Abstract
Viruses are abundant in the ocean and a major driving force in plankton ecology and evolution. It has been assumed that most of the viruses in seawater contain DNA and infect bacteria, but RNA-containing viruses in the ocean, which almost exclusively infect eukaryotes, have never been quantified. We compared the total mass of RNA and DNA in the viral fraction harvested from seawater and using data on the mass of nucleic acid per RNA- or DNA-containing virion, estimated the abundances of each. Our data suggest that the abundance of RNA viruses rivaled or exceeded that of DNA viruses in samples of coastal seawater. The dominant RNA viruses in the samples were marine picorna-like viruses, which have small genomes and are at or below the detection limit of common fluorescence-based counting methods. If our results are typical, this means that counts of viruses and the rate measurements that depend on them, such as viral production, are significantly underestimated by current practices. As these RNA viruses infect eukaryotes, our data imply that protists contribute more to marine viral dynamics than one might expect based on their relatively low abundance. This conclusion is a departure from the prevailing view of viruses in the ocean, but is consistent with earlier theoretical predictions.
Keywords: RNA viruses, DNA viruses, virioplankton, marine, seawater, abundance, metagenome
Introduction
Microbial ecologists have devoted considerable effort to understanding the nature of the viruses in seawater, because viruses have key roles in the evolution, ecology and mortality of marine plankton (Rohwer and Vega Thurber, 2007; Suttle, 2007). For at least the past two decades, researchers have assumed that the pool of viruses in the ocean is dominated by bacteriophages with DNA genomes (Steward et al., 1992; Breitbart et al., 2002; Weinbauer, 2004; Comeau et al., 2010; Sullivan et al., 2010). Perhaps as a consequence, studies of the molecular diversity of marine viruses have most commonly (exclusively, before 2003) focused on DNA viruses (Edwards and Rohwer, 2005; Kristensen et al., 2010). However, evidence that RNA viruses are important contributors to marine plankton ecology has been steadily accumulating (Lang et al., 2009).
The isolation of a positive-sense, single-stranded RNA (ssRNA) virus that infects the raphidophyte Heterosigma akashiwo (HaRNAV) was the first recorded instance of an RNA virus infecting a marine protist (Tai et al., 2003). This was followed by reports of similar picorna-like ssRNA viruses infecting diatoms (Nagasaki et al., 2004; Shirai et al., 2008; Tomaru et al., 2009), and a thraustochytrid (Takao et al., 2005). All of these viruses are now classified as members of the order Picornavirales. Molecular surveys using degenerate primers to target the RNA-dependent RNA polymerase gene of picorna-like viruses have shown that, in addition to the handful of isolates, a very diverse pool of uncultivated picornavirads exist in seawater (Culley et al., 2003; Culley and Steward, 2007).
Other novel RNA viruses infecting marine protists have also been isolated. A double-stranded RNA virus infecting the abundant marine prymnesiophyte Micromonas pusilla represents a new genus in the family Reoviridae (Brussaard et al., 2004). A positive-sense, ssRNA virus that infects the dinoflagellate Heterocapsa circularisquama (Tomaru et al., 2004) is only distantly related to existing viral families (Nagasaki et al., 2005) and may represent a new family. Metagenomic surveys of the RNA viral fraction of seawater are consistent with the view provided by the limited number of isolates available so far, and suggest that the positive-sense, single-stranded picornavirads dominate the marine RNA virus pool, but other diverse RNA viruses, including some containing double-stranded RNA, are present as well (Culley et al., 2006).
Despite the emerging evidence that marine RNA viruses are diverse and infect ecologically important members of the marine planktonic food web, there have been no reports that satisfactorily address the question of whether these viruses constitute a substantial fraction of the total virioplankton. This question is of considerable ecological interest, because, unlike the pool of DNA viruses in seawater, which is composed predominantly of bacteriophage-like sequences (Edwards and Rohwer, 2005), marine RNA viruses are almost exclusively comprised of those that infect eukaryotes (Lang et al., 2009).
Directly quantifying the abundance of RNA viruses in a mixed viral assemblage has proven difficult, because of technical limitations. Differences in counts using a DNA-specific stain vs a nonspecific nucleic acid stain have been reported (Weinbauer and Suttle, 1997; Guixa-Boixereu et al., 1999; Bettarel et al., 2000), but the results are of uncertain significance, because the stains differ in their sensitivity (Weinbauer, 2004). In particular, the small genomes of single-stranded DNA and RNA viruses make the individual virions difficult to detect even with appropriate stains (Brussaard et al., 2000; Tomaru and Nagasaki, 2007; Holmfeldt et al., 2012).
Given the difficulties of obtaining reliable direct counts of RNA viruses, we took a different approach. In the work reported here, we measured the relative masses of RNA and DNA in natural assemblages of viruses purified from tropical coastal seawater and coupled this with estimates of the mass of nucleic acid per RNA or DNA virion to obtain the first estimates of the relative abundance of RNA viruses in seawater.
Materials and methods
Study site
The samples for this study were collected from a pier in the southern portion of Kāne'ohe Bay on the windward side of O'ahu, Hawai‘i (21° 25′ 46.80″ N, 157° 47′ 31.51″ W). This tropical embayment is characterized by year-round warm temperatures (22 °C to 28 °C) and salinities ranging from 32 to 35, except during periods of heavy rain, when freshwater plumes from stream runoff can transiently suppress salinity to <30 (Drupp et al., 2011). Concentrations of chlorophyll a in surface waters are typically low with average values recorded over a 2.5-year period ranging from 0.5 to 1.1 μg l−1 in summer and from 1.3 to 3.4 μg l−1 in the winter depending on location (Drupp et al., 2011). Following heavy rainfall, blooms have been reported with transient increases in chlorophyll a to ca 6 μg l−1 (Hoover et al., 2006; De Carlo et al., 2007).
