Abstract
Methanol biogeochemistry and its importance as a carbon source in seawater is relatively unexplored. We report the first microbial methanol carbon assimilation rates (k) in productive coastal upwelling waters of up to 0.117±0.002 d−1 (∼10 nmol l−1 d−1). On average, coastal upwelling waters were 11 times greater than open ocean northern temperate (NT) waters, eight times greater than gyre waters and four times greater than equatorial upwelling (EU) waters; suggesting that all upwelling waters upon reaching the surface (⩽20 m), contain a microbial population that uses a relatively high amount of carbon (0.3–10 nmol l−1 d−1), derived from methanol, to support their growth. In open ocean Atlantic regions, microbial uptake of methanol into biomass was significantly lower, ranging between 0.04–0.68 nmol l−1 d−1. Microbes in the Mauritanian coastal upwelling used up to 57% of the total methanol for assimilation of the carbon into cells, compared with an average of 12% in the EU, and 1% in NT and gyre waters. Several methylotrophic bacterial species were identified from open ocean Atlantic waters using PCR amplification of mxaF encoding methanol dehydrogenase, the key enzyme in bacterial methanol oxidation. These included Methylophaga sp., Burkholderiales sp., Methylococcaceae sp., Ancylobacter aquaticus, Paracoccus denitrificans, Methylophilus methylotrophus, Methylobacterium oryzae, Hyphomicrobium sp. and Methylosulfonomonas methylovora. Statistically significant correlations for upwelling waters between methanol uptake into cells and both chlorophyll a concentrations and methanol oxidation rates suggest that remotely sensed chlorophyll a images, in these productive areas, could be used to derive total methanol biological loss rates, a useful tool for atmospheric and marine climatically active gas modellers, and air–sea exchange scientists.
Keywords: methanol uptake, oligotrophic gyres, upwelling, chlorophyll a, methylotrophic bacteria, loss rates
Introduction
We have only just begun to understand methanol biogeochemistry and its significance in marine microbial carbon cycling in the oceans, and it represents one of the emerging scientific research areas. Recent analytical developments (for example, Beale et al., 2011) coupled with proven published methods utilising 14C-labelled methanol (Dixon et al., 2011a, 2011b, 2012) have begun to uncover high seawater concentrations up to 429 nℳ (Beale et al., 2011) coupled with biological turnover times as low as 1 day in the surface waters of the oligotrophic North Atlantic (Dixon et al., 2011b). The occurrence of methylotrophic bacteria (that is, bacteria that grow on methanol and other one carbon compounds) in the marine environment has been known for many years (for example, see Murrell et al., 1992 and references therein), but there has been relatively little research on marine methylotrophs until recently. For example, using cultivation-independent methods such as DNA stable isotope probing, it was shown that methylotrophs and, in particular, Methylophaga species are prevalent in coastal waters (Neufeld et al., 2007, 2008). More recently, Giovannoni and colleagues, through cultivation-based studies, have provided conclusive evidence that SAR 11 Alphaproteobacteria, which are some of the most abundant heterotrophs in the oceans, use one-carbon compounds like methanol for energy generation, and thus microbial oxidation of one carbon compounds could represent a significant in situ source of CO2 in the upper ocean (Sun et al., 2011). Using elegant genomics and environmental proteomics methods they have also shown that Betaproteobacteria of the OM43 clade, which are commonly found in productive coastal ocean ecosystems, also metabolise methanol and other one carbon compounds (Giovannoni et al., 2008; Sowell et al., 2011; Halsey et al., 2012). All of this research indicates the potential for significant turnover of methanol in the marine environment, and thus it is important to understand how different ocean systems contribute to the cycling of methanol.
Upwelling regions of the global oceans inject inorganic nutrients to often impoverished waters, and are thus normally highly biologically productive regions, despite their limited geographical extent. The coastal Mauritanian upwelling area is one of two major coastal upwellings in the Atlantic Ocean, and globally one of the most biologically productive systems (Pauly and Christensen, 1995). However, this area remains relatively unexplored (Rees et al., 2011). In contrast, oligotrophic oceanic gyres, which are the oceans least productive waters, cover ∼40% of the planet's surface and are thought to be expanding due to global warming (McClain et al., 2004; Polovina et al., 2008). The cyanobacterium Prochlorococcus and the SAR11 group of Alphaproteobacteria numerically dominate the oligotrophic gyres (Chisholm et al., 1988; Morris et al., 2002), while very small plastidic protists are the most numerous of the eukaryotes (Hartmann et al., 2012 and references therein). Thus, the aim of this study was to investigate the microbial demand for methanol as a carbon source for growth along a gradient of productivity from chlorophyll-rich coastal upwelling waters to oligotrophic oceanic gyres in the Atlantic Ocean.
Materials and methods
Sample collection
Seawater samples were collected from the Atlantic ocean (Figure 1) during two research cruises (a) SOLAS ICON (Surface Ocean Lower Atmosphere Study, UK ‘The impact of coastal upwellings on the production of climatically active gases') aboard the RRS Discovery (D338, 15 April–27 May 2009) and (b) AMT19 (Atlantic Meridonal Transect cruise number 19) aboard RRS James Cook (JC039, 13 October–1 December 2009). Samples were collected with 20 l Niskin bottles deployed on a rosette equipped with a Seabird conductivity, temperature and depth sensors. The seawater was collected directly into acid washed polycarbonate bottles that were in black plastic bags, and experiments were started within <1 h of sampling.
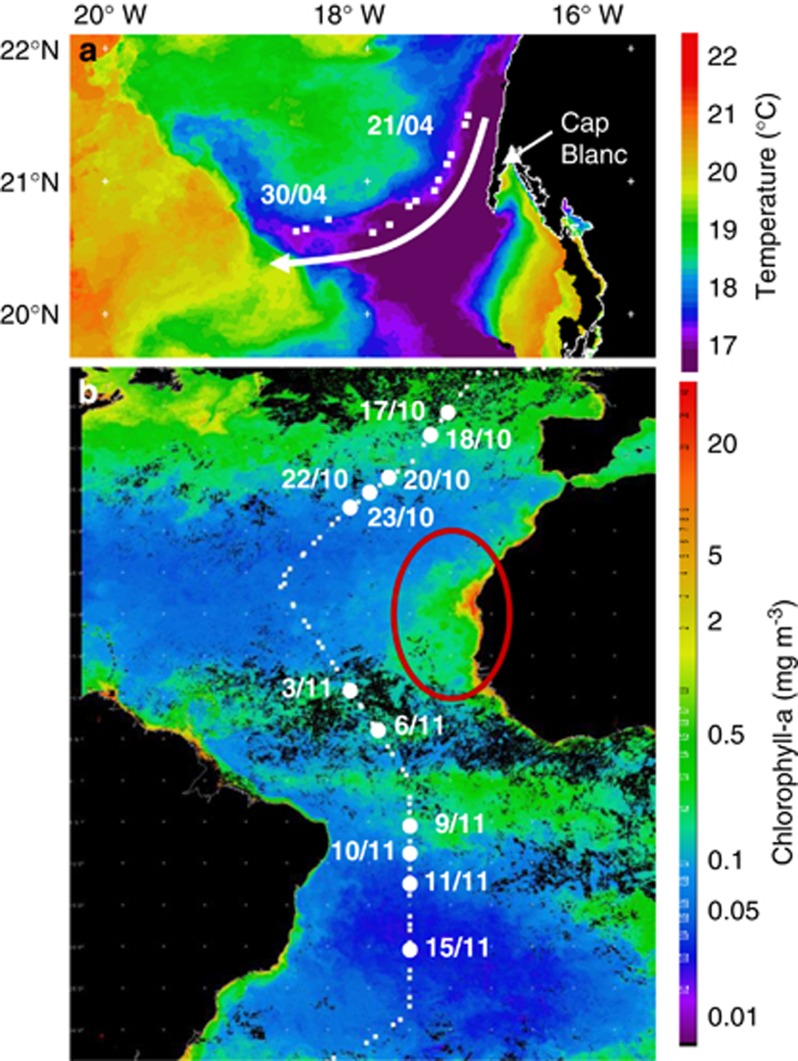
Figure 1.
Study area showing (a) AVHRR image of sea surface temperature showing the cold water filament, which was sampled during 21–30 April 2009 where the arrow indicates the direction of filament flow away from the coast, and (b) remotely sensed MODIS—Aqua chlorophyll a composite image from November 2009. The red oval shows the high concentrations associated with the Mauritanian upwelling. White squares show station positions, while white circles and corresponding station numbers (sampling date) show location of methanol uptake experiments during AMT19. These images are courtesy of NEODAAS.
