers typical of biogeochemical systems (Table 2). Spatial distributions of the infrared absorption intensities (from univariate analysis) of these molecular markers are shown in Figure 5a.ii. Notice that the infrared absorption intensity ratio of CH2 to CH3 was ∼30% higher in the biofilm regions occupied by large, filamentous-shaped, Beggiatoa-like bacteria, compared with the surrounding Archaea-dominated area.
Figure 5.
(a) SR-FTIR images of Bacteria in the Archaea-dominated biofilm. SR-FTIR images (220 μm by 180 μm) showing the distribution of microorganisms and biogeochemical products in an Archaea-dominated biofilm. (a i and a ii) Distribution heatmap (from univariate analysis) of the relative abundance of total proteins (based on the peak area centered at ∼1548 cm−1), of bacterial lipids (the ratio of the peak area of CH2 centered at ∼2852 cm−1 to the peak area of CH3 at ∼2872 cm−1), carbohydrates (the peak area centered at ∼1089 cm−1), sulfur/carbon biochemical cycling products (S=O from organic sulfate products, centered at ∼1240 cm−1, S=O of inorganic sulfate centered at ∼1130 cm−1 and CO32− groups of carbonate minerals in the 880–840 cm−1 region. The OH of clay centered at ∼3695 cm−1 is not shown here). The white circles with numbers (1–6) in the bright field and in the SR-FTIR images (a ii) correspond to the transflectance spectra (a i). The circles represent pixels where the spectra were recorded. Note: Filamentous bacterial structures in the biofilm were rarely observed but specifically presented here in order to illustrate the lipid signatures of Bacteria and Archaea (for more samples please see Supplementary Figure S9). Scale bars=25 μm. (b) Multivariate curve resolution analysis to differentiate Archaea and Bacteria. (b i) Spectra of the three components extracted from the MCR, in red component 1 (Archaea), in green component 2 (Bacteria), in blue component presenting sulfate spectral features, with arrows pinpointing the spectral markers used in the analysis, in the panels below highlighted the spectral region of lipids, important region for the distinction of Bacteria and Archaea since their different membrane composition, and protein region where a shift is observable in Amide I band, index of a different protein content in Bacteria and Archaea. (b ii): Relative concentration images (220 μm by 180 μm) of Archaea (component 1) and Bacteria (component 2) recovered by the MCR analysis and the chemical distribution maps of organic sulfate (C-S=O) in blue. Merging the relative bacterial concentration image (in green color) with the organic sulfate distribution map (in blue) reveals the co-localization of bacteria and organic sulfate. Scale bars=50 μm.
Table 2. Tentative band assignments of the fundamental vibrational modes used in SR-FTIR spectromicroscopy.
| Molecules | Frequency (cm−1) | Assignment | References |
|---|---|---|---|
| Calcium carbonates | ∼1798 | Multiphonon band | White, 1974; Beniash et al., 1997 |
| ∼874, ∼845, ∼710 | Coupling between CO32− groups in adjacent layers of calcite | Huang and Kerr, 1960; White, 1974; Beniash et al., 1997 | |
| Carbohydrates | 1200–900 | C−O−C, C−O ring vibrations of carbohydrates | Nauman, 2000 |
| Clay | ∼3695, ∼3620 | OH groups in the inner-surface of kaolinite | Ledoux and White, 1964 |
| Fatty acids/ lipids | ∼2959, ∼2872 | C−H asym and sym stretching modes of −CH3 in fatty acids | Sinclair et al., 1952; Nauman, 2000; Whittaker et al., 2003 |
| ∼2920, ∼2852 | C−H asymmetric and symmetric stretching modes of 〉CH2 in fatty acids | Sinclair et al., 1952; Nauman, 2000; Whittaker et al., 2003 | |
| ∼1468, ∼1420 | C–H deformation of 〉CH2 | Parker, 1983; Nauman, 2000 | |
| ∼1378 | C–H deformation of −CH3 | Parker, 1983 | |
| Proteins | 1695–1600 | Protein amide I | Nauman, 2000 |
| 1580–1510 | Protein amide II | Nauman, 2000 | |
| Sulfate: inorganic | ∼1150, ∼1130, ∼1105 | S−O of the SO42− | Adler and Kerr, 1965; Peak et al., 1999; Smith, 1999 |
| 800–850 | C–O–S vibration of sulfate in carbonates | Takano, 1985 | |
| Sulfate: organic | ∼1250, ∼1240 | S=O stretching vibration of sulfate ester | Percival and Wold, 1963; Mayers et al., 1969; Goren, 1970; Asker et al., 2007 |
Abbreviation: SR-FTIR, synchrotron radiation-based Fourier transform infrared.
