Abstract
Dispersal limitation is generally considered to have little influence on the spatial structure of biodiversity in microbial metacommunities. This notion derives mainly from the analysis of spatial patterns in the field, but experimental tests of dispersal limitation using natural communities are rare for prokaryotes and, to our knowledge, non-existent for viruses. We studied the effects of dispersal intensity (three levels) and patch heterogeneity (two levels) on the structure of replicate experimental metacommunities of bacteria and viruses using outdoor mesocosms with plankton communities from natural ponds and lakes. Low levels of dispersal resulted in a decrease in the compositional differences (beta diversity) among the communities of both bacteria and viruses, but we found no effects of patch heterogeneity. The reductions in beta diversity are unlikely to be a result of mass effects and only partly explained by indirect dispersal-mediated interactions with phytoplankton and zooplankton. Our results suggest that even a very limited exchange among local communities can alter the trajectory of bacterial and viral communities at small temporal and spatial scales.
Keywords: viruses, bacteria, metacommunity, experiment, dispersal limitation, patch heterogeneity
Introduction
A metacommunity is defined as a group of local communities connected via dispersal of interacting species (Leibold et al., 2004). Metacommunity theory integrates the study of interactions between organisms and their local biotic and abiotic environment with spatial dynamics at the regional scale. The theory makes a variety of predictions about the spatial structure of biodiversity that depend on patch connectivity, patch heterogeneity and niche differences among species. In neutral metacommunities, where organisms are assumed to have identical niches and local habitat patches are the same, increased connectivity among the patches will counteract the effects of drift and lead to metacommunity homogenization, resulting in a reduction in compositional differences among communities in different patches (that is, ‘beta diversity'). In niche-based metacommunities, patch quality is not spatially homogenous, species occupy different niches and spatial patch heterogeneity can enhance beta diversity (Chesson, 2000). However, in such cases, the impact of patch heterogeneity on metacommunity structure will depend on the degree of dispersal among communities. At very low rates of dispersal, species may not reach suitable habitat patches, resulting in species-poor communities with vacant niches and a weak match between species composition and environmental conditions. Higher dispersal rates may therefore increase beta diversity in a heterogeneous landscape. At very high dispersal rates, mass effects can lead to the predominance of regionally superior competitors and result in a decline of beta diversity (Mouquet and Loreau, 2003). Dispersal-mediated ecological interactions can also add an additional level of complexity because patch connectivity and heterogeneity may differentially affect metacommunity structure of organism groups belonging to different but interacting trophic levels (Verreydt et al., 2012).
The prevailing view is that the geographic distribution and community composition of microbial organisms is largely unaffected by dispersal limitation, which is consistent with the idea that everything is everywhere, and the environment selects. The small size of microbes allows for wide dispersal and large population sizes make extinction unlikely at local scales (Fenchel and Finlay, 2004). Furthermore, some populations can recruit from dormant cells, allowing them to grow rapidly to high densities when suitable conditions arise (Jones and Lennon, 2010). Such characteristics allow local dynamics to have a dominant role in structuring microbial metacommunities and reduce the likelihood that dispersal limitation will influence spatial patterns of microbial diversity (Van der Gucht et al., 2007; De Bie et al., 2012). These predictions are consistent with the wide distribution patterns of many microbial taxa (Short and Suttle, 2002; Fenchel and Finlay, 2004; Short and Suttle, 2005) and strong associations between community composition and environmental gradients (Beisner et al., 2006; De Bie et al., 2012). However, studies covering a broad range of spatial scales, varying from widely separated extreme habitats (Papke et al., 2003; Whitaker et al., 2003), stream networks (Heino et al., 2010), rock pool clusters (Langenheder and Ragnarsson, 2007), and individual salt marshes (Martiny et al., 2011), suggest that microbial dispersal can be too slow to counteract community differentiation resulting from ecological drift. In light of such mixed evidence from field studies, there is a strong need for experimental tests of dispersal limitation in microbial metacommunities.
In the current study, we tested whether low levels of dispersal can influence the compositional changes of microbial metacommunities in response to spatial variation in environmental conditions. In a neutral scenario, we would expect no effects of dispersal or environmental heterogeneity on beta diversity. In a niche-based scenario, however, we would expect an interaction between dispersal and patch heterogeneity, whereby dispersal enhances community differentiation in metacommunities with heterogeneous compared with homogenous environments. To test these ideas, we performed an outdoor mesocosm experiment using plankton from natural ponds and lakes in British Colombia (Canada). We created replicate experimental metacommunities with different levels of patch heterogeneity by adding different amounts of nutrients to mesocosms. We analyzed the changes in the metacommunity structure of both bacteria and viruses in response to different levels of dispersal intensity. We chose very low exchange rates of organisms among communities in the dispersal treatments to ensure that dispersal effects on the trajectories of community composition reflect demographic responses of dispersed phylotypes to environmental conditions rather than merely being the result of metacommunity homogenization due to mixing. We also tested if changes in metacommunity structure of bacteria and viruses that we observe in response to the dispersal treatments can be attributed to direct dispersal effects or to indirect interactions with other organisms (for example, phytoplankton and zooplankton) that may also be constrained by dispersal themselves (Verreydt et al., 2012).
