Abstract
Members of the genus Tetrasphaera are considered to be putative polyphosphate accumulating organisms (PAOs) in enhanced biological phosphorus removal (EBPR) from wastewater. Although abundant in Danish full-scale wastewater EBPR plants, how similar their ecophysiology is to ‘Candidatus Accumulibacter phosphatis' is unclear, although they may occupy different ecological niches in EBPR communities. The genomes of four Tetrasphaera isolates (T. australiensis, T. japonica, T. elongata and T. jenkinsii) were sequenced and annotated, and the data used to construct metabolic models. These models incorporate central aspects of carbon and phosphorus metabolism critical to understanding their behavior under the alternating anaerobic/aerobic conditions encountered in EBPR systems. Key features of these metabolic pathways were investigated in pure cultures, although poor growth limited their analyses to T. japonica and T. elongata. Based on the models, we propose that under anaerobic conditions the Tetrasphaera-related PAOs take up glucose and ferment this to succinate and other components. They also synthesize glycogen as a storage polymer, using energy generated from the degradation of stored polyphosphate and substrate fermentation. During the aerobic phase, the stored glycogen is catabolized to provide energy for growth and to replenish the intracellular polyphosphate reserves needed for subsequent anaerobic metabolism. They are also able to denitrify. This physiology is markedly different to that displayed by ‘Candidatus Accumulibacter phosphatis', and reveals Tetrasphaera populations to be unusual and physiologically versatile PAOs carrying out denitrification, fermentation and polyphosphate accumulation.
Keywords: enhanced biological phosphorus removal, Tetrasphaera, metabolic model, genomic sequencing
Introduction
Enhanced biological phosphorus removal (EBPR) is an environmentally friendly and cost-effective process encouraged in many wastewater treatment plants (Oehmen et al., 2007). These systems exploit the ability of polyphosphate accumulating organisms (PAOs) to assimilate large amounts of phosphorus and store it intracellularly as polyphosphate granules (Fuhs and Chen, 1975). The dominance of these PAO populations in EBPR communities is achieved by cycling the biomass through alternating anaerobic (feast) and aerobic (famine) phases (Barnard, 1975).
The many models explaining PAO metabolism have been the subject of much debate. Their general features are that PAOs take up volatile fatty acids like acetate and store them as polyhydroxyalkanoates (PHAs) during the anaerobic phase. This reserve polymer is metabolized in the subsequent aerobic phase, where readily metabolizable exogenous carbon and energy sources are no longer available, to supply the energy needed for PAO to assimilate phosphate from the mixed liquor and store it as intracellular polyphosphate granules. It now appears that the energy needed for the anaerobic uptake of volatile fatty acids and PHA synthesis is derived from hydrolysis of the intracellular polyphosphate stores, while reducing power and additional energy is generated from glycolysis of intracellular glycogen stores (Mino et al., 1987), anaerobic operation of the tricarboxylic acid (TCA) cycle (Comeau et al., 1986; Louie et al., 2000) or both (Pereira et al., 1996; Wexler et al., 2009; Zhou et al., 2009).
‘Candidatus Accumulibacter phosphatis' (subsequently referred to as Accumulibacter) is the organism thought to be largely responsible for EBPR in both laboratory- and full-scale plants (Crocetti et al., 2000; and reviewed by He and McMahon, 2011). Data from laboratory-scale studies repeatedly show that this organism possesses the phenotype expected of a PAO (Pereira et al., 1996; Mino et al., 1998; Hesselmann et al., 2000; Liu et al., 2001). A metabolic model was reconstructed for the dominant Accumulibacter strain (Clade IIA str. UW-1) in a metagenome from two laboratory-scale EBPR plants enriched in it (García Martín et al., 2006). The model displayed the mechanisms thought to account for phosphate accumulation, and has since provided a basis for analysis of the transcription of key genes (He et al., 2010). A metagenomic study of a full-scale EBPR plant has also confirmed the presence of Accumulibacter populations although their genomes show substantial differences to that of the reference genome (Albertsen et al., 2011).
Members of the Actinobacterial genus Tetrasphaera were suggested to be putative PAOs (Maszenan et al., 2000; Kong et al., 2005), but with a markedly different physiology to Accumulibacter. Both could take up phosphate aerobically and store it intracellularly as polyphosphate, and assimilate a range of substrates under anaerobic conditions. However, in situ staining has failed to show that Tetrasphaera store these in the form of intracellular PHA in EBPR systems, in contrast to the situation with Accumulibacter (Kong et al., 2005; Nguyen et al., 2011). Furthermore, Tetrasphaera appears able to ferment glucose (Kong et al., 2008; Nguyen et al., 2011), but like Accumulibacter, Tetrasphaera also assimilates phosphate into polyphosphate granules under aerobic conditions only if in a previous anaerobic phase organic substrates have been available to them. Thus, the ecophysiology of Tetrasphaera seems to be more versatile than that of Accumulibacter. Tetrasphaera-related PAOs can be detected in high relative abundances in many full-scale EBPR plants where they account for up to 30% of the total bacterial biovolume (Kong et al., 2005; Nguyen et al., 2011). They are also phylogenetically diverse, comprising three distinct clades, where cells within each clade exhibit a range of morphologies (Nguyen et al., 2011).
Currently, the genus Tetrasphaera consists of six species cultured from activated sludge; T. australiensis (strains Ben 109 and Ben 110), T. japonica (strain T1-X7) (Maszenan et al., 2000), T. elongata (strain Lp2) (Hanada et al., 2002), T. elongata (strain ASP12) (Onda and Takii, 2002), T. jenkinsii, T. veronensis and T. vanveenii (McKenzie et al., 2006), but no genome sequence data exist for any of these isolates. As these organisms seem to have an important role in phosphorus removal in EBPR plants (Kong et al., 2005; Nguyen et al., 2011), a metabolic model for Tetrasphaera is needed to clarify what their function in these systems is, and how their ecophysiology might differ from that of Accumulibacter. Thus, the aim of this study was to sequence the genomes of four Tetrasphaera isolates (T. australiensis, T. elongata, T. japonica and T. jenkinsii), and to interpret these data to develop a metabolic model for them. These models were confirmed experimentally, where possible, and a metabolic model for the Tetrasphaera present in EBPR configured systems is proposed.
Materials and methods
Bacterial strains
Four Tetrasphaera strains (T. australiensis str. Ben110, DSM17519; T. elongata str. Lp2, DSM14184; T. japonica str. T1-X7, DSM13192; T. jenkinsii str. Ben74, DSM17519) isolated from activated sludge were used. The strains were grown in GS or R2A medium at 26 °C. Genomic DNA from each isolate was extracted using FastDNA SPIN kit for soil (MP Biomedicals, Seven Hills, NSW, Australia) according to manufacturer's instructions.
Genome sequencing and assembly
From 0.5 to 1 μg of DNA, a library for Illumina paired-end sequencing was constructed using the Paired-end DNA Sample Prep Kit (PE-102-1001; Illumina, CA, USA) according to manufacturer's instructions (Part # 1005063 Rev. A), but with minor modifications. The genomic DNA was fragmented at 32 p.s.i. for 8 min, and the adaptor-modified DNA fragments were enriched by 14 PCR cycles. The purified library was sequenced using an Illumina GAII with a paired-end module. Up to 200 000 clusters were generated per tile with paired-end reads of a length of 36 bp for T. australiensis and T. elongata. For T. japonica, paired-end reads of both 36 bp and 72 bp were obtained, and for T. jenkinsii, the paired-end reads were 72 bp long.