Assignments were used for identification of bio- and geochemical molecules (of dominance) measured in the investigated biofilms.
MCR analysis confirmed that Bacteria can be distinguished from Archaea by their spectral features (Figure 5b.i versus Figures 1a and b; Figure 5b.ii versus Supplementary Figure S3, Supplementary Figure S2 versus Figure 1b). Furthermore, specific metabolites as well as biogeochemical materials were found associated with these prokaryotic groups (Figure 5b). For example, in Figure 5b.ii, a combined univariate and MCR SR-FTIR analysis revealed strikingly overlapping infrared signals of organic sulfate products (R-S=O) and carbonate minerals with Bacteria-rich areas. Similar results were observed in other samples collected during this field experimental period (Supplementary Figures S9 and S10). This implies the presence of microscale mixtures of bacteria that are involved in subsurface sulfur and carbon turnover.
Discussion
Life in the subsurface is highly diverse and comprises an enormous fraction of Earth's biomass. However, the microbial community living in this extreme environment remains largely mysterious, as subsurface biotopes are hardly accessible, which makes it also difficult to understand ongoing geochemical processes in these environments (Onstott et al., 2009). The Muehlbacher Schwefelquelle, however, provides an extraordinary window to the subsurface and allowed the discovery of a highly unusual, Archaea-dominated microbial community (the SM1 euryarchaeal biofilm), which is continuously being washed up from the subsurface and can be harvested from the spring water. As the dominant SM1 Euryarchaeon still resists efforts to be cultivated and metabolically understood, this study focused on the bacterial minority thriving in this type of biofilms. Using a combined approach of molecular techniques and SR-FTIR spectromicroscopy, we demonstrated that the interplay between the underrepresented bacterial fraction and geologically important chemicals could be analyzed in order to obtain insights into a possible ecological role of this extraordinary microbial community.
The dominance of one specific archaeon, the SM1 Euryarchaeon, was revisited and confirmed in this study by qPCR techniques and FISH, proving the constancy of this subsurface system over several years (Henneberger et al., 2006). The sensitive PhyloChip technology also confirmed the abundance of the SM1 Euryarchaeon, but additionally identified the presence of other archaea disproving the initial statement of an ‘archaeal monospecies biofilm' (Henneberger et al., 2006). Nevertheless, because none of these alternate archaea was visualized either in FISH or in the archaeal clone library, it can be assumed that they represent only a very minor fraction of the biofilm.
We demonstrated that the spectral features of membrane lipids can be used to distinguish Archaea from Bacteria even in complex samples without using either a MS- or a nucleic acid-based approach (Sprott, 1992; Elvert et al., 2000; Sturt et al., 2004). Furthermore, we also could use SR-FTIR to map the distribution of biogeochemical compounds, and to relate this molecular information to certain dominant ecological functions of even underrepresented microbial groups.
Raman microspectroscopy has often been used to characterize spatial distribution and molecular composition of biological samples (Wagner, 2009; Beier et al., 2010; Hall et al., 2011; Li et al., 2012). However, to date, Raman microscopy has often been used together with FISH (Raman-FISH) to differentiate microbial populations such as Bacteria and Archaea (Huang et al., 2007). On the contrary, SR-FTIR does not require cell labeling. SR-FTIR is also non-destructive, and therefore allows additional in situ studies of chemical composition changes in microbes on the same sample (Holman et al., 2010). As demonstrated in this study SR-FTIR spectromicroscopy imaging could associate the distribution of Archaea and Bacteria with biogeochemical compounds, giving us the opportunity to gain a more in-depth insight of the underpinning biogeochemical processes. In Figure 5, for example, large, filamentous bacterial cells were observed along with increases in organic sulfate intensities, which suggests that these bacteria could belong to an sulfur-accumulating and -oxidizing bacterium such as Beggiatoa (Larkin and Strohl, 1983), a genus also identified in the biofilm core microbiome.