Sample collection and processing
Seawater was collected in acid-washed, polycarbonate carboys on 1 August 2009 (35 liters) and on 3 June 2010 (80 liters) during non-storm conditions. Seawater was transported immediately to the laboratory (<1 h) and filtered through 0.22 μm pore-size filters (Sterivex, Millipore, Billerica, MA, USA). The sample from June was split and processed as two parallel subsamples (40 liters each). Viruses in the filtrates were concentrated by iron flocculation (John et al., 2011) then concentrated further in a centrifugal 30 kDa ultrafiltration device (Amicon 15, Millipore). Viruses in each concentrate were purified by a two-step process in CsCl buoyant density gradients (Lawrence and Steward, 2010). Fractions (ca 0.5 ml each) were collected from the final gradient for analysis with an Auto Densi-Flow (Labconco, Kansas City, MO, USA).
Fraction analysis
The density of each fraction was measured using a micropipet and an analytical balance (Lawrence and Steward, 2010). CsCl was exchanged with buffer (10 mℳ Tris, 1 mℳ EDTA, pH 8) by centrifugal ultrafiltration (Steward, 2001) and total nucleic acids in a subsample of each fraction was extracted using spin columns (QIAamp MinElute Viral Spin Kit, Qiagen, Valencia, CA, USA) according to the manufacturer's instructions. Samples for RNA analysis were treated twice with DNase (TURBO DNase, Life Technologies, Carlsbad, CA, USA) to avoid nonspecific signal. RNA and DNA contents of each fraction were measured separately in parallel subsamples by fluorometry (Quant-iT DNA and RNA assays, Life Technologies) in a cuvette fluorometer (TD-700, Turner Designs, Sunnyvale, CA, USA). The RNA content of the individual fractions from the June sample was low, so we also measured the RNA content of the pooled putative viral fractions to obtain a more accurate estimate. The nucleic acid masses (±95% confidence interval (CI)) were calculated by inverse prediction from the standard curves (Zar, 1996).
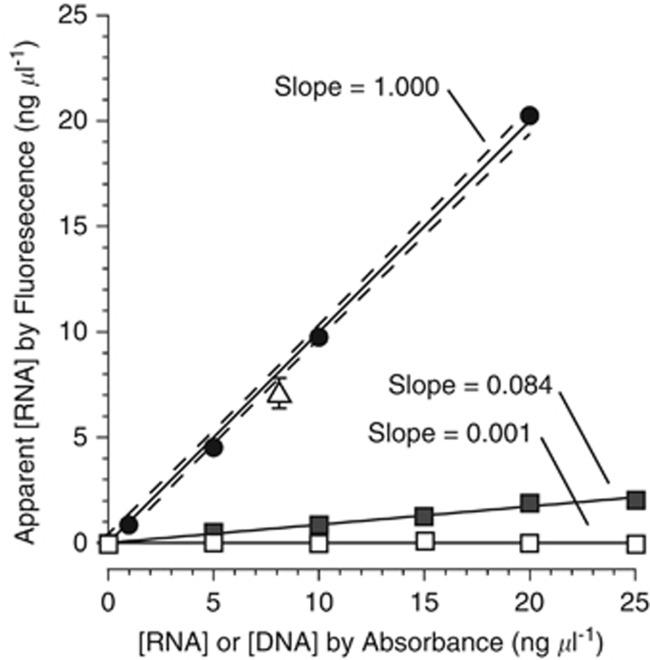
The fluorometric assay was necessary to achieve the required sensitivity. To ensure that we did not overestimate viral RNA with the fluorometric assay, we compared the measurements of the mass of RNA obtained from a purified RNA virus by fluorometry and by spectrophometry (Model DU 800, Beckman Coulter, Inc., Brea, CA, USA). The virus was a positive-sense, single-stranded, picornavirad that infects a marine diatom and has a genome of 8800 nucleotides (nt) (Schvarcz et al., unpublished results). We also tested for cross-reaction of DNA in the RNA fluorescence assay with and without the DNase digestion procedure described above (Figure 1). The DNA used was double-stranded genomic DNA from Enterobacteria phage lambda. The fluorometer was calibrated with the Quant-iT kit standards (Escherichia coli RNA). The RNA standards were then read as samples along with undigested and digested DNA standards and the purified viral RNA. Apparent RNA concentration for each sample as determined by fluorometry was plotted as a function of the given RNA or DNA concentrations (Quant-iT kit standards) or, in the case of the viral RNA, the concentration determined by absorption at 260 nm (Sambrook and Russell, 2001). Model I regression lines were calculated and a two-tailed t-test used to determine if the slopes were significantly different from zero (Zar, 1996).
Figure 1.
Test of the fluorescence-based RNA assay. The x axis represents concentrations of DNA or RNA as determined by absorbance of purified nucleic acid solutions at 260 nm. The y axis represents the apparent RNA concentration based on fluorometric signal after calibration with the RNA standards in the Quant-iT kit (Life Technologies). Closed circles are the E. coli RNA standards provided with the kit and which were used to calibrate the fluorometer. The open triangle is purified genomic RNA from a ssRNA virus that infects a marine diatom (mean±95% CI). Squares are dilutions of Enterobacteria phage lambda genomic DNA (double-stranded DNA (dsDNA)). Closed squares are undigested lambda DNA, and the open squares lambda DNA digested with TURBO DNase. Slopes of the model I linear regressions (solid lines) are noted above each line. Dashed lines represent the 95% confidence bands for the RNA standards.
