Abstract
Sediment-hosting hydrothermal systems in the Okinawa Trough maintain a large amount of liquid, supercritical and hydrate phases of CO2 in the seabed. The emission of CO2 may critically impact the geochemical, geophysical and ecological characteristics of the deep-sea sedimentary environment. So far it remains unclear whether microbial communities that have been detected in such high-CO2 and low-pH habitats are metabolically active, and if so, what the biogeochemical and ecological consequences for the environment are. In this study, RNA-based molecular approaches and radioactive tracer-based respiration rate assays were combined to study the density, diversity and metabolic activity of microbial communities in CO2-seep sediment at the Yonaguni Knoll IV hydrothermal field of the southern Okinawa Trough. In general, the number of microbes decreased sharply with increasing sediment depth and CO2 concentration. Phylogenetic analyses of community structure using reverse-transcribed 16S ribosomal RNA showed that the active microbial community became less diverse with increasing sediment depth and CO2 concentration, indicating that microbial activity and community structure are sensitive to CO2 venting. Analyses of RNA-based pyrosequences and catalyzed reporter deposition-fluorescence in situ hybridization data revealed that members of the SEEP-SRB2 group within the Deltaproteobacteria and anaerobic methanotrophic archaea (ANME-2a and -2c) were confined to the top seafloor, and active archaea were not detected in deeper sediments (13–30 cm in depth) characterized by high CO2. Measurement of the potential sulfate reduction rate at pH conditions of 3–9 with and without methane in the headspace indicated that acidophilic sulfate reduction possibly occurs in the presence of methane, even at very low pH of 3. These results suggest that some members of the anaerobic methanotrophs and sulfate reducers can adapt to the CO2-seep sedimentary environment; however, CO2 and pH in the deep-sea sediment were found to severely impact the activity and structure of the microbial community.
Keywords: CO2 seep, low pH, anaerobic oxidation of methane, acidophilic sulfate reduction
Introduction
Since the discovery of natural liquid CO2 accumulation and leakage in the Izena Hole hydrothermal field in the Okinawa Trough in 1990 (Sakai et al., 1990), similar phenomena have been observed in other hydrothermal systems at Yonaguni Knoll IV and Hatoma Knoll in the Okinawa Trough (Inagaki et al., 2006; Konno et al., 2006; Shitashima et al., 2008) and in volcanic arc hydrothermal vents of the northwest Eifuku seamount in the Mariana arc (Lupton et al., 2006, 2008). Geochemical data indicate that the CO2 is derived from magmatic chambers via liquid–vapor phase separation of subseafloor hydrothermal fluids (Konno et al., 2006; Lupton et al., 2006). Vapor-phase fluids stagnate in the sediment, cool and accumulate liquid CO2 below the surface seafloor. CO2 hydrates and sulfur-rich crusts form in the overlying sediments, which may act as a cap preventing CO2 migration to the hydrosphere (Inagaki et al., 2006; Konno et al., 2006; Suzuki et al., 2008).
Numerous studies have demonstrated that pH is one of the most important factors influencing microbial energy respiration, physiology and growth because of its direct effect on enzyme activity and hence cellular metabolism. However, accurately measuring pH in deep-sea CO2-seep environments is difficult because the pressure decrease associated with sample recovery will change the phase of CO2 from the liquid and/or dissolved forms to the gaseous form. Therefore, in order to understand the geochemical and geophysical nature of liquid CO2-seep environments, accurate measurements of pH (and other geochemical variables) require the use of in situ microsensors or in situ sampling with high-pressure gas-tight vessels. However, even previous onboard (ex situ) analyses of porewater from gassy Yonaguni Knoll IV CO2-seep sediments recovered with standard push corers showed a pH of ∼6.5, which is significantly lower than the ambient bottom water pH of about 7.2–7.5 (Inagaki et al., 2006).
Given the extremely high concentration of CO2 in sediments around hydrothermal CO2 vents, the pH of the porewater and the overlying seawater, where CO2-rich fluids reach the bottom waters and get dispersed with the currents, would be expected to reach extreme values. The results of direct CO2 deep-sea injection experiments and ecological surveys of elevated CO2 environments around volcanic vents suggest that changes in seawater chemistry associated with high concentrations of CO2 may reduce the biodiversity of benthic ecosystems (Barry et al., 2004; Hall-Spencer et al., 2008; Tunnicliffe et al., 2009; Fabricius et al., 2011).
The high-CO2 and low-pH deep-sea environment is an ideal natural laboratory for addressing some important ecological questions, such as: how do microbial and benthic ecosystems associated with natural CO2 reservoirs respond and adapt to this extreme environment? Can we detect significant biological processes that convert CO2 to reduced carbon compounds (for example, biomass)? Which types of microorganisms are metabolically active under extremes of CO2 and pH, and what are their biogeochemical roles in the natural environment? This knowledge is critical for determining the potential ecological impact of CO2 sequestration in deep-sea sediments (Onstott, 2005; House et al., 2006).
Previous DNA- and lipid-based molecular ecological studies at the Yonaguni Knoll IV hydrothermal field identified a relatively diverse array of bacterial and archaeal 16S ribosomal RNA (rRNA) genes, as well as some functional genes and 13C-depleted lipid biomarkers, suggesting that one-carbon (that is, CO2 and CH4) biogeochemical processes can occur in CO2-rich sedimentary environments (Inagaki et al., 2006; Nunoura et al., 2010). In this study, we focused on metabolically active microbial communities in CO2-seep sediment samples obtained from the Yonaguni Knoll IV hydrothermal field. To characterize metabolically active microbial populations in the CO2-seep sedimentary habitat, we used RNA-based molecular ecological techniques. Some important geochemical characteristics, such as pH and the concentration of CO2, were measured in situ using microsensors, and potential metabolic sulfate reduction at various pH conditions was assessed using rate measurements of radiotracer turnover. These activity-sensitive analyses expand our knowledge of microbial life and deep-sea ecosystems under high-CO2 and low-pH extremes.
Materials and methods
Sample collection
Sediment samples were obtained from the Yonaguni Knoll IV hydrothermal field of the southern Okinawa Trough during the SO196 cruise of the German RV Sonne in March 2008 (Rehder et al., 2008; Schenke et al., 2008). Using a video-guided multiple-corer system as well as push corers deployed on the ROV Quest 4000 (MARUM, Univ. Bremen), CO2-seep sediment core samples were taken from the seafloor near the Swallow Chimney (MUC8: 24° 50.838′E, 122° 41.992′E, 1362 m below sea level (mbsl)) and the Abyss vent (MUC10 located 30 m away from the Abyss Vent: 24° 50.791′N, 122° 42.020′E, 1392 mbsl; Dive 201 PC1, PC5 and PC28, located about 1 m off Abyss vent: 24° 50.781′N, 122° 42.027′E, 1382 mbsl), where CO2-rich hydrothermal activity was previously observed (Inagaki et al., 2006; Konno et al., 2006; Nunoura et al., 2010). Sediment cores were also retrieved from a low-CO2-seepage site about 15 m off the Abyss vent (Dive 203 PC8 and PC11: 24° 50.784′N, 122° 42.036′E, ∼1380 mbsl) and from hydrothermally unaffected marine sediment (MUC23: 24° 50.355′N, 122° 41.736′E, 1324 mbsl) (see Supplementary Figure S1).
After samples were recovered onboard, the sediment cores (in which CO2 bubbling was still observed) were immediately subsampled at 4-cm depth intervals using 2.5 ml-sterilized tip-cut syringes for subsequent microbiological and geochemical processing. A portion of the sediment was mixed with RNAlater (Ambion, Austin, TX, USA) and stored at −80 °C for subsequent shore-based analysis of microbial RNA. For microscopic observations, such as determination of microbial numbers and fluorescence in situ hybridization, cells were fixed with 3% paraformaldehyde for 3 h at 4 °C, washed twice with phosphate-buffered saline (PBS) (pH 7.6) and then stored in PBS/ethanol (1:1 (v/v)) at −20 °C. For radiotracer incubation experiments, 2.5 cm3 of mini-core sediment was obtained aseptically using a tip-cut syringe and placed anaerobically in glass vials containing argon in the headspace. Anaerobic sediment samples were kept at 4 °C until analyzed in the laboratory.
Porewater geochemistry
Porewater was extracted from sediment subsamples using a low-pressure squeezer (argon at 1–4 bar) in a cold room (∼4 °C) onboard the research vessel. While squeezing, the porewater was filtered through 0.2-μm Nuclepore filters. An aliquot of the extract was immediately fixed with zinc acetate and gelatin for subsequent H2S analysis. The H2S concentrations were measured onboard photometrically as methylene blue (Grasshoff et al., 1999) using a Hitachi UV/VIS spectrophotometer (Hitachi High-Technologies Co., Tokyo, Japan). Total alkalinity (TA) was determined by titration with 0.02 N HCl using a mixture of methyl red and methylene blue as an indicator. The titration vessel was bubbled with argon to strip any CO2 and H2S produced during the titration. The IAPSO (International Association for the Physical Sciences of the Ocean) seawater standard was used for method calibration. The porewater sulfate content was determined using ion chromatography at the IFM-GEOMAR laboratories. Again, the IAPSO seawater standard was used for calibration. Porewater samples were stored frozen until ion chromatography analysis.