The sequences obtained for each genome were subjected to de novo assembly using both CLC Genomics Workbench version 4.5.1 (CLC bio, Aarhus, Denmark) and ABySS (Simpson et al., 2009) (n=10, k=27 for the 36-bp reads, k=47 for the 72-bp reads from T. japonica and k=49 for the reads obtained from T. jenkinsii). When assembling the reads with CLC Genomics Workbench, default settings were used as the assembly parameters. The minimum and maximum distances between the paired-end reads were set individually for each assembly, depending on the approximate size of the fragment sequenced, and for the 72-bp reads, a similarity of 0.8 and length fraction of 0.5 were used. The resulting contigs from CLC Genomics Workbench version 4.5.1 and ABySS were merged into one set of contigs using Minimus from the AMOS software package (Sommer et al., 2007).
Comparative genome analysis
The contigs were annotated using the web interface Magnifying Genomes (MaGe) of the MicroScope platform from GenoScope (Vallenet et al., 2006, 2009). The automatic annotations provided by MaGe were curated manually to validate the presence or absence of a particular gene involved in selected pathways of interest. Based on the annotations for each genome, a statistical distribution of the protein encoding genes was determined according to their classification within the Cluster of Orthologous Groups functional categories.
The unique and conserved genes in the four Tetrasphaera genomes were determined by comparing the protein sequences from each Tetrasphaera genome against a complete Tetrasphaera database using blastP. The resulting genes that were >50% identical over a minimum of 50% of the length of the protein with one or more genes from the other Tetrasphaera genomes were considered as non-unique genes. All genes not having a hit by these criteria in the other Tetrasphaera genomes were considered as unique genes.
Pure culture validation experiments
Two isolates (T. elongata and T. japonica) were cultured in Erlenmeyer flasks in an orbital incubator at 22 °C in R2A broth (which includes glucose) without starch and sodium pyruvate, to generate enough biomass for biochemical studies. When an adequate amount had been produced (typically after 7 days), cells were harvested by centrifugation and resuspended in the chemically defined MSV media (Williams and Unz, 1989) without any carbon source. Biomass suspensions were incubated aerobically for 4 h to exhaust any storage polymers and residual carbon sources before experiments were initiated. All experiments were performed in duplicate at 20–22 °C.
To determine the ability of the Tetrasphaera strains to assimilate glucose and release phosphate under anaerobic conditions, biomass was incubated anaerobically in R2A with 1 mℳ 13C-glucose, but without starch, sodium pyruvate and potassium dihydrogen phosphate. To ensure anaerobic conditions, cells were incubated in vials that were capped and sealed before being flushed sequentially with nitrogen gas and vacuum. During incubation, samples were removed for analyses of phosphate, 13C-glucose and 13C-labeled fermentation products. Cell biomass was also sampled for glycogen and PHA analyses. To determine the ability of Tetrasphaera strains to take up phosphate under subsequent aerobic conditions, biomass was incubated anaerobically with glucose, then harvested and washed after 3 h with MSV media under anaerobic conditions, before being incubated under aerobic conditions for a further 3 h in MSV with 0.5 mℳ of phosphate, but without any exogenous carbon source. Samples were taken for phosphate uptake measurements and cell contents of glycogen and PHA were measured. All experiments were performed in duplicate.
Denitrification
Inocula were obtained as above by growing cultures in shake flasks in R2A broth lacking starch and sodium pyruvate. When a sufficient inoculum was obtained, cells were harvested by centrifugation and resuspended in fresh R2A broth without starch and sodium pyruvate. To ensure that the cultures could tolerate nitrate or nitrite, low concentrations of nitrate and nitrite were added to the flasks during aerobic growth (0.25 mℳ of NaNO3 and 0.1 mℳ of NaNO2, respectively). To assess whether the Tetrasphaera isolates could denitrify, a final concentration of either 2 mℳ NaNO3 or 0.5 mℳ NaNO2 was added to each culture. Residual oxygen was removed as described earlier for the anaerobic incubations. Nitrate and nitrite levels were monitored. All experiments were performed in duplicate.
Anaerobic growth
Isolates were grown in R2A broth without starch and sodium pyruvate under the anaerobic conditions described above. The cell numbers were determined after 0, 1, 7, 14 and 21 days by counting DAPI-stained cells (see later).
Chemical analyses
Glycogen and PHA
Intracellular glycogen levels were determined according to the method described by Bond et al. (1999) with minor modifications. Ten micrograms of lyophilized biomass was resuspended in 5 ml of 0.9 M HCl and digested at 100 °C for 5 h. Glucose equivalents in the supernatant were quantified using HPLC equipped with the ICS-5000 (Dionex, Thermo Fisher Denmark A/S, Hvidovre, Denmark) column. PHA was analyzed by gas chromatography as described by Braunegg et al. (1978) with minor modifications. Ten micrograms of lyophilized biomass was resuspended in 2 ml chloroform, 2 ml methanol with 3% or 10% [v/v] H2SO4 and a hepta-decane internal standard, and digested for 6 h at 100 °C. One microliter of milliQ water was added and the sample was left to phase separate for at least 1 h. In all, 0.5 g of sodium sulfate was added to the chloroform phase to remove excess water. The samples were analyzed using an Agilent Technologies 7890A GC system (Horsholm, Denmark) equipped with a HP-5 column.
Fermentation products
In total, 10 ml aliquots of supernatant from the anaerobic cultures were lyophilized and resuspended in 600 μl of D2O with 0.075% [w/v] of sodium 2,2,3,3-tetradeutero, 3-(trimethylsilyl) propionate, pH 7. The spectra were acquired with a Bruker AVIII-600 Spectrometer (Fällanden, Switzerland) equipped with a 5-mm TXI (H/C/N) probe operating at 600.13 MHz for 1H and 150.9 MHz for 13C. Fermentation products were identified by acquiring HSQC (heteronuclear single quantum coherence) and 2D-HSQC-TOCSY spectra and comparing the shifts found with the Human Metabolome Database (Wishart et al., 2009) and BRUKER's BBIOREFCODE spectral database (Karlsruhe, Germany).
Orthophosphate
The samples were filtered through a 0.45-μm cellulose acetate filter, stored in 0.04 M H2SO4 at 4 °C and analyzed spectrophotometrically according to the ascorbic acid standard method 4500-P E (Murphy and Riley, 1962; APHA et al, 1995).
Biomass determination
T. elongata was grown in Erlenmeyer flasks in an orbital incubator at 22 °C in R2A broth without starch and sodium pyruvate for 7 days. Every day, 2 ml of the culture was fixed in a final concentration of 2% [w/v] formaldehyde for cell number determination. The fixed sample was homogenized and stained for 10 min with 5 μg ml−1 of DAPI, filtered through a 0.2-μm black polycarbonate filter and the stained cells counted using an epifluorescence microscope. On day 7, the suspended solids were also determined by filtering an aliquot through a 0.2-μm cellulose ester membrane filter that was dried overnight at 60 °C (APHA et al., 1995).
Nucleotide sequence accession numbers
These sequence data have been submitted to the DDBJ/EMBL/GenBank databases under the accession numbers CAIZ01000001-CAIZ01000173, CAJA01000001-CAJA01000528, CAJB01000001-CAJB01000435 and CAJC01000001-CAJC01000215 for T. elongata, T. australiensis, T. japonica and T. jenkinsii, respectively.