Metagenomic analysis
To determine the composition of the RNA in the selected RNA peaks, we created and analyzed metagenomes from this material for the August sample and from one of the duplicate subsamples (replicate 1) from June. Purified, DNase-treated RNA from the indicated fractions (Figure 2) of each sample was pooled and amplified using random priming-mediated sequence-independent single-primer amplification as described previously (Djikeng et al., 2008; Culley et al., 2010). Tests of this method on RNA viral genomes indicated that it results in coverage and redundancy similar to the ideal values that are predicted by the Lander–Waterman model (Djikeng et al., 2008), which suggests amplification biases are limited. Sequence libraries from the resulting amplified complementary DNA samples were produced by pyrosequencing (GS FLX Titanium, 454 Life Sciences, Branford, CT, USA). Sequences that were >2 s.d. from the mean length or had an average phred score of <15 were discarded using the MG-RAST QC pipeline (http://metagenomics.anl.gov/) and sequence-independent single-primer amplification primers were trimmed from the remaining reads. Artificial (or technical) replicates (Gomez-Alvarez et al., 2009) were removed using the online tool MG-RAST (Meyer et al., 2008).
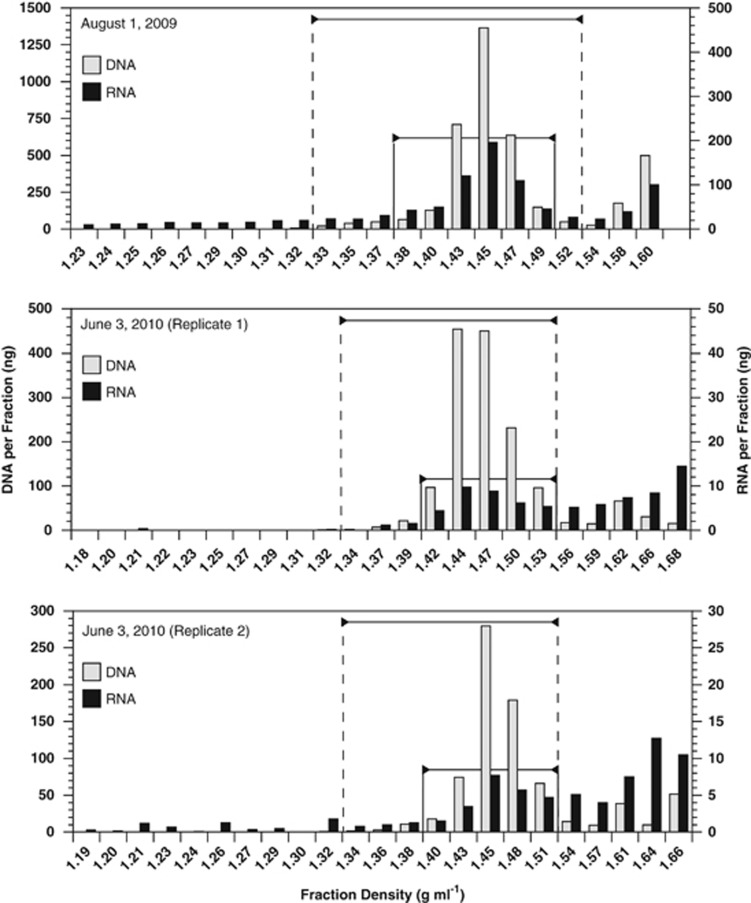
Figure 2.
Distribution of nucleic acids after separation of viral concentrates in CsCl buoyant density gradients. DNA and RNA per fraction for samples collected on 1 August 2009 (top panel) and the duplicate samples from 3 June 2010 (middle and bottom panels) is shown. DNA in the fractions between the dashed lines and RNA in the fractions between the solid lines was considered viral.
Processed sequences were assembled with CLC Genomics Workbench version 5.0 (CLCbio, Cambridge, MA, USA) using global alignment with automatic word and bubble sizes, a minimum contig length of 200, mismatch, insertion and deletion costs set to 3, length fraction set to 0.5, and the similarity threshold set to 0.8. The community composition of each sample was analyzed with MEGAN (Huson et al., 2011) using the output from blastx comparisons (Altschul et al., 1990) of the assembled metagenomes (contigs plus singletons) with the non-redundant NCBI sequence database. The threshold E-value for considering a hit to be significant was 10−5. The taxonomic assignment for a given contig was applied to all reads comprising that contig. MEGAN assignments were manually checked and some reassignments made in the case of annotation errors. As an independent check on possible contamination from cellular RNA, sequences were compared with the SILVA database (Pruesse et al., 2007) using blastn to search for ribosomal RNA contamination, which is expected to be the major source of cellular RNA contamination (Karpinets et al., 2006). The mass of RNA in the pooled viral fraction for each library was adjusted downward by the percentage of reads identified as being cell derived.
