Abstract
Since human embryonic stem cells (hESCs) were first differentiated to beating cardiomyocytes a decade ago, interest in their potential applications has increased exponentially. This has been further enhanced over recent years by the discovery of methods to induce pluripotency in somatic cells, including those derived from patients with hereditary cardiac diseases. Human pluripotent stem cells have been among the most challenging cell types to grow stably in culture but advances in reagent development now mean that most laboratories can expand both embryonic and induced pluripotent stem cells robustly using commercially available products. However, differentiation protocols have lagged behind and, in many cases, only produce the cell types required with low efficiency. Cardiomyocyte differentiation techniques were also initially inefficient and not readily transferable across cell lines, but there are now a number of more robust protocols available. Here we review the basic biology underlying the differentiation of pluripotent cells to cardiac lineages and describe current state-of-the-art protocols as well as ongoing refinements. This should provide a useful entry for laboratories new to this area to start their research. Ultimately, efficient and reliable differentiation methodologies are essential to generate desired cardiac lineages in order to realize the full promise of human pluripotent stem cells for biomedical research, drug development, and clinical applications.
Keywords: human embryonic stem cells, human induced pluripotent stem cells, cardiomyogenesis, differentiation, cardiac disease models
The successful isolation of human embryonic stem cells (hESCs) and, more recently, the generation of induced pluripotent stem cells (hiPSCs) has ushered in a new era of opportunities for cardiovascular research and therapies.1–4 These human pluripotent stem cells (hPSCs) can undergo differentiation in vitro to generate derivatives of the three primary germ layers and hence potentially all the cell types present in the body. However, to take advantage of the promise of these cell sources, reproducible and efficient differentiation protocols to form the cell types of interest are essential. Protocols for different cell lineages have been described that exhibit variable success. In most cases, the in vitro differentiation recapitulates the stepwise stages of embryological development for the cell type of interest. In this review, we focus on differentiation of hPSCs to cardiomyocytes (CMs).
The generation of hPSC-derived CMs is of growing interest for multiple applications. First, access to an in vitro model of human development permits the study of human heart development in ways not otherwise possible. Secondly, stem cell-derived CMs serve as a human cardiac model that can be used for diverse basic research studies ranging from cellular electrophysiology to protein biochemistry. Furthermore, the ability to generate hiPSCs from patients with inherited cardiac diseases provides unprecedented opportunities for studying disease in human CMs.5–7 Access to abundant populations of human CMs is of particular interest to the pharmaceutical industry as a tool to develop new cardioactive compounds, and perhaps more importantly, to screen compounds for potential cardiotoxicity, such as drug-induced QT prolongation.8, 9 Finally, in the long-term, clinical applications using hPSC-derived CMs might provide a powerful approach to repair the injured heart, although the challenges will take time to overcome.10, 11 Regardless of the use of hPSC-derived CMs, efficient and reproducible differentiation protocols are needed.
Here we review current best methods for differentiating hPSC to CMs and describe the underlying biology. There is still room for further improvement, since even the most successful laboratories are continuing to refine their protocols. Compared to just a few years ago, however, it is now possible to determine whether cells have the capacity to differentiate to cardiomyocytes on the basis of just a few principle protocols. Some of the protocols require that the stem cells have a particular “history” or have been pre-adapted to a particular starting condition as undifferentiated cells. Some protocols can be scaled-up, others are more limited in this respect. We indicate the particular merits and caveats for each protocol discussed.