Microbial methanol uptake
Seawater samples (320 ml) were incubated with ∼4.4 nℳ (final concentration) of 14C labelled methanol (14CH3OH) for between 4.5–10 h in the dark at in situ temperature, to determine the microbial uptake of methanol into particulate cell biomass. For full details of experimental procedures please refer to Dixon et al. (2011a). Typical seawater methanol concentrations range between 27–212 nℳ (Beale, 2011, personal communication) and 100–429 nℳ (Williams et al., 2004; Beale et al., 2011; Dixon et al., 2011a, 2011b) in the Mauritanian upwelling and the wider Atlantic ocean respectively. Thus the radiotracer addition typically represents between 2–16% and 1–4% of in situ concentrations in upwelling and Atlantic waters, respectively. The data are reported as apparent rate constants, k (h−1) calculated from a ratio of the sample 14C counts (from the particles on the filter, nCi ml−1 h−1) divided by total 14CH3OH added to the sample (nCi ml−1). Evaluation of control samples suggests that ⩽0.3% is recovered on the filters. Methanol uptake rates in nmol l−1 h−1 were calculated by multiplying the methanol counts obtained from the filter (nCi ml−1 h−1) by the specific activity of 14C methanol (57.1 mCi mmol−1).
Bacterial production
Heterotrophic bacterial production rates were determined using 1.7 ml seawater samples by measuring the incorporation of 3H-leucine (20 nℳ final concentration) into bacterial protein synthesis following the method of Smith and Azam (1992). The relative error based on three replicates was on average 7.1%. Leucine incorporation rates were multiplied by carbon to leucine conversion factors. For the Mauritanian upwelling samples, an empirically derived conversion factor of 0.7 kg C mol leu−1 from Alonso-Sáez et al. (2007) was used, which matches with upwelling location and sampling season. However, for an upper estimate, the theoretical conversion factor of 1.55 kg C mol leu−1 derived from Simon and Azam (1989) and used notably by Hoppe et al. (2002) was applied. An average empirical conversion factor of 0.73 kg C mol leu−1 (Morán et al., 2002, 2004; Dixon, 2008; Calvo-Díaz and Morán 2009; Dixon et al., 2011b) was also applied for all samples. For Atlantic transect samples, where surface chlorophyll a levels were relatively low (0.02–0.68 mg m−3, Table 1), a lower estimate of bacterial carbon production was estimated using a conversion factor of 0.1 kg C mol leu−1 (Alonso-Sáez et al., 2007; Dixon et al., 2011b).
Table 1. Summary of parameters in the euphotic zone (down to 1% PAR depth).
| Methanol uptake (d−1) | Chl a (mg m−3) | PP (mg C m−3 h−1) | BN ( × 106 cells ml−1) | BPa(pM Leu d−1) | Nitrate (μmol l−1) | Phosphate (μmol l−1) | Syns ( × 103cells ml−1) | Pros ( × 105cells ml−1) | Peuksb ( × 103cells ml−1) | Nanoc ( × 103cells ml−1) | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Coastal upwelling | |||||||||||
| Av ± 1s.d. | 0.033 ± 0.020 | 2.8 ± 2.0 | 114 ± 112d | 1.2 ± 0.3 | 1465 ± 495 | 6.6 ± 1.5 | 0.47 ± 0.10 | 0.35 ± 0.20 | ND | 0.54 ± 1.1 | 1.1 ±0.7 |
| Range | 0.004–0.117 | 0.5–6.5 | 5–338d | 0.5–1.8 | 190–2279 | 4.5–13.3 | 0.35–0.81 | 0.12–1.3 | ND | 0.03–5.3 | 0.2–2.4 |
| n | 43 | 43 | 43 | 43 | 43 | 42 | 42 | 43 | 43 | 43 | |
| Northern Temperate | |||||||||||
| Av ± 1s.d. | 0.003 ± 0.001 | 0.18 ± 0.15 | 4.7 ± 2.7 | NA | 203 ± 112 | 0.09 ± 0.005 | 0.06 ± 0.03 | 20.2 ± 19.2 | 1.6 ± 0.8 | 5.1 ± 3.8 | 0.62 ±0.44 |
| Range | 0.001–0.004 | 0.02–0.35 | 1.4–8.3 | 34–341 | 0.08–0.09 | 0.03–0.13 | 2.3–41.9 | 0.02–2.2 | 1.1–11.3 | 0.18–1.3 | |
| n | 8 | 8 | 8 | 8 | 8 | 8 | 8 | 8 | 8 | 8 | |
| Northern Gyre | |||||||||||
| Av ± 1s.d. | 0.004 ± 0.003 | 0.15 ± 0.16 | 2.7 ± 1.4 | NA | 344 ± 369 | 0.28 ± 0.69e | 0.06 ± 0.05e | 2.4 ± 2.0 | 1.3 ± 0.4 | 1.6 ± 1.7 | 0.22 ± 0.07 |
| Range | 0.001–0.010 | 0.06–0.59 | 1.2–5.3 | 124–1488 | 0.01–2.5e | 0.04–0.21e | 0.1–5.6 | 0.5–1.8 | 0.8–6.5 | 0.05–0.36 | |
| n | 12 | 12 | 12 | 12 | 12 | 12 | 12 | 12 | 12 | 12 | |
| Equatorial upwelling | |||||||||||
| Av ± 1s.d. | 0.008 ± 0.004 | 0.27 ± 0.18 | 10.5 ± 6.0 | NA | 391 ± 175 | 0.08 ± 0.07 | 0.02 ± 0.03 | 30.9 ± 24.1 | 2.5 ± 1.1 | 3.4 ± 4.1 | 0.9 ± 1.4 |
| Range | 0.003–0.015 | 0.13–0.68 | 1.2–19.8 | 135–635 | 0.02–0.20 | 0.01–0.08 | 1.0–81 | 1.1–3.6 | 1.5–13.3 | 0.3–4.3 | |
| n | 8 | 8 | 8 | 8 | 8 | 8 | 8 | 8 | 8 | 8 | |
| Southern Gyre | |||||||||||
| Av ± 1s.d. | 0.004 ± 0.002 | 0.13 ± 0.12 | 3.2 ± 2.2 | NA | 525 ± 757 | 0.18 ± 0.41f | 0.15 ± 0.07f | 2.0 ± 1.2 | 2.2 ± 1.1 | 2.5 ± 2.9 | 0.28 ± 0.16 |
| Range | 0.001–0.007 | 0.03–0.45 | 0.7–7.9 | 51–3217 | 0.02–1.6f | 0.06–0.29f | 0.01–3.7 | 0.7–3.8 | 0.6–11.5 | 0.16–0.77 | |
| n | 16 | 16 | 16 | 16 | 16 | 16 | 16 | 16 | 16 | 16 | |
Abbreviations: BN, bacterial numbers; ND, not detected; NA, not available; PP, primary productivity; Pros, Prochlorococcus sp; Syns, Synechococcus sp.
Bacterial leucine incorporation.
Picophytoplankton (<2 μm).
Nanophytoplankton (∼2–12 μm).
For coastal Mauritanian upwelling primary productivity was carried out over 24 h period hence units of mgC m−3 d−1, whereas for all other regions incubations were from dawn to dusk and are shown in mg C m−3 h−1.
For one depth profile on 20 October 2009 the 1% PAR equivalent depth of 57 m was within the start of the nitricline, where concentration of nitrate was 2.5 μmol l-1 and phosphate was 0.21 μmol l−1
The 1% PAR equivalent depth was within the beginning of the nitricline.
Primary production and Chlorophyll a concentrations
Primary production (PP) was estimated using the standard method of 14C-bicarbonate incorporation (for example, Joint and Pomroy, 1993; Dixon, 2008), except that during the ICON upwelling cruise, samples were incubated for ∼24 h (between 0600–0600), and for Atlantic Meridonal Transect (AMT) stations, samples were incubated from dawn to dusk. Chlorophyll a concentrations were determined by fluorometric analysis of acetone-extracted pigments (Holm-Hansen et al., 1965).
Community composition
The numbers of bacterioplankton cells were determined by flow cytometry on SYBR Green I DNA-stained cells from 1.8 ml seawater samples fixed in paraformaldehyde (5%, final concentration). Numbers of Prochlorococcus sp. and Synechococcus sp. were analysed on unstained samples by flow cytometry. Water samples (250 ml) for plankton identification were collected pre-dawn and fixed in 2% acid Lugol's solution. Preserved samples were stored in cool, dark conditions until they were analysed in the laboratory by inverted settlement microscopy (Utermöhl, 1958). The plankton assemblage was characterised as detailed previously in Widdicombe et al. (2002).
Nutrient concentrations
Inorganic phosphate and nitrate concentrations were determined on discrete seawater samples using a 5-channel Technicon segmented flow autoanalyser for micromolar determinations (Woodward and Rees, 2001).