Calculation of DNA and RNA genome copies
To estimate the number of RNA virus genomes, we first summed the RNA mass within a narrow buoyant density range (between the solid vertical lines, Figure 2). Although RNA viruses can have densities outside of that range, we used this conservative window because we analyzed the RNA in only those fractions by metagenomic analysis. Using the relative representation of different taxa in the metagenomic library and the average genome sizes for those taxa derived from data in the Ninth report of the International Committee on the Taxonomy of Viruses (King et al., 2012), we calculated a weighted average RNA mass per virion. For our prima facie estimates of the RNA virus contribution, we assumed that the sequences having no significant hit to any sequences in GenBank (either directly or by association with other sequences in a contig) had the same taxonomic distribution as the collection of sequences that did have significant BLAST hits. The portion of the total RNA determined to be viral was then divided by the weighted average mass of RNA per virion to obtain the number of RNA-containing viruses. Errors were calculated (and propagated) as the 95% CI of the inverse predictions of RNA mass from the standard curves.
To estimate abundance of DNA viruses, we summed the DNA content in the fractions encompassing the main DNA peak in the viral buoyant density range (between the dashed vertical lines, Figure 2) and converted the total DNA mass to numbers of DNA viruses assuming an average DNA content per virion of 5.5 × 10−17 g (equivalent to 50 kb double-stranded DNA), an average that was found to be similar in a wide range of environments (Steward et al., 2000). This value is conservative compared with some other estimates (Brum, 2005; Angly et al., 2009) and therefore potentially overestimates the DNA virus abundance.
We also calculated the contribution of RNA viruses using more extreme assumptions to get a sense of how much higher and lower the percentage might be. For the high estimate, we assumed that the DNA per virion was twofold greater (100 kb, double-stranded DNA) as calculated for marine viral metagenomic data (Angly et al., 2009). For the low estimate, we assumed an average DNA content per virion twofold lower than expected (25 kb, double-stranded DNA), and conservatively assumed that any RNA sequences with no significant match in GenBank were not viral (which reduced the total mass of viral RNA by about half).
Results and Discussion
To achieve the sensitivity necessary to assay the nucleic acids in our CsCl-purified viral fractions, we had to employ fluorometry. We performed simple tests of the RNA assay kit to ensure that it would provide reasonable estimates of viral RNA concentration, and that our measurements would not be overestimated by cross-reaction from DNA. With the fluorometer calibrated to the kit standards, the standards themselves result in a linear curve with a significant slope of 1.000 as expected (P<0.001). The assay of a purified ssRNA virus falls very close to the calibration line (low by 14%) suggesting that the kit standards (dilutions of E. coli RNA) are reasonably accurate for quantifying a ssRNA viral genome (Figure 1). DNA was found to have a limited cross-reaction with the RNA stain with a significant (P<0.001) slope of 0.084, which suggests a cross-reaction of 8.4% on average. However, this cross-reaction was effectively removed (slope statistically indistinguishable from zero; P>0.5) by the DNase treatment that we applied to all of our samples before RNA assay (Figure 1).
The majority of DNA (83–90%, depending on the sample) fell within a range of buoyant densities from ⩾1.33 to ⩽1.53 g ml−1 with a peak around 1.45 g ml−1 (Figure 2). This buoyant density range and the peak location are similar to previous observations for DNA-containing viruses from other marine environments (Steward et al., 2000) and is well within the range for all known viruses (1.16–1.6; King et al., 2012). RNA concentrations displayed a local peak at the same density with 32–69% of the total in the same viral range. DNA and RNA were also found in higher density fractions nearer to, or at, the bottom of the gradient. As these fractions are outside the known range of viral buoyant densities, data from them were not included in subsequent analyses. This resulted in the exclusion of a higher percentage of the RNA than DNA.
For the metagenomic libraries prepared from RNA within the viral density range (Figure 2), the number of quality-controlled, de-replicated reads was 139 801 and 110 140 for the 2009 and 2010 samples, respectively (Table 1). The majority of reads (69–78%) in both libraries formed contigs with maximum lengths of 9378 bp for August and 9480 bp for June. We found that, after assembly and classification of the sequences in the RNA viral metagenomes, roughly half (50–57%) of the reads derived from the designated viral fractions were most similar to known eukaryote-infecting RNA viruses, with the majority matching positive-sense, ssRNA viruses in the order Picornavirales (Table 2). The percentage of sequences matching known double-stranded RNA viruses was very small (0.02–1.2%). The calculated weighted average mass of RNA per virion was 5.38 × 10−18 g in August and 5.25 × 10−18 g in June (Table 2), which translate into weighted average genome sizes of 9528 nt and 9301 nt, respectively. The average of these (9414 nt) is very similar to the maximum contig lengths observed in each library and somewhat larger than the three complete RNA virus genomes (ranging from 4449 to 9212 nt) assembled from a sample from coastal British Columbia (Culley et al., 2007).
Table 1. Number and length (in nucleotides, nt) of initial quality-controlled reads and of contigs after assembly for the August 2009 and the June 2010 libraries, % G+C content of each library, and the percentage of reads in each library that were matched with at least one other sequence to form a contig.
|
Length (nt) |
G+C (%) | Matched (%) | |||||
|---|---|---|---|---|---|---|---|
| Count | Mean | Min | Max | Total nt | |||
| 2009 | |||||||
| Reads | 139 801 | 349 | 54 | 591 | 48 817 678 | 42.8 | 69 |
| Contigs | 3552 | 578 | 47 | 9378 | 2 054 043 | ||
| 2010 | |||||||
| Reads | 110 140 | 361 | 134 | 555 | 39 811 849 | 43.8 | 78 |
| Contigs | 1738 | 600 | 136 | 9480 | 1 043 483 | ||
Table 2. The percentage of reads in each of two libraries (August 2009 and June 2010) that matched known viruses, that had no significant match in the sequence databases, or were most similar to sequences derived from cells.