The concentration of dissolved methane was determined using gas chromatography via the headspace technique: 3 ml of wet sediment was placed into a glass vial containing 9 ml of 0.1 M NaOH solution, tightly crimped and the sediment was suspended by vigorous shaking for 1 h. Headspace vials were stored refrigerated before analysis.
The analytical precision and accuracy of all methods described above was well below 5%.
In situ microsensor measurements
For in situ investigation of the geochemical and geophysical characteristics of the Yonaguni Knoll IV hydrothermal field, we deployed an autonomous microsensor profiler using the ROV Quest 4000 during the SO196 cruise. The profiler module (Wenzhöfer et al., 2000; de Beer et al., 2006) was equipped with microsensors for dissolved oxygen, temperature (Pt100, UST Umweltsensortechnik GmbH, Geschwenda, Germany), pH, H2S, redox and pCO2 (Microelectrodes Ltd, Ottawa, Canada). The temperature and CO2 minisensors had a diameter of 2 mm. The sensors were calibrated before deployment. The CO2 sensor was calibrated in acidified seawater, stripped with N2, to which aliquots of CO2 saturated seawater were added. The other sensors had a tip diameter of 50–100 μm, and were made and calibrated as described previously (Revsbech and Ward, 1983; Jeroschewski et al., 1996; de Beer et al., 1997). Positioned at the sediment surface, the profiler gradually moves the sensors downward in 250-μm increments over a total distance of 12.5 cm. The sensor recordings are stored internally. Readings of known O2 concentrations in the bottom water and anoxic sediment were used to crosscheck O2 sensor calibration curves.
RNA extraction
Total RNA from MUC8 and Dive 203 PC8 was extracted from 4 cm3 of RNAlater-containing frozen sediment using a RNA PowerSoil Total RNA Isolation Kit (MO BIO Laboratories, Carlsbad, CA, USA), according to the manufacturer's protocol. Immediately after resuspension of the RNA, DNA was removed by treatment with DNase I (TURBO DNA-free Kit, Ambion), and DNA removal was confirmed by negative amplification of archaeal and bacterial 16S rRNA genes using the Bac27F-Uni1492R and Arc21F-Uni1492R primer sets, respectively (DeLong, 1992). The DNA amplification conditions were as follows: 40 cycles at 98 °C for 10 s, 52 °C for 30 s and 72 °C for 120 s. The RNA concentration determined by Quant-iT RNA Assay Kit (Invitrogen, Carlsbad, CA, USA) was less than ca. 0.1 μg ml−1 of RNAlater-fixed frozen sediment. Parallel processing of sediment-free negative controls resulted in no visible amplification from the RNA extracts.
Reverse transcription and amplification of archaeal and bacterial 16S rRNA, and phylogenetic analysis
Reverse transcription (RT) was carried out to obtain 16S rRNA complementary DNA using the SuperScript III One-Step RT-PCR System with Platinum Taq DNA Polymerase (Invitrogen). The primers for the RT reaction were Bac27F, Arc21F and Uni1492R (DeLong, 1992). The RT-PCR program was as follows: 30 min at 50 °C and 2 min at 94 °C, followed by 40 cycles of 15 s at 94 °C, 30 s at 50 °C and 90 s at 68 °C. The final elongation step was increased to 5 min. The amplified 16S crDNA products were gel-purified, cloned and sequenced as described previously (Yanagawa et al., 2011).
Similarity among the 16S rRNA sequences was assessed using the FAST Group II web-based program (Yu et al., 2006). Sequences that were more than 97% similar were classified as identical phylotypes. Homology of the representative phylotypes was compared using the FASTA program against sequences in the DDBJ/EMBL/GenBank databases. The sequences were aligned with closely related sequences using the CLUSTAL-W program, followed by manual alignment. Aligned sequences were checked for chimeras with the Bellerophon v. 3 (Huber et al., 2004). Phylogenetic affiliations were then assigned by neighbor-joining analysis using ARB software (Ludwig et al., 2004). Bootstrap analyses for 1000 trial replicates were performed to assign confidence levels to the tree topology.
Pyrosequencing of 16S crDNA
Extracted RNA was reverse-transcribed and amplified with the primers EUB27F (Amann et al., 1990) and EUB338Rmix (I: 5′-GCTGCCTCCCGTAGGAGT-3′, II: 5′-GCAGCCACCCGTAGGTGT-3′, III: 5′-GCTGCCACCCGTAGGTGT-3′) (Frank et al., 2008) for bacterial 16S rRNA, and UNIV530Fmix (I: 5′-GTGCCAGCMGCCGCGG-3′, II: 5′-GTGTCAGCCGCCGCGG-3′) (Hoshino et al., 2011) and ARC912Rmix (I: 5′-CCCCCGCCAATTCCTTTAA-3′, II: 5′-CCCCCGTCAATTCCTTCAA-3′, III: 5′-CCCCCGCCAATTTCTTTAA-3′) (Miyashita et al., 2009) for archaeal 16S rRNA. The 5′-end of the forward primers contained the 454 Life Sciences Adapter (454 Life Sciences, Branford, CT, USA). The PCR conditions were as follows: initial denaturation at 94 °C for 3 min; 30 cycles of 94 °C for 30 s, 57 °C for 45 s, 72 °C for 1 min and a final 2-min extension at 72 °C. The products were pooled after cycling and cleaned using a MiniElute PCR purification kit (Qiagen, Valencia, CA, USA). Only sharp, distinct amplification products with a total yield of 200 ng were subjected to deep sequencing analysis using a GS FLX pyrosequencer (454 Life Sciences). Purification of the amplified products, quality checks and sequencing were conducted by Takara Bio Inc. (Shiga, Japan).
All reads, including sample identifier tags and primer sequences, were first processed with the Pipeline Initial Process (http://pyro.cme.msu.edu/init/form.spr), which is a part of the Ribosomal Database Project (Cole et al., 2009). Parameters for the pipeline initial process were: forward primer maximum edit distance=2, maximum number of N=0, minimum average experiment quality score=20, reverse primer maximum edit distance=0 and minimum sequence length=150. Taxonomic classification for each processed read was assigned using BLAST with a customized computer script using the ARB SILVA sequence package (Pruesse et al., 2007) as the database. The Mothur Utility package (Schloss et al., 2009) was used for statistical analyses of the 16S rRNA sequence data, and the operational taxonomic unit (OTU) at 97% cutoff, Chao-1 estimate (Chao, 1987) and Shannon diversity index (Krebs, 1989) were calculated. The evenness of the community structure was estimated by calculating the Pielou's evenness index (J′) using the equation (Pielou, 1966):
 |
where H′ represents the number derived from the Shannon diversity index and S represents the total number of OTUs.
Microscopic determination of microbial numbers in CO2-seep sediment
The number of microbes in CO2-seep sediment samples was determined using fluorescence microscopic image analysis with SYBR Green I stain (Morono et al., 2009). Paraformaldehyde-fixed slurry samples were treated with 1% hydrogen fluoride solution and gently sonicated at 5 W for 1 min with a UH-50 ultrasonic homogenizer (SMT Co. Ltd., Tokyo, Japan). An aliquot was filtered with a 0.2-μm polycarbonate filter (Isopore, Millipore, Billerica, MA, USA) and the number of SYBR Green I-stained cells was determined microscopically using an automated slide-loader system (Morono and Inagaki, 2010). The acquired images (>140 microscopic fields for each sample) were processed using MetaMorph software (Molecular Devices, Downingtown, PA, USA).
The number of cell aggregates was determined by manual counting on a BX61 fluorescence microscope (Olympus, Tokyo, Japan). Paraformaldehyde-fixed slurry samples were diluted with 0.01 M sodium pyrophosphate and gently sonicated for 30 s (UT-104, 100 W Sharp, Osaka, Japan). The samples were centrifuged and the supernatants were filtered with 0.2-μm polycarbonate filters. Microbial cells were immobilized by dipping the polycarbonate filter in 0.2% low-gelling-temperature agarose, followed by drying at 45 °C. Filters were then stained with SYBR Green I (diluted 250-fold) for 10 min at room temperature. After rinsing with pure water, each filter was mounted on a glass slide with Prolong antifade solution (Molecular Probes, Eugene, OR, USA) to prevent photobleaching. The cells on the filter were observed under a fluorescence microscope and the number of cell aggregates was determined by manual counting of >30 microscopic fields for each sample.
Catalyzed reporter deposition-fluorescence in situ hybridization
Fixed slurry samples of the MUC8 sediment core were used for catalyzed reporter deposition-fluorescence in situ hybridization (CARD-FISH) analysis. Cells were detached from the sediment matrix using a protocol described by Kallmeyer et al. (2008) with slight modifications. Briefly, a 100-μl aliquot of fixed slurry was diluted with 350 μl of NaCl solution, mixed with 50 μl of detergent mix (100 mℳ ethylenediaminetetraacetic acid, 100 mℳ sodium pyrophosphate, 1% (v/v) Tween 80 and 3% NaCl) and sonicated for 1 min. Fixed microbial cells were separated from sediment particles by layering a cushion of 500 μl of 50% (w/v) Nycodenz below the slurry with a needle and centrifuging at 4000 g for 15 min. The supernatant, including the slurry and Nycodenz layer interface, was carefully removed, transferred to a separate vial and filtered through 0.2-μm polycarbonate filters to trap microbial cells.