Results and discussion
Tetrasphaera genome characteristics
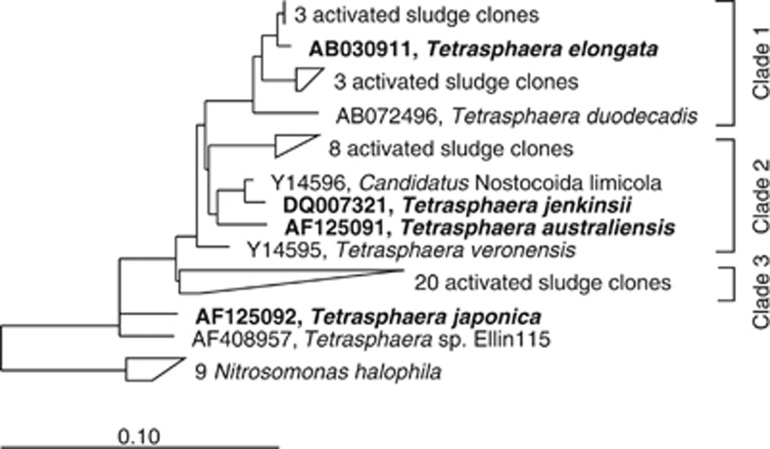
Of the phylogenetic clades proposed for the genus Tetrasphaera (Nguyen et al., 2011), T. elongata belongs to clade 1, T. australiensis and T. jenkinsii to clade 2, while T. japonica does not belong to any of the described clades (Figure 1). The morphology of these isolates varies, with T. australiensis and T. japonica always growing as cocci in tetrads or clusters (Maszenan et al., 2000), T. elongata as rod or oval unicells (Hanada et al., 2002), while the morphology of T. jenkinsii depends on the medium on which it is cultured; being filamentous on GS media and growing as irregularly sized cocci in clusters on R2A media (McKenzie et al., 2006).
Figure 1.
Phylogenetic tree of the Tetrasphaera genus showing the grouping of Tetrasphaera isolates in three clades described by Nguyen et al. (2011). Names of isolates used to construct the Tetrasphaera genomes are indicated with boldface font. The outgroup used to construct this tree was nine strains of Nitrosomonas halophila.
To reconstruct the genomes of these four Tetrasphaera species, the reads were assembled de novo and annotated using the MicroScope platform. The assembly resulted in genomes comprising 173–526 contigs (Table 1). The genome size varied from 3.15 to 5.16 Mbp with an average genome coverage ranging from 316 × to 1327 × . According to Farrer et al. (2009), a sequence depth above 30 × should be sufficient to assemble >96% of the genome de novo with the remaining unassembled part of the genome likely to be mainly composed of non-coding RNA and mobile genetic elements. Based on the presence of 106 essential genes, the completeness of the genomes is above 95%. These four Tetrasphaera genomes consist of 3258–5003 coding sequences, constituting up to 92.37% of their total genomes. An analysis of the Clusters of Orthologous Groups classification distribution of the coding sequences was conducted, and in general, none of the genomes is distinctly different from the others (Supplementary Material; Supplementary Figure S1). For all isolates, the coverage of the 16S rRNA gene was 1.3–1.5 times higher than the average coverage of the genome, which indicates that each isolate contains 1 or 2 copies of the rRNA operon.
Table 1. Genome characteristics of four sequenced Tetrasphaera isolates.
| Organism | Sequence length (bp) | Genome completeness (%)a | mol% GC | Coverage | Number of contigs | N50 | Number of CDS | Protein coding density (%) |
|---|---|---|---|---|---|---|---|---|
| T. australiensis | 4 282 171 | 96.2 | 69.01 | 366 | 526 | 13 130 | 4748 | 89.40 |
| T. elongata | 3 157 491 | 95.3 | 68.99 | 316 | 173 | 35 110 | 3258 | 92.37 |
| T. japonica | 5 162 321 | 98.1 | 71.51 | 737 | 435 | 19 833 | 5003 | 87.83 |
| T. jenkinsii | 3 809 986 | 100 | 68.24 | 1327 | 215 | 29 678 | 3983 | 91.78 |
Abbreviation: CDS, coding sequences.
Based on percentage essential genes present.
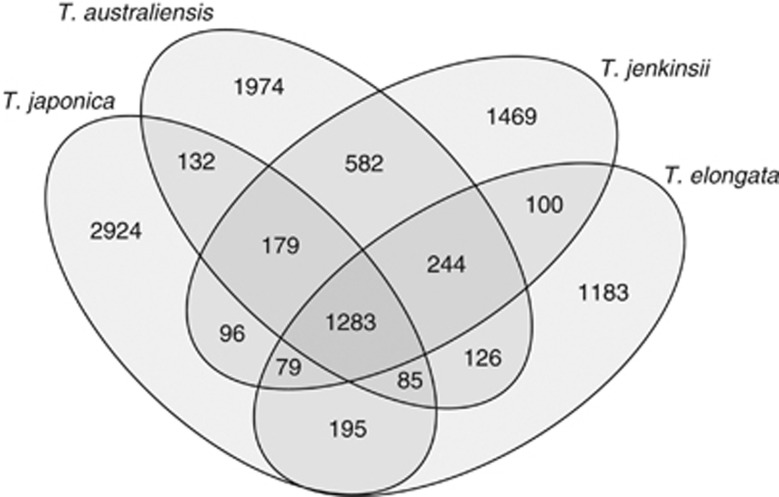
Conserved and unique genes were identified in the four Tetrasphaera isolates. Recognizing whether an individual gene is unique or conserved depends heavily on the criteria used. As no definitions exist for determining which represent core genes, it is difficult to elucidate the core genome (Bentley, 2009). In this study, we have recognized the genes involved in central metabolic pathways as being conserved genes, and those involved in a pathway unique to a single isolate as unique genes. With these criteria, the conserved genes from all four genomes constitute 1283 sequences (Figure 2). They include those involved in the central metabolic pathways possessed by all four isolates, and include polyphosphate metabolism, glycolysis and the TCA cycle (see below). The unique genes constitute from 1183 to 2924 sequences, and for example are genes involved in pathways for PHA synthesis, and assimilatory nitrate reduction to ammonia (see below).
Figure 2.
Venn diagram of unique and conserved genes for four Tetrasphaera isolates (T. australiensis, T. elongata, T. japonica and T. jenkinsii). T. japonica has 2924 unique genes and T. elongata has 1183 unique genes. T. australiensis and T. jenkinsii, both belonging to clade 2, have 1974 and 1469 unique genes, respectively, and share 582 genes. The conserved genes constitute 1283 genes of each genome.
Metabolic reconstruction
The annotation of the four Tetrasphaera genomes showed that many of the genes involved in the central metabolic pathways in Accumulibacter are also found in Tetrasphaera. These pathways include the TCA cycle, glycolysis, gluconeogenesis and polyphosphate metabolism. However, as detailed below, they differ fundamentally in several important respects, especially in the ability of these Tetrasphaera species to ferment.