In other biofilm samples, such as those presented in Supplementary Figures S9 and S10, a majority of the areas that exhibited infrared spectral signatures of bacterial cells coincides with signals that are indicative of an accumulation of organic sulfate. The increasing sulfate signals could imply the presence of compounds such as adenosine-5′-phosphosulfate), 3′phosphoadenosine-5′phosphosulfate or sulfolipids (Goren, 1970). However, adenosine-5′-phosphosulfate is a typical intermediate of metabolically active either assimilatory or dissimilatory SRB and sulfur-oxidizing bacteria.
qPCR assays were able to detect a high amount of dsrB genes of Deltaproteobacteria, which were also identified by the PhyloChip G3 technology to be highly enriched in the biofilm. In FISH analyses of multiple biofilms a vast majority of the bacteria showed a positive signal after hybridization with two different (sets of) SRB-directed probes (the 385 probe and the Delta495a/b/c probe mix). This observation was supported by the CTC-FISH assay, which showed metabolic activity of bacteria stained with the Delta495 probe mix (see Supplementary Figure S7). These investigations confirmed that the bacterial microbiome of the SM1 Euryarchaeon biofilm is comprised mostly of Deltaproteobacteria, involved in sulfate reduction.
The fact that little sulfate signals were detected in the Archaea-rich regions implies two possible scenarios for the samples taken. In the first scenario, the supposed sulfate-reducing SM1 Euryarchaeon might be alive but metabolically inactive, having already reduced most of the sulfate compounds in its direct vicinity of the biofilm. In the second scenario, the SM1 Euryarchaeon might not be capable of sulfate reduction, a conclusion which is in stark contrast to the previous hypothesis (Moissl et al., 2002).
A number of metabolic pathways of SRB-associated archaea have already been reported in literature. For instance, in the AMO consortium SRB have a key role for archaeal, anaerobic methane oxidation (Orphan et al., 2001). However, it still remains unclear if the SM1 Euryarchaeon is capable of methane-oxidation or methanogenesis, or if it performs a completely different metabolism. Nevertheless it can be speculated, that a classical methanogen would quickly be outcompeted by SRB for hydrogen or organic substrates in sulfate-rich, anoxic environments such as the Muehlbacher Schwefelquelle (Lovley and Klug, 1983). Previous investigations have failed to detect F420, a key co-enzyme for methanogenesis, showing no positive amplification of the according gene, nor a positive chemical detection based on chromatography (Moissl et al., 2003).
Possible metabolic functions of the SM1 Euryarchaeon remain speculative but may be responsible for the environmental success of this organism. As the SM1 Euryarchaeon is currently the only known archaeon to absolutely predominate one specific biotope, combined with its appearance in hot spots in Europe and maybe even beyond (Rudolph et al., 2004), a larger (ecological) role can be assumed, which is currently still mysterious. However, a metagenomic study of the biofilm is currently performed, for which the knowledge about the microbial diversity is an important and very helpful prerequisite. This approach may reveal the metabolic capabilities of the SM1 Euryarchaeon in the biofilm.
Although a broad diversity of microbes is detectable in the Muehlbacher Schwefelquelle biotope, the accumulation of SRB, which represent the overwhelming majority of the minor bacterial part, appears to not be an accident; rather it is clear that these bacteria provide a valuable function within the biofilm as their presence in the biofilm was monitored as its discovery more than 8 years ago. However, the open question is if and how the SM1 Euryarchaeon influences the (bacterial) diversity in the biofilm. Does it—as it seems to be obvious for living together with (selected) filamentous sulfide-oxidizers in surface waters—actively recruit SRB to the biofilm, or is this phenomenon a passive enrichment? How and why does the SM1 Euryarchaeon switch from biofilm to string-of-pearls community status and are transition states detectable? These and many more questions will have to be answered in future studies and promise astonishing insights into this fascinating natural archaeal system.
Acknowledgments
Technical assistance by Lauren Tom as well as review and discussion provided by Robert Huber and Reinhard Wirth are much appreciated. Work at University of Regensburg was performed under the DFG grant MO19773–1 given to Christine Moissl-Eichinger. Phylogenetic work at Lawrence Berkeley National Laboratory was performed under the auspices of the U.S. Department of Energy under contract no. DE-AC02–05CH11231. The SR-FTIR and associated imaging work were performed under the Berkeley Synchrotron Infrared Structural Biology (BSISB) Program and the Subsurface Science Scientific Focus Area funded by the U.S. Department of Energy, Office of Science and Office of Biological and Environmental Research through contracts DE-AC02–05CH11231. The Advanced Light Source is supported by the Director, Office of Science, Office of Basic Energy Sciences, of the U.S. Department of Energy under contract no. DE-AC02–05CH11231. The authors are grateful to PreSens (Germany, Regensburg) for providing the oxygen dipping probe PSt6 and the Fibox 3, LCD trace. Alexander J Probst was supported by the German National Academic Foundation (Studienstiftung des deutschen Volkes).