Next, CARD-FISH was performed using the hybridization procedure described by Pernthaler et al. (2002), with slight modifications. Horseradish peroxidase-labeled oligonucleotide probes targeting the general archaea (ARCH915) (Amann et al., 1995), Desulfosarcina/Desulfococcus bacteria (DSS658) (Manz et al., 1998) and the SEEP-SRB2 group (SEEP2-658: 5′-TCCACTTCCCTCTCCGGT-3′) (Kleindienst et al., 2012) were used, according to the published protocols, with slight modifications as described below.
Cells immobilized on polycarbonate filters were permeabilized with lysozyme (10 mg ml−1 in 0.05 M ethylenediaminetetraacetic acid and 0.1 M Tris-HCl (pH 7.5)) for 70 min at 37 °C and then digested with achromopeptidase (60 U ml−1 in 0.01 M NaCl and 0.01 M Tris-HCl (pH 8.0)) for 30 min at 37 °C. The filters were then treated with 1% H2O2 in methanol for 30 min at room temperature to deactivate endogenous peroxidase. Microbial cells were hybridized for 6 h at 46 °C in hybridization buffer containing an horseradish peroxidase-labeled oligonucleotide probe (final concentration: 0.1 pmol μl−1) and 35% (ARCH915) or 45% (DSS658 and SEEP2-658) formamide solution. The filter was then treated in a washing buffer (5 mℳ ethylenediaminetetraacetic acid (pH 8.0), 20 mℳ Tris-HCl (pH 7.5) and 0.01% sodium dodecyl sulfate) for 10 min at 48 °C, and subsequently in 1 × PBS for 15 min at room temperature. The filter was incubated in 1/50 fluorochrome-labeled tyramide solution (TSA direct; PerkinElmer, Waltham, MA, USA) for 15 min at 46 °C in the dark. Alexa488- and Cy3-labeled tyramides were used for the first and second signal amplification, respectively. The filter was then washed in 1 × PBS and dehydrated with ethanol. Fluorescence microscopic images were collected with an epifluorescent microscope (BX51; Olympus) equipped with a cooled CCD camera (DP-72; Olympus) or with an LSM 510-META laser scanning confocal microscope (Carl Zeiss, Jene, Germany).
Potential sulfate reduction rate
Potential metabolic sulfate reduction in the presence and absence of methane was examined at various pH conditions through radiotracer incubation experiments in glass tubes sealed with butyl-rubber stoppers and screw caps. Slurry samples used for potential sulfate reduction rate (pSRR) measurement were prepared in an anoxic glove chamber filled with N2. Sediment samples (2.5 cm3) were amended with 5 ml of anoxic artificial seawater containing 10 mℳ sodium sulfate. To obtain different pH values, HCl was added to aliquots of the medium. The adjusted pH values remained constant throughout the experiment. Next, 35S-labeled sodium sulfate (1 MBq) was injected into the slurry samples, and the samples were incubated horizontally for 35 days at 8 °C with 200 kPa of methane or nitrogen gas in the headspace. Sulfate reduction was stopped by adding 20 ml of 20% (w/v) zinc acetate and the slurry samples were mixed with 20 ml of a 50% (v/v) ethanol–water solution and stored at −20 °C until cold distillation of Cr(II)-reduced sulfur compounds was performed as described elsewhere (Kallmeyer et al., 2004). Reduced sulfur compounds were stripped from the sediment as H2S and carried to tubes containing 7 ml of 5% (w/v) zinc acetate by a flow of N2. The radioactivity of trapped sulfide as Zn35S was determined using liquid scintillation counting by mixing scintillation cocktail (Lumasafe Plus, PerkinElmer) with the ZnS (2:1). Potential activity was calculated from the ratio of radioactive sulfide to total radioactive sulfate (Fossing and Jørgensen, 1989).
Nucleotide sequence accession numbers
The 16S crDNA gene nucleotide sequences determined in this study were deposited in the DDBJ/EMBL/GenBank nucleotide sequence databases under the following accession numbers: AB663253–AB663295 (bacterial 16S crDNA) and AB663296–AB663305 (archaeal 16S crDNA).
Results
In situ microsensor measurements and porewater geochemistry
To measure in situ geochemical characteristics associated with CO2 seeps, we deployed a microsensor profiler close to the Abyss vent of the Yonaguni Knoll IV hydrothermal field. The microsensor data of Dives 201 were taken at about 1 and 10 m distance from the vent. Microsensor measurements of redox potential (ORP), H2S concentration and O2 concentration showed that CO2-seep sediment is a highly reduced environment and that oxygen from the overlying seawater does not penetrate more than 1 millimeter into the sediment. Concentrations of dissolved inorganic carbon and CO2 increased substantially with depth. However, we were unable to precisely determine the concentration of in situ CO2 using the microsensor unit owing to extraordinarily high CO2 levels, which presumably reach approximately 1000 to 1700 mℳ in a few centimeters below the seafloor (cmbsf). This is ∼2 orders of magnitude above the possible calibration range at 1 atm, thus in situ readings are unreliable. The pH decreased strongly with depth, reaching a value of 4.6 at a sediment depth of 6 cm (Table 1). This decrease in pH is owing to the increasing CO2 concentration in those sediment horizons. In the bottom seawater overlying the sediment, the in situ pH directly at the CO2-venting site (pH∼5.5) was significantly lower than at the low-CO2-seep site (pH∼6.6).
Table 1. Sample location, in situ pH and temperature.
| Core | Site | Location | Latitude | Longitude | Water depth (mbsl) | Core length (cmbsf) |
pH in porewatera |
Temperature |
|||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 cmbsf | 6 cmbsf | Core bottom | 0 cmbsf | 6 cmbsf | |||||||
| MUC 8 | Swallow Chimney | CO2 seep with thick pavement | 24°50.838'N | 122°41.992'E | 1362 | 30 | ND (ND) | ND (ND) | ND (ND) | ND | ND |
| Dive 201 PC28 | Abyss Vent | About 1 m off hydrothermal vent | 24°50.781'N | 122°42.027'E | 1382 | 16 | 5.5 (7.1)b | 4.6 (7.4)b | ND (7.1)b | 3.5 | 8 |
| Dive 203 PC8 | Abyss Vent | About 15 m off hydrothermal vent | 24°50.784'N | 122°42.036'E | ∼1380 | 20 | 6.6 (7.6)c | 4.9 (7.7)c | ND (7.7)c | 3.0 | 5.5 |
| MUC 10 | Abyss Vent | About 30 m off hydrothermal vent | 24°50.791'N | 122°42.020'E | 1392 | 26 | ND (7.0) | ND (6.4) | ND (6.2) | ND | ND |
| MUC 23 | — | Hydrothermally unaffected sediment | 24°50.355'N | 122°41.736'E | 1324 | 26 | ND (8.5) | ND (7.8) | ND (7.9) | ND | ND |
Abbreviations: cmbsf, centimeter below the seafloor; mbsl, meter below sea level; ND, not determined.
Onboard measurement data of pH are shown in parenthesis.
The data were obtained from adjacent core Dive201 PC5.
The data were obtained from adjacent core Dive203 PC11.