DNA extraction and bacterial identification
Seawater samples were only collected during AMT19 from the surface, 33% and 1% photosynthetically active radiation (PAR) equivalent depths and 200 m. Samples of 20 l were filtered through 0.2 μm Sterivex filters (Millipore, Watford, UK) using a peristaltic pump. DNA samples were incubated with 1.4 ml of RNAlater (Life Technologies, Paisley, UK) overnight at 6 °C. The RNAlater was removed before storage at −80 °C for the remainder of the cruise. Samples were transported back to the UK on dry ice and stored at −20 °C.
DNA was extracted from the Sterivex filters using the method previously described in Neufeld et al. (2007). Centrifugal filter devices (Amicon, Millipore) were used to clean the extracted DNA. PCR (35 cycles) amplification was carried out with functional gene primers for mxaF (encoding the large subunit of methanol dehydrogenase, found in Gram-negative methylotrophic bacteria)1003f and 1555r (McDonald and Murrell 1997; Neufeld et al., 2007) with an annealing temperature of 55 °C. PCR products of the appropriate size (∼550 bp) were purified from agarose gels using the QIAquick gel extraction kit (Qiagen, Crawley, UK) and cloned into JM109 high efficiency competent cells using the pGEM-T easy vector system (Promega, Southampton, UK). Colonies were selected for colony PCR using M13 primers, PCR products were cleaned using a QIAquick PCR purification kit (Qiagen) and DNA sequenced (using M13f as a sequencing primer). Nucleotide sequences retrieved were trimmed in Chromas Lite (2.01) and BLAST (Basic Local Alignment Search Tool, NCBI) was used to search for similar sequences within the nucleotide collection (nr/nt) database using the Blastn algorithm.
Results
Mauritanian upwelling filament
The upwelling filament was tracked using SF6 from its upwelling in surface waters close to the Mauritanian coast for 10 days (21–30 April 2009), over a distance of over 200 km. The top 200 m of this filament was 50–80% dominated by North Atlantic Central waters (Rees et al., 2011). Microbial uptake of methanol into the particulate phase was initially 0.086±0.008 d−1 in the most recently upwelled surface waters (Figure 2a), with maximum rates determined in subsurface waters (15 m) of 0.117±0.002 d−1. Rates thereafter generally decreased in the euphotic zone until the 26 April, when methanol uptake rates stabilised to an average of 0.026±0.007 d−1 (Figure 2a). The overall range in methanol uptake rate constants found in Mauritanian upwelling waters was between 0.004–0.117 d−1 (Table 1), which equates to 0.2–9.8 nmol l−1 d−1 (calculated using the specific activity of added 14C methanol). If the methanol uptake rate constants are multiplied by the range in methanol concentrations measured in the upwelling filament, methanol uptake varies between 0.1–24.8 nmol l−1 d−1. Depth profiles consistently showed subsurface maxima between 8–20 m, with rates generally decreasing at depths >20 m. On average, during the filament, 91–97% of the microbial assimilation of methanol carbon was associated with the 0.2–2.0 μm size fraction.
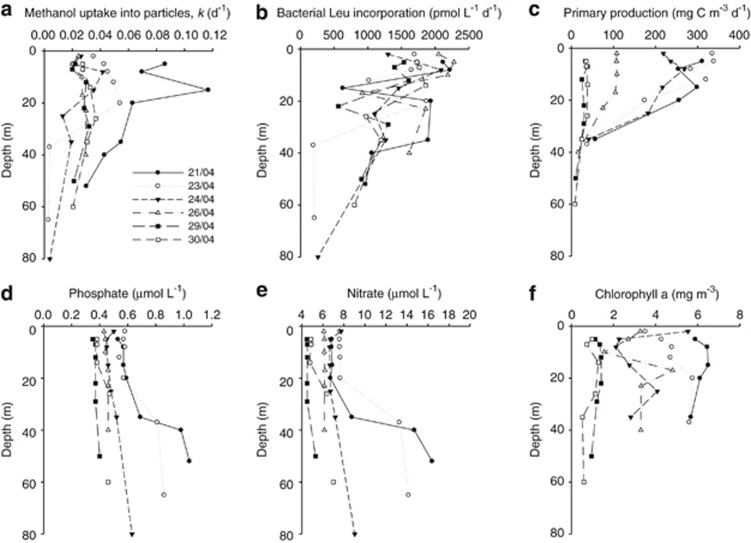
Figure 2.
Depth profiles (97–1% PAR equivalent depth) of biogeochemical parameters (a) methanol uptake into particles, (b) bacterial leucine incorporation, (c) rates of primary production and concentrations of (d) phosphate, (e) nitrate and (f) chlorophyll a determined while tracking an upwelling filament of water originating off Cap Blanc in Mauritanian waters.
Surface bacterial leucine incorporation rates did not show any progressive changes with filament age. Overall leucine uptake rates varied between 190–2279 pmol Leu l−1 d−1 (Table 1) and typically displayed subsurface minima between 12–22 m (Figure 2b). Surface rates decreased from 2116 to 1299 pmol Leu l−1 d−1 in the initial 3 days of filament progression, but peaked on day 4 at 2279 pmol Leu l−1 d−1 before gradually decreasing until the end of the experiment (Figure 2b). Rates of PP were exceptionally high during the first 3 days of the filament ranging between 218–338 mg C m−3 d−1 in surface waters, before reducing to 107 and to 35 mg C m−3 d−1 5 and 9 days later (Figure 2c). Rates of PP typically decreased significantly below 20 m (∼7% PAR). On average 90% of the PP in the top 20 m was in large cells >2 μm during first 4–5 days of bloom before decreasing slightly to 77%. Phytoplankton abundance was largely dominated (51–82%) by flagellates and diatoms (13–22%), although their contribution decreased slightly in the last 2 days of sampling the filament. The diatoms were numerically dominated (on average 52% ranging between 31–78%) by small Chaetoceros species. However, in terms of carbon biomass, diatoms comprised 74–80% of the total phytoplankton carbon for most of the filament, except during the final 2 days, when their dominance dropped to 51–61%. Chlorophyll a concentrations were maximum at 6.5 mg m−3 in the most recently upwelled waters, and decreased to a minimum of 0.5 mg m−3 10 days later (Figure 2f and Table 1), with depth profiles often showing subsurface maxima (Figure 2f). Concentrations of the inorganic nutrients nitrate and phosphate ranged between 6.1–7.7 μℳ and 0.43–0.58 μℳ respectively, in surface waters (down to 20 m) during the first 5 to 6 days of the filament, before reducing to between 4.5–4.9 μℳ and 0.35–0.38 μℳ in the last 2 days (Figures 2d and e, Table 1). The N:P ratio was on average 15 for the most part of the filament development, but dropped to 13 in the last 2 days. Typically the nitricline was between 20–40 m, below which maximum concentrations of nitrate and phosphate were 16.4 and 1.0 μℳ respectively. Prochlorococcus cells were notably absent from the Mauritanian upwelling waters, while Synechococcus numbers ranged between 0.12–1.3 × 103 cells ml−1 (Table 1). Pico-plankton and nano-phytoplankton abundance ranged between 0.03–5.3 and 0.2–2.4 × 103 cells ml−1 (Table 1).
Methanol uptake rates (calculated using the specific activity of 14C methanol) were integrated to the 1% PAR light equivalent depth and were initially 2.6 ng C m−2 d−1 before decreasing to an average of 1.1±0.2 ng C m−2 d−1 on all subsequent sampling days (Table 2). Surface chlorophyll a was also the highest initially at 6.2 mg m−3, but decreased to an average of 3.5 and 1.2 mg m−3 between 2–6 and 7–9 days later, respectively. Integrated methanol carbon rates were compared with calculated bacterial carbon production and suggest that in the most recently upwelled waters between 3–6% of the carbon assimilated by heterotrophic bacteria could be derived from methanol. This percentage reduced to between 1–4% as the upwelled filament aged (Table 2).