|
Viral classification |
% Of all sequences |
% Of viral sequences |
Genome length (nt) | RNA mass per virion (ag) |
Weighted RNA mass per virion (ag) |
|||||
|---|---|---|---|---|---|---|---|---|---|---|
| N A type | Order | Family | 2009 | 2010 | 2009 | 2010 | 2009 | 2010 | ||
| dsRNA | Unassigned | Reoviridae | 1.2 | 0.02 | 2.4 | 0.0 | 25.6 | 28.9 | ||
| (+) ssRNA | Picornavirales | Dicistroviridae | 6.5 | 8.8 | 12.9 | 15.3 | 9.3 | 5.24 | ||
| Marnaviridae | 1.5 | 0.1 | 3.0 | 0.2 | 8.6 | 4.85 | 5.38 | 5.25 | ||
| Unassigneda | 6.9 | 6.3 | 13.7 | 11.0 | 9.3 | 5.23 | ||||
| Unclassifiedb | 23.7 | 22.8 | 47.2 | 39.7 | 9.6 | 5.42 | ||||
| Unculturedc | 10.4 | 19.4 | 20.7 | 33.8 | 9.0 | 5.08 | ||||
| All other virusesd | 0.3 | 0.6 | ||||||||
| Virus total | 50.5 | 58.0 | ||||||||
| No hits | 42.8 | 41.1 | ||||||||
| Cells | 6.7 | 0.8 | ||||||||
Abbreviations: dsRNA, double-stranded RNA; NA, nucleic acid; ssRNA, single-stranded RNA.For each virus group, the mean genome length and the corresponding mass of RNA per virion is presented along with the weighted average mass per virion in attograms (10−18 g) based on the percent representation of each group in the libraries. Only those viral families that were matched by ⩾1% of the sequences in at least one of the libraries are included in the calculation of RNA mass per virion.
This group represents diatom viruses in the new genus Bacillarnavirus, which has not yet been assigned to a family.
Sequences in this group are unclassified members of the order Picornavirales. The genome length used is an average for all members of the order.
Sequences were assigned to this group if they matched sequences from uncultivated marine picorna-like viruses JP-A and JP-B or the related RNA-dependent RNA polymerase gene sequences amplified from seawater by PCR.
Includes hits to dsRNA viruses (members of the Picobirnaviridae, Totiviridae families and the Saccharomyces cerevisiae virus L-A) and to ssRNA viruses (members of the Avernaviridae, Caliciviridae, Hepeviridae, Nodaviridae, Tombusviridae, Virgaviridae families, and members of the genera Ourmiavirus, Sobemovirus).
Less than half of the reads (41–43%) had no significant matches in GenBank. We presume that most of these unidentified sequences are viral as well, because many otherwise unidentifiable sequences were found to assemble with virus-like sequences and the fraction of reads identified as cellular was small (Table 2). Estimates of cellular contamination in each library based on blastn comparisons with the Silva database (6.1% and 0.7%) were nearly identical to those based on blastx comparisons with GenBank nr database (6.7% and 0.8% of the sequences). The reads identified as cell derived by the former method were a subset of those identified by the latter. The identification of all of the putative ribosomal RNA reads by blastx was somewhat surprising, but appears to be a result of mis-annotations of ribosomal RNA genes as hypothetical proteins in GenBank. As the blastx annotations used for MEGAN were more comprehensive in the assignment of cell-derived reads, the larger percentages from that method were accepted as correct. After adjusting the RNA mass estimates to discount the contribution from cellular RNA, our estimates put the contribution of RNA viruses to the total number of viruses at 38–63% (Table 3). By applying more conservative and more liberal assumptions, we calculated extreme high and low estimates that ranged from 15% to 77% (Table 3).
Table 3. Relative abundance of RNA viruses in coastal waters.
| Date | Viral genome copies ( × 109) | RNA viruses (%) | ||
|---|---|---|---|---|
| |
RNA |
DNA |
Mean (± err) |
Range |
| 01 August 2009a | 97.7 (±3.5) | 58.6 (±5.4) | 63 (±6) | 31–77% |
| 03 June 2010b | 11.4 (±0.4) | 18.5 (±8.9) | 38 (±18) | 15–55% |
| Avg. (±s.d.) | 50 (±18) | |||
The estimated numbers of DNA and RNA viruses recovered from 35- to 40-liter samples of seawater after concentration and gradient purification are presented along with the calculated percent contributions of RNA viruses to the total viruses. Ranges are the extreme low and high estimates calculated as described in the Materials and methods section.
Errors are analytical errors propagated as±95% confidence interval (CI).
Errors are±s.d. of duplicate samples processed in parallel.