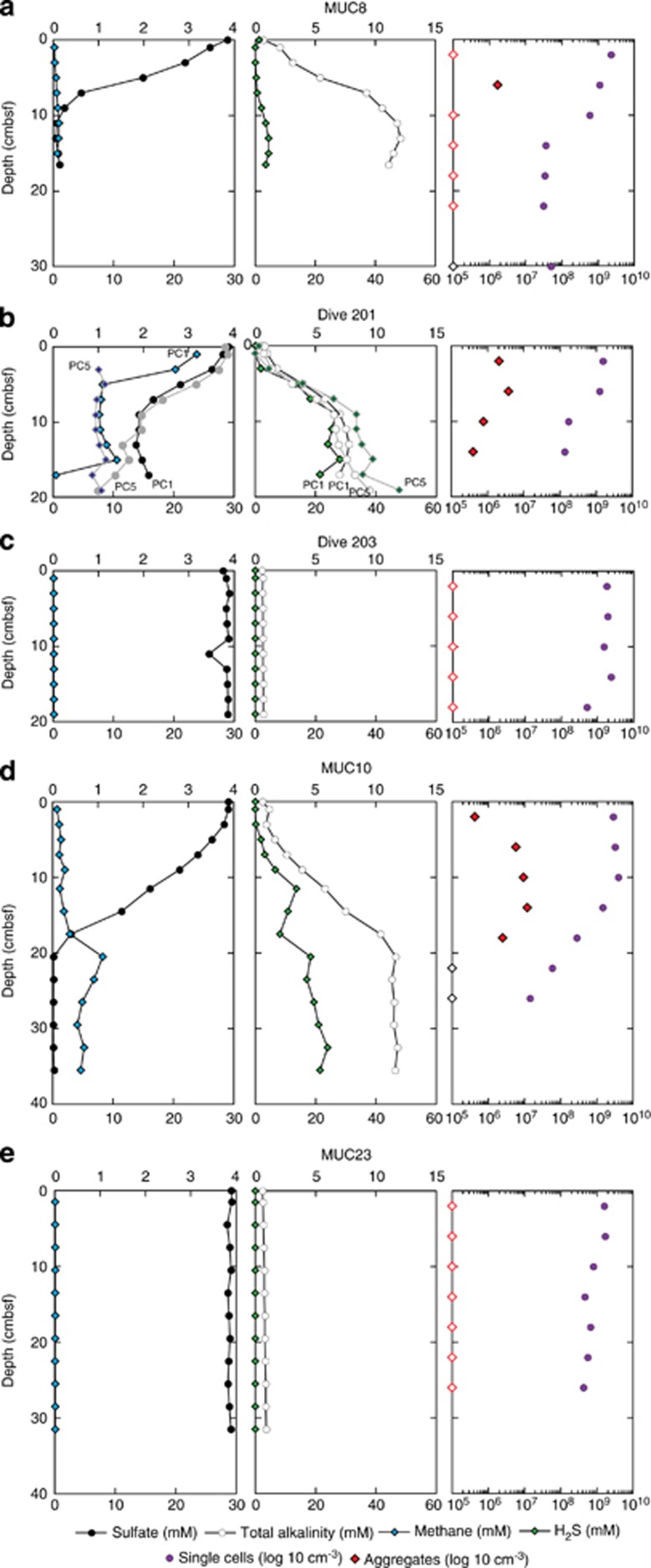
After recovery of cores from the CO2-seep sites, concentrations of dissolved methane, sulfate, H2S and TA in the porewater were determined onboard (Figure 1). The profiles from MUC8 and MUC10 showed that the concentrations of methane and sulfate increase and decrease with depth, respectively, as far down as tens of centimeters. The concentration of H2S and TA generally increase with increasing depth. MUC 8 and 10 show a depletion of sulfate in the top 10–20 cm, which is not reflected in the H2S profiles and cannot fully be explained by the anaerobic oxidation of methane with sulfate, but may indicate a strong upward transport of sulfate-depleted vent fluids. Also the presence of dissolved methane and the high TA in the sulfate reduction zone near the seafloor (Dive 201, PC1 and PC5) are indicative of upward fluid and gas transport. These geochemical characteristics in the CO2-seep areas around Swallow and Abyss Vent are clearly distinct from those in the reference sites (for example, MUC23), where the concentrations of methane and sulfate and TA were nearly constant at background seawater levels throughout the cored sediment samples, and the concentration of H2S was below the detection limit. The geochemical profile of the low-seepage core, taken about 15 m from the Abyss vent (Dive 203), was similar to the MUC23 profile.
Figure 1.
Depth profiles of porewater methane, sulfide, sulfate, TA and number of microbial cells around CO2 seepage from the Swallow Chimney (a, MUC8), Abyss vent (b, Dive 201; c, Dive 203; and d, MUC10) and reference site (e, MUC23). Microbial cell number profiles for Dive 201 and Dive 203 were obtained from PC28 and PC8, respectively. Single cells and cell aggregates were detected using SYBR Green I staining. The two geochemical profiles for Dive 201 (about 1 m from the Abyss vent) were obtained from PC1 and PC5, both of which were adjacent to PC28. The geochemical data from Dive 203 (about 10 m from the Abyss Vent) were based on PC11, which was adjacent to PC8. Black circles, sulfate; blue diamonds, methane; green diamonds, sulfide; white circles, TA; purple circles, number of single cells; and red diamonds, number of cell aggregates. Red open diamonds on left axis denote number of cell aggregates below the detection limit.
Number of microbial cells and CARD-FISH
In MUC8, MUC10 and Dive 201 PC28 samples from CO2-seep sites, the number of microbes in the upper 10 cm of sediment was generally above 109 cells per cm3, which was consistent with the number found in samples from MUC23 at the reference site and Dive 203 PC8 at the low-CO2-seep site (Figure 1). The abundance of microbes below 10 cm in the CO2-seep sediment cores (MUC8 and Dive 201 PC28) was markedly lower (107 cells per cm3), and was consistent with previous cell count data from CO2-rich sediments of the Yonaguni Knoll IV hydrothermal field (Inagaki et al., 2006).
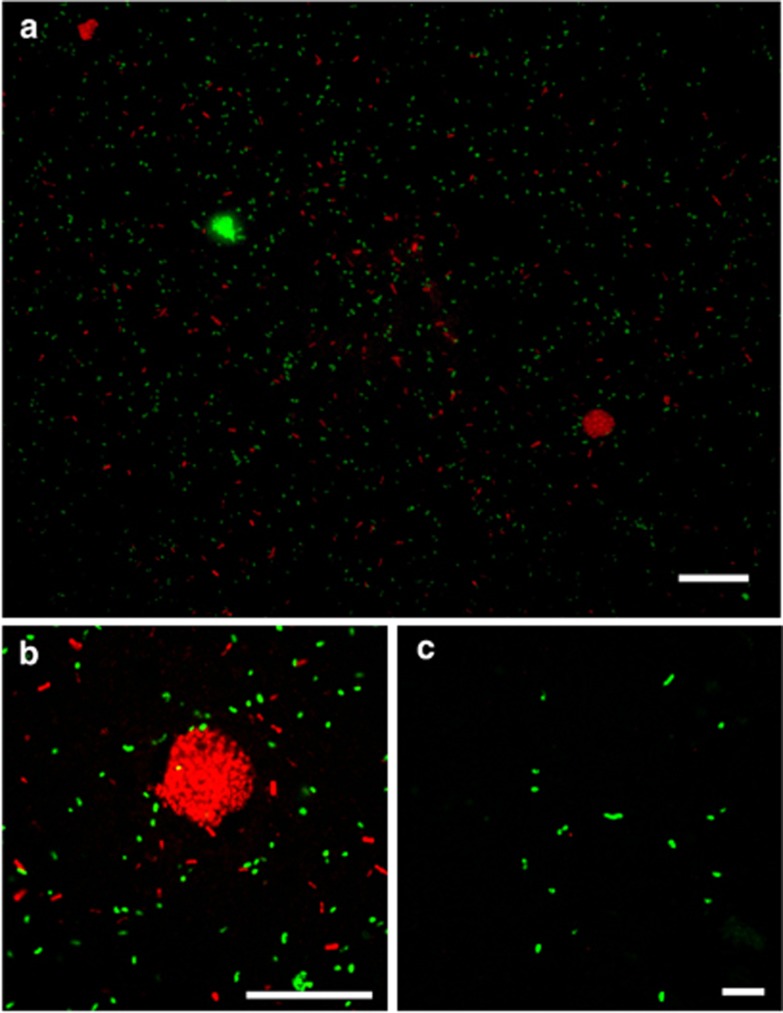
We also evaluated the number of cell aggregates in the cored sediment, which may indicate the presence of AOM consortia (Figure 1). Aggregates were frequently observed in MUC8, MUC10 and Dive 201 PC28 samples from CO2-seep sites, especially in the near-surface sediments where sulfate was abundant (Figure 1). The aggregates ranged from 4–20 μm in diameter (Figures 2a and b), and were morphologically similar to the anaerobic methanotrophic archaea (ANME)-sulfate-reducing bacteria (SRB) consortium that mediates anaerobic oxidation of methane (Boetius et al., 2000; Orphan et al., 2002; Knittel and Boetius, 2009). The number of aggregates was 2–3 orders of magnitude lower than the total number of cells (Figure 1), but each aggregate contained 210–3700 cells and an average of 1300 cells.
Figure 2.
Fluorescent microscopic images of cell aggregates from CO2-seeping sediment of the Swallow Chimney (MUC8). (a, b) Microbial cells in the sediment at a depth of 6 cm were stained using CARD-FISH. ARCH915- and SEEP2-658-positive cells are shown in red and green, respectively. Varied forms of microbial cells and cell aggregates were observed. Bar indicates 30 μm. (c) Single-type SEEP2-658-positive cells were detected from the deep zone at 30 cmbsf. Bar indicates 10 μm.
The aggregated cells were visualized using CARD-FISH with either the ARCH915 or SEEP2-658 probes, but no direct physical connections characteristic of ANME-SRB consortia were observed in the aggregate structures (Figure 2 and Supplementary Table S1). Many single cells that hybridized with either the ARCH915 or SEEP2-658 probe were observed. Single SEEP-SRB2-hybridized cells occurred together with the aggregates composed of either SEEP-SRB2- or ARC915-hybridized cells in samples collected from 6 cmbsf. At a the depth of 30 cm, at which no cell aggregates were observed, all of the SEEP-SRB2-positive cells were detected as single cells (Figure 2c). SEEP-SRB1 existed as single cells hybridized with DSS658, though they were previously found as AOM consortia partner of several ANME types (Knittel and Boetius, 2009). The commonly used probes for ANME-2 were invalid because of several mismatches with the majority of ANME-2 sequences detected in Yonaguni Knoll IV sediment (Supplementary Table S1).