Table 2. Summary of % contribution of microbial methanol uptake to bacterial carbon production.
| Cruise/date | Zonea | Surface Chl a (mg m−3) | Methanol uptake Σ1%b (ng C m−2 d−1) | Bacterial leucine incorporation (pmol Leu L−1 d−1) | Bacterial carbon productionc Σ1% (ng C m−2 d−1) s-CF Min-CF Max-CF | Methanol % contribution to Bacterial carbon production | ||
|---|---|---|---|---|---|---|---|---|
| Mauritania filament | ||||||||
| 21 April 09 | U | 6.2 | 2.6 | 1760±578 (626–2217) | 44.4 | 42.5 | 94.2 | 3–6 |
| 23 April 09 | U | 3.9 | 1.3 | 1358±645 (190–1870) | 35.0 | 33.6 | 74.3 | 2–4 |
| 24 April 09 | U | 3.9 | 0.93 | 1488±361 (1101–2087) | 36.2 | 34.8 | 77.0 | 1–3 |
| 26 April 09 | U | 3.0 | 0.97 | 1817±503 (913–2279) | 51.3 | 49.2 | 109 | 1–2 |
| 27 April 09 | U | 3.2 | 1442±472d | |||||
| 28 April 09 | U | 1.1 | 1228±612d | |||||
| 29 April 09 | U | 1.2 | 1.2 | 1217±405 | 42.8 | 41.0 | 90.9 | 1–3 |
| 30 April 09 | U | 0.86 | 1.6 | 1504±393 | 55.8 | 53.5 | 119 | 1–3 |
| Atlantic transect | ||||||||
| 17 October 09 | NT | 0.31 | 0.19 | 289±74 (182–341) | 13.3 | 1.82 | 28.2 | 1–10 |
| 18 October 09 | NT | 0.16 | 0.09 | 117±63 (34–183) | 4.01 | 0.55 | 8.53 | 1–9 |
| 20 October 09 | NG | 0.06 | 0.17 | 261±42 (207–295) | 10.4 | 1.44 | 22.3 | 1–17 |
| 22 October 09 | NG | 0.08 | 0.19 | 159±30 (124–197) | 7.65 | 1.05 | 16.3 | 1–19 |
| 23 October 09 | NG | 0.07 | 0.27 | 611±589 (216–1488) | 59.6 | 8.17 | 127 | 0.2–3 |
| 03 November 09 | EqU | 0.14 | 0.18 | 487±137 (307–635) | 14.4 | 1.96 | 30.5 | 1–9 |
| 06 November 09 | EqU | 0.14 | 0.27 | 294±168 (135–445) | 17.3 | 2.37 | 36.8 | 1–13 |
| 09 November 09 | SG | 0.08 | 0.19 | 249±28 (220–283) | 17.0 | 2.33 | 36.1 | 1–10 |
| 10 November 09 | SG | 0.10 | 0.24 | 1188 ±1364 (324–3217) | 111 | 15.1 | 235 | 0.1–2 |
| 11 November 09 | SG | 0.04 | 0.19 | 536±319 (256–995) | 57.3 | 7.85 | 122 | 0.2–2 |
| 15 November 09 | SG | 0.03 | 0.07 | 128±68 (21–187) | 13.4 | 1.84 | 28.4 | 0.3–4 |
U, Mauritanian upwelling; NT, north east temperate Atlantic; NG, North Atlantic gyre; EqU, Equatoral upwelling; SG, southern gyre.
Missing data due to incomplete depth profile, for Atlantic transect dark incubation data shown to compare with upwelling filament.
Integrated (to 1% PAR) bacterial production has been calculated three ways: Max-CF using the theoretical leucine to carbon conversion factor of 1.55 kg C mol leu−1 (Simon and Azam, 1989; Hoppe et al., 2002), which is also the maximum conversion factor found, Min-CF is the lowest. empirical conversion factor found in the literature matched to season and place where possible, 0.7 kg C mol leu−1 has been applied to Mauritanian upwelling waters and 0.1 kgC mol leu−1 has been applied to low chlorophyll a Atlantic transect samples (Alonso-Sáez et al., 2007) s-CF using a conversion factor of 0.73 kg C mol leu−1 applied to all the samples (Morán et al., 2002, 2004; Dixon, 2008; Calvo-Díaz and Morán, 2009; Dixon et al., 2011b).
Only two depths sampled in the top 1–97% PAR depths therefore only average±1 s.d. shown.
Microbial methanol uptake in regions of the Atlantic Ocean
We have used the MODIS–Aqua surface chlorophyll a composite image from November 2009 (Figure 1b) to broadly split the Atlantic Ocean into four regions: Northern temperate (NT, Chl ⩾0.15 mg m−3), Northern Gyre (NG, Chl <0.15 mg m−3), Equatorial upwelling (EU, Chl ⩾0.15 mg m−3), Southern Gyre (SG, Chl <0.15 mg m−3), using a similar approach taken by Hill et al. (2011). Microbial uptake of methanol into the particulate phase during the meridonal transect of the Atlantic Ocean was overall relatively low (0.001–0.015 d−1) compared with the highly productive Mauritanian upwelling waters (Table 1 and Figure 3a). This range is equivalent to between 0.04–0.68 nmol l−1 d−1 (using the specific activity of added 14C methanol). Samples taken in NT waters showed the lowest microbial methanol uptake rates (0.001–0.004 d−1), with an average of 0.003±0.001 d−1, which is ∼10 times lower than Mauritanian upwelling waters (Table 1). The highest rates (during AMT19) were found in EU waters (0.003–0.015 d−1) followed by NG (0.001–0.010 d−1) and SG (0.001–0.007 d−1). Sub surface peaks in the microbial uptake of methanol into the cells were found in waters receiving ∼33% PAR light (equivalent to 10–25 m). Below which methanol uptake rates generally decreased with depth (Figure 3a). On average, during the Atlantic Ocean transect, 82–95% of the microbial uptake of methanol was associated with the 0.2–2.0 μm size fraction.
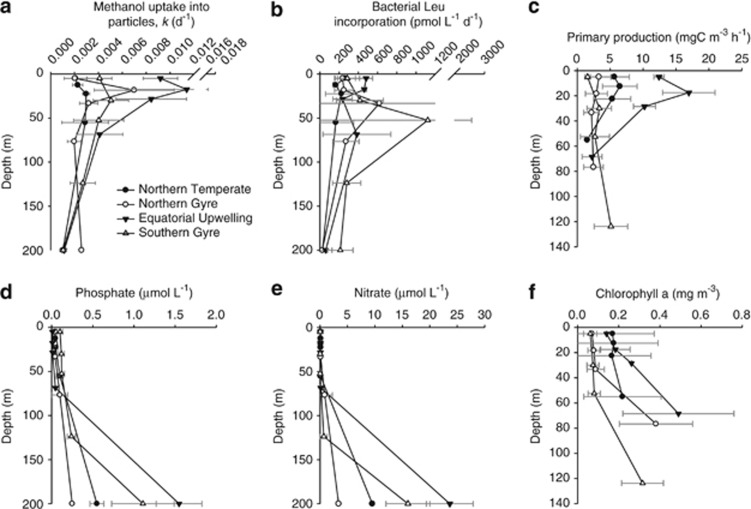
Figure 3.
Depth profiles (97–1% PAR equivalent) of biogeochemical parameters (a) methanol uptake into particles, (b) bacterial leucine incorporation, (c) rates of primary production and concentrations of (d) phosphate, (e) nitrate and (f) chlorophyll a determined in different provinces of the Atlantic Ocean. The profiles represent the average found in each province where for NT n=2, NG n=3, EU n=2, SG n=4. For each individual profile the number of replicate at each depth were ⩾3.
The average bacterial leucine incorporation rates found in NT, NG, EU and SG waters were between 3–7 times lower than incorporation rates in Mauritanian upwelling waters. Overall average rates of bacterial leucine uptake were lowest in NT waters, where they varied between 34–341 pℳ Leu d−1, with an average of 203 pℳ Leu d−1(Table 1). Average values were highest in SG waters, but depth profiles reveal two hotspots of anomalously high activity (3217 and 995 pℳ Leu d−1) associated with two profiles (out of four) at depths equivalent to 14% PAR (45–55 m, Figure 3b). If these are excluded from calculations, then average bacterial leucine rates decrease to 299±179 pM Leu d−1. Similarly, in NG waters, one depth profile (out of three) also showed a large hotspot of bacterial activity at the 14% PAR light level (1488 pℳ Leu d−1 at 45 m), and if this is excluded from calculations, then the average bacterial leucine incorporation rates decreases from 344±369 to 240±85 pℳ Leu d−1, which are similar to rates found in SG depth profiles. If these outliers are ignored, the EU waters show the highest average bacterial leucine uptake rates of 391±175 pℳ Leu d−1. However, for each hotspot in bacterial leucine incorporation shown in gyre water from between 45–55 m, three replicate samples were obtained with good precision (1–5%), so although the data seem anomalous when compared with the whole data set, it is difficult to justify their complete removal as they could represent hot spots in activity associated with detrital material (Azam et al., 1994).
Rates of PPwere characteristically low in gyre waters (Figure 3c), with an average of 2.7±1.4 mg C m−3 h−1 and 3.2±2.2 mg C m−3 h−1 in NG and SG depth profiles, respectively. PP was on average slightly higher in NT profiles (4.7±2.7 mg C m−3 h−1), but not surprisingly showed the highest rates in EU waters (10.5±6.0 mg C m−3 h−1). In NT, NG and EU waters, on average (for each depth profile) between 67–75% of the PP was associated with the 0.2–2 μm fraction. However for SG profiles, this decreased to 58%.