Polyphosphate metabolism
Polyphosphate metabolism has been detailed in many bacteria including Escherichia coli. Orthologous genes involved in polyphosphate metabolism are also possessed by each of these Tetrasphaera species (Table 2; Supplementary Table S1). These include the genes important in both polyphosphate degradation and synthesis, and in transporting inorganic phosphate across the membrane using a low-affinity inorganic phosphate transporter (Pit) system and a high-affinity phosphate ABC transporter (Pst) system (Willsky and Malamy, 1980). E. coli is thought to synthesize polyphosphate using a reversible PPK1 (polyphosphate kinase 1), which transfers high-energy phosphate groups from ATP to a growing polyphosphate chain (Akiyama et al., 1992). Intracellularly stored polyphosphate can be degraded to yield energy in reactions catalyzed by a polyphosphate kinase 2 (Ishige et al., 2002), exophosphatase (PPX) (Akiyama et al., 1993), PAP (polyphosphate AMP phosphotransferase) (Bonting et al., 1991), or the combined enzymatic activity of ADK (adenylate kinase) and PPK1 (Shiba et al., 2000). The PAP/ADK reaction may also serve as an additional mechanism for polyphosphate synthesis (García Martín et al., 2006).
Table 2. Key metabolic pathways as predicted from the genomes from four Tetrasphaera isolates.
| Pathway | Accumulibacter | T. australiensis | T. elongata | T. jenkinsii | T. japonica |
|---|---|---|---|---|---|
| Glycolysis | + | + | + | + | + |
| Gluconeogenesis | + | + | + | + | + |
| Glycogen synthesis | + | + | + | + | + |
| Glycogenolysis | + | + | + | + | + |
| TCA cycle | + | + | + | + | + |
| Glyoxylate shunt | + | − | − | − | − |
| PHA synthesisa | + | − | − | − | + |
| Polyphosphate metabolism | + | + | + | + | + |
| Acetate uptake | + | + | + | + | + |
| Glucose uptake | − | + | + | + | + |
| Glutamate/aspartate uptake | − | + | + | + | + |
Abbreviations: PHA, polyhydroxyalkanoate; TCA, tricarboxylic acid.
+, Candidate genes coding for all enzymes involved in the pathway have been located in the genome; −, Not all candidate genes for the enzymes involved in the pathway have been found in the genome.
Based on the presence of phaC gene.
These observations suggest that Tetrasphaera may share much of their polyphosphate metabolic machinery with Accumulibacter (García Martín et al., 2006). Thus, intracellular polyphosphate in Tetrasphaera may be degraded to Pi during the EBPR anaerobic phase, generating energy, with the Pi transported out of the cell, and the energy generated supporting substrate uptake and conversion into storage reserve material. In the subsequent aerobic phase, polyphosphate storage levels are replenished by cells assimilating Pi from the bulk liquid.
Substrate uptake and storage polymers
Tetrasphaera appears able to assimilate a range of substrates, including acetate, propionate, glucose, glutamate and aspartate. Previous in situ ecophysiological investigations have shown that they utilize glucose and amino acids, but controversy exists regarding their ability to utilize acetate (Kong et al., 2004; Nguyen et al., 2011). All four Tetrasphaera genomes contain an acetate transporter gene (actP) and genes involved in acetate activation (acs, ackA and pta). Thus, they clearly have the potential to take up and utilize acetate. Furthermore, the genes encoding a general sugar transporter and glucokinase are present in all four Tetrasphaera genomes, as are the genes encoding for a glutamate/aspartate transporter. Thus, it seems that Tetrasphaera is versatile in the substrates it can potentially utilize. The genome for Accumulibacter lacks genes encoding for assimilation of glucose, agreeing with in situ observations that unlike Tetrasphaera which can utilize glucose, Accumulibacter cannot (Kong et al., 2004; Burow et al., 2008; Nguyen et al., 2011). This major metabolic difference between Tetrasphaera and Accumulibacter might have a key role in deciding which of these PAO populations outcompete the other in EBPR plants.
All genes encoding for enzymes involved in the forward TCA cycle, glycolysis, gluconeogenesis, glycogen synthesis and glycogenolysis were present in all four Tetrasphaera genomes. However, those encoding for enzymes involved in the glyoxylate shunt (isocitrate lyase (icl) and malate synthase (mas)) indicating that Tetrasphaera does not seem to bypass the steps in the TCA cycle where carbon is lost as CO2 as Accumulibacter has been proposed to do (García Martín et al., 2006). The absence of genes for the glyoxylate shunt may explain the poor growth when acetate is provided as the sole carbon source, despite the presence of the genes for acetate transport and activation.
Both PHA and glycogen are important bacterial storage polymers. Candidate genes for both acetyl-CoA acetyltransferase (phaA) and acetoacetyl-CoA reductase (phaB) were identified in all four Tetrasphaera genomes, but PHA synthase (phaC) was only found in the T. japonica genome (Table 2). Thus, only T. japonica appears to have the potential to synthesize PHA as a storage compound. This partly disagrees with previous published data. Of the four isolates studied here, T. jenkinsii was the only isolate shown previously by histochemical staining to contain ‘PHA', although the positive staining reaction was reported to be inconsistent for this organism (Blackall et al., 2000; McKenzie et al., 2006) and other lipidic material may also have given a positive signal with the applied Nile blue A staining method (Serafim et al., 2002). On the other hand, all four isolates have the potential to transform substrate into glycogen, another common carbon and energy storage polymer produced by bacteria. In Accumulibacter, glycogen is synthesized in the aerobic EBPR phase, and hydrolyzed in the subsequent anaerobic phase to provide energy and reducing power for synthesis of PHA from assimilated volatile fatty acids. However, glycogen and glycogen-like inclusions can also be synthesized under anaerobic conditions, as in Methanothrix str. FE (Pellerin et al., 1987). Thus, it is possible that glycogen is synthesized as the anaerobic storage polymer in Tetrasphaera, and used under subsequent aerobic conditions as an energy source for Pi assimilation and polyP synthesis. No ecophysiological studies have investigated this possibility because of the lack of a staining method for the specific detection of glycogen in situ (Serafim et al., 2002).
Any discussion on which source of reducing power Accumulibacter uses to synthesize PHA anaerobically from acetate (glycolysis, TCA, split TCA or a combination, reviewed by He and McMahon, 2011) is not relevant for Tetrasphaera clade 1 and 2 members because no PHA is formed, although their ability to ferment (see below) would provide an alternative source of energy.
Fermentation
In situ studies of the Tetrasphaera populations have shown that they may possess a fermentative metabolism (Kong et al., 2008; Nguyen et al., 2011). The genes important in glucose fermentation in E. coli (Clark, 1989) and Bacillus subtilis (Nakano et al., 1997) are indeed present in these four Tetrasphaera genomes (Table 3). This includes the candidate gene encoding an alanine dehydrogenase (ald), which suggests that all isolates may produce alanine as an end product of glucose fermentation, whereas the genes coding for ethanol production (acetaldehyde dehydrogenase adhE and alcohol dehydrogenase (adh)) have only been observed in T. australiensis. The gene responsible for lactate formation (lactate dehydrogenase (ldh)) was detected only in T. elongata and T. japonica. However, the lactate dehydrogenase in T. japonica (1.1.2.3 and 1.1.2.4) is theoretically metabolically irreversible, catalyzing the conversion of lactate into pyruvate, while that present in the T. elongata genome (1.1.1.27) is theoretically reversible (Stansen et al., 2005). From this observation, we hypothesize that only T. elongata has the potential to produce lactate as an end product of glucose fermentation. Acetate and succinate may also be produced as fermentation end products from glucose in all four isolates. If the annotated succinate dehydrogenase is capable of operating in reverse, then the observed anaerobically produced succinate (see later) may be a by-product of fermentation via the forward and/or reverse operation of the TCA cycle. The ability of these Tetrasphaera to generate energy from fermentation is an additional key metabolic feature distinguishing them from Accumulibacter.