Materials and methods
Experimental design
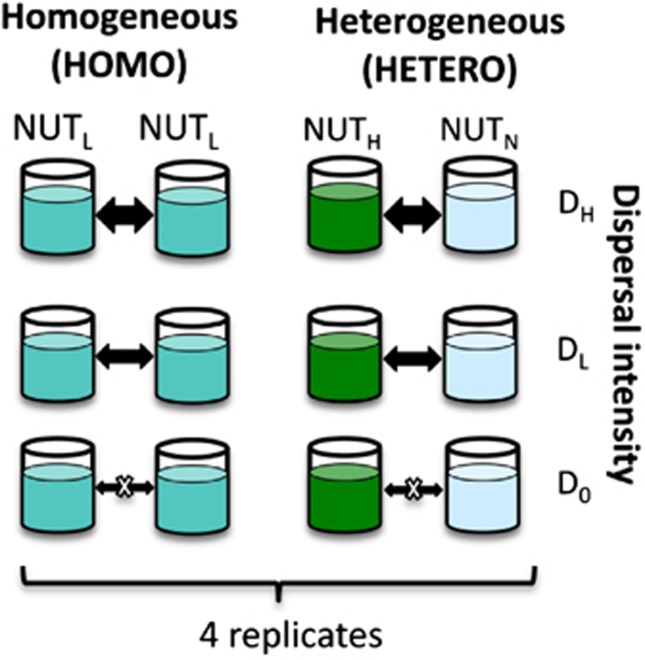
‘Metacommunities' consisted of pairs of plastic containers (280 l) outdoors that were connected through the dispersal of planktonic organisms. The experimental design consisted of the cross-factorial combination of ‘patch heterogeneity' and ‘dispersal intensity' (Figure 1). Patch heterogeneity was created through the addition of nutrients in either the same (homogenous metacommunities) or different amounts (heterogeneous metacommunities) between the two containers. Homogenous metacommunities (HOMO) received an identical, low amount of nutrients only at the start of the experiment (NUTL). Heterogeneous metacommunities (HETERO) were created by the repeated addition of a high amount of nutrients to one of the containers (NUTH), while no nutrients were added to the other (NUTN). The dispersal treatment consisted of three levels of exchange rates of planktonic organisms among containers: no dispersal (D0), low dispersal (DL) and high dispersal (DH) rates. All multifactorial combinations of dispersal intensity and nutrient heterogeneity were replicated four times, according to a randomized block design (Figure 1), with blocks corresponding to the spatial grouping of the containers on the experimental terrain.
Figure 1.
Schematic representation of the randomized block experimental design. This scheme represents one block consisting of six pairs of two containers (‘metacommunities') connected by dispersal. The experimental treatments consist of the six cross-factorial combinations of two factors: ‘nutrient heterogeneity' (two levels) and ‘dispersal intensity' (three levels). Three metacommunities received identical amounts of nutrients at low concentrations (NUTL) and are referred to as metacommunities with homogenous nutrient additions (HOMO). Heterogeneous metacommunities (HETERO) were created by repeatedly adding high amounts of nutrients to one of the containers (NUTH) and by adding no nutrients to the other container (NUTN). The factor dispersal consisted of three levels of exchange rates of planktonic organisms among containers, with no (D0), low (DL) and relatively high dispersal rates (DH). Blocks were replicated four times with identical heterogeneity and dispersal treatment combinations but inoculations from different lakes (see Supplementary Appendix 1 for a more detailed explanation of inoculation assignment).
Microbial and zooplankton communities were introduced into containers at the beginning of the experiment by adding water and sediments from lakes near Vancouver, Kelowna, Squamish and Victoria, BC, Canada. Within blocks, all NUTH and half of the NUTL containers were inoculated with a water and sediment mixture of three lakes, whereas all NUTN and the remaining NUTL containers received inoculations of three other lakes (see Supplementary Appendix 1 for details on the inoculation design).
The bottom of the containers was covered with coarse sand and filled with municipal drinking water on 28 March 2008. Lake water and sediments were collected during April 2008. The lake water was added to the containers as soon as it was brought from the field. Sediment samples were kept in the dark at 4 °C until 200 g from each lake was added to each container on 9 May 2008. On 16 May 2008, we added nutrients (nitrogen and phosphorus) according to the Redfield ratio to the NUTL (0.34 μℳ KH2PO4 and 5.48 μℳ NaNO3 final concentrations) and NUTH containers (17 μℳ KH2PO4 and 274 μℳ NaNO3 final concentrations). Due to experimental problems, we had to restart the NUTH containers on 16 June. The NUTH containers were emptied, cleaned, refilled with tap water and nutrients were added again (same concentrations as initially). On 1 July, we re-inoculated these tanks with water from the corresponding NUTL tanks with the aim of providing them with the same initial plankton community composition as in the NUTL tanks. Nutrient additions were then repeated weekly with 3.4 μℳ KH2PO4 and 55 μℳ NaNO3 to the NUTH containers and 0.17 μℳ KH2PO4 and 2.74 μℳ NaNO3 to the NUTL containers, until 20 August 2008. The dispersal treatment was started on 13 July, and continued on a weekly basis until 19 September.
We exchanged 0.025 l among DL containers and 1 l among DH containers, corresponding to 0.009% and 0.35% of the container volume, respectively. These volumes also contained zooplankton collected from 8 and 80 l, respectively, with a Schindler-Patalas trap (mesh size: 64 μm). To keep the same level of physical disturbance in all treatments, the same actions were repeated for D0, but without exchanging organisms or water among containers. Containers were covered by a mosquito net (mesh size: 1 mm) during the entire experiment to prevent insects and birds from entering.
Sampling and sample analysis
We took samples of bacterial, viral and zooplankton communities at two occasions, once just before the start of the dispersal treatment (10–11 July 2008) and once at the end of the experiment (29 September–1 October). In addition, during April 2008, we sampled the microbial communities (bacteria and viruses) of all the lakes that we used as inocula for the experiment (the ‘source lakes').