The absolute numbers for both types of viruses in our final samples are minimum estimates, because of losses during processing. Assuming typical viral concentrations for these waters based on epifluorescence microscopy to be on the order of 0.5 to 1 × 1010 per liter, this implies overall final yields of DNA viruses on the order of 17–33% (June) and 5–9% (August), but these estimates have considerable uncertainty. In the absence of data to the contrary, we assume these losses to be similar for RNA and DNA viruses. If DNA viruses were preferentially lost, this would have led to overestimates of the contribution of RNA viruses. Some of the largest viruses (especially those >0.2 μm in diameter) will be lost by the initial 0.2 μm filtration and all of the largest viruses known so far contain double-stranded DNA (King et al., 2012), suggesting that the purification procedure will have some bias against DNA viruses. However, the numerical contribution of viruses >0.2 μm to the total virioplankton appears to be low based on quantitative surveys using electron microscopy (Bratbak et al., 1992; Cochlan et al., 1993; Wommack and Colwell, 2000; Auguet et al., 2006). The viral concentration method we used has not been tested specifically on RNA viruses, but is reported to be exceptionally efficient for DNA viruses (John et al., 2011). We were also more conservative in our assignment of RNA as viral compared with DNA. From the above considerations, we feel it is unlikely that we have grossly overestimated the contribution of RNA viruses, but we cannot conclusively rule out the possibility that our procedure is significantly biased against DNA viruses.
If RNA viruses are as abundant as our data suggest, this would have important consequences for our understanding of marine viral ecology. The data imply, for example, that eukaryotic viruses can be just as abundant as bacteriophages in coastal ocean waters, despite the fact that the eukaryotic plankton concentrations are orders of magnitude lower than those of bacteria. It seems that the much larger burst sizes of eukaryotic RNA viruses (thousands to tens-of-thousands; Lang et al., 2009) relative to those of bacteriophages (tens to hundreds; Wommack and Colwell, 2000), compensates for the lower host abundances. This is consistent with earlier theoretical work based on mass transport calculations (Murray and Jackson, 1992). At the time of that report, only DNA viruses with relatively small burst sizes were known to infect marine protists. As a consequence, the authors tentatively concluded that the viruses in seawater primarily infect bacteria. They point out, however, that small viruses having a large burst size (that is, those like the RNA viruses that we now know exist), could have a large contribution to protistan mortality.
As RNA viruses and single-stranded DNA viruses are not reliably detected with the current routine methods for viral direct counts, our data also suggest that many rate estimates that depend on fluorescence-based viral direct counts, such as viral turnover times, virus–host contact rates and viral production rates, may be in need of revision. Development of new methods to directly count even the smallest viruses would be helpful in better constraining the rates of these important processes. In the meantime, the approach we described here provides a means to estimate the relative contribution of RNA viruses in natural aquatic habitats to determine whether our observations are more broadly representative of marine and freshwater habitats.
Acknowledgments
This work was supported by NSF grants to GFS and AIC (OCE 08-26650) and the Center for Microbial Oceanography Research and Education (EF 04-24599). Metagenome sequencing was supported by the Gordon and Betty Moore Foundation and conducted at the Broad Institute. Bioinformatics analyses were supported in part through the use of the COBRE/INBRE Bioinformatics core, supported jointly by NIH Grants from the National Institute of General Medical Sciences (P20GM103516 and P20GM103466). We acknowledge GO Walker for assistance with sampling, CR Schvarcz for providing an RNA virus isolate for assay testing, and Shulei Sun and the Broad Institute for assistance with the processing and sequencing of the metagenomes. Metagenomic data referred to in this paper are available at the CAMERA web site (http://camera.calit2.net/) under the accession number CAM_PROJ_BROADPHAGE and sample names CAM_SMPL_000815 (1 August 2009) and CAM_SMPL_000824 (3 June 2010).
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Angly FE, Willner D, Prieto-Davó A, Edwards RA, Schmieder R, Vega-Thurber R, et al. The GAAS metagenomic tool and its estimations of viral and microbial average genome size in four major biomes. PLoS Comput Biol. 2009;5:e1000593. doi: 10.1371/journal.pcbi.1000593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auguet JC, Montanié H, Lebaron P. Structure of virioplankton in the Charente Estuary (France): transmission electron microscopy versus pulsed field gel electrophoresis. Microb Ecol. 2006;51:197–208. doi: 10.1007/s00248-005-0043-0. [DOI] [PubMed] [Google Scholar]
- Bettarel Y, Sime-Ngando T, Amblard C, Laveran H. A comparison of methods for counting viruses in aquatic systems. Appl Environ Microbiol. 2000;66:2283–2289. doi: 10.1128/aem.66.6.2283-2289.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bratbak G, Haslund OH, Heldal M, Nœss A, Røeggen T. Giant marine viruses. Mar Ecol Progr Ser. 1992;85:201–202. [Google Scholar]