Footnotes
Supplementary Information accompanies the paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Adler HH, Kerr PF. Variations in infrared spectra, molecular symmetry and site symmetry of sulfate minerals. Am Mineralogist. 1965;50:132–147. [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Amann RI, Binder BJ, Olson RJ, Chisholm SW, Devereux R, Stahl DA. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl Environ Microbiol. 1990a;56:1919–1925. doi: 10.1128/aem.56.6.1919-1925.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amann RI, Krumholz L, Stahl DA. Fluorescent-oligonucleotide probing of whole cells for determinative, phylogenetic, and environmental studies in microbiology. J Bacteriol. 1990b;172:762–770. doi: 10.1128/jb.172.2.762-770.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashelford KE, Chuzhanova NA, Fry JC, Jones AJ, Weightman AJ. At least 1 in 20 16S rRNA sequence records currently held in public repositories is estimated to contain substantial anomalies. Appl Environ Microbiol. 2005;71:7724–7736. doi: 10.1128/AEM.71.12.7724-7736.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asker MMS, Mohamed SF, Ali FM, El-Sayed OH. Chemical structure and antiviral activity of water-soluble sulfated polysaccharides from Surgassum Iatifolium. J Appl Sci Res. 2007;3:1178–1185. [Google Scholar]
- Beier BD, Quivey RG, Berger AJ. Identification of different bacterial species in biofilms using confocal Raman microscopy. J Biomed Opt. 2010;15:06001-1–06001-5. doi: 10.1117/1.3505010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beniash E, Aizenberg J, Addadi L, Weiner S. Amorphous calcium carbonate transforms into calcite during sea-urchin larval spicule growth. Proc R Soc Lond. 1997;B:461–465. [Google Scholar]
- Budevska BO, Sum ST, Jones TJ. Application of multivariate curve resolution for analysis of FT-IR microspectroscopic images of in situ plant tissue. Appl Spectrosc. 2003;57:124–131. doi: 10.1366/000370203321535015. [DOI] [PubMed] [Google Scholar]
- Burggraf S, Olsen GJ, Stetter KO, Woese CR. A phylogenetic analysis of Aquifex pyrophilus. Syst Appl Microbiol. 1992;15:352–356. doi: 10.1016/S0723-2020(11)80207-9. [DOI] [PubMed] [Google Scholar]
- Carr GL, Reffner JA, Williams GP. Performance of an Infrared Microspectrometer at the Nsls. Rev Sci Instrum. 1995;66:1490–1492. [Google Scholar]
- Cole JR, Wang Q, Cardenas E, Fish J, Chai B, Farris RJ, et al. The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 2009;37:D141–D145. doi: 10.1093/nar/gkn879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekas AE, Poretsky RS, Orphan VJ. Deep-sea archaea fix and share nitrogen in methane-consuming microbial consortia. Science. 2009;326:422–426. doi: 10.1126/science.1178223. [DOI] [PubMed] [Google Scholar]
- DeLong EF. Everything in moderation: archaea as 'non-extremophiles'. Curr Opin Genet Dev. 1998;8:649–654. doi: 10.1016/s0959-437x(98)80032-4. [DOI] [PubMed] [Google Scholar]
- DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, et al. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol. 2006;72:5069–5072. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumas P, Miller LM, Tobin MJ. Challenges in biology and medicine with synchrotron infrared light. Acta Phys Pol A. 2009;115:446–454. [Google Scholar]
- Elvert M, Suess E, Greinert J, Whiticar MJ. Archaea mediating anaerobic methane oxidation in deep-sea sediments at cold seeps of the eastern Aleutian subduction zone. Org Geochem. 2000;31:1175–1187. [Google Scholar]