We estimated prokaryotic and viral abundances with flow cytometry. We determined phytoplankton chlorophyll a concentrations fluorometrically and measured dissolved organic carbon by filtering a water sample through ashed GF/F filters (Whatman), which were then analyzed on a Shimadzu 5000 TOC analyzer (Shimadzu, Tokyo, Japan). We assessed bacterial community composition using terminal restriction fragment length polymorphism (Moeseneder et al., 1999) and viral community composition with a randomly amplified polymorphic DNA PCR (Winget and Wommack, 2008). Samples of crustacean zooplankton were counted and identified to the genus level using a stereo microscope. We refer to Supplementary Appendix 2 for a more detailed description of sampling protocols and analysis procedures.
Data analysis
The central aim of our study was to investigate the response of beta diversity in bacterial and viral metacommunities to nutrient heterogeneity and dispersal intensity. For a metacommunity at a given sampling day, we quantified spatial beta diversity with the Jaccard dissimilarity index. Jaccard dissimilarity is the complement of Jaccard similarity, the latter equaling the ratio of the number of operational taxonomic units (OTUs) occurring in both patches of a metacommunity and the total number of OTUs observed in that metacommunity. The Jaccard dissimilarity thus reflects the degree to which communities differ in their observed OTU composition and varies from 0 (equal OTU composition) to 1 (no OTUs shared). We used two-way ANOVA to evaluate the interaction effect of nutrient heterogeneity and dispersal intensity on spatial metacommunity beta diversity. In case of significant effects, we applied paired t-tests to further explore differences among treatment levels.
Treatment effects on phytoplankton and zooplankton communities in our experiment may have indirectly affected the metacommunity structure of bacteria and viruses (Verreydt et al., 2012). For example, heterogeneous nutrient addition may have caused more heterogeneous metacommunities of interacting species (for example, zooplankton, phytoplankton and viruses) that indirectly increased bacterial beta diversity relative to the homogenous nutrient addition regime. Similarly, the dispersal treatment may have homogenized the biotic environment, contributing to a stronger decrease in beta diversity of bacteria than would have been expected from the physical exchange of these organisms alone. To evaluate the importance of such indirect effects (Verreydt et al., 2012), we measured the degree of biotic heterogeneity in metacommunities (i) by calculating ΔCHLA, as log10 (|chla1-chla2|+1), where chla1 and chla2 represent the chlorophyll a concentrations in two containers of the same metacommunity at 19 September 2008 and (ii) by calculating the Bray-Curtis distance using the zooplankton species abundance matrix (BCzoop). We incorporated ΔCHLA and BCzoop in general linear models explaining the bacterial and viral beta diversity to evaluate the direct effects of the experimentally manipulated dispersal and nutrient heterogeneity independently of any potential indirect effects.
We used two-way ANOVA to test for the effect of nutrient addition and dispersal intensity on abundances of prokaryotes, viruses and zooplankton, and on concentrations of chlorophyll a and dissolved organic carbon. Similarly, we applied distance-based redundancy analysis (Supplementary Appendix 4) to evaluate the effect of the experimental treatments on the community composition of the bacterial and viral communities.
With Mantel tests, we investigated pair-wise associations in the community composition of bacteria, viruses and zooplankton. In these analyses, we used Jaccard dissimilarity matrices derived from the presence-abundance matrices of bacteria and viruses and the Bray-Curtis dissimilarity matrix calculated from the zooplankton species abundance matrix.
To evaluate the strength of the temporal dynamics of bacterial and viral community composition, we calculated temporal beta diversity for communities within each mesocosm by calculating the Jaccard dissimilarity index on community data from the start and end of the experiment (July and September).
Results
Effects of dispersal and nutrient addition on metacommunity structure of bacteria and viruses
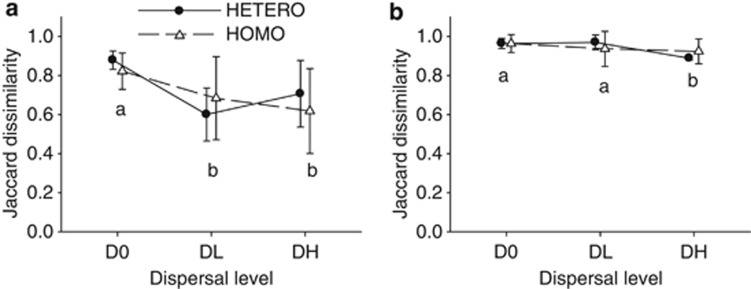
The dispersal treatment affected the beta diversity of both bacterial and viral metacommunities. At the end of September, beta diversity of bacteria was significantly lower in DL and DH than in D0 treatments (Figure 2a; Table 1). Thirty percent of the observed OTUs were shared by both mesocosms in the DL and DH metacommunities, compared with 15% in D0 metacommunities (an average of, respectively, 3 and 1.5 OTUs on a total of c. 10 OTUs). Beta diversity of viruses was lower in the DH treatment than in the D0 and DL treatments (Figure 2b; Table 1): mesocosms of the DH treatment shared 9.5% of the observed OTUs, compared with 4% in D0 and DL treatments (an average of, respectively, 1.6 and 0.8 OTUs on a total of c. 18 OTUs). Heterogeneity in nutrient additions had no effect on the beta diversity of bacteria or virus metacommunities (Figure 2; Table 1). Blocks significantly explained variation in bacterial and viral beta diversity (Table 1).
Figure 2.