Concentrations of nitrate and phosphate in NT, SG, EU and SG profiles were also very low compared with Mauritanian upwelling coastal waters (Table 1 and Figures 3d and e). Nitrate concentrations in NT profiles were consistently low, ranging between 0.08–0.09 μℳ. The ranges shown for gyre waters are slightly extended because on occasion the 1% PAR equivalent depth was within the start of the nitricline (Table 1). Phosphate concentrations were on depth average 0.06±0.03 μℳ, 0.06±0.05 μℳ and 0.02±0.03 μℳ in NT, NG and EU profiles waters, respectively, but showed a relatively higher average concentration of 0.15±0.07 μℳ in SG waters. The resulting N:P ratios were very low and ranged between 0.9–3.3, 2–2.5, 2–13.2 and 0.2–5.8 for NT, NG, EU and SG depth profiles, respectively. For SG profiles, the exceptionally low N:P values of 0.2–0.4 were due to relatively more P compared with other regions, while in three out of four profiles, N was relatively enriched at the 1% PAR depth (95–125 m) due to incursion within the nitricline resulting in N:P ratios of between 2.5–5.8.
Numbers of Synechococcus were characteristically low in the gyres (2.4±2.0 cells ml−1 and 2.0±1.2 × 103 cells ml−1 for NG and SG, respectively), but were on average, two orders of magnitude higher in NT and EU waters (20.2±19.2 cells ml−1 and 30.9±24.1 × 103 cells ml−1 for NT and EU, respectively) compared with Mauritanian upwelling waters (Table 1). Although NT and EU profiles had relatively high variability due largely to a rapid decrease in numbers below 14% PAR equivalent depth, when light starts to become limiting. Prochlorococcus cells favoured EU and SG waters showing average numbers of 2.5±1.1 × 105 cells ml−1 and 2.2±1.1 × 105 cells ml−1, respectively. They showed lower averages of 1.6±0.8 × 105 cells ml−1 and 1.3±0.4 × 105 cells ml−1 in NT and NG waters, respectively. Pico-plankton numbers throughout the Atlantic open ocean regions were on average higher than in coastal upwelling waters and varied between 1.6–5.1 × 103 cells ml−1 across the regions, while nano-phytoplankton abundance was on average lower in the open ocean compared with coastal upwelling ranging between 0.22–0.9 × 103 cells ml−1 (Table 1).
Integrated methanol uptake rates in open ocean waters were lower than their coastal upwelling counterparts, ranging between 0.09–0.19 ng C m−2 d−1, 0.17–0.27 ng C m−2 d−1, 0.18–0.27 ng C m−2 d−1 and 0.07–0.24 ng C m−2 d−1 for NT, NG, EU and SG regions, respectively. However, because integrated bacterial carbon production is generally lower in the open ocean compared with the chlorophyll a-rich eutrophic coastal upwelling area (except when integrated rates have been elevated due to the measured ‘hot spots' of bacterial leucine incorporation as discussed above), the contribution that methanol carbon could make to bacterial carbon incorporation can be as high as 10%, 19%, 13% and 10% in NT, NG, EU and SG regions, respectively, although the ranges shown in Table 2 suggest that methanol contribution to bacterial carbon incorporation is relatively less important in the SG compared with the other Atlantic regions.
Preliminary characterisation of methylotrophic bacteria in regions of the Atlantic Ocean
Using cultivation-independent molecular biological methods, based on retrieval of mxaF, the gene encoding the large subunit of methanol dehydrogenase, a key enzyme in methanol metabolism in Gram-negative methylotrophs, the presence of several methylotrophic bacteria was noted in open ocean water samples of the Atlantic Ocean (Table 3). Of the 14 mxaF sequence types (operational taxonomic units) retrieved by PCR from DNA from these samples, seven of them had no significant taxonomic affiliation and were assigned as mxaF clones from uncultivated methylotrophic bacteria. However, it was obvious from sequence analysis that they were bona fide mxaF sequences (82–87% identity at the amino acid level to mxaF from Methylophaga spp. over 145 amino acyl residues). The mxaF phylotype 33–7_A02 was found in all samples (NT, NG, EU and SG) and was identified as mxaF from Methylophaga (Neufeld et al., 2007, 2008). A total of 89 clones were screened (19 from NT, 20 from NG, 26 from EgU, 19 from SG and 5 from ST Atlantic region).
Table 3. Occurrence of mxaF genes in DNA samples from different Atlantic Ocean areas.
| Phylogenetic affiliation of mxaF clones | Atlantic area | Light level (% PAR) | Genbank accession number | Reference | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| |
NT |
NG |
EqU |
SG |
ST |
97% |
33% |
1% |
200 m (dark) |
|
|
| Ancylobacter aquaticus | * | * | AB455976 | Suzuki et al., 2009 | |||||||
| Uncultured bacterium 33–7_A02 | * | * | * | * | * | * | EU001854 | Neufeld et al., 2007 | |||
| Burkholderiales bacterium RZ18–153 | * | * | * | * | * | * | EU548067 | Kalyuzhnaya et al., 2008 | |||
| Methylococcaceae bacterium ET-SHO | * | * | * | * | * | * | * | AB453966 | Nakamura et al. unpublished. | ||
| Uncultured clone L4_E01 | * | * | * | * | EU001809 | Neufeld et al., 2007 | |||||
| Methylophilus methylotrophus | * | * | EU194905 | Madhaiyan et al., 2009 | |||||||
| Paracoccus denitrificans | * | * | U34346 | Ras et al., 1991 | |||||||
| Uncultured clone C04 | * | * | * | * | JQ254993 | Irvine et al., 2012 | |||||
| Uncultured Methylophaga sp. | * | * | * | EU031899 | Neufeld et al., 2007 | ||||||
| Uncultured clone C7 | * | * | * | * | JN665049 | Antony et al., 2012 | |||||
| Methylophaga sp. DMS021 | * | * | * | * | EU001861 | Neufeld et al., 2007 | |||||
| Uncultured clone 24–7_F06 | * | * | * | EU001823 | Neufeld et al., 2007 | ||||||
| Methylobacterium oryzae NBHP3 | * | * | HQ221359 | Meena et al., 2012 | |||||||
| Hyphomicrobium sp. B294 | * | * | Y08081 | Fesefeldt and Gliesche, 1997 | |||||||
| Methylosulfonomonas methylovora | * | * | U70525 | McDonald and Murrell, 1997 | |||||||
| Uncultured bacterium clone C23 | * | * | EF212333 | Chen et al., 2007 | |||||||
Abbreviations: NT, north east temperate Atlantic; NG, North Atlantic gyre; EqU, Equatoral upwelling; SG, southern gyre; ST, Southern temperate.*indicates presence of mxaF genes.
While some mxaF clones, identified as originating from members of the Burkholderiales (RZ18–153), Methylococcaceae (ET-SHO) and also Methylophaga sp. (DMS021), were present in DNA samples from several open ocean Atlantic regions, other mxaF sequences, such as those identified as belonging to the methylotrophs Ancylobacter aquaticus, Paracoccus denitrificans, Methylophilus methylotrophus, Methylobacterium oryzae, Hyphomicrobium sp. and Methylosulfonomonas methylovora (Table 3 and references therein), were only identified in DNA from seawater samples in specific regions and under particular light.
Discussion
We measured the highest ever microbial methanol uptake rates (k) of 0.117±0.002 d−1 in the most recently upwelled coastal waters, which approximately equates to 10 nmol l−1 d−1. However, this microbial incorporation of methanol carbon into cells reduced and stabilised to an average of 2 nmol l−1 d−1 in the euphotic zone within 5 days of following the upwelled waters. This maximum rate is >100-fold higher than that reported for North Atlantic oligotrophic waters (Dixon et al., 2011b), and about nine times higher than the average found in surface waters of the NE Atlantic (55.2° N 12.7 °W) where chlorophyll a concentrations ranged between 0.7–1.0 mg m−3 (Dixon et al., 2011a). On average coastal upwelling waters were 11 times greater than NT waters, eight times greater than gyre waters and four times greater than EU waters in this study. These results suggest that coastal and EU waters upon reaching the surface (⩽20 m), contain a microbial population that use a relatively high amount of carbon (0.3–10 nmol l−1 d−1), derived from methanol, to support their growth. Preliminary characterisation of methylotrophic bacteria in the equatorial region of the Atlantic Ocean reveal a number of uncultivated methylotrophic bacteria, Methylococcaceae (ET-SHO), Methylophaga sp. (DMS021) and Methylosulfonomonas methylovora, the latter is a marine methanesulfonate degrading methylotroph that also uses methanol for growth (Holmes et al., 1997). When the use of methanol for incorporation into cell biomass was compared with the total amount of methanol used (for energy and uptake into cellular structures), then analysis of coastal upwelling samples revealed up to 57% of the total methanol was used to assimilate carbon into cells (average of 32% over all depths and days), which compares with an average of 12% in the EU, 1% in NT and NG regions, and 8% in the SG. This agrees with previously published results, which also suggest that ⩽4% of methanol is used for assimilation in open ocean gyre waters (Dixon et al., 2011b), but between 12–50% for more seasonally eutrophic shelf and coastal areas of the North-East Atlantic (Dixon et al., 2011a, 2011b).