- Engel AS, Lee N, Porter ML, Stern LA, Bennett PC, Wagner M. Filamentous "Epsilonproteobacteria" dominate microbial mats from sulfidic cave springs. Appl Environ Microbiol. 2003;69:5503–5511. doi: 10.1128/AEM.69.9.5503-5511.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel AS, Porter ML, Stern LA, Quinlan S, Bennett PC. Bacterial diversity and ecosystem function of filamentous microbial mats from aphotic (cave) sulfidic springs dominated by chemolithoautotrophic "Epsilonproteobacteria". FEMS Microbiol Ecol. 2004;51:31–53. doi: 10.1016/j.femsec.2004.07.004. [DOI] [PubMed] [Google Scholar]
- Frols S, Ajon M, Wagner M, Teichmann D, Zolghadr B, Folea M, et al. UV-inducible cellular aggregation of the hyperthermophilic archaeon Sulfolobus solfataricus is mediated by pili formation. Mol Microbiol. 2008;70:938–952. doi: 10.1111/j.1365-2958.2008.06459.x. [DOI] [PubMed] [Google Scholar]
- Geets J, Borremans B, Diels L, Springael D, Vangronsveld J, van der Lelie D, et al. DsrB gene-based DGGE for community and diversity surveys of sulfate reducing bacteria. J Microbiol Methods. 2006;66:194–205. doi: 10.1016/j.mimet.2005.11.002. [DOI] [PubMed] [Google Scholar]
- Good IJ. The population frequencies of species and the estimation of population parameters. Biometrika. 1953;40:237–264. [Google Scholar]
- Goren MB. Sulfolipid I of Mycobarterium tuberculosis, strain H37Rv II. Structural studies. Biochem Biophys Acta. 1970;210:127–138. doi: 10.1016/0005-2760(70)90068-8. [DOI] [PubMed] [Google Scholar]
- Hall EK, Singer GA, Pölzl M, Hämmerle I, Schwarz C, Daims H, et al. Looking inside the box: using Raman microspectroscopy to deconstruct microbial biomass stoichiometry one cell at a time. ISME J. 2011;5:196–208. doi: 10.1038/ismej.2010.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatzenpichler R, Lebedeva EV, Spieck E, Stoecker K, Richter A, Daims H, et al. A moderately thermophilic ammonia-oxidizing crenarchaeote from a hot spring. Proc Natl Acad Sci USA. 2008;105:2134–2139. doi: 10.1073/pnas.0708857105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazen TC, Dubinsky EA, DeSantis TZ, Andersen GL, Piceno YM, Singh N, et al. Deep-sea oil plume enriches indigenous oil-degrading bacteria. Science. 2010;330:204–208. doi: 10.1126/science.1195979. [DOI] [PubMed] [Google Scholar]
- Heidelberg JF, Seshadri R, Haveman SA, Hemme CL, Paulsen IT, Kolonay JF, et al. The genome sequence of the anaerobic, sulfate-reducing bacterium Desulfovibrio vulgaris Hildenborough. Nat Biotechnol. 2004;22:554–559. doi: 10.1038/nbt959. [DOI] [PubMed] [Google Scholar]
- Henneberger R, Moissl C, Amann T, Rudolph C, Huber R. New insights into the lifestyle of the cold-loving SM1 euryarchaeon: natural growth as a monospecies biofilm in the subsurface. Appl Environ Microbiol. 2006;72:192–199. doi: 10.1128/AEM.72.1.192-199.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holman HY, Bechtel HA, Hao Z, Martin MC. Synchrotron IR spectromicroscopy: chemistry of living cells. Anal Chem. 2010;82:8757–8765. doi: 10.1021/ac100991d. [DOI] [PubMed] [Google Scholar]
- Holman HY, Wozei E, Lin Z, Comolli LR, Ball DA, Borglin S, et al. Real-time molecular monitoring of chemical environment in obligate anaerobes during oxygen adaptive response. Proc Natl Acad Sci USA. 2009;106:12599–12604. doi: 10.1073/pnas.0902070106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang CK, Kerr PF. Infrared study of the carbonate minerals. The American Mineralogist. 1960;45:311–324. [Google Scholar]