Beta diversity of bacterial (a) and viral (b) metacommunities in response to dispersal and nutrient addition treatments. Beta diversity was calculated as the Jaccard dissimilarity index among communities within metacommunities. Different letters at symbols indicate significant differences among dispersal levels. Error bars represent the standard deviation.
Table 1. Results of ANOVA testing for the effects of dispersal intensity and metacommunity type (heterogeneous versus homogeneous nutrient addition regime) on the beta diversity (Jaccard dissimilarity) of bacterial and viral metacommunities.
| Factor |
Bacteria |
Viruses |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| df | SS | MS | F | P | SS | MS | F | P | |
| Dispersal intensity (Disp) | 2 | 0.211 | 0.106 | 6.62 | 0.009 | 0.0151 | 0.008 | 4.36 | 0.032 |
| Metacommunity type (Mctype) | 1 | 0.003 | 0.003 | 0.15 | 0.699 | 0.000 | 0.000 | 0.00 | 0.964 |
| Disp × Mctype | 2 | 0.033 | 0.017 | 1.05 | 0.375 | 0.005 | 0.002 | 1.35 | 0.298 |
| Blocks | 3 | 0.212 | 0.071 | 4.42 | 0.020 | 0.023 | 0.008 | 4.46 | 0.020 |
| Residuals | 15 | 0.240 | 0.016 | 0.026 | 0.002 | ||||
Abbreviations: Blocks, randomized block design; df, degrees of freedom; MS, mean squares; P, significance level; SS, sum of squares.Significant P-values are in bold.
Abundances of prokaryotes and viruses were higher under conditions of strong nutrient enrichment compared with low or no nutrient additions (Supplementary Appendix 3). Redundancy analyses revealed effects of nutrient addition, dispersal and inoculation origin on the community composition of bacteria (Supplementary Appendix 4). Viral community composition was affected by a dispersal × nutrient effect and an inoculation effect (Supplementary Appendix 4).
Effects of treatments on phytoplankton and zooplankton heterogeneity
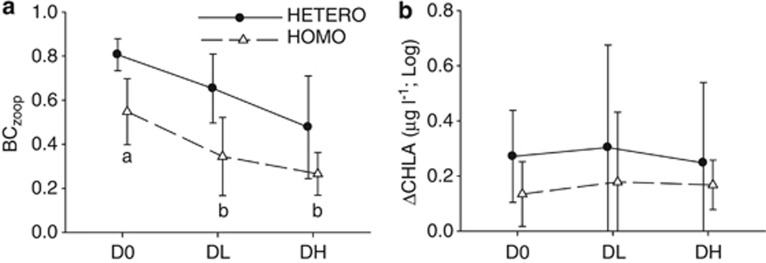
The experimental treatments also affected the heterogeneity of other biota in the metacommunities. Bray-Curtis distances calculated from zooplankton abundance data (BCzoop) were lower in the DL and DH than in the D0 treatments and were higher in heterogeneous than in homogenous metacommunities at the end of the experiment (Figure 3a; Table 2; see also Supplementary Appendix 3 for more details on treatment effects on population densities). We found no effects of dispersal intensity and heterogeneity in nutrient addition on ΔCHLA values (Figure 3b; Table 2).
Figure 3.
Biotic heterogeneity in metacommunities in response to dispersal and nutrient addition treatments. (a) Bray-Curtis distances among the pairs of zooplankton communities (BCzoop). (b) Differences in chlorophyll a concentration (log-transformed absolute values) (ΔCHLA). Different letters at symbols indicate significant differences among dispersal levels. Error bars represent the standard deviation.
Table 2. Results of ANOVA testing for the effects of dispersal intensity and metacommunity type (heterogeneous versus homogeneous nutrient addition regime) on the heterogeneity found for other trophic levels.
| Factor |
BCzoop |
ΔCHLA |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| df | SS | MS | F | P | SS | MS | F | P | |
| Dispersal intensity (Disp) | 2 | 0.349 | 0.174 | 8.870 | 0.003 | 0.045 | 0.023 | 0.084 | 0.920 |
| Metacommunity type (Mctype) | 1 | 0.344 | 0.344 | 17.503 | 0.001 | 0.536 | 0.536 | 1.988 | 0.179 |
| Disp × Mctype | 2 | 0.010 | 0.005 | 0.246 | 0.785 | 0.200 | 0.100 | 0.370 | 0.697 |
| Blocks | 3 | 0.162 | 0.054 | 2.746 | 0.082 | 3.079 | 1.026 | 3.804 | 0.033 |
| Residuals | 14 | 0.275 | 0.020 | 4.047 | 0.270 | ||||
Abbreviations: ΔCHLA, difference in chlorophyll a concentrations among the mesocosms of the same metacommunity; BCzoop, Bray-Curtis distance among the zooplankton communities of metacommunities; Blocks, randomized block design; df, degrees of freedom; MS, mean squares; P, significance level; SS, sum of squares.Significant P-values are in bold.
Assessment of indirect treatment effects through trophic interactions
BCzoop was positively correlated with beta diversity in metacommunities of bacteria (r=0.47, P=0.02, Supplementary Appendix 5) but not of viruses (r=0.32, P=0.137, Supplementary Appendix 5). ΔCHLA was positively correlated with beta diversity of bacteria (r=0.44, P=0.031, Supplementary Appendix 5), and a similar but marginal trend was also observed for viruses (r=0.36, P=0.058, Supplementary Appendix 5). According to General Linear Model analyses, the effect of the dispersal treatment on the beta diversity of bacteria and viruses was significant even with the inclusion of ΔCHLA and BCzoop in the model (Table 3). Beta diversity of bacteria did not contribute to the explanation of variation in the beta diversity of viruses or vice versa (Table 3).