- Breitbart M, Salamon P, Andresen B, Mahaffy JM, Segall AM, Mead D, et al. Genomic analysis of uncultured marine viral communities. Proc Natl Acad Sci USA. 2002;99:14250–14255. doi: 10.1073/pnas.202488399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brum JR. Concentration, production and turnover of viruses and dissolved DNA pools at Stn ALOHA, North Pacific Subtropical Gyre. Aquat Microb Ecol. 2005;41:103–113. [Google Scholar]
- Brussaard CPD, Marie D, Bratbak G. Flow cytometric detection of viruses. J Virol Meth. 2000;85:175–182. doi: 10.1016/s0166-0934(99)00167-6. [DOI] [PubMed] [Google Scholar]
- Brussaard CPD, Noordelos AAM, Sandaa RA, Heldal M, Bratbak G. Discovery of a dsRNA virus infecting the marine phtosynthetic protist Micromonas pusilla. Virol. 2004;319:280–291. doi: 10.1016/j.virol.2003.10.033. [DOI] [PubMed] [Google Scholar]
- Cochlan WP, Wikner J, Steward GF, Smith DC, Azam F. Spatial distribution of viruses, bacteria and chlorophyll-a in neritic, oceanic and estuarine environments. Mar Ecol Progr Ser. 1993;92:77–87. [Google Scholar]
- Comeau AM, Arbiol C, Krisch HM. Gene network visualization and quantitative synteny analysis of more than 300 marine T4-like phage scaffolds from the GOS metagenome. Mol Biol Evol. 2010;27:1935–1944. doi: 10.1093/molbev/msq076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culley AI, Lang AS, Suttle CA. High diversity of unknown picorna-like viruses in the sea. Nature. 2003;424:1054–1057. doi: 10.1038/nature01886. [DOI] [PubMed] [Google Scholar]
- Culley AI, Lang AS, Suttle CA. Metagenomic analysis of coastal RNA virus communities. Science. 2006;312:1795–1798. doi: 10.1126/science.1127404. [DOI] [PubMed] [Google Scholar]
- Culley AI, Lang AS, Suttle CA. The complete genomes of three viruses assembled from shotgun libraries of marine RNA virus communities. Virol J. 2007;4:69. doi: 10.1186/1743-422X-4-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culley AI, Steward GF. New genera of RNA viruses in subtropical seawater inferred from polymerase gene sequences. Appl Environ Microbiol. 2007;73:5937–5944. doi: 10.1128/AEM.01065-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culley AI, Suttle CA, Steward GF.2010Characterization of the diversity of marine RNA virusesIn: Wilhelm SW, Weinbauer MG, Suttle CA, (eds)Manual of Aquatic Viral Ecology American Society of Limnology and Oceanography: Waco, TX; 193–201. [Google Scholar]
- De Carlo EH, Hoover DJ, Young CW, Hoover RS, Mackenzie FT. Impact of storm runoff from tropical watersheds on coastal water quality and productivity. Appl Geochem. 2007;22:1777–1797. [Google Scholar]
- Djikeng A, Halpin R, Kuzmickas R, Depasse J, Feldblyum J, Sengamalay N, et al. Viral genome sequencing by random priming methods. BMC Genomics. 2008;9:5. doi: 10.1186/1471-2164-9-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drupp P, De Carlo EH, Mackenzie FT, Bienfang P, Sabine CL. Nutrient inputs, phytoplankton response, and CO2 variations in a semi-enclosed subtropical embayment, Kaneohe Bay, Hawaii. Aquat Geochem. 2011;17:473–498. [Google Scholar]
- Edwards RA, Rohwer F. Viral metagenomics. Nat Rev Microbiol. 2005;3:504–510. doi: 10.1038/nrmicro1163. [DOI] [PubMed] [Google Scholar]
- Gomez-Alvarez V, Teal TK, Schmidt TM. Systematic artifacts in metagenomes from complex microbial communities. ISME J. 2009;3:1314–1317. doi: 10.1038/ismej.2009.72. [DOI] [PubMed] [Google Scholar]
- Guixa-Boixereu N, Lysnes K, Pédros-Alió C. Viral lysis and bacterivory during a phytoplankton bloom in a coastal water microcosm. Appl Environ Microbiol. 1999;65:1949–1958. doi: 10.1128/aem.65.5.1949-1958.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmfeldt K, Odić D, Sullivan MB, Middelboe M, Riemann L. Cultivated single-stranded DNA phages that infect marine Bacteroidetes prove difficult to detect with DNA-binding stains. Appl Environ Microbiol. 2012;78:892–894. doi: 10.1128/AEM.06580-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoover RS, Hoover D, Miller M, Landry MR, DeCarlo EH, Mackenzie FT. Zooplankton response to storm runoff in a tropical estuary: bottom-up and top-down controls. Mar Ecol Progr Ser. 2006;318:187–201. [Google Scholar]
- Huson DH, Mitra S, Ruscheweyh H-J, Weber N, Schuster SC. Integrative analysis of environmental sequences using MEGAN4. Genome Res. 2011;21:1552–1560. doi: 10.1101/gr.120618.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- John SG, Mendez CB, Deng L, Poulos B, Kauffman AKM, Kern S, et al. A simple and efficient method for concentration of ocean viruses by chemical flocculation. Environ Microbiol Rep. 2011;3:195–202. doi: 10.1111/j.1758-2229.2010.00208.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpinets TV, Greenwood DJ, Sams CE, Ammons JT. RNA: protein ratio of the unicellular organism as a characteristic of phosphorous and nitrogen stoichiometry and of the cellular requirement of ribosomes for protein synthesis. BMC Biol. 2006;4:30. doi: 10.1186/1741-7007-4-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ.(eds) (2012Virus Taxonomy. Ninth report of the International Committee on Taxonomy of Viruses Elsevier: New York; 1327pp. [Google Scholar]