- Huang WE, Stoecker K, Griffiths R, Newbold L, Daims H, Whiteley AS, et al. Raman-FISH: combining stable-isotope Raman spectroscopy and fluorescence in situ hybridization for the single cell analysis of identity and function. Environ Microbiol. 2007;9:1878–1889. doi: 10.1111/j.1462-2920.2007.01352.x. [DOI] [PubMed] [Google Scholar]
- Karner MB, DeLong EF, Karl DM. Archaeal dominance in the mesopelagic zone of the Pacific Ocean. Nature. 2001;409:507–510. doi: 10.1038/35054051. [DOI] [PubMed] [Google Scholar]
- Klappenbach JA, Saxman PR, Cole JR, Schmidt TM. rrndb: the ribosomal RNA operon copy number database. Nucleic Acids Res. 2001;29:181–184. doi: 10.1093/nar/29.1.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kodama Y, Watanabe K. Sulfuricurvum kujiense gen. nov., sp. nov., a facultatively anaerobic, chemolithoautotrophic, sulfur-oxidizing bacterium isolated from an underground crude-oil storage cavity. Int J Syst Evol Microbiol. 2004;54:2297–2300. doi: 10.1099/ijs.0.63243-0. [DOI] [PubMed] [Google Scholar]
- Lane DJ.199116S/23S rRNA sequencingIn: Stackebrandt E, Goodfellow M (eds)Nucleic Acid Techniques in Bacterial Systematics John Wiley & Sons: Chichester; 115–175. [Google Scholar]
- Lapaglia C, Hartzell PL. Stress-Induced production of biofilm in the hyperthermophile Archaeoglobus fulgidus. Appl Environ Microbiol. 1997;63:3158–3163. doi: 10.1128/aem.63.8.3158-3163.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin JM, Strohl WR. Beggiatoa, Thiothrix, and Thioploca. Annu Rev Microbiol. 1983;37:341–367. doi: 10.1146/annurev.mi.37.100183.002013. [DOI] [PubMed] [Google Scholar]
- Ledoux RL, White JL. Infrared study of the OH groups in expanded kaoline. Science. 1964;143:244–246. doi: 10.1126/science.143.3603.244. [DOI] [PubMed] [Google Scholar]
- Lee ZM, Bussema C, Schmidt TM. rrnDB: documenting the number of rRNA and tRNA genes in bacteria and archaea. Nucleic Acids Res. 2009;37:D489–D493. doi: 10.1093/nar/gkn689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic I, Bork P. Interactive Tree Of Life (iTOL): an online tool for phylogenetic tree display and annotation. Bioinformatics. 2007;23:127–128. doi: 10.1093/bioinformatics/btl529. [DOI] [PubMed] [Google Scholar]
- Li M, Canniffe DP, Jackson PJ, Davison PA, FitzGerald S, Dickman MJ, et al. Rapid resonance Raman microspectroscopy to probe carbon dioxide fixation by single cells in microbial communities. ISME J. 2012;6:875–885. doi: 10.1038/ismej.2011.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovley DR, Klug MJ. Sulfate reducers can outcompete methanogens at freshwater sulfate concentrations. Appl Environ Microbiol. 1983;45:187–192. doi: 10.1128/aem.45.1.187-192.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loy A, Lehner A, Lee N, Adamczyk J, Meier H, Ernst J, et al. Oligonucleotide microarray for 16S rRNA gene-based detection of all recognized lineages of sulfate-reducing prokaryotes in the environment. Appl Environ Microbiol. 2002;68:5064–5081. doi: 10.1128/AEM.68.10.5064-5081.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig W, Strunk O, Westram R, Richter L, Yadhukumar MeierH, et al. ARB: a software environment for sequence data. Nucleic Acids Res. 2004;32:1363–1371. doi: 10.1093/nar/gkh293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancuso CA, Nichols PD, White DC. A method for the separation and characterization of archaebacterial signature ether lipids. J Lipid Res. 1986;27:49–56. [PubMed] [Google Scholar]