Table 3. Results of General Linear Model analyses explaining variation of the beta diversity (Jaccard dissimilarity) in bacterial and viral metacommunities.
|
Bacteria |
Viruses |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| df | SS | MS | F | P | SS | MS | F | P | |
| Dispersal intensity (Disp) | 2 | 0.177 | 0.088 | 7.584 | 0.009 | 0.015 | 0.007 | 3.990 | 0.050 |
| Metacommunity type (Mctype) | 1 | 0.001 | 0.001 | 0.117 | 0.739 | 0.000 | 0.000 | 0.001 | 0.981 |
| Disp × Mctype | 2 | 0.055 | 0.028 | 2.360 | 0.140 | 0.006 | 0.003 | 1.508 | 0.264 |
| ΔCHLA | 1 | 0.160 | 0.160 | 13.738 | 0.003 | 0.010 | 0.010 | 5.501 | 0.039 |
| BCzoop | 1 | 0.073 | 0.073 | 6.295 | 0.029 | 0.005 | 0.005 | 2.625 | 0.133 |
| βvir | 1 | 0.001 | 0.001 | 0.047 | 0.833 | ||||
| βbact | 0.000 | 0.000 | 0.042 | 0.842 | |||||
| Blocks | 3 | 0.064 | 0.021 | 1.818 | 0.202 | 0.013 | 0.004 | 2.328 | 0.131 |
| Residuals | 11 | 0.128 | 0.012 | 0.020 | 0.002 | ||||
Dispersal intensity and metacommunity type (heterogeneous versus homogeneous nutrient addition regime) represent the experimental treatments.
Abbreviations: ΔCHLA, difference in chlorophyll a concentrations among the mesocosms of the same metacommunity; BCzoop, Bray-Curtis distance among the zooplankton communities of metacommunities; Blocks, randomized block design; df, degrees of freedom; MS, mean squares; P, significance level; SS, sum of squares.
We also included bacterial beta diversity (βbact) as explanatory variable in the model for the beta diversity of viruses (βvir) and vice versa. Significant P-values are in bold.
Community concordance patterns
Mantel correlation analyses revealed no association between the bacterial and viral communities in either July or September, but a significant association of bacteria with zooplankton community composition in both July (r=0.015, P=0.001) and September (r=0.04, P=0.001).
Distribution of bacterial and viral OTU's across time and metacommunities
Bacteria and viruses exhibited contrasting patterns of diversity across the array of mesocosms. The number of distinct OTUs observed for bacteria (N=88 OTUs) was much lower than for viruses (N=140 OTUs). The number of detected bacterial OTUs declined by 65% over 2 months (from 82 OTUs in July to 29 OTUs in September), leaving a nested subset of the original species pool at the end of the experiment, with only six new OTUs found in September. On both dates, several of the bacterial OTUs were widespread across the experiment, with more than half of the OTUs occurring in >5 mesocosms (Supplementary Appendix 6). In contrast, the total number of detected viral OTUs increased by 30% (from 99 OTUs in July to 124 in September, with 40 new OTUs in September), with viral OTUs being more sparsely distributed than those for bacteria. At both time points, <20% of the original viral OTUs occurred in five or more mesocosms, and no OTU occurred in >12 of the mesocosms (Supplementary Appendix 6).
We observed strong temporal dynamics in bacterial and viral community composition within local patches, as indicated by low Mantel correlations between July and September communities (bacteria: rSP=0.1; P=0.021; viruses: non-significant). The average number of bacterial OTUs per local community was 21 in July and 6 in September, and temporal beta diversity amounted to 0.85. The average number of viral OTUs per container was 6.6 in July and 9.6 in September, and temporal beta diversity equaled 0.96. Despite strong temporal dynamics, the bacterial communities within each patch shared a considerable proportion of OTUs with the original inocula from the source lakes. Across all sampled lakes, 54 bacterial OTUs were detected in total, and 29 and 17 of these were shared with July and September communities, respectively. For these shared OTUs, there was a positive correlation between their frequency of occurrence in mesocosms during September and the source lakes (r=0.49; P=0.007). No such correlation was found for viruses, and of the 101 viral OTUs found in the source lakes, only 6 were detected in the experiment.
Discussion
Despite strong effects of nutrient additions on the abundance of prokaryotes, viruses and zooplankton, we observed no effects of environmental heterogeneity on the metacommunity structure of bacteria and viruses. The fact that beta diversity in metacommunities with heterogeneous nutrient addition treatments showed no difference from metacommunities with homogenous patches is consistent with our predictions of a neutral scenario. However, multivariate analyses (distance-based redundancy analysis) demonstrated effects of nutrient additions on community composition, indicating that niche–environment interactions partly steered the compositional trajectories of the communities of bacteria and viruses. These niche–environment interactions, however, may not have been strong enough to affect compositional differentiation among patches at the metacommunity scale. Contrary to our predictions, dispersal did not enhance community differentiation along our established environmental gradient. Instead, the weekly exchange of very small volumes of water among containers reduced beta diversity of both bacterial and viral metacommunities. For example, the fraction of bacterial OTUs shared by patches within metacommunities was more than twice as high in those with dispersal as in isolated communities.