Diversity of metabolically active microbial communities
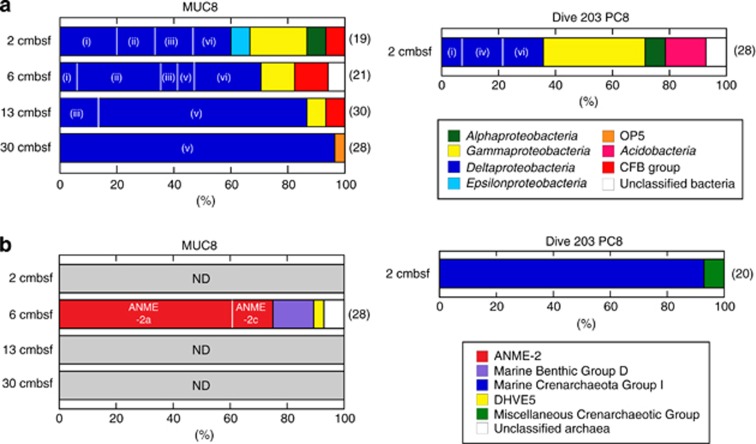
We evaluated the metabolic activity of microbial communities in MUC8 and Dive 203 PC8 samples using RNA-based molecular ecological approaches. RNA-based approach generally reflects active and living population, because ribosomes are labile and continuously turn over in cells. Clone library analysis of reverse-transcribed 16S rRNA (16S crDNA) showed that, at the CO2-seep site (that is, MUC8), the surface sediment (top 6 cm), which consists of elemental sulfur-rich altered clay and CO2 hydrates (Suzuki et al., 2008), harbors a diverse array of metabolically active bacteria that include members of the Delta-, Gamma- and Epsilon-proteobacteria and the Cytophaga-Flavobacterium-Bacteroidetes (Figure 3a). In deeper sediments collected at 13 and 30 cmbsf, the bacterial 16S crDNA clone library data clearly showed that members of the SEEP-SRB2 group (previously designated as the Eel-2 group) predominate within the Deltaproteobacteria, comprising 70–96% of the total number of sequence reads. However, also a substantial proportion of SEEP-SRB1 sequences were detected in the shallow depth, which was consistent with CARD-FISH detection of DSS658-positive single cells. As for archaeal communities, we were able to obtain archaeal 16S rRNA only from a 6-cm-deep horizon of MUC8 sediment and a surface sediment sample from Dive 203 PC8. The sequence analysis showed that ANME-2a and -2c dominated the clone library from MUC8 (60 and 15% of the total number of clone sequences, respectively), while members of Nitrosopumilales within Marine Crenarchaeota Group-I were predominantly detected in the surface sediment from Dive 203 PC8 (Figure 3b). Several phylotypes within Marine Benthic Group-D were also detected as relatively minor archaeal components in MUC8 samples. Detailed phylogenetic positions of these bacterial and archaeal 16S crDNA sequences are shown in Supplementary Figures S2 and S3.
Figure 3.
Phylogenetic community structures based on 16S crDNA clone libraries of domains. (a) Bacteria and (b) archaea from MUC8 and Dive 203 PC8 samples. The number of clones examined at each depth is indicated to the right in parentheses. The phylogenetic affiliation of each clone sequence was determined based on the trees shown in the Supplementary Figures S2 and S3. The Deltaproteobacteria were classified into six major subgroups: (i) Desulfobulbaceae, (ii) SEEP-SRB1 (DSC/DSS), (iii) Desulfobacteriaceae (excluding SEEP-SRB1), (iv) NB1-j, (v) SEEP-SRB2 and (vi) unclassified Deltaproteobacteria. ND, not detected.
Community structure analysis of metabolically active microbial communities based on pyrosequence data
We determined the structures of the CO2-seep archaeal and bacterial communities by obtaining complementary DNA sequences for over 40 000 16S rRNA amplicons and compared the results with those for the low-seepage site. The 1245 bacterial 16S rRNA sequences from the CO2-seep sediment at 6 cmbsf (MUC8) clustered into 575 OTUs at the 3% distance threshold. Chao-1 and Shannon diversity indices showed that bacterial communities from surface sediments at the CO2-seep site are generally less diverse than those in the low-seepage sediments from Dive 203 PC8 (Table 2). Significant differences in richness and evenness scores were observed between shallower and deeper sediments at the CO2-seep site. In the deeper layers, the number of OTUs was nearly the same as in the surface sediment; over 14 000 reads of 16S rRNA fragments provided a significantly lower diversity index for the deeper layers than for the surface or low-seepage samples. The asymptotic shape of the rarefaction curve indicated that most of the amplifiable sequence diversities from the CO2-seep sediment were covered by pyrosequencing of 16S crDNA (Supplementary Figure S4).
Table 2. Summary of of 16S crDNA-tag sequencing analysis of the metabolically active microbial community in the CO2-seep area of the Yonaguni Knoll IV hydrothermal field.
| Library | Sample | Depth (cmbsf) | Location | Number of sequences read | Number of OTUa | Chao 1b | Shannon index (H') | Pielou's index (J) |
|---|---|---|---|---|---|---|---|---|
| Bacteria | MUC8 | 6 | CO2 seep | 1245 | 575 | 936 (845–1057) | 5.95 | 0.94 |
| 30 | CO2 seep | 14 176 | 682 | 737 (718–766) | 3.91 | 0.60 | ||
| Dive 203 PC8 | 2 | Low seepage | 2476 | 1274 | 2232 (2066–2432) | 6.77 | 0.95 | |
| Archaea | MUC8 | 6 | CO2 seep | 14 941 | 877 | 921 (905–944) | 4.94 | 0.73 |
| Dive 203 PC8 | 2 | Low seepage | 10 496 | 698 | 722 (712–740) | 5.06 | 0.77 |
Abbreviations: cmbsf, centimeter below the seafloor; OTUs, operational taxonomic units.
OTUs defined at 97% sequence similarity.
The scores were determined by 97% sequence similarly. The numbers in parenthesis represent 95% confidence interval.
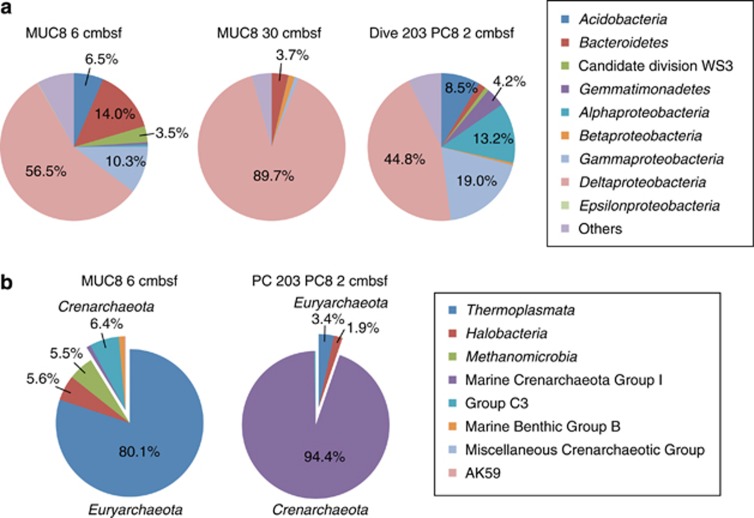
Microbial community composition at the phylum or class level was determined from pyrosequencing of 16S crDNA fragments. Major archaeal groups in surface sediments from the CO2-seep and low-seepage sites were assigned into Thermoplasmata and Crenarchaeota Group-I, respectively (Figure 4). The Marine Benthic Group-D-related Thermoplasmata represented a higher proportion of archaeal pyrosequencing fragments than in the 16S crDNA clone library (Figure 3), while Methanomicrobia, including ANME, were in a minority group (5.5%). The ANME-2a population in the pyrosequence-based community might be underestimated because their sequences from the clone library analysis had at least 1-bp mismatch with ARC912R primer. In the sample from MUC8, a single sequence phylotype within the Deltaproteobacteria dominated the bacterial community. This uneven bacterial community structure is consistently reflected in the lower Pielou's evenness index in Table 2. Phylogenetic analysis showed that more than 90% of the bacterial community sequences were composed of the SEEP-SRB2 group that represents a cluster of SRB within the Deltaproteobacteria. These sequences are closely related to OT-B08.16 (AB252432), which was previously identified in samples from the Yonaguni CO2-seep site using DNA-based molecular analysis (Inagaki et al., 2006). The representative sequence was found to comprise >95% of all delta proteobacterial sequences from MUC8 CO2-seep samples.
Figure 4.
Phylum- and class-level taxonomic classifications of (a) bacterial and (b) archaeal 16S crDNA sequences.