- Manders EEM, Verbeek FJ, Aten JA. Measurement of co-localization of objects in dual-colour confocal images. J Microsc. 1993;169:375–382. doi: 10.1111/j.1365-2818.1993.tb03313.x. [DOI] [PubMed] [Google Scholar]
- Mayers GL, Pausada M, Haines TH. Microbial Sulfolipids. III. The dislfate of (+)-1,14-Docosanediol in Ochromonas danica. Biochemistry. 1969;8:2981–2986. doi: 10.1021/bi00835a045. [DOI] [PubMed] [Google Scholar]
- Moissl C, Rachel R, Briegel A, Engelhardt H, Huber R. The unique structure of archaeal 'hami', highly complex cell appendages with nano-grappling hooks. Mol Microbiol. 2005;56:361–370. doi: 10.1111/j.1365-2958.2005.04294.x. [DOI] [PubMed] [Google Scholar]
- Moissl C, Rudolph C, Huber R. Natural communities of novel archaea and bacteria with a string-of-pearls-like morphology: molecular analysis of the bacterial partners. Appl Environ Microbiol. 2002;68:933–937. doi: 10.1128/AEM.68.2.933-937.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moissl C, Rudolph C, Rachel R, Koch M, Huber R. In situ growth of the novel SM1 euryarchaeon from a string-of-pearls-like microbial community in its cold biotope, its physical separation and insights into its structure and physiology. Arch Microbiol. 2003;180:211–217. doi: 10.1007/s00203-003-0580-1. [DOI] [PubMed] [Google Scholar]
- Moissl-Eichinger C. Archaea in artificial environments: their presence in global spacecraft clean rooms and impact on planetary protection. ISME J. 2011;5:209–219. doi: 10.1038/ismej.2010.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moissl-Eichinger C, Huber H. Archaeal symbionts and parasites. Curr Opin Microbiol. 2011;14:364–370. doi: 10.1016/j.mib.2011.04.016. [DOI] [PubMed] [Google Scholar]
- Nauman D. Infrared Spectroscopy in Microbiology. John Wiley & Sons, Ltd.: Chichester; 2000. [Google Scholar]
- Onstott TC, Colwell FS, Kieft TL, Murdoch L, Phelps TJ. New horizons for deep subsurface microbiology. Microbe. 2009;4:499–505. [Google Scholar]
- Orphan VJ, House CH, Hinrichs KU, McKeegan KD, DeLong EF. Methane-consuming archaea revealed by directly coupled isotopic and phylogenetic analysis. Science. 2001;293:484–487. doi: 10.1126/science.1061338. [DOI] [PubMed] [Google Scholar]
- Palmer AN. Origin and morphology of limestone caves. Geol Soc Am Bull. 1991;103:1–21. [Google Scholar]
- Parker FS.1983Applications of Infrared, Raman, and Resonance Raman Spectroscopy in Biochemistry Plenum Press: New York; 550pp. [Google Scholar]
- Peak D, Ford RG, Sparks DL. An in situ ATR-FTIR investigation of sulfate bonding mechanisms on goethite. J Colloid Interface Sci. 1999;218:289–299. doi: 10.1006/jcis.1999.6405. [DOI] [PubMed] [Google Scholar]
- Percival E, Wold JK. The acid polysaccharide from the green seaweed Ulva lactuca. Part II. The site of the ester sulphate. J Chem Soc. 1963. pp. 5459–5468.
- Pruesse E, Quast C, Knittel K, Fuchs BM, Ludwig W, Peplies J, et al. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 2007;35:7188–7196. doi: 10.1093/nar/gkm864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudolph C.2003. Molekularbiologische Untersuchungen zur Verbreitung und Physiologie neuartiger, unkultivierter Archaeen in kalten Schwefelquellen. Dissertation, University of Regensburg.