The observed reductions of beta diversity for both bacteria and viruses were surprising, particularly given dispersal treatments that consisted of weekly exchanges ranging from 0.009% to 0.35% of mesocosm volumes in the low and high dispersal treatments, respectively. From a purely neutral perspective, such low levels of dispersal relative to the population sizes of the resident community would unlikely lead to metacommunity homogenization due to mixing. Given the relatively high detection limits of our screening methods, it is therefore unlikely that neutral dynamics have resulted in a convergence of community profiles in the dispersal treatments. Metacommunity homogenization caused by mass effects is even less likely, given that the sustenance of populations of mal-adapted phylotypes would require exchange rates higher than in a neutral scenario. Indeed, previous experiments (Jones and McMahon, 2009; Lindström and Ostman, 2011) and field surveys (Logue and Lindström, 2010) have shown that dispersal rates need to be very high to overwhelm the local dynamics of species sorting in bacterial communities. A more likely explanation for the observed reduction in beta diversity is that the dispersal treatment has alleviated dispersal limitation in the metacommunities and has thereby facilitated the matching of community composition to environmental conditions in our experimental mesocosms. This explanation makes sense if the environmental conditions in the mesocosms generate a selection regime that favors only a subset of the original microbial communities. The observation of higher numbers of shared OTUs in metacommunities with dispersal suggests that local communities have enriched each other with such OTUs.
Experiments demonstrating dispersal limitation in natural microbial communities are scarce, and to our knowledge none have examined the effect of dispersal on viral communities. Lindström and Ostman (2011) studied the influence of dispersal treatments on bacterial community composition of three lakes for a wide range of dispersal rates. They found evidence of mass effects at very high dispersal rates, but no indications of dispersal limitation at low dispersal rates. In a series of incubation experiments with microbial communities from rainwater pools in a woodland, Bell (2010) found evidence for dispersal limitation, but only over short time scales (a few days). Langenheder et al. (2006) incubated bacterial communities of eight different aquatic habitats under identical environmental conditions and found no increase in the similarity among communities during the course of the experiment. Although their experiment did not involve a dispersal treatment, their result could have occurred because of dispersal limitation from source communities. Verreydt et al. (2012) showed stronger compositional shifts of bacterial communities in response to a nutrient gradient when dispersal was present, which was largely explained by dispersal-mediated trophic interactions, in which effects of dispersal on zooplankton and phytoplankton communities translated into apparent dispersal effects in bacterial communities, probably through modified grazing and nutrient recycling regimes (Langenheder and Jürgens, 2001; Degans et al., 2002; Jürgens and Matz, 2002). Such dispersal-induced homogenization of zooplankton metacommunities may also have contributed to the observed reduction of bacterial beta diversity in our experiment. Indeed, we detected significant Mantel correlations between zooplankton and bacterial communities and a positive association between zooplankton and bacterial beta diversity. However, General Linear Model analysis showed a significant effect of the dispersal treatment on the beta diversity of bacteria and viruses, independent of zooplankton beta diversity, indicating that the dispersal treatment also exerted unique effects. Viruses are typically highly host specific such that a decrease in bacterial community differentiation could lead to a reduction in the beta diversity of the viral metacommunities, but our results show no evidence for this. Mantel correlation analyses revealed no association between bacterial and viral communities, and our General Linear Model analyses showed no effects of bacterial beta diversity on viral beta diversity.
Our results show that very low exchange rates have enabled microbial communities to affect each other's compositional trajectories. Although this seems to be at odds with the idea that prokaryotes and viruses are ubiquitously distributed in nature, our results do not allow us to make statements about distribution patterns of OTUs in natural lakes at larger spatial scales, given the high detection limits of terminal restriction fragment length polymorphism and randomly amplified polymorphic DNA and the possibility that our initial inoculums may have only represented a fraction of the microbial communities present in the lakes. Our results, however, do suggest that the compositional changes of bacterial and viral communities in response to new environmental circumstances can be constrained by dispersal limitation, at least over time scales corresponding up to a few months.
A considerable share of the bacterial OTUs (35% in July and 49% in September) was also found in the source lakes, and in July we found a positive correlation between the frequency of occurrence of OTUs across containers in the experiment and the lakes supplying the corresponding inocula. In contrast, viral communities shared fewer OTUs with the source lakes and we detected a large fraction of new viral OTUs in September that had not been observed during July or in the lake survey. The majority of viruses in freshwater environments are viruses infecting prokaryotes (phages) and these have been shown to exhibit strong host specificity (Wommack and Colwell, 2000). The Bank model (Breitbart and Rohwer, 2005), for example, suggests that the active viral community composition is strongly determined by the composition of the hosts. Our results provide no support for this prediction as responses of viral and bacterial composition to the experimental treatments differed qualitatively (for example, response to nutrient addition in bacteria but not in viruses) and Mantel tests provided no evidence for overall compositional associations. Similarly, Shurin et al. (2012) found that the response of viruses to experimental treatments of warming, fish predators and eutrophication was distinct from those of bacteria. Weak associations between bacteria and viruses can be expected if a large fraction of the viruses infect other organisms, such as phytoplankton cells. In most cases, however, bacteriophages are much more abundant than other viruses, because of the numerical dominance of bacteria in plankton communities. Furthermore, viruses infecting other organisms than prokaryotes tend to be considerably larger than bacteriophages. Our sampling procedures may have partially excluded such viruses, given that samples were filtered over 0.22 μm filters and then shock frozen, which facilitates the break-up of large viruses. Patterns of association may also be reduced if viral communities show low host specificity (Sano et al., 2004; Holmfeldt et al., 2007). Prokaryotic hosts may also be infected by multiple viruses (Holmfeldt et al., 2007) and, given the high reproduction potential of viruses, viral communities can show rapid compositional turnover rates leading to poor associations between bacterial and viral communities. Finally, host–virus interactions may also have been obscured by limitations in the resolution of our molecular screening techniques (for example, Holmfeldt et al., 2007).