Potential sulfate reduction rates
As previous work suggested that AOM coupled with sulfate reduction occurs in Yonaguni CO2-seep sediment (Inagaki et al., 2006), we determined the pSRRs with and without methane in the slurry headspace at various pH conditions. Radiotracer incubation experiments using 35S-labeled sulfate showed that the pSRR in the presence of methane was consistently higher than in its absence (tested with N2 in the headspace). This suggests that methane can fuel sulfate reduction in this environment, and that organoclastic SRR from sedimented organic matter has a minor role. Other potential SR substrates in the vent fluids may include hydrogen, but were not tested here.
We evaluated the pSRR with methane in the headspace under a wide range of pH conditions in sediment samples from MUC10 (close to Abyss vent) collected at depths of 10 and 18 cm. The pSRR at 18 cmbsf was significantly lower than at 10 cmbsf, though the trend with respect to pH was generally similar. The data indicate that pSRR fueled by methane increases with decreasing pH (Figure 5a). A maximum sulfate reduction activity of 81 nmol cm−3 per day was observed at pH 3 in 10-cm-deep sediment.
Figure 5.
pSRRs determined via the cold chromium-distillation method (Kallmeyer et al., 2004). (a) Methane-dependent sulfate reduction at different pH conditions in MUC10 sediment at 10 cm (circles) and 18 cmbsf (triangles). (b) Sediment slurries from MUC8, MUC10 and Dive 201 PC28 were incubated in a 0.2-MPa methane atmosphere (closed red circles, pH 4.5; closed blue triangles, pH 7.5) and in the absence of methane (open red circles, pH 4.5; open blue triangles, pH 7.5).
The pSRR in the presence and absence of methane was determined at pH 4.5 and 7.5 for different samples of Swallow vent (MUC 8) and Abyss vent (Dive 201 PC28—15 m; MUC10—30 m). Methane-dependent sulfate reduction was observed at both pH values, and the activity at pH 4.5 was similar to or slightly higher than that observed at pH 7.5 (Figure 5b).
The pSRRs determined for MUC8 samples from Swallow vent were significantly lower than those determined for Dive 201 PC28 and MUC10 from Abyss vent, which is consistent with the environmental H2S concentration and cell aggregate number data (Figure 1). In MUC10 samples 30 m off Abyss vent, a peak in the pSRR at pH 7.5 was observed at 6 cmbsf (46 nmol cm−3 per day), whereas at pH 4.5 the peak pSRR occurred at ∼14 cmbsf (63 nmol cm−3 per day). In both samples closer to the vents (MUC8 and Dive 201 PC28), the pSRR maxima were observed at sediment surface, and barely any activity was observed >10 cm.
Discussion
One of the key features of the deep-sea environment uncovered by this study is the presence of a metabolically active microbial community in the high-CO2 low-pH sedimentary habitat. The density, diversity and metabolic activity of the sediment microbes decrease toward the CO2-rich deeper zone, which is composed of liquid CO2, CO2 hydrate and/or supercritical CO2. This indicates that CO2 and pH extremes are critical geochemical constraints for biomes in the marine sedimentary habitat (Nealson, 2006). Our RNA- and biogeochemical-based assessments of microbial composition clearly indicated that one-carbon compounds, such as methane and CO2 as well as seawater-derived electron acceptors, could serve as major carbon and energy sources in the CO2-seep environment.
Using microsensors in the deep-sea hydrothermal environment (Table 1), we first demonstrated that the in situ pH of the bottom seawater and sediment porewater at the CO2-seep area of the Yonaguni Knoll IV hydrothermal field is markedly lower (∼4.6) than previously measured onboard, when samples degassed during recovery (Inagaki et al., 2006). Considering the range of CO2 concentrations that can be measured with a microsensor, the in situ CO2 concentrations in sediment are exceptionally high, with several tens of mol m−3. In this study, we selected relatively soft, non-altered sedimentary areas associated with CO2 seepage for microsensor measurements above and beneath the seafloor. However, visible patches of yellowish elemental sulfur were observed in the examined sediment cores (for example, MUC8), and the seafloor sediment at the Yonaguni Knoll IV hydrothermal field is widely covered by a CO2-altered pavement structure that contains significant concentrations of elemental sulfur (Suzuki et al., 2008). The accumulation of a hard crust of elemental sulfur has also been observed in other CO2-seep hydrothermal environments, such as the Izena Hole and the Hatoma Knoll in the Okinawa Trough and upon Eifuku and Nikko seamounts in the Mariana volcanic arc (K Nakamura et al., unpublished data). Using microsensors, we observed a linear profile of diffusive sulfide flux down to 4 cmbsf, indicative of no or very low sulfate-reducing activity in the CO2-seep uppermost sediment. This result was inconsistent with the high pSRR determined in samples near the top of the sediment from Dive 201 PC28 collected near the Abyss vent. One conceivable explanation is that the microbial community in the CO2-seep sedimentary environment does not kinetically produce sulfide as the sulfur metabolism metabolic end product, but instead produces other elemental sulfur compounds. In addition to sulfate reducers, chemolithoautotrophic sulfur oxidizers, such as members of the Epsilonproteobacteria, were detected in the RNA-based sequence library constructed from sample collected at the top of the sediment. The reduced sulfur compounds may support some sulfur oxidizers (Inagaki et al., 2002, 2003) that were not detected in the deeper sediment, possibly owing to the lack of available electron acceptors (for example, O2 and nitrate) in the CO2-rich low-pH sediment.
The seafloor sediment in the Yonaguni Knoll IV hydrothermal field harbors a remarkable number and diversity of microbes (Inagaki et al., 2006; Nunoura and Takai, 2009; Nunoura et al., 2010). Our RNA-based molecular ecological study demonstrated that the deeper sedimentary habitat, which is directly influenced by CO2 leakage, is an extremely harsh environment for many microbes, and thus harbors only specialized sedimentary acidophilic microbes. Compared with the microbial diversity at the methane-seep sediment in the Nankai Trough, very low diversity scores for 16S rRNA-tagged sequences were obtained from the CO2-rich sediment (Nunoura et al., 2012). These results suggest that only life forms that can physiologically adapt to high-CO2 and low-pH conditions can survive in this environment.
The existence of metabolically active ANMEs and sulfate reducers (for example, SEEP-SRB1, SEEP-SRB2 and Desulfobulbuceae within the Deltaproteobacteria) in upper sediments (∼6 cm in depth) was confirmed by multiple RNA-based approaches. In addition, radiotracer incubation experiments indicated that methane-dependent acidophilic sulfate reduction potentially occurs in the CO2-seep sedimentary environment. Previous studies of ANME communities from methane seeps found that AOM-related sulfate reduction is inhibited under acidic conditions (Nauhaus et al., 2005), indicating that AOM consortia adapted to high CO2 inhabit the sediments of Yonaguni Knoll IV.
In this context, we found a high sequence abundance of sulfate reducers within the SEEP-SRB2 group of the Deltaproteobacteria, which are commonly present as a relatively minor component of anoxic methane-seep sediment communities (for example, Teske et al., 2002; Knittel et al., 2003; Yanagawa et al., 2011). The dominance of SEEP-SRB2 group was only reported from surface sediment underlying Beggiatoa mat in a Gulf of Mexico hydrocarbon seep (Lloyd et al., 2010). Our results suggested that they acquired specific metabolic functions that enable them to adapt to the high-CO2 and low-pH sedimentary habitat. Using CARD-FISH, we found, however, that the SEEP-SRB2 bacteria exist as single cells or single cell aggregates in the shallow depths at which AOM occurs, and that they are missing the archaeal partners (Figure 2). We hypothesize that SEEP-SRB2 bacteria may use multiple electron donors in the high-CO2 and low-pH sedimentary habitat, for example, these bacteria use not only methane via the syntrophic reaction with ANMEs, but probably also hydrogen or other vent fluid constituents. In the Yonaguni Knoll IV hydrothermal field, a wide range of hydrogen concentrations (up to 5.2 mmol kg−1) have been measured in venting fluids and sediments (Konno et al., 2006), representing the heterologous structure of fluid–gas stagnation that influences the availability of hydrogen for indigenous microbial activity. This hypothesis is also supported by the detection of potentially hydrogenotrophic microbes such as Epsilonproteobacteria and Hydrogenovibrio in such sediments (Inagaki et al., 2006; Nunoura et al., 2010). In contrast, no molecular signals (for example, 16S rRNA and methyl co-enzyme M reductase) related to methanogenic archaea have been detected at the CO2-impacted sites.
Anaerobic oxidation of methane may occur under a wide range of geochemical and geophysical conditions, for example, at temperatures ranging from the freezing point of seawater to 60 °C or more (Kallmeyer and Boetius, 2004; Lloyd et al., 2006; Niemann et al., 2006; Holler et al., 2011; Biddle et al., 2012). Also it was reported to occur at pH values ranging from neutral to 9–11 at the Lost City hydrothermal field (Brazelton et al., 2006). In this study, we found that AOM occurs under conditions of high CO2 causing low pH of as low as 3–4.5, mediated by novel types of ANME-2a and sulfate-reducing partners (SEEP-SRB2) not associating in cell consortia.