- Rudolph C, Moissl C, Henneberger R, Huber R. Ecology and microbial structures of archaeal/bacterial strings-of-pearls communities and archaeal relatives thriving in cold sulfidic springs. FEMS Microbiol Ecol. 2004;50:1–11. doi: 10.1016/j.femsec.2004.05.006. [DOI] [PubMed] [Google Scholar]
- Rudolph C, Wanner G, Huber R. Natural communities of novel archaea and bacteria growing in cold sulfurous springs with a string-of-pearls-like morphology. Appl Environ Microbiol. 2001;67:2336–2344. doi: 10.1128/AEM.67.5.2336-2344.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol. 2009;75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinclair RG, McKay AF, Myers GS, Norman Jones R. The infrared absorption spectra of unsaturaed fatty acids and esters. J Am Chem Soc. 1952;74:2578–2585. [Google Scholar]
- Smith B. Infrared Spectral Interpretation - A Systematic Approach. CRC Press: Washington, DC; 1999. [Google Scholar]
- Sprott GD. Structures of archaebacterial membrane lipids. J Bioenerg Biomembr. 1992;24:555–566. doi: 10.1007/BF00762348. [DOI] [PubMed] [Google Scholar]
- Stellmach J. Fluorescent redox dyes 1. Production of fluorescent formazan by unstimulated and phorbal esteror digitonin-stimulated Ehrlich ascites tumor cells. Histochemistry. 1984;80:137–143. doi: 10.1007/BF00679987. [DOI] [PubMed] [Google Scholar]
- Stellmach J, Severin E. A fluorescent redox dye. Influence of several substrates and electron carriers on the tetrazolium salt-formazan reaction of Ehrlich ascites tumor cells. Histochem J. 1986;19:21–26. doi: 10.1007/BF01675289. [DOI] [PubMed] [Google Scholar]
- Sturt HF, Summons RE, Smith K, Elvert M, Hinrichs KU. Intact polar membrane lipids in prokaryotes and sediments deciphered by high-performance liquid chromatography/electrospray ionization multistage mass spectrometry--new biomarkers for biogeochemistry and microbial ecology. Rapid Commun Mass Spectrom. 2004;18:617–628. doi: 10.1002/rcm.1378. [DOI] [PubMed] [Google Scholar]
- Takano B. Geochemical implications of sulfate in sedimentary carbonates. Chem Geol. 1985;49:393–403. [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Tillett D, Neilan BA. Xanthogenate nucleic acid isolation from cultured and environmental cyanobacteria. J Phycol. 2000;36:251–258. [Google Scholar]
- Tourna M, Stieglmeier M, Spang A, Konneke M, Schintlmeister A, Urich T, et al. Nitrososphaera viennensis, an ammonia oxidizing archaeon from soil. Proc Natl Acad Sci USA. 2011;108:8420–8425. doi: 10.1073/pnas.1013488108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyson GW, Chapman J, Hugenholtz P, Allen EE, Ram RJ, Richardson PM, et al. Community structure and metabolism through reconstruction of microbial genomes from the environment. Nature. 2004;428:37–43. doi: 10.1038/nature02340. [DOI] [PubMed] [Google Scholar]
- Vaneechoutte M, Rossau R, De Vos P, Gillis M, Janssens D, Paepe N, et al. Rapid identification of bacteria of the Comamonadaceae with amplified ribosomal DNA-restriction analysis (ARDRA) FEMS Microbiol Lett. 1992;72:227–233. doi: 10.1111/j.1574-6968.1992.tb05102.x. [DOI] [PubMed] [Google Scholar]
- Wagner M. Single-cell ecophysiology of microbes as revealed by raman microspectroscopy or secondary ion mass spectrometry imaging. Annu Rev Microbiol. 2009;63:411–429. doi: 10.1146/annurev.micro.091208.073233. [DOI] [PubMed] [Google Scholar]
- Wagner M, Roger AJ, Flax JL, Brusseau GA, Stahl DA. Phylogeny of dissimlatory sulfite reductases supports an early origin of sulfate respiration. J Bacteriol. 1998;180:2975–2982. doi: 10.1128/jb.180.11.2975-2982.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker CB, de la Torre JR, Klotz MG, Urakawa H, Pinel N, Arp DJ, et al. Nitrosopumilus maritimus genome reveals unique mechanisms for nitrification and autotrophy in globally distributed marine crenarchaea. Proc Natl Acad Sci USA. 2010;107:8818–8823. doi: 10.1073/pnas.0913533107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallner G, Amann R, Beisker W. Optimizing fluorescent in situ hybridization with rRNA-targeted oligonucleotide probes for flow cytometric identification of microorganisms. Cytometry. 1993;14:16–143. doi: 10.1002/cyto.990140205. [DOI] [PubMed] [Google Scholar]
- White WB. The Carbonated Minerals. Mineralogical Society: London; 1974. [Google Scholar]
- Whittaker P, Mossoba MM, Al-Khaldi S, Fry FS, Dunkel VC, Tall BD, et al. Identification of foodborne bacteria by infrared spectroscopy using cellular fatty acid methyl esters. J Microbiol Methods. 2003;55:709–716. doi: 10.1016/j.mimet.2003.07.005. [DOI] [PubMed] [Google Scholar]
- Yoshida N, Hiraishi A. An improved redox dye-staining method using 5-cyano-2,3-ditoryl tetrazolium chloride for detection of metabolically active bacteria in activated sludge. Microbes Environ. 2004;19:61–70. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.