Six months after the start of the experiment, we were still able to find signatures of the original lake inoculations in the composition of bacterial communities, and in the beta diversity of bacterial and viral metacommunities. This indicates persistent effects of the initial distribution of taxa within the original source communities. Such lasting differences between bacterial communities of different origin suggest dispersal limitation, although it should be noted that our inoculations also included lake sediments. Inoculation effects may therefore also have been consolidated by the presence of large reservoirs of bacterial spores (Jones and Lennon, 2010) and persistent effects of sediments on water quality.
Although our results demonstrate that low levels of dispersal can affect the compositional trajectory of microbial communities, we found no interaction effects of environmental heterogeneity and dispersal on the beta diversity of microbial metacommunities. The lack of such an interaction may be the result of several specific features of our experiment. First, each of the patches in our experimental metacommunities was initially inoculated with communities from a mixture of three different locations. If compositional similarity among the source lakes was already low, then increased nutrient heterogeneity may not have been able to decrease the similarity among these communities any further. Second, dispersal failed to enhance compositional differentiation in response to nutrient heterogeneity. Possibly, the initial inoculums were too species rich and functionally saturated so as to be constrained in their response to a nutrient enrichment gradient. Third, the ‘regional' species pool in each community consisted of only two communities, which limits the potential for new functional types to establish in the communities. Therefore, the probability that dispersal will interact with patch heterogeneity in determining metacommunity structure of microbial organisms will likely increase when functionally poor communities are connected with a relatively rich pool of potential immigrants along pronounced environmental gradients. Fourth, we used a relatively coarse method for quantifying microbial diversity, because the detection limits of terminal restriction fragment length polymorphism and randomly amplified polymorphic DNA are high and only allow detecting relatively abundant OTUs. This methodological limitation may have obscured responses of the less abundant OTUs to the experimental treatments. Overall, such limitations have also made it impossible to identify the original sources of individual OTUs and track their spatial dynamics throughout the experiment. We therefore see great potential in the combination of high-throughput sequencing technology with similarly designed experiments to study microbial metacommunity dynamics.
Conclusions
Very low dispersal rates among local communities within replicate metacommunities resulted in a decrease in the compositional differences (beta diversity) in the metacommunities of both bacteria and viruses. Given the relatively small numbers of individuals that were exchanged, this response cannot be attributed to community homogenization or to mass effects. Beta diversity of bacteria and to lesser extent viruses was also associated with the heterogeneity in phytoplankton and zooplankton communities. Nevertheless, the effects of the dispersal treatment could not be explained by indirect effects of dispersal on these other trophic levels. While we found no evidence for interactive effects between dispersal intensity and patch heterogeneity on metacommunity beta diversity, our results demonstrate that a very limited exchange of organisms among local communities can alter the species sorting trajectory of bacterial and viral communities at temporal spatial scales of a few months.
Acknowledgments
We acknowledge the Natural Sciences and Engineering Research Council of Canada and the Tula Foundation for financial support. SD acknowledges a postdoctoral grant and a travel grant to UBC, Vancouver, Canada from the Research Foundation Flanders (FWO). BM acknowledges SNF grant number 31003A-125006: "Food-web and ecosystem responses to global change". We would like to thank Anita Norman and Michaela Martin for helping us with the maintenance and sampling of the mesocosms.
Footnotes
Supplementary Information accompanies the paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Beisner BE, Peres PR, Lindström ES, Barnett A, Longhi ML. The role of environmental and spatial processes in structuring lake communities from bacteria to fish. Ecology. 2006;87:2985–2991. doi: 10.1890/0012-9658(2006)87[2985:troeas]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Bell T. Experimental tests of the bacterial distance-decay relationship. ISME J. 2010;4:1357–1365. doi: 10.1038/ismej.2010.77. [DOI] [PubMed] [Google Scholar]
- Breitbart M, Rohwer F. Here a virus, there a virus, everywhere the same virus. Trends Microbiol. 2005;13:278–284. doi: 10.1016/j.tim.2005.04.003. [DOI] [PubMed] [Google Scholar]
- Chesson P. Mechanisms of maintenance of species diversity. Annu Rev Ecol Syst. 2000;31:343–366. [Google Scholar]
- De Bie T, De Meester L, Brendonck L, Martens K, Goddeeris B, Ercken D, et al. Body size and dispersal mode as key traits determining metacommunity structure of aquatic organisms. Ecol Lett. 2012;15:740–747. doi: 10.1111/j.1461-0248.2012.01794.x. [DOI] [PubMed] [Google Scholar]
- Degans H, Zollner E, Van der Gucht K, De Meester L, Jürgens K. Rapid Daphnia-mediated changes in microbial community structure: an experimental study. FEMS Microbiol Ecol. 2002;42:137–149. doi: 10.1111/j.1574-6941.2002.tb01003.x. [DOI] [PubMed] [Google Scholar]
- Fenchel T, Finlay BJ. The ubiquity of small species: Patterns of local and global diversity. Bioscience. 2004;54:777–784. [Google Scholar]