Table 3. Genes coding for enzymes involved in fermentation.
| Gene | Protein | EC no. | Accumulibacter | T. australiensis | T. elongata | T. jenkinsii | T. japonica |
|---|---|---|---|---|---|---|---|
| pdc | Pyruvate decarboxylase | 4.1.1.1 | − | − | − | − | − |
| mhpF | Acetaldehyde dehydrogenase | 1.2.1.10 | − | + | − | − | − |
| aldA | Aldehyde dehydrogenase | 1.2.1.3 | − | + | + | + | + |
| adh | Alcohol dehydrogenase | 1.1.1.1 | − | + | + | + | + |
| ldh | Lactate dehydrogenase | 1.1.1.27 | − | − | + | − | + |
| pfl | Pyruvate formate lyase | 2.3.1.54 | − | − | + | − | − |
| gcvP | Glycine dehydrogenase | 1.4.4.2 | + | + | + | + | + |
| gcvT | Aminomethyltransferase | 2.1.2.10 | + | + | + | + | + |
| lpd | Dihydrolipoyl dehydrogenase | 1.8.1.4 | + | + | + | + | + |
| gcvH | Gcv-system lipoylprotein | + | + | + | + | + | |
| ltaE | Threonine aldolase | 4.1.2.5 | − | + | + | + | + |
| tdh | Threonine 3-dehydrogenase | 1.1.1.103 | − | + | + | + | − |
| kbl | Glycine acetyl transferase | 2.3.1.29 | − | + | + | + | − |
+, Candidate gene present in the genome; −, a candidate gene has not been identified in the genome.
Denitrification
All four Tetrasphaera isolates possess the potential to utilize nitrate and/or nitrite as terminal electron acceptors in anaerobic respiration or denitrification. Genes involved in the reduction of nitrate to nitrite were detected in each species. In all four genomes nirK, the defining gene for denitrification was similar phylogenetically to nirK sequences in other Gram-positive bacteria (data not shown). However, sets of genes allowing complete denitrification were not found (Table 4). Interestingly, those encoding the nitrate reductases responsible for reducing nitrate to ammonia (nirB and nirD) were detected in the genome of T. japonica (Table 4), suggesting that this organism can reduce nitrate by dissimilatory nitrate reduction to ammonia. Thus, Tetrasphaera may be able to couple nitrate and/or nitrite reduction to phosphorus uptake, which is similar to the behavior described previously for Accumulibacter clade IA members (Flowers et al., 2009).
Table 4. Genes coding for enzymes involved in denitrification and dissimilatory nitrate reduction to ammonia.
| Gene | Protein | EC no. | Accumulibacter | T. australiensis | T. elongata | T. jenkinsii | T. japonica |
|---|---|---|---|---|---|---|---|
| narI | Nitrate reductase (γ subunit) | 1.7.99.4 | − | + | + | + | + |
| narJ | Nitrate reductase (δ subunit) | 1.7.99.4 | − | + | + | + | + |
| narH | Nitrate reductase (β subunit) | 1.7.99.4 | − | + | + | + | + |
| narG | Nitrate reductase (α subunit) | 1.7.99.4 | − | + | + | + | + |
| napA | Nitrate reductase | 1.9.6.1 | + | − | − | − | + |
| nirB | Nitrite reductase, large subunit | 1.7.1.4 | + | − | − | − | + |
| nirD | Nitrite reductase, small subunit | 1.7.1.4 | + | − | − | − | + |
| nirK | Copper-containing nitrite reductase | 1.7.2.1 | − | + | + | + | + |
| norB | Nitric oxide reductase, large subunit | 1.7.99.7 | − | +a | − | − | +a |
| norC | Nitric oxide reductase, small subunit | 1.7.99.7 | − | − | − | − | − |
| norZ | Nitric oxide reductase | 1.7.99.7 | + | +a | − | − | +a |
| nosZ | Nitrous oxide reductase | 1.7.99.6 | + | − | − | − | − |
+, Candidate gene present in the genome; −, a candidate gene has not been identified in the genome.
norB and norZ have amino-acid identities of 90%. Therefore, this sequence may be either of these sequences.
Experimental metabolic investigations
The genomic investigations of the four Tetrasphaera isolates provided insight into the metabolic potential of Tetrasphaera isolates and how they may survive and compete successfully in EBPR plants. The hypotheses developed and discussed above were validated through experimental investigations of their ability to consume glucose and acetate, to ferment, to synthesize different storage compounds, to denitrify and to actively take up and release P. However, only T. elongata and T. japonica were studied as the other isolates grew too slowly in all the media tested. T. elongata represents the most abundant clade (clade 1) observed in full-scale plants in Denmark (based on FISH (fluorescent in situ hybridization)), while T. japonica has not been found so far in any Danish full-scale plants (Nguyen et al., 2011).
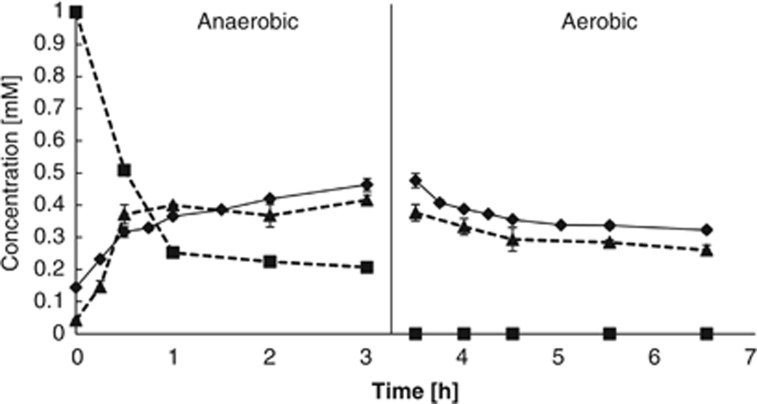
All the potential pathways revealed by the genome sequencing were shown to operate in these two species. Both consumed acetate and glucose, but as they grew very poorly on acetate, all subsequent experiments were carried out with glucose in R2A media. T. elongata grew at 20 °C with a doubling time of ∼4 and 19 h under aerobic and anaerobic conditions, respectively. During an anaerobic-aerobic cycle, T. elongata demonstrated a typical PAO phenotype (Figure 3). Thus, it consumed glucose and released Pi during the anaerobic phase, and then assimilated Pi during the subsequent aerobic conditions. In the absence of glucose in the anaerobic phase, no Pi was assimilated aerobically (data not shown). A fraction of the glucose was stored as glycogen (Figure 3) and a fraction was fermented to succinate, lactate, alanine, and acetate as determined by 13C-labeling (data not shown). Glycogen was only produced as stored Pi was released, which is similar to the pattern observed in Accumulibacter for the anaerobic formation of PHA from acetate (Oehmen et al., 2007), suggesting that the energy required is sourced from the hydrolysis and release of intracellular polyphosphate. In the aerobic phase, the glycogen was slowly consumed with time (Figure 3) and no fermentation products were observed (data not shown).
Figure 3.
The ability of T. elongata to release Pi and consume glucose in the anaerobic phase (0–3 h) and take up Pi in the aerobic phase (3.5–6.5 h), and to produce and consume glycogen in the anaerobic and aerobic phases, respectively. The average concentration and standard deviation from duplicate experiments are shown for the Pi concentration of the media (diamonds) and glycogen content of the biomass (triangles). Media glucose concentration is also shown (squares).