Dissolved organic carbon production by phytoplankton is believed to be the primary organic carbon source for marine bacteria (Dixon et al., 2011b). However there is current debate over whether apparent mismatches can, or do occur, that is, where bacterial carbon demand exceeds that apparently available from phytoplankton production (for example, del Giorgio and Duarte, 2002; Duarte et al., 2001; Carlson et al., 2007; López-Sandoval et al., 2011). Dixon et al. (2011b) have previously suggested that for oligotrophic regions, methanol could be a significant, perhaps non-phytoplankton derived, organic carbon and energy source for bacteria. These current results further suggest that, even in highly productive, nutrient rich coastal upwelling regions, methanol could also be a significant carbon source for microbial anabolism (and energy), which is perhaps counter intuitive in such phytoplankton-rich waters. However, what remains uncertain is where the methanol originates, that is, whether the methanol is produced in situ, perhaps from photochemical degradation of dissolved organic carbon compounds, ultimately derived from phytoplankton or from air-sea transfer.
Relationships between the microbial assimilation of methanol carbon and other biogeochemical variables (including bacterial production, PP, cellular abundance and inorganic nutrient concentrations) have been explored in order to try and understand what controls, or determines the uptake of methanol into particulate cellular material. Statistically significant relationships are summarised in Table 4. We did not find a significant relationship between methanol uptake and bacterial leucine incorporation for this data, in contrast to previous experiments, which were from more limited geographical and biogeochemical areas (Dixon et al., 2011a, 2012). Methanol assimilation into the particulate phase for both the coastal upwelling and open ocean Atlantic samples (NT, NG, EU and SG combined) showed positive statistically significant relationships with rates of primary productivity (r=0.611, n=43, P⩽0.001 and r=0.676, n=44, P⩽0.001, respectively). For open ocean samples, when carbon fixation is dominated by unicellular cyanobacteria such as Synechococcus and Prochlorococcus sp. (and small plastidic protists, Hartmann et al., 2012), statistically significant relationships are also shown between their abundance and the uptake of methanol into cellular biomass (r=0.518 and 0.611, respectively, for n=50, P⩽0.001), in agreement with previously published results from diel experiments in the subtropical region of the NG, and in waters west of the Iberian peninsula located within the NT region (Dixon et al., 2011b; 2012). However, these relationships were not shown for more shelf waters of the north-east Atlantic (Dixon et al., 2011a). For the Mauritanian coastal upwelling waters where PP was dominated by cells >2 μm, a statistically significant relationship, albeit weaker, was shown between methanol uptake and the numbers of nano-phytoplankton (2–20 μm, r=0.381 n=43, P⩾0.05), rather than cyanobacterial cells. The link between the uptake of methanol into microbial cells and rates of carbon fixation by photosynthesis is unlikely to be direct in the coastal upwelling because 91–97% of the methanol uptake was in the 0.2–2 μm particle fraction, whereas on average 90% of the PP, was associated with particles >2 μm (nanoplankton). Thus, we suggest that mixotrophy of large plastidic protists, that is, preying on methylotrophic bacteria could have been occurring. Mixotrophy in natural populations of nano-plankton has been previously documented in other coastal upwelling regions, for example, North West Iberian upwelling system (Crespo et al., 2011; Vargas et al., 2011; Hartmann et al., 2012 and references therein) and in both temperate and oligotrophic regions of the Atlantic ocean (Zubkov and Tarran, 2008; Hartmann et al., 2012). Alternatively, the correlation found in Mauritanian upwelling waters between microbial methanol assimilation and primary productivity could be an indirect effect, of possibly a distinct group of methylotrophs associated with upwelling waters that uniquely use up to 57% of the methanol for cellular incorporation. This percentage is much higher than previously reported in oligotrophic and temperate coastal waters (⩽4%, Dixon et al., 2011a, 2011b; 2012), but more similar to that predicted in Murrell and McDonald (2000).
Table 4. Summary of statistically significant relationships between methanol uptake and biogeochemical variables.
|
Methanol uptake |
Primary production |
Chlorophyll a |
Synechococcus sp. |
Prochlorococcus sp. |
Nanophytoplankton |
| Mauritanian upwelling (D338) | 0.611** | 0.610** | NS | NA | 0.381* |
| Open Ocean (AMT19) | 0.676** | NS | 0.518** | 0.611** | NS |
Abbreviation: NS, not significant; *P⩾0.05; **P⩽0.001; NA, not applicable because the upwelling did not have any Prochlorococcus cells.
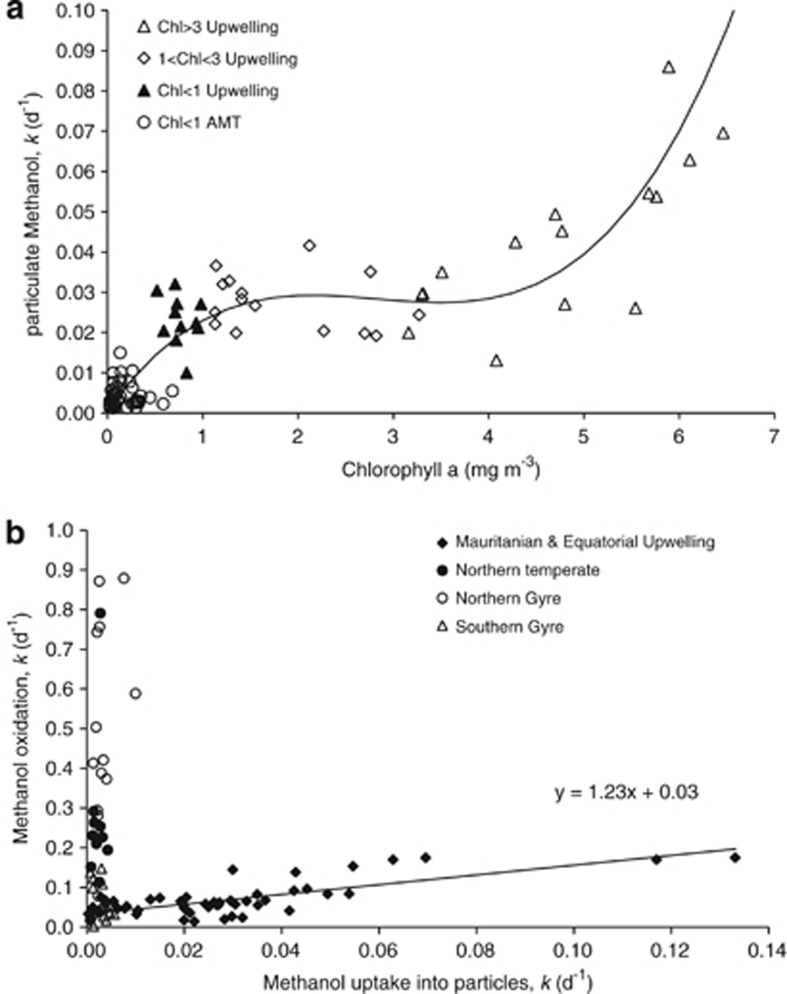
Because the two primary productivity data sets cannot be combined (different incubation lengths), microbial methanol uptake as a function of chlorophyll a concentrations for all the data is shown in Figure 4a. The relationship is best described by a third-order polynomial (r=0.915, n=86, P<0.001), although no statistically significant linear relationship between methanol uptake and chlorophyll a is apparent for the open ocean data only (Table 4). However, these results do suggest that for upwelling waters, the rate of methanol incorporated in cellular biomass could be estimated from chlorophyll a concentrations. The relationship between methanol oxidation (used as an energy source) and uptake into particles is shown in Figure 4b. The upwelling data from the Mauritanian coast and Equator have been combined, and show a statistically significant linear relationship (r=0.755, n=54, P⩽0.001). No relationship was found in NT or gyre waters where methanol oxidation rates tend to be much higher, and ⩾97% of methanol is used by microbes for obtaining energy (Dixon et al., 2011b). The relationships found for upwelling waters suggest that remotely sensed chlorophyll a images of the sea surface could be used to predict the total amount of methanol loss due to biological activity, which could potentially be a very useful tool when trying to determine global methanol budgets (for example, Jacob et al., 2005).