- Heino J, Bini LM, Karjalainen SM, Mykra H, Soininen J, Vieira LCG, et al. Geographical patterns of micro-organismal community structure: are diatoms ubiquitously distributed across boreal streams. Oikos. 2010;119:129–137. [Google Scholar]
- Holmfeldt K, Middelboe M, Nybroe O, Riemann L. Large variabilities in host strain susceptibility and phage host range govern interactions between lytic marine phages and their Flavobacterium hosts. Appl Environ Microbiol. 2007;73:6730–6739. doi: 10.1128/AEM.01399-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones SE, McMahon KD. Species-sorting may explain an apparent minimal effect of immigration on freshwater bacterial community dynamics. Environ Microbiol. 2009;11:905–913. doi: 10.1111/j.1462-2920.2008.01814.x. [DOI] [PubMed] [Google Scholar]
- Jones SE, Lennon JT. Dormancy contributes to the maintenance of microbial diversity. Proc Nat Acad Sci USA. 2010;107:5881–5886. doi: 10.1073/pnas.0912765107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jürgens K, Matz C. Predation as a shaping force for the phenotypic and genotypic composition of planktonic bacteria. Anton Leeuwenhoek Int J Gen M. 2002;81:413–434. doi: 10.1023/a:1020505204959. [DOI] [PubMed] [Google Scholar]
- Langenheder S, Jürgens K. Regulation of bacterial biomass and community structure by metazoan and protozoan predation. Limnol Oceanogr. 2001;46:121–134. [Google Scholar]
- Langenheder S, Lindström ES, Tranvik LJ. Structure and function of bacterial communities emerging from different sources under identical conditions. Appl Environ Microbiol. 2006;72:212–220. doi: 10.1128/AEM.72.1.212-220.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langenheder S, Ragnarsson H. The role of environmental and spatial factors for the composition of aquatic bacterial communities. Ecology. 2007;88:2154–2161. doi: 10.1890/06-2098.1. [DOI] [PubMed] [Google Scholar]
- Leibold MA, Holyoak M, Mouquet N, Amarasekare P, Chase JM, Hoopes MF, et al. The metacommunity concept: a framework for multi-scale community ecology. Ecol Lett. 2004;7:601–613. [Google Scholar]
- Lindström ES, Ostman O. The importance of dispersal for bacterial community composition and functioning. PLoS ONE. 2011;6:25883. doi: 10.1371/journal.pone.0025883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logue JB, Lindström ES. Species sorting affects bacterioplankton community composition as determined by 16S rDNA and 16S rRNA fingerprints. ISME J. 2010;4:729–738. doi: 10.1038/ismej.2009.156. [DOI] [PubMed] [Google Scholar]
- Martiny JBH, Eisen JA, Penn K, Allison SD, Horner-Devine MC. Drivers of bacterial beta-diversity depend on spatial scale. Proc Nat Acad Sci USA. 2011;108:7850–7854. doi: 10.1073/pnas.1016308108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moeseneder MM, Arrieta JM, Muyzer G, Winter C, Herndl GJ. Optimization of terminal-restriction fragment length polymorphism analysis for complex marine bacterioplankton communities and comparison with denaturing gradient gel electrophoresis. Appl Environ Microbiol. 1999;65:3518–3525. doi: 10.1128/aem.65.8.3518-3525.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouquet N, Loreau M. Community patterns in source-sink metacommunities. Am Nat. 2003;162:544–557. doi: 10.1086/378857. [DOI] [PubMed] [Google Scholar]
- Papke RT, Ramsing NB, Bateson MM, Ward DM. Geographical isolation in hot spring cyanobacteria. Environ Microbiol. 2003;5:650–659. doi: 10.1046/j.1462-2920.2003.00460.x. [DOI] [PubMed] [Google Scholar]
- Sano E, Carlson S, Wegley L, Rohwer F. Movement of viruses between biomes. Appl Environ Microbiol. 2004;70:5842–5846. doi: 10.1128/AEM.70.10.5842-5846.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Short CM, Suttle CA. Nearly identical bacteriophage structural gene sequences are widely distributed in both marine and freshwater environments. Appl Environ Microbiol. 2005;71:480–486. doi: 10.1128/AEM.71.1.480-486.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Short SM, Suttle CA. Sequence analysis of marine virus communities reveals that groups of related algal viruses are widely distributed in nature. Appl Environ Microbiol. 2002;68:1290–1296. doi: 10.1128/AEM.68.3.1290-1296.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shurin JB, Clasen JL, Greig HS, Kratina P, Thompson PL. Warming shifts top-down and bottom-up control of pond food web structure and function. Philos Trans R Soc Lond B Biol Sci. 2012;367:3008–3017. doi: 10.1098/rstb.2012.0243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Gucht K, Cottenie K, Muylaert K, Vloemans N, Cousin S, Declerck S, et al. The power of species sorting: Local factors drive bacterial community composition over a wide range of spatial scales. Proc Nat Acad Sci USA. 2007;104:20404–20409. doi: 10.1073/pnas.0707200104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verreydt D, De Meester L, Decaestecker E, Villena MJ, Van der Gucht K, Vannormelingen P, et al. Dispersal-mediated trophic interactions can generate apparent patterns of dispersal limitation in aquatic metacommunities. Ecol Lett. 2012;15:218–226. doi: 10.1111/j.1461-0248.2011.01728.x. [DOI] [PubMed] [Google Scholar]
- Whitaker RJ, Grogan DW, Taylor JW. Geographic barriers isolate endemic populations of hyperthermophilic archaea. Science. 2003;301:976–978. doi: 10.1126/science.1086909. [DOI] [PubMed] [Google Scholar]
- Winget DM, Wommack KE. Randomly amplified polymorphic DNA PCR as a tool for assessment of marine viral richness. Appl Environ Microbiol. 2008;74:2612–2618. doi: 10.1128/AEM.02829-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wommack KE, Colwell RR. Virioplankton: viruses in aquatic ecosystems. Microbiol Mol Biol Rev. 2000;64:69–114. doi: 10.1128/mmbr.64.1.69-114.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.