The amount of phosphate released after 3 h incubation was ∼19 mgP g−1 dry biomass and the amount taken up was ∼9 mgP g−1 dry biomass. A sample was also taken after 24 h of aerobic incubation which showed that the isolate had taken up ∼16 mgP g−1 dry biomass (corresponding to ∼1.4 × 10−16 mol per cell). It is difficult to extrapolate results from pure cultures to the complex conditions of full-scale plants where the P content may be even higher when considering the experiments only covered a single cycle, and P-uptake capacity may increase over time. The experimental design was intended to demonstrate the ability of Tetrasphaera to release/uptake P and not to determine the maximum uptake capacity.
T. japonica could ferment glucose, generating acetate and alanine as end products. T. japonica also produced intracellular PHA anaerobically from glucose. Following uptake, the glucose is likely activated and degraded to pyruvate through the glycolysis pathway. The produced pyruvate may be further converted into PHA as polyhydroxybutyrate via acetyl-CoA and polyhydroxyvalerate through propionyl-CoA.
Both T. japonica and T. elongata can potentially denitrify, using both nitrate and nitrite as electron acceptors (Supplementary Figure S2). Both isolates reduced the nitrate supplied completely within 48 h. During nitrate reduction, nitrite accumulated in the cultures and was not reduced until the nitrate was completely removed. Whether nitrite reduction proceeded completely to N2 was not pursued.
Developing a metabolic model for Tetrasphaera
Based on the genomic sequence data from these four Tetrasphaera isolates and experimental validation of pathways of special interest with T. elongata and T. japonica, we propose the following model for their metabolism in EBPR systems.
The genome sequence of T. japonica suggests it has the potential to synthesize PHA, glycogen and polyphosphate, and to reduce nitrate and nitrite, thus sharing key features of its metabolism with Accumulibacter (García Martín et al., 2006; He and McMahon, 2011). Furthermore, T. japonica has a fermentative ability, and thus can grow anaerobically. However, this species has not been detected by FISH in full-scale plants in Denmark (Nguyen et al., 2011), and so more data are needed from plants in other countries before its importance in EBPR can be properly assessed.
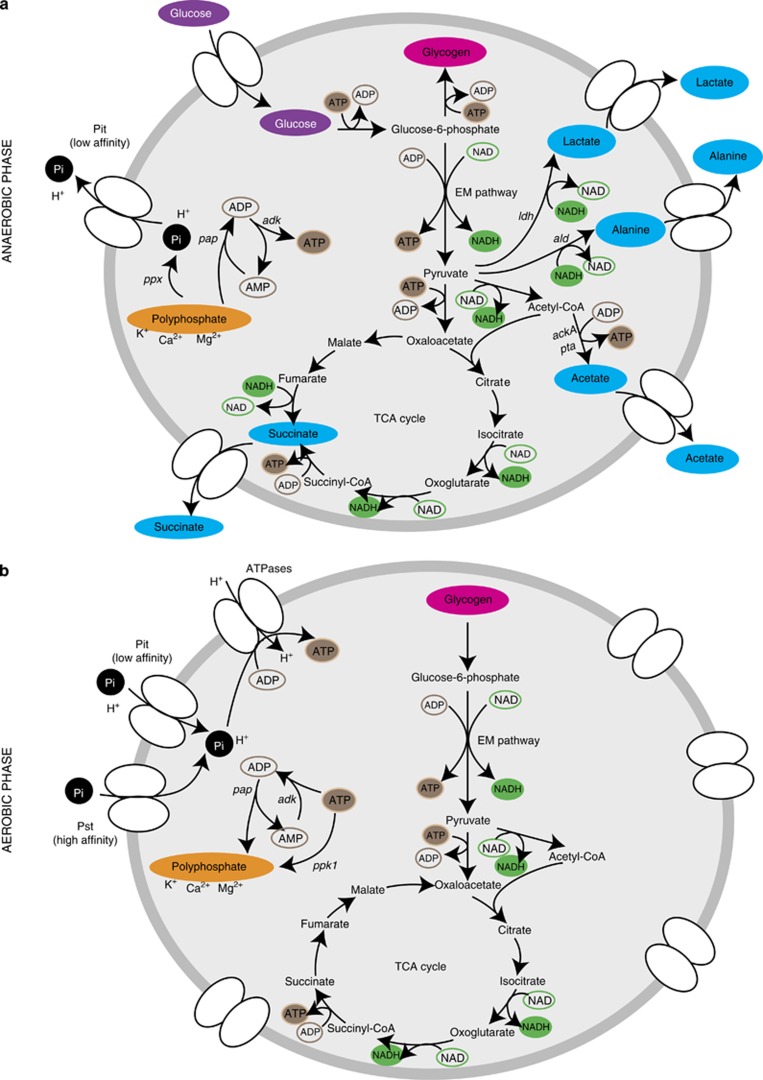
The dominant Tetrasphaera in Danish full-scale EBPR plants belonging to clades 1 and 2 (T. australiensis, T. elongata and T. jenkinsii) seem unable to synthesize PHA anaerobically, but instead produce glycogen and polyphosphate in situ. They can also denitrify and ferment. These attributes agree with the analysis presented in this study and imply that these Tetrasphaera species are metabolically highly versatile. Consequently, they may be active during all stages of EBPR, being involved in key processes that contribute to efficient nutrient removal (Nielsen et al., 2012). We propose in the model that these Tetrasphaera can grow and ferment glucose and produce glycogen under anaerobic conditions, and that the energy required for these anabolic reactions is obtained from fermentation and polyphosphate degradation (Figure 4). During the aerobic phase, where exogenous energy and carbon sources are considered to be in short supply, the stored glycogen is degraded to provide carbon and energy for growth, phosphate assimilation and polyphosphate formation. The fermentation capability that allows Tetrasphaera to grow under anaerobic conditions and the ability, like Accumulibacter, to store substrate under the carbon-rich anaerobic conditions, for later use in the carbon-deficient anoxic/aerobic periods, are the key physiological traits that potentially allow them to be so successful in EBPR plants.
Figure 4.
Metabolic model for T. elongata. The key metabolic pathways enabling Tetrasphaera to compete in full-scale EBPR plants are shown. (a) In the anaerobic phase, glucose is taken up and either stored as glycogen or fermented to acetate, lactate, succinate and alanine (highlighted in blue circles). The energy required for glycogen synthesis is supplied by fermentation and polyphosphate degradation to orthophosphate (Pi). (b) In the aerobic phase, the stored glycogen is degraded, supplying energy for growth and replenishing the polyphosphate stores.
The metabolic model proposed in this study should also provide a strong basis for future investigations into regulation of gene expression and the niche ecology of Tetrasphaera PAOs in EBPR systems. More detailed kinetic studies, expanding on the findings of this study, will be important in understanding the competition between the Tetrasphaera and other key organisms in activated sludge; such as the Accumulibacter PAO and the glycogen accumulating organisms. Although, the demonstrated ability of the Tetrasphaera PAO to ferment glucose and utilize amino acids indicates that they occupy a slightly different niche. Incorporation of such information into more comprehensive process models will also markedly advance our understanding of the structure and population dynamics of the EBPR community, and its functional stability, which is important if these biotechnological processes are to be operated more efficiently.