Figure 4.
Microbial uptake of methanol into cellular particles (a) as a function of chlorophyll a concentration for all data, which is best described by the third order polynomial y=0.0015x3–0.0127x2+0.034x+0.004 (r=0.915, n=86, P<0.001) and (b) relationship with methanol oxidation.
As a first attempt to assess the robustness of the relationships described in Figure 4, we have estimated the concentration of chlorophyll a from remotely sensed MODIS Aqua images (to coincide for when we have in situ methanol oxidation rates for the upwelling D338 cruise we have used a composite image from 21 to 27 April 2009 of the Mauritanian upwelling area, and for the EU we have used a composite AMT19 image from October 2009). For the Mauritanian upwelling we estimate chlorophyll a concentrations between 2–4 mg m−3 from the image. This equates to between 0.028–0.029 d−1 for methanol uptake into particles and consequently between 0.065–0.066 d−1 for methanol oxidation (using the relationships in Figures 4a and b, respectively). We have five surface in situ measurements over the image time giving an average 14C-derived methanol oxidation of 0.088±0.054 d−1 (n=5). For the EU area we estimate chlorophyll a from the image to be between 0.3–0.7 mg m−3, which equates to between 0.009–0.018 d−1 particulate methanol uptake, and between 0.042–0.053 d−1 for methanol oxidation. This compares with in situ oxidation measurements using 14C-labelled methanol to between 0.033–0.052 d−1 (n=2).
This research offers the first ever microbial methanol assimilation rates from highly productive coastal upwelling waters, which sharply contrasts with oligotrophic oceanic gyre water. Maximum microbial methanol carbon incorporation rates up to 10 nmol l−1 d−1 have been shown, which suggest that up to 57% of the total methanol is assimilated into carbon biomass. In comparison, microbes in oligotrophic gyre waters use ⩾97% methanol as an energy source (Dixon et al., 2011b). The statistically significant correlations for upwelling water between methanol uptake into cells and both chlorophyll a and methanol oxidation rates suggest that remotely sensed chlorophyll a images could be used in these productive areas to derive total methanol biological loss rates, a potentially useful tool for atmospheric and marine climatically active gas modellers and air–sea exchange scientists. Although we have identified some methylotrophic bacterial species from the open Atlantic Ocean, further research should identify which microbial species are utilising methanol, in addition to further development and testing of methanol production and loss processes parameterisations.
Acknowledgments
We thank all the scientists, officers and crew of the RRS Discovery and James Cook during D338 and JC029. In particular, we thank R Beale for methanol concentrations, G Tarran for the flow cytometry data, Y Chen for advice on use of mxaF primers, C Widdicombe for PP rates, chlorophyll a and phytoplankton species analysis, and EMS Woodward, A Rees and C Harris for the nutrient data. Satellite data were processed by the NERC Earth Observation Data Acquisition and Analysis Service (NEODAAS) at Plymouth Marine Laboratory (http://www.neodaas.ac.uk). This study was supported by the UK Natural Environmental Research Council (NERC) via UK SOLAS and Oceans 2025 funding for Plymouth Marine Laboratory.
References
- Alonso-Sáez L, Gasol JM, Arístegui J, Vilas JC, Vaqué D, Duarte CM. Large scale variability in surface bacterial carbon demand and growth efficiency in the subtropical northeast Atlantic Ocean, Limnol. Oceanogr. 2007;52:533–546. [Google Scholar]
- Antony CP, Doronina NV, Boden R, Trotsenko Y, Shouche YS, Murrell JC. Methylophaga lonarensis sp. nov., a novel moderately haloalkaliphilic methylotroph isolated from the soda lake sediments of a meteorite impact crater. Int J Syst Evol Microbiol. 2012;62:1613–1618. doi: 10.1099/ijs.0.035089-0. [DOI] [PubMed] [Google Scholar]
- Azam F, Smith DC, Steward GF, Hagström Å. Bacteria-organic matter coupling and its significance for oceanic carbon cycling. Microb Ecol. 1994;28:167–179. doi: 10.1007/BF00166806. [DOI] [PubMed] [Google Scholar]
- Beale R, Liss PS, Dixon JL, Nightingale PD. Quantification of oxygenated volatile organic compounds in seawater by membrane inlet-proton transfer reaction/mass spectrometry. Anal Chim Acta. 2011;706:128–134. doi: 10.1016/j.aca.2011.08.023. [DOI] [PubMed] [Google Scholar]
- Calvo-Díaz A, Morán XAG. Empirical leucine-to-carbon conversion factors for estimating heterotrophic bacterial production: Seasonality and predictability in a temperate coastal ecosystem. Appl. Environ. Microbiol. 2009;75:3216–3221. doi: 10.1128/AEM.01570-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson CA, Del Giorgio PA, Herndl GJ. Microbes and the dissipation of energy and respiration: from cells to ecosystems. Oceanography. 2007;20:89–100. [Google Scholar]
- Chen Y, Dumont MG, Cebron A, Murell JC. Identification of active methanotrophs in a landfill cover soil through detection of expression of 16S rRNA and functional genes. Environ Microbiol. 2007;9:2855–2869. doi: 10.1111/j.1462-2920.2007.01401.x. [DOI] [PubMed] [Google Scholar]
- Chisholm SW, Olson RJ, Zettler ER, Goericke R, Waterbury JB, Welschmeyer NA. A novel free-living prochlorophyte abundant in the oceanic euphotic zone. Nature. 1988;334:340–343. [Google Scholar]
- Crespo BG, Espinoza-Gonzalez O, Teixeira IG, Castro CG, Figueiras FG. Possible mixotrophy of pigmented nanoflagellates: Microbial plankton biomass, primary production and phytoplankton growth in the NW Iberian upwelling in spring. Est Coast Shelf Sci. 2011;94:172–191. [Google Scholar]
- Del Giorgio PA, Duarte CM. Respiration in the open ocean. Nature. 2002;420:379–384. doi: 10.1038/nature01165. [DOI] [PubMed] [Google Scholar]
- Dixon JL. Macro and micro nutrient limitation of microbial productivity in oligotrophic sub tropical Atlantic waters. Environ. Chem. 2008;5:135–142. [Google Scholar]
- Dixon JL, Beale R, Nightingale PD. Microbial methanol uptake in northeast Atlantic waters. ISME J. 2011a;5:704–716. doi: 10.1038/ismej.2010.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon JL, Beale R, Nightingale PD. Rapid biological oxidation of methanol in the tropical Atlantic: significance as a microbial carbon source. Biogeosciences. 2011b;8:2707–2716. [Google Scholar]
- Dixon JL, Nightingale PD. Fine scale variability in methanol uptake and oxidation in the micro-layer and near-surface waters of the Atlantic. Biogeosciences. 2012;9:2961–2972. [Google Scholar]
- Duarte CM, Agustí S, Arístegui J, González N, Anadón R. Evidence for a heterotrophic subtropical northeast Atlantic. Limnol Oceanogr. 2001;46:425–428. [Google Scholar]
- Fesefeldt A, Gliesche CG. Identification of Hyphomicrobium spp. using PCR-amplified fragments of the mxaF gene as a molecular marker. Syst Appl Microbiol. 1997;20:387–396. [Google Scholar]
- Giovannoni SJ, Hayakawa DH, Tripp J, Sting U, Givan SA, Cho J-C, et al. The small genome of an abundant coastal ocean methylotroph. Environ Microbiol. 2008;10:1771–1782. doi: 10.1111/j.1462-2920.2008.01598.x. [DOI] [PubMed] [Google Scholar]
- Halsey HH, Carter AE, Giovannoni SJ. Synergistic metabolism of a broad range of C1 compounds in the marine methylotrophic bacterium HTCC2181. Environ Microb. 2012;14:630–640. doi: 10.1111/j.1462-2920.2011.02605.x. [DOI] [PubMed] [Google Scholar]
- Hartmann M, Grob C, Tarran GA, Martin AP, Burkhill PH, Scanlan DJ, et al. Mixotrophic basis of Atlantic oligotrophic ecosystems. Proc Natl Acad Sci USA. 2012;109:5756–5760. doi: 10.1073/pnas.1118179109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill PG, Mary I, Purdie DA, Zubkov MV. Similarity in microbial amino acid uptake in surface waters of the North and South Atlantic (sub-)tropical gyres. Prog Oceanogr. 2011;91:437–446. [Google Scholar]