Acknowledgments
We thank the shipboard science party of the SO196 cruise and the crew and operation teams of the RV Sonne and ROV Quest 4000 for their support in sample collection. We also thank Noriaki Masui for technical assistance. We are grateful to Katrin Knittel and Sara Kleindienst for helpful discussion regarding CARD-FISH using the SEEP2-658 probe. This work was supported in part by a Grant-in-Aid from the Japan Society for the Promotion of Science (JSPS) Fellows (20-10764, to KY), a Grant-in-Aid for Scientific Research: Project TAIGA (New Scientific Research on Innovative Areas, 20109003, to MS and TU), the JSPS Funding Program for the Next Generation of World-Leading Researchers (NEXT Program, to FI), the JSPS Strategic Fund for Strengthening Leading-edge Research and Development (to JAMSTEC), the Max Planck Society (MPG project funds) and project SUMSUN (grant no. 03G0196B), funded by the German Federal Ministry of Education and Research.
Footnotes
Supplementary Information accompanies the paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Amann RI, Binder BJ, Olson RJ, Chisholm SW, Devereux R, Stahl DA. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl Environ Microbiol. 1990;56:1919–1925. doi: 10.1128/aem.56.6.1919-1925.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amann RI, Ludwig W, Schleifer KH. Phylogenetic identification and in-situ detection of individual microbial-cells without cultivation. Microbiol Rev. 1995;59:143–169. doi: 10.1128/mr.59.1.143-169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barry JP, Buck KR, Lovera CF, Kuhnz L, Whaling PJ, Peltzer ET, et al. Effects of direct ocean CO2 injection on deep-sea meiofauna. J Oceanogr. 2004;60:759–766. [Google Scholar]
- Biddle JF, Cardman Z, Mendlovitz H, Albert DB, Lloyd KG, Boetius A, et al. Anaerobic oxidation of methane at different temperature regimes in Guaymas Basin hydrothermal sediments. ISME J. 2012;6:1018–1031. doi: 10.1038/ismej.2011.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boetius A, Ravenschlag K, Schubert CJ, Rickert D, Widdel F, Gieseke A, et al. A marine microbial consortium apparently mediating anaerobic oxidation of methane. Nature. 2000;407:623–626. doi: 10.1038/35036572. [DOI] [PubMed] [Google Scholar]
- Brazelton WJ, Schrenk MO, Kelley DS, Baross JA. Methane- and sulfur-metabolizing microbial communities dominate the Lost City hydrothermal field ecosystem. Appl Environ Microbiol. 2006;72:6257–6270. doi: 10.1128/AEM.00574-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao A. Estimating the population-size for capture-recapture data with unequal catchability. Biometrics. 1987;43:783–791. [PubMed] [Google Scholar]
- Cole JR, Wang Q, Cardenas E, Fish J, Chai B, Farris RJ, et al. The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 2009;37:D141–D145. doi: 10.1093/nar/gkn879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Beer D, Sauter E, Niemann H, Kaul N, Foucher J, Witte U, et al. In situ fluxes and zonation of microbial activity in surface sediments of the Håkon Mosby Mud Volcano. Limnol Oceanogr. 2006;51:1315–1331. [Google Scholar]
- de Beer D, Schramm A, Santegoeds CM, Kühl M. A nitrite microsensor for profiling environmental biofilms. Appl Environ Microbiol. 1997;63:973–977. doi: 10.1128/aem.63.3.973-977.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLong EF. Archaea in coastal marine environments. Proc Natl Acad Sci USA. 1992;89:5685–5689. doi: 10.1073/pnas.89.12.5685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabricius KE, Langdon C, Uthicke S, Humphrey C, Noonan S, De'ath G, et al. Losers and winners in coral reefs acclimatized to elevated carbon dioxide concentrations. Nat Clim Chang. 2011;1:165–169. [Google Scholar]
- Fossing H, Jørgensen BB. Measurement of bacterial sulfate reduction in sediments—evaluation of a single-step chromium reduction method. Biogeochemistry. 1989;8:205–222. [Google Scholar]
- Frank JA, Reich CI, Sharma S, Weisbaum JS, Wilson BA, Olsen GJ. Critical evaluation of two primers commonly used for amplification of bacterial 16S rRNA genes. Appl Environ Microbiol. 2008;74:2461–2470. doi: 10.1128/AEM.02272-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grasshoff K, Ehrhardt M, Kremling K. Methods of Seawater Analysis. Wiley-VCH: Weinheim; 1999. p. 600. [Google Scholar]
- Hall-Spencer JM, Rodolfo-Metalpa R, Martin S, Ransome E, Fine M, Turner SM, et al. Volcanic carbon dioxide vents show ecosystem effects of ocean acidification. Nature. 2008;454:96–99. doi: 10.1038/nature07051. [DOI] [PubMed] [Google Scholar]
- Holler T, Widdel F, Knittel K, Amann R, Kellermann MY, Hinrichs K-U, et al. Thermophilic anaerobic oxidation of methane by marine microbial consortia. ISME J. 2011;5:1946–1956. doi: 10.1038/ismej.2011.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshino T, Morono Y, Terada T, Imachi H, Ferdelman TG, Inagaki F. Comparative study of subseafloor microbial community structures in deeply buried coral fossils and sediment matrices from the Challenger Mound in the Porcupine Seabight. Front Microbiol. 2011;2:231. doi: 10.3389/fmicb.2011.00231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- House KZ, Schrag DP, Harvey CF, Lackner KS. Permanent carbon dioxide storage in deep-sea sediments. Proc Natl Acad Sci USA. 2006;103:12291–12295. doi: 10.1073/pnas.0605318103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber T, Faulkner G, Hugenholtz P. Bellerophon: a program to detect chimeric sequences in multiple sequence alignments. Bioinformatics. 2004;20:2317–2319. doi: 10.1093/bioinformatics/bth226. [DOI] [PubMed] [Google Scholar]
- Inagaki F, Kuypers MMM, Tsunogai U, Ishibashi J, Nakamura K, Treude T, et al. Microbial community in a sediment-hosted CO2 lake of the southern Okinawa Trough hydrothermal system. Proc Natl Acad Sci USA. 2006;103:14164–14169. doi: 10.1073/pnas.0606083103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inagaki F, Sakihama Y, Inoue A, Kato C, Horikoshi K. Molecular phylogenetic analysis of reverse-transcribed bacterial rRNA obtained from deep-sea cold seep sediments. Environ Microbiol. 2002;4:277–286. doi: 10.1046/j.1462-2920.2002.00294.x. [DOI] [PubMed] [Google Scholar]
- Inagaki F, Takai K, Nealson KH, Horikoshi K. Sulfurimonas autotrophica gen. nov., sp. nov., a sulfur and thiosulfate oxidizing epsilon proteobacterium isolated from the Okinawa Trough. Int J Syst Evol Microbiol. 2003;53:1801–1805. doi: 10.1099/ijs.0.02682-0. [DOI] [PubMed] [Google Scholar]
- Jeroschewski P, Steuckart C, Kühl M. An amperometric microsensor for the determination of H2S in aquatic environments. Anal Chem. 1996;68:4351–4357. [Google Scholar]
- Kallmeyer J, Boetius A. Effects of temperature and pressure on sulfate reduction and anaerobic oxidation of methane in hydrothermal sediments of Guaymas Basin. Appl Environ Microbiol. 2004;70:1231–1233. doi: 10.1128/AEM.70.2.1231-1233.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallmeyer J, Ferdelman TG, Weber A, Fossing H, Jørgensen BB. A cold chromium distillation procedure for radiolabeled sulfide applied to sulfate reduction measurements. Limnol Oceanogr: Methods. 2004;2:171–180. [Google Scholar]
- Kallmeyer J, Smith DC, Spivack AJ, D'Hondt S. New cell extraction procedure applied to deep subsurface sediments. Limnol Oceanogr: Methods. 2008;6:236–245. [Google Scholar]
- Kleindienst S, Ramette A, Amann R, Knittel K. Distribution and in situ abundance of sulfate-reducing bacteria in diverse marine hydrocarbon seep sediments. Environ Microbiol. 2012;14:2689–2710. doi: 10.1111/j.1462-2920.2012.02832.x. [DOI] [PubMed] [Google Scholar]
- Knittel K, Boetius A. Anaerobic oxidation of methane: progress with an unknown process. Annu Rev Microbiol. 2009;63:311–334. doi: 10.1146/annurev.micro.61.080706.093130. [DOI] [PubMed] [Google Scholar]
- Knittel K, Boetius A, Lemke A, Eilers H, Lochte K, Pfannkuche O, et al. Activity, distribution, and diversity of sulfate reducers and other bacteria in sediments above gas hydrate (Cascadia margin, Oregon) Geomicrobiol J. 2003;20:269–294. [Google Scholar]