Acknowledgments
This study has been funded by Danish wastewater treatment plants (MiDas-DK, The Microbial Database), Viborg Energy, Aalborg University and the Danish Strategic Research Council (EcoDesign-MBR).
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Akiyama M, Crooke E, Kornberg A. The polyphosphate kinase gene of Escherichia coli. Isolation and sequence of the ppk gene and membrane location of the protein. J Biol Chem. 1992;267:22556–22561. [PubMed] [Google Scholar]
- Akiyama M, Crooke E, Kornberg A. An exopolyphosphatase of Escherichia coli. The enzyme and its ppx gene in a polyphosphate operon. J Biol Chem. 1993;268:633–639. [PubMed] [Google Scholar]
- Albertsen M, Hansen LBS, Saunders AM, Nielsen PH, Nielsen KL. A metagenome of a full-scale microbial community carrying out enhanced biological phosphorus removal. ISME J. 2011;6:1094–1106. doi: 10.1038/ismej.2011.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- APHA, AWWA, WPCF . Standard Methods for the Examination of Water and Wastewater. Port City Press: Baltimore; 1995. [Google Scholar]
- Barnard JL. Biological nutrient removal without the addition of chemicals. Water Res. 1975;9:485–490. [Google Scholar]
- Bentley S. Sequencing the species pan-genome. Nature Rev Microbiol. 2009;7:258–259. doi: 10.1038/nrmicro2123. [DOI] [PubMed] [Google Scholar]
- Blackall LL, Seviour EM, Bradford D, Rossetti S, Tandoi V, Seviour RJ. ‘Candidatus Nostocoida limicola', a filamentous bacterium from activated sludge. Int J Syst Evol Microbiol. 2000;50:703–709. doi: 10.1099/00207713-50-2-703. [DOI] [PubMed] [Google Scholar]
- Bond PL, Erhart R, Wagner M, Keller J, Blackall LL. Identification of some of the major groups of bacteria in efficient and nonefficient biological phosphorus removal activated sludge systems. Appl Environ Microbiol. 1999;65:4077–4084. doi: 10.1128/aem.65.9.4077-4084.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonting CF, Kortstee GJ, Zehnder AJ. Properties of polyphosphate: AMP phosphotransferase of Acinetobacter strain 210A. J Bacteriol. 1991;173:6484–6488. doi: 10.1128/jb.173.20.6484-6488.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braunegg G, Sonnleitner B, Lafferty RM. A rapid gas chromatographic method for the determination of poly-β-hydroxybutyric acid in microbial biomass. Eur J Appl Microbiol Biotechnol. 1978;6:29–37. [Google Scholar]
- Burow LC, Mabbett AN, Blackall LL. Anaerobic glyoxylate cycle activity during simultaneous utilization of glycogen and acetate in uncultured Accumulibacter enriched in enhanced biological phosphorus removal communities. ISME J. 2008;2:1040–1051. doi: 10.1038/ismej.2008.45. [DOI] [PubMed] [Google Scholar]
- Clark DP. The fermentation pathways of Escherichia coli. FEMS Microbiol Rev. 1989;5:223–234. doi: 10.1016/0168-6445(89)90033-8. [DOI] [PubMed] [Google Scholar]
- Comeau Y, Hall KJ, Hancock REW, Oldham WK. Biochemical model for enhanced biological phosphorus removal. Water Res. 1986;20:1511–1521. [Google Scholar]
- Crocetti GR, Hugenholtz P, Bond PL, Schuler A, Keller J, Jenkins D, et al. Identification of polyphosphate-accumulating organisms and design of 16S rRNA-directed probes for their detection and quantitation. Appl Environ Microbiol. 2000;66:1175–1182. doi: 10.1128/aem.66.3.1175-1182.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrer RA, Kemen E, Jones JDG, Studholme DJ. De novo assembly of the Pseudomonas syringae pv.syringae B728a genome using Illumina/Solexa short sequence reads. FEMS Microbiol Lett. 2009;291:103–111. doi: 10.1111/j.1574-6968.2008.01441.x. [DOI] [PubMed] [Google Scholar]
- Flowers JJ, He S, Yilmaz S, Noguera DR, McMahon KD. Denitrification capabilities of two biological phosphorus removal sludges dominated by different 'Candidatus Accumulibacter' clades. Environ Microbiol Rep. 2009;1:583–588. doi: 10.1111/j.1758-2229.2009.00090.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuhs GW, Chen M. Microbiological basis of phosphate removal in the activated sludge process for the treatment of wastewater. Microb Ecol. 1975;2:119–138. doi: 10.1007/BF02010434. [DOI] [PubMed] [Google Scholar]
- García Martín H, Ivanova N, Kunin V, Warnecke F, Barry KW, McHardy AC, et al. Metagenomic analysis of two enhanced biological phosphorus removal (EBPR) sludge communities. Nat Biotechnol. 2006;24:1263–1269. doi: 10.1038/nbt1247. [DOI] [PubMed] [Google Scholar]
- Hanada S, Liu WT, Shintani T, Kamagata Y, Nakamura K. Tetrasphaera elongata sp. nov., a polyphosphate-accumulating bacterium isolated from activated sludge. Int J Syst Evol Microbiol. 2002;52:883. doi: 10.1099/00207713-52-3-883. [DOI] [PubMed] [Google Scholar]
- He S, Kunin V, Haynes M, Martin HG, Ivanova N, Rohwer F, et al. Metatranscriptomic array analysis of 'Candidatus Accumulibacter phosphatis'-enriched enhanced biological phosphorus removal sludge. Environ Microbiol. 2010;12:1205–1217. doi: 10.1111/j.1462-2920.2010.02163.x. [DOI] [PubMed] [Google Scholar]
- He S, McMahon K. ‘Candidatus Accumulibacter' gene expression in response to dynamic EBPR conditions. ISME J. 2011;5:329–340. doi: 10.1038/ismej.2010.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hesselmann RPX, Von Rummell R, Resnick SM, Hany R, Zehnder AJB. Anaerobic metabolism of bacteria performing enhanced biological phosphate removal. Water Res. 2000;34:3487–3494. [Google Scholar]
- Ishige K, Zhang H, Kornberg A. A polyphosphate kinase (PPK2) widely conserved in bacteria. Proc Natl Acad Sci USA. 2002;99:16678–16683. doi: 10.1073/pnas.262655199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong Y, Nielsen JL, Nielsen PH. Microautoradiographic study of Rhodocyclus-related polyphosphate-accumulating bacteria in full-scale enhanced biological phosphorus removal plants. Appl Environ Microbiol. 2004;70:5383–5390. doi: 10.1128/AEM.70.9.5383-5390.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong Y, Nielsen JL, Nielsen PH. Identity and ecophysiology of uncultured actinobacterial polyphosphate-accumulating organisms in full-scale enhanced biological phosphorus removal plants. Appl Environ Microbiol. 2005;71:4076–4085. doi: 10.1128/AEM.71.7.4076-4085.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong Y, Xia Y, Nielsen PH. Activity and identity of fermenting microorganisms in full-scale biological nutrient removing wastewater treatment plants. Environ Microbiol. 2008;10:2008–2019. doi: 10.1111/j.1462-2920.2008.01617.x. [DOI] [PubMed] [Google Scholar]