- Holm-Hansen O, Lorenzen CJ, Holmes RW, Strickland JDH. Fluorometric determinations of chlorophyll. Journal du Conseil Permenant International pour l'Exploration de la Mer. 1965;30:3–15. [Google Scholar]
- Holmes AJ, Kelly DP, Baker SC, Thompson AS, De Marco P, Kenna EM, et al. Methylosulfonomonas methylovora gen. nov. and Marinosulfonomonas methylotropha gen. nov.: novel methylotrophs able to grow on methanesulfonic acid. Arch Microbiol. 1997;167:46–53. doi: 10.1007/s002030050415. [DOI] [PubMed] [Google Scholar]
- Hoppe H-G, Gocke K, Koppe R, Begler C. Bacterial growth and primary production along a north-south transect of the Atlantic Ocean. Nature. 2002;416:168–171. doi: 10.1038/416168a. [DOI] [PubMed] [Google Scholar]
- Irvine IC, Vivanco L, Bentley PN, Martiny JB. The effect of nitrogen enrichment on c(1)-cycling microorganisms and methane flux in salt marsh sediments. Front Microbiol. 2012;3:90. doi: 10.3389/fmicb.2012.00090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob DJ, Field BD, Li Q, Blake DR, de Gouw J, Warneke C, et al. Global budget of methanol: Constraints from atmospheric observations. J Geophys Res. 2005;110:D08303. [Google Scholar]
- Joint I, Pomroy A. Phytoplankton biomass and production in the southern North Sea, Mar. Ecol Prog Ser. 1993;99:169–182. [Google Scholar]
- Kalyuzhnaya MG, Hristova KR, Lidstrom ME, Chistoserdova L. Characterization of a novel methanol dehydrogenase in representatives of Burkholderiales: implications for environmental detection of methylotrophy and evidence for convergent evolution. J Bacteriol. 2008;190:3917–3823. doi: 10.1128/JB.00180-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Sandoval DC, Fernández A, Maranón E. Dissolved and particulate primary production along a longitudinal gradient in the Mediterranean Sea. Biogeosciences. 2011;8:815–825. [Google Scholar]
- Madhaiyan M, Poonguzhali S, Kwon SW, Sa TM. Methylophilus rhizosphaerae sp. nov., a restricted facultative methylotroph isolated from rice rhizosphere soil. Int J Syst Evol Microbiol. 2009;59:2904–2908. doi: 10.1099/ijs.0.009811-0. [DOI] [PubMed] [Google Scholar]
- McClain CR, Signorini SR, Christian JR. Subtropical gyre variability observed by ocean-colour satellites. Deep- Sea Res II. 2004;51:281–301. [Google Scholar]
- McDonald IR, Murrell JC. The methanol dehydrogenase structual gene mxaF and its use as a functional gene probe for methanotrophs and methylotrophs. Appl Environ Microbiol. 1997;63:3218–3224. doi: 10.1128/aem.63.8.3218-3224.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meena KK, Kumar M, Kalyuzhnaya MG, Yandigeri MS, Singh DP, Saxena AK, et al. Epiphytic pink-pigmented methylotrophic bacteria enhance germination and seedling growth of wheat (Triticum aestivum) by producing phytohormone. Antonie Van Leeuwenhoek. 2012;101:777–786. doi: 10.1007/s10482-011-9692-9. [DOI] [PubMed] [Google Scholar]
- Morán XAG, Estrada M, Gasol JM, Pedrós-Alió C. Dissolved primary production and the strength of phytoplankton-bacterioplankton coupling in contrasting marine regions. Microb Ecol. 2002;44:217–223. doi: 10.1007/s00248-002-1026-z. [DOI] [PubMed] [Google Scholar]
- Morán XAG, Fernández E, Pérez V. Size-fractionated primary production, bacterial production and net community production in subtropical and tropical domains of the oligotrophic NE Atlantic in autumn. Mar Ecol Prog Ser. 2004;274:17–29. [Google Scholar]
- Morris RM, Rappé1 MS, Connon SA, Vergin KL, Siebold WA, Carlson CA, et al. SAR11 clade dominates ocean surface bacterioplankton communities. Nature. 2002;420:806–810. doi: 10.1038/nature01240. [DOI] [PubMed] [Google Scholar]
- Murrell JC, McGowan V, Cardy DLN. Detection of methylotrophic bacteria in natural samples by molecular probing techniques. Chemosphere. 1992;26:1–11. [Google Scholar]
- Murrell JC, McDonald IR.2000MethylotrophyIn: Lederberg J, (eds). Encyclopedia of Microbiologysecond edn.Academic Press: London; 245–255. [Google Scholar]
- Neufeld JD, Schäfer H, Cox MJ, Boden R, McDonald IR, Murrell JC. Stable-isotope probing implicates Methylophaga spp. and novel Gammaproteobacteria in marine methanol and methylamine metabolism. ISME J. 2007;1:480–491. doi: 10.1038/ismej.2007.65. [DOI] [PubMed] [Google Scholar]
- Neufeld JD, Chen Y, Dumont MG, Murrell JC. Marine methylotrophs revealed by stable-isotope probing, multiple displacement amplification and metagenomics. Environ. Microbiol. 2008;10:1526–1535. doi: 10.1111/j.1462-2920.2008.01568.x. [DOI] [PubMed] [Google Scholar]
- Pauly D, Christensen V. Primary production required to sustain global fisheries. Nature. 1995;374:255–257. [Google Scholar]
- Polovina JJ, Howell EA, Abecassis M. Oceans's least productive waters are expanding. Geophys Res Letts. 2008;35:L03618. [Google Scholar]
- Ras J, Reijnders WN, Van Spanning RJ, Harmis N, Oltmann LF, Stouthamer AH. Isolation, sequencing, and mutagenesis of the gene encoding cytochrome c553i of Paracoccus denitrificans and characterization of the mutant strain. J Bacteriol. 1991;173:6971–6979. doi: 10.1128/jb.173.21.6971-6979.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rees AP, Brown IJ, Clark D, Torres R. The Lagrangian progression of nitrous oxide within filaments formed in the Mauritanian upwelling. Geophys Res Letts. 2011;38:L21606. [Google Scholar]
- Simon M, Azam F. Protein content and protein synthesis rates of planktonic marine bacteria. Mar Ecol Prog Ser. 1989;51:201–213. [Google Scholar]
- Smith DC, Azam F. A simple, economical method for measuring bacterial protein synthesis rates in seawater using 3H-leucine. Mar Microb Food Webs. 1992;6:107–114. [Google Scholar]
- Sowell SM, Abraham PE, Shah M, Verberkmoes NC, Smith DP, Barofsky DF, et al. Environmental proteomics of microbial plankton in a highly productive coastal upwelling system. ISME J. 2011;5:856–865. doi: 10.1038/ismej.2010.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Steindler L, Thrash JC, Halsey KH, Smith DP, Carter AE, et al. One carbon metabolism in SAR11 pelagic marine bacteria. PLoS ONE. 2011;6:e23973. doi: 10.1371/journal.pone.0023973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki R, Lisdiyantic P, Komagata K, Uchimura T. MxaF gene, a gene encoding alpha subunit of methanol dehydrogenase in and false growth of acetic acid bacteria on methanol. J Gen Appl Microbiol. 2009;55:101–110. doi: 10.2323/jgam.55.101. [DOI] [PubMed] [Google Scholar]
- Utermöhl H. Zur vervollkommnung der quantitativen phytoplankton methodik. Mitteilongen der Internationalen Vereinigung fűf Limnologie. 1958;9:1–38. [Google Scholar]
- Vargas CA, Contreras PY, Iriarte JL. Relative importance of phototrophic, heterotrophic, and mixotrophic nanoflagellates in the microbial food web of a river-influenced coastal upwelling area. Aquat Micro Ecol. 2011;65:233–248. [Google Scholar]
- Widdicombe CE, Archer SD, Burkhill PH, Widdicombe S. Diversity and structure of the microplankton community during a coccolithophore bloom in the stratified northen North Sea. Deep-sea Res II. 2002;49:2887–2903. [Google Scholar]
- Williams J, Holzinger R, Gros V, Xu X, Atlas E, Wallace DWR. Measurements of organic species in air and seawater from the reopical Atlantic. Geophys Res Letts. 2004;31:LS23S06. [Google Scholar]
- Woodward EMS, Rees AP. Nutrient distributions in an anticyclonic eddy in the northeast Atlantic Ocean, with reference to nanomolar ammonium concentrations. Deep- Sea Res II. 2001;48:775–793. [Google Scholar]
- Zubkov MV, Tarran GA. High bacterivory by the smallest phytoplankton in the the North Atlantic Ocean. Nature. 2008;455:224–226. doi: 10.1038/nature07236. [DOI] [PubMed] [Google Scholar]