- Kristensen DM, Mushegian AR, Dolja VV, Koonin EV. New dimensions of the virus world discovered through metagenomics. Trends Microbiol. 2010;18:11–19. doi: 10.1016/j.tim.2009.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang AS, Rise ML, Culley AI, Steward GF. RNA viruses in the sea. FEMS Microbiol Rev. 2009;33:295–323. doi: 10.1111/j.1574-6976.2008.00132.x. [DOI] [PubMed] [Google Scholar]
- Lawrence JE, Steward GF.2010Purification of viruses by centrifugationIn: Wilhelm SW, Weinbauer MG, Suttle CA, (eds)Manual of Aquatic Viral Ecology American Society of Limnology and Oceanography: Waco, TX; 166–181. [Google Scholar]
- Meyer F, Paarmann D, D'Souza M, Olson R, Glass EM, Kubal M, et al. The metagenomics RAST server–a public resource for the automatic phylogenetic and functional analysis of metagenomes. BMC Bioinformatics. 2008;9:386. doi: 10.1186/1471-2105-9-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray AG, Jackson GS. Viral dynamics: a model of the effects of size, shape, motion and abundance of single-celled planktonic organisms and other particles. Mar Ecol Progr Ser. 1992;89:103–116. [Google Scholar]
- Nagasaki K, Tomaru Y, Katanozaka N, Shirai Y, Nishida K, Itakura S, et al. Isolation and characterization of a novel single-stranded RNA virus infecting the bloom-forming diatom Rhizosolenia setigera. Appl Environ Microbiol. 2004;70:704–711. doi: 10.1128/AEM.70.2.704-711.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagasaki K, Shirai Y, Takao Y, Mizumoto H, Nishida K, Tomaru Y. Comparison of genome sequences of single-stranded RNA viruses infecting the bivalve-killing dinoflagellate Heterocapsa circularisquama. Appl Environ Microbiol. 2005;71:8888–8894. doi: 10.1128/AEM.71.12.8888-8894.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruesse E, Quast C, Knittel K, Fuchs BM, Ludwig W, Peplies J, et al. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucl Acids Res. 2007;35:7188–7196. doi: 10.1093/nar/gkm864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohwer F, Vega Thurber R. Viruses manipulate the marine environment. Nature. 2007;459:207–212. doi: 10.1038/nature08060. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Russell DW.2001Molecular Cloning: A laboratory manual3rd edn.Cold Spring Harbor Laboratory Press: Cold Spring Harbor; 2.56–52.58. [Google Scholar]
- Shirai Y, Tomaru Y, Takao Y, Suzuki H, Nagumo T, Nagasaki K. Isolation and characterization of a single-stranded RNA virus infecting the marine planktonic diatom Chaetoceros tenuissimus Meunier. Appl Environ Microbiol. 2008;74:4022–4027. doi: 10.1128/AEM.00509-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steward GF, Wikner J, Cochlan WP, Smith DC, Azam F. Estimation of virus production in the sea: II. Field results. Mar Microb Food Webs. 1992;6:79–90. [Google Scholar]
- Steward GF, Montiel JL, Azam F. Genome size distributions indicate variability and similarities among marine viral assemblages from diverse environments. Limnol Oceanogr. 2000;45:1697–1706. [Google Scholar]
- Steward GF.2001Fingerprinting viral assemblages by pulsed field gel electrophoresisIn: Paul JH, (ed)Marine Microbiology Academic Press, Ltd.: London; 85–103. [Google Scholar]
- Sullivan MB, Huang KH, Ignacio-Espinoza JC, Berlin AM, Kelly L, Weigele PR, et al. Genomic analysis of oceanic cyanobacterial myoviruses compared with T4-like myoviruses from diverse hosts and environments. Environ Microbiol. 2010;12:3035–3056. doi: 10.1111/j.1462-2920.2010.02280.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suttle CA. Marine viruses—major players in the global ecosystem. Nat Rev Microbiol. 2007;5:801–812. doi: 10.1038/nrmicro1750. [DOI] [PubMed] [Google Scholar]
- Tai V, Lawrence JE, Lang AS, Chan AM, Culley AI, Suttle CA. Characterization of HaRNAV, a single-stranded RNA virus causing lysis of Heterosigma akashiwo (Raphidophyceae) J Phycol. 2003;39:343–352. [Google Scholar]
- Takao Y, Nagasaki K, Mise K, Okuno T, Honda D. Isolation and characterization of a novel single-stranded RNA Virus infectious to a marine fungoid protist, Schizochytrium sp. (Thraustochytriaceae, Labyrinthulea) Appl Environ Microbiol. 2005;71:4516–4522. doi: 10.1128/AEM.71.8.4516-4522.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomaru Y, Katanozaka N, Nishida K, Shirai Y, Tarutani K, Yamaguchi M, et al. Isolation and characterization of two distinct types of HcRNAV, a single-stranded RNA virus infecting the bivalve-killing microalga Heterocapsa cicularisquama. Aquat Microb Ecol. 2004;34:207–218. [Google Scholar]
- Tomaru Y, Nagasaki K. Flow cytometric detection and enumeration of DNA and RNA viruses infecting marine eukaryotic microalgae. J Oceanogr. 2007;63:215–221. [Google Scholar]
- Tomaru Y, Takao Y, Suzuki H, Nagumo T, Nagasaki K. Isolation and characterization of a single-stranded RNA virus infecting the bloom-forming diatom Chaetoceros socialis. Appl Environ Microbiol. 2009;75:2375–2381. doi: 10.1128/AEM.02580-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinbauer MG, Suttle CA. Comparison of epifluorescence and transmission electron microscopy for counting viruses in natural marine waters. Aquat Microb Ecol. 1997;13:225–232. [Google Scholar]
- Weinbauer MG. Ecology of prokaryotic viruses. FEMS Microbiol Rev. 2004;28:127–181. doi: 10.1016/j.femsre.2003.08.001. [DOI] [PubMed] [Google Scholar]
- Wommack KE, Colwell RR. Virioplankton: viruses in aquatic ecosystems. Microbiol Mol Biol Rev. 2000;64:69–114. doi: 10.1128/mmbr.64.1.69-114.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zar J.1996Biostatistical Analysis3rd edn.Prentice Hall: Upper Saddle River, NJ [Google Scholar]