- Liu JR, McKenzie CA, Seviour EM, Webb RI, Blackall LL, Saint CP, et al. Phylogeny of the filamentous bacterium 'Nostocoida limicola' III from activated sludge. Int J Syst Evol Microbiol. 2001;51:195–202. doi: 10.1099/00207713-51-1-195. [DOI] [PubMed] [Google Scholar]
- Louie TM, Mah TJ, Oldham W, Ramey WD. Use of metabolic inhibitors and gas chromatography/mass spectrometry to study poly-[beta]-hydroxyalkanoates metabolism involving cryptic nutrients in enhanced biological phosphorus removal systems. Water Res. 2000;34:1507–1514. [Google Scholar]
- Maszenan AM, Seviour RJ, Patel BKC, Schumann P, Burghardt J, Tokiwa Y, et al. Three isolates of novel polyphosphate-accumulating Gram-positive cocci, obtained from activated sludge, belong to a new genus, Tetrasphaera gen. nov., and description of two new species, Tetrasphaera japonica sp. nov. and Tetrasphaera australiensis sp. nov. Int J Syst Evol Microbiol. 2000;50:593–603. doi: 10.1099/00207713-50-2-593. [DOI] [PubMed] [Google Scholar]
- McKenzie CM, Seviour EM, Schumann P, Maszenan AM, Liu JR, Webb RI, et al. Isolates of ‘Candidatus Nostocoida limicola' Blackall et al. (2000) should be described as three novel species of the genus Tetrasphaera, as Tetrasphaera jenkinsii sp. nov. Tetrasphaera vanveenii sp. nov. and Tetrasphaeraveronensis sp. nov. Int J Syst Evol Microbiol. 2006;56:2279–2290. doi: 10.1099/ijs.0.63978-0. [DOI] [PubMed] [Google Scholar]
- Mino T, van Loosdrecht MCM, Heijnen JJ. Microbiology and biochemistry of the enhanced biological phosphate removal process. Water Res. 1998;32:3193–3207. [Google Scholar]
- Mino T, Tsuzuki Y, Matsuo T. Effect of phosphorus accumulation on acetate metabolism in the biological phosphorus removal process. Adv Water Pollut Control. 1987;27:27–38. [Google Scholar]
- Murphy J, Riley JP. A modified single solution method for the determination of phosphate in natural waters. Anal Chim Acta. 1962;27:31–36. [Google Scholar]
- Nakano MM, Dailly YP, Zuber P, Clark DP. Characterization of anaerobic fermentative growth of Bacillus subtilis: identification of fermentation end products and genes required for growth. J Bacteriol. 1997;179:6749–6755. doi: 10.1128/jb.179.21.6749-6755.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen HT, Le VQ, Hansen AA, Nielsen JL, Nielsen PH. High diversity and abundance of putative polyphosphate-accumulating Tetrasphaera-related bacteria in activated sludge systems. FEMS Microbiol Ecol. 2011;76:256–267. doi: 10.1111/j.1574-6941.2011.01049.x. [DOI] [PubMed] [Google Scholar]
- Nielsen PH, Saunders AM, Hansen AA, Larsen P, Nielsen JL. Microbial communities involved in enhanced biological phosphorus removal from wastewater—a model system in environmental biotechnology. Curr Opin Biotechnol. 2012;23:452–459. doi: 10.1016/j.copbio.2011.11.027. [DOI] [PubMed] [Google Scholar]
- Oehmen A, Lemos PC, Carvalho G, Yuan Z, Keller J, Blackall LL, et al. Advances in enhanced biological phosphorus removal: from micro to macro scale. Water Res. 2007;41:2271–2300. doi: 10.1016/j.watres.2007.02.030. [DOI] [PubMed] [Google Scholar]
- Onda S, Takii S. Isolation and characterization of a Gram-positive polyphosphate-accumulating bacterium. J Gen Appl Microbiol. 2002;48:125–133. doi: 10.2323/jgam.48.125. [DOI] [PubMed] [Google Scholar]
- Pellerin P, Gruson B, Prensier G, Albagnac G, Debeire P. Glycogen in Methanothrix. Arch Microbiol. 1987;146:377–381. [Google Scholar]
- Pereira H, Lemos PC, Reis MA, Crespo JPS, Carrondo MJ, Santos H. Model for carbon metabolism in biological phosphorus removal processes based on in vivo 13C-NMR labelling experiments. Water Res. 1996;30:2128–2138. [Google Scholar]
- Serafim LS, Lemos PC, Levantesi C, Tandoi V, Santos H, Reis MA. Methods for detection and visualization of intracellular polymers stored by polyphosphate-accumulating microorganisms. J Microbiol Methods. 2002;51:1–18. doi: 10.1016/s0167-7012(02)00056-8. [DOI] [PubMed] [Google Scholar]
- Shiba T, Tsutsumi K, Ishige K, Noguchi T. Inorganic polyphosphate and polyphosphate kinase: their novel biological functions and applications. Biochemistry (Mosc) 2000;65:315–323. [PubMed] [Google Scholar]
- Simpson JT, Wong K, Jackman SD, Schein JE, Jones SJM, Birol I. ABySS: A parallel assembler for short read sequence data. Genome Res. 2009;19:1117–1123. doi: 10.1101/gr.089532.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer DD, Delcher AL, Salzberg SL, Pop M. Minimus: a fast, lightweight genome assembler. BMC Bioinformatics. 2007;8:64. doi: 10.1186/1471-2105-8-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stansen C, Uy D, Delaunay S, Eggeling L, Goergen JL, Wendisch VF. Characterization of a Corynebacterium glutamicum lactate utilization operon induced during temperature-triggered glutamate production. Appl Environ Microbiol. 2005;71:5920–5928. doi: 10.1128/AEM.71.10.5920-5928.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallenet D, Labarre L, Rouy Z, Barbe V, Bocs S, Cruveiller S, et al. MaGe: a microbial genome annotation system supported by synteny results. Nucleic Acids Res. 2006;34:53–65. doi: 10.1093/nar/gkj406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallenet D, Engelen S, Mornico D, Cruveiller S, Fleury L, Lajus A, et al. 2009MicroScope: a platform for microbial genome annotation and comparative genomics Database (Oxford)2009:doi: 10.1093/database/bap021 [DOI] [PMC free article] [PubMed]
- Wexler M, Richardson DJ, Bond PL. Radiolabelled proteomics to determine differential functioning of Accumulibacter during the anaerobic and aerobic phases of a bioreactor operating for enhanced biological phosphorus removal. Environ Microbiol. 2009;11:3029–3044. doi: 10.1111/j.1462-2920.2009.02007.x. [DOI] [PubMed] [Google Scholar]
- Williams TM, Unz RF. The nutrition of Thiothrix, Type 021N, Beggiatoa and Leucothrix strains. Water Res. 1989;23:15–22. [Google Scholar]
- Willsky GR, Malamy MH. Characterization of two genetically separable inorganic phosphate transport systems in Escherichia coli. J Bacteriol. 1980;144:356–365. doi: 10.1128/jb.144.1.356-365.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wishart DS, Knox C, Guo AC, Young N, Gautam B, Hau DD, et al. HMDB: a knowledgebase for the human metabolome. Nucleic Acids Res. 2009;37:D603–D610. doi: 10.1093/nar/gkn810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Pijuan M, Zeng RJ, Yuan Z. Involvement of the TCA cycle in the anaerobic metabolism of polyphosphate accumulating organisms (PAOs) Water Res. 2009;43:1330–1340. doi: 10.1016/j.watres.2008.12.008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.