- Konno U, Tsunogai U, Nakagawa F, Nakaseama M, Ishibashi J, Nunoura T, et al. Liquid CO2 venting on the seafloor: Yonaguni knoll IV hydrothermal system, Okinawa Trough. Geophys Res Lett. 2006;33:L16607. [Google Scholar]
- Krebs CJ.1989Ecological Methodology2nd ednBenjamin/Cummings: Menlo Park, CA, USA [Google Scholar]
- Lloyd KG, Albert DB, Biddle JF, Chanton JP, Pizarro O, Teske A. Spatial structure and activity of sedimentary microbial communities underlying a Beggiatoa spp. mat in a Gulf of Mexico hydrocarbon seep. PLoS ONE. 2010;5:e8738. doi: 10.1371/journal.pone.0008738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd KG, Lapham L, Teske A. Anaerobic methane-oxidizing community of ANME-1b archaea in hypersaline Gulf of Mexico sediments. Appl Environ Microbiol. 2006;72:7218–7230. doi: 10.1128/AEM.00886-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig W, Strunk O, Westram R, Richter L, Meier H, Yadhukumar, et al. ARB: a software environment for sequence data. Nucleic Acids Res. 2004;32:1363–1371. doi: 10.1093/nar/gkh293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupton J, Butterfield D, Lilley M, Evans L, Nakamura K, Chadwick W, et al. Submarine venting of liquid carbon dioxide on a Mariana Arc volcano. Geochem Geophy Geosy. 2006;7:Q08007. [Google Scholar]
- Lupton J, Lilley M, Butterfield D, Evans L, Embley R, Massoth G, et al. Venting of a separate CO2-rich gas phase from submarine arc volcanoes: examples from the Mariana and Tonga-Kermadec arcs. J Geophys Res. 2008;113:B8S12. [Google Scholar]
- Manz W, Eisenbrecher M, Neu TR, Szewzyk U. Abundance and spatial organization of Gram-negative sulfate-reducing bacteria in activated sludge investigated by in situ probing with specific 16S rRNA targeted oligonucleotides. FEMS Microbiol Ecol. 1998;25:43–61. [Google Scholar]
- Miyashita A, Mochimaru H, Kazama H, Ohashi A, Yamaguchi T, Nunoura T, et al. Development of 16S rRNA gene-targeted primers for detection of archaeal anaerobic methanotrophs (ANMEs) FEMS Microbiol Lett. 2009;297:31–37. doi: 10.1111/j.1574-6968.2009.01648.x. [DOI] [PubMed] [Google Scholar]
- Morono Y, Inagaki F. Automatic slide-loader fluorescence microscope for discriminative enumeration of subseafloor life. Sci Drilling. 2010;9:32–36. [Google Scholar]
- Morono Y, Terada T, Masui N, Inagaki F. Discriminative detection and enumeration of microbial life in marine subsurface sediments. ISME J. 2009;3:503–511. doi: 10.1038/ismej.2009.1. [DOI] [PubMed] [Google Scholar]
- Nauhaus K, Treude T, Boetius A, Krüger M. Environmental regulation of the anaerobic oxidation of methane: a comparison of ANME-I and ANME-II communities. Environ Microbiol. 2005;7:98–106. doi: 10.1111/j.1462-2920.2004.00669.x. [DOI] [PubMed] [Google Scholar]
- Nealson KH. Lakes of liquid CO2 in the deep sea. Proc Natl Acad Sci USA. 2006;103:13903–13904. doi: 10.1073/pnas.0606709103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niemann H, Lösekann T, de Beer D, Elvert M, Nadalig T, Knittel K, et al. Novel microbial communities of the Haakon Mosby mud volcano and their role as a methane sink. Nature. 2006;443:854–858. doi: 10.1038/nature05227. [DOI] [PubMed] [Google Scholar]
- Nunoura T, Oida H, Nakaseama M, Kosaka A, Ohkubo SB, Kikuchi T, et al. Archaeal diversity and distribution along thermal and geochemical gradients in hydrothermal sediments at the Yonaguni Knoll IV hydrothermal field in the southern Okinawa Trough. Appl Environ Microbiol. 2010;76:1198–1211. doi: 10.1128/AEM.00924-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunoura T, Takai K. Comparison of microbial communities associated with phase-separation-induced hydrothermal fluids at the Yonaguni Knoll IV hydrothermal field, the Southern Okinawa Trough. FEMS Microbiol Ecol. 2009;67:351–370. doi: 10.1111/j.1574-6941.2008.00636.x. [DOI] [PubMed] [Google Scholar]
- Nunoura T, Takaki Y, Kazama H, Hirai M, Ashi J, Imachi H, et al. 2012Microbial diversity in deep-sea methane seep sediments presented by SSU rRNA gene tag sequencing Microb Environdoi: 10.1264/jsme2.ME12032 [DOI] [PMC free article] [PubMed]
- Onstott TC.2005Impact of CO2 injections on deep subsurface microbial ecosystems and potential ramifications for the subsurface biosphereIn: Thomas DC, Benson SM, (eds)Carbon Dioxide Capture for Storage in Deep Geologic Formation-Results from the CO2 Capture Project: Vol. 2, Geologic Storage of Carbon Dioxide with Monitoring and Verification Elsevier: London; 1207–1239. [Google Scholar]
- Orphan VJ, House CH, Hinrichs KU, McKeegan KD, DeLong EF. Multiple archaeal groups mediate methane oxidation in anoxic cold seep sediments. Proc Natl Acad Sci USA. 2002;99:7663–7668. doi: 10.1073/pnas.072210299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pernthaler A, Pernthaler J, Amann R. Fluorescence in situ hybridization and catalyzed reporter deposition for the identification of marine bacteria. Appl Environ Microbiol. 2002;68:3094–3101. doi: 10.1128/AEM.68.6.3094-3101.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pielou EC. Species-diversity and pattern-diversity in study of ecological succession. J Theor Biol. 1966;10:370–383. doi: 10.1016/0022-5193(66)90133-0. [DOI] [PubMed] [Google Scholar]
- Pruesse E, Quast C, Knittel K, Fuchs BM, Ludwig WG, Peplies J, et al. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 2007;35:7188–7196. doi: 10.1093/nar/gkm864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehder G, Schneider von Deimling J, Cruise participants 2008. RV Sonne Cruise Report SO 196, SUMSUN 2008, Suva Guam Okinawa Trough Manila, 19 February —26 March, 2008, Leibniz Institut für Ostseeforschung: Sektion Meereschemie, Warnemünde.
- Revsbech NP, Ward DM. Oxygen microelectrode that is insensitive to medium chemical composition: use in an acid microbial mat dominated by Cyanidium caldarium. Appl Environ Microbiol. 1983;45:755–759. doi: 10.1128/aem.45.3.755-759.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai H, Gamo T, Kim ES, Tsutsumi M, Tanaka T, Ishibashi J, et al. Venting of carbon-dioxide rich fluid and hydrate formation in mid-Okinawa Trough backarc basin. Science. 1990;248:1093–1096. doi: 10.1126/science.248.4959.1093. [DOI] [PubMed] [Google Scholar]
- Schenke, Werner H, Rehder G.2008Swath sonar bathymetry during Sonne cruise SO196 with links to multibeam raw data files Alfred Wegener Institute for Polar and Marine Research: Bremerhaven; doi: 10.1594/PANGAEA.701955 [DOI] [Google Scholar]
- Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol. 2009;75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shitashima K, Maeda Y, Koike Y, Ohsumi T. Natural analogue of the rise and dissolution of liquid CO2 in the ocean. Int J Greenhouse Gas Control. 2008;2:95–104. [Google Scholar]
- Suzuki R, Ishibashi J, Nakaseama M, Konno U, Tsunogai U, Gena K, et al. Diverse range of mineralization induced by phase separation of hydrothermal fluid: a case study of the Yonaguni Knoll IV hydrothermal field in the Okinawa Trough back-arc basin. Res Geol. 2008;58:267–288. [Google Scholar]
- Teske A, Hinrichs K-U, Edgcomb V, Gomez AD, Kysela D, Sylva SP, et al. Microbial diversity of hydrothermal sediments in the Guaymas Basin: evidence for anaerobic methanotrophic communities. Appl Environ Microbiol. 2002;68:1994–2007. doi: 10.1128/AEM.68.4.1994-2007.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tunnicliffe V, Davies KTA, Butterfield DA, Embley RW, Rose JM, Chadwick WW. Survival of mussels in extremely acidic waters on a submarine volcano. Nature Geosci. 2009;2:344–348. [Google Scholar]
- Yanagawa K, Sunamura M, Lever MA, Morono Y, Hiruta A, Ishizaki O, et al. Niche separation of methanotrophic archaea (ANME-1 and -2) in methane-seep sediments of the eastern Japan Sea offshore Joetsu. Geomicrobiol J. 2011;28:118–129. [Google Scholar]
- Yu YN, Breitbart M, McNairnie P, Rohwer F. FastGroupII: a web-based bioinformatics platform for analyses of large 16S rDNA libraries. BMC Bioinformatics. 2006;7:57. doi: 10.1186/1471-2105-7-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenzhöfer F, Holby O, Glud RN, Nielsen HK, Gundersen JK. In situ microsensor studies of a shallow water hydrothermal vent at Milos, Greece. Mar Chem. 2000;69:43–54. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.