Lessons from embryonic cardiac development
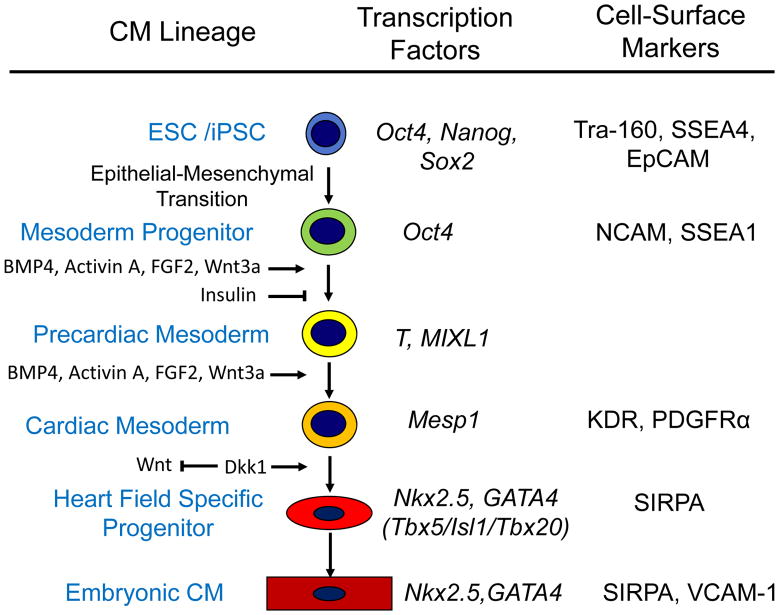
Because in vitro differentiation of stem cells to CMs mimics the sequential stages of embryonic cardiac development, a brief description of the key steps and factors in cardiac development are highlighted. However, readers are referred to more comprehensive reviews on cardiac development for detailed information.12–14 The heart is one of the first identifiable tissues to develop in vertebrate embryos. It forms soon after gastrulation from anterior migrating mesodermal cells that intercalate between the ectoderm and endoderm cell layers in the primitive streak. Heart forming- or cardiac progenitor cells are primarily localized in the mid-streak. Signals from adjacent cell populations promote induction of cardiac mesoderm, and the endoderm in particular, appears to have a highly conserved instructive function in cardiogenesis.15
Three families of protein growth factors are thought to control these early stages of mesoderm formation and cardiogenesis: bone morphogenetic proteins (BMPs), which are members of the transforming growth factor β superfamily; the Wingless/INT proteins (WNTs); and the fibroblast growth factors (FGFs). These factors, or their inhibitors, are expressed in the endoderm. Genetic disruption of their signaling has dramatic effects on cardiac development (reviewed by Olson and Schneider).14 In vertebrates, BMP signaling generally promotes cardiogenesis whilst Wingless in Drosophila and related Wnt proteins in vertebrates are involved in cardiac specification. Wnt action is complex, however, and may inhibit or promote differentiation depending on spatiotemporal context and whether the canonical signaling pathway (acting via β-catenin/GSK3 to repress cardiogenesis) or the non-canonical pathway (acting via PKC/JNK to promote cardiogenesis) is activated.14, 16 Finally, studies in chick and zebrafish have implicated a cardioinductive role for FGFs, although in Drosophila, FGFs appear to provide positional cues to cells for specification. BMP2 can upregulate FGF8 ectopically and BMP2 and FGF8 probably synergize to drive mesodermal cells into myocardial differentiation.15 Furthermore, conditional knockouts of Fgf8 and FgfR1 result in impaired outflow tract development due in part to proliferation defects in cells from the second heart field.17, 18 Overall, it appears that the timing and relative expression of different growth factor combinations induce then pattern the cardiogenic mesoderm.
Once anteriorally migrated mesoderm cells have received appropriate signals, they switch on a highly conserved heart-specific combination of transcription factors that establish the cardiac transcriptional program. Initially, the mesodermal precursor cells in the primitive streak express transcription factors such as the T-box factor Brachyury (T) and the homeodomain protein, Mixl1. Prior to migration from the streak, these cells transiently activate the basic helix-loop-helix transcription factor mesoderm posterior 1 (Mesp1) to enter a 'precardiac' mesoderm stage of development.19, 20 A subset of the Mesp1+ cell population then begins to express the homeodomain transcription factor Nkx2-5, the T-box protein Tbx5 and Isl1, a LIM homeodomain transcription factor, which are early markers of the cardiac lineage that are activated during the formation of the heart fields. In mice, Nkx2-5 is essential for the interpretation of patterning signals within the primitive heart tube and is likely to act in concert with Tbx5 during formation of both the atrial and left ventricular compartments to positively regulate transcription. Nkx2-5 and Tbx5 associate with members of the GATA family of zinc finger transcription factors (GATA4/5/6) and with serum response factor (SRF) to activate cardiac structural genes, such as actin, myosin light chain, myosin heavy chain (MHC), troponins and desmin. Tbx5 can also cooperate with Nkx2-5 to activate expression of ANF and the junctional protein connexin 40. GATA4, which has an early role in heart tube formation through regulation of extra-embryonic endoderm and embryo folding21, 22, is essential for proepicardium formation23 and muscle development as cardiomyocyte specific ablation of GATA4 results in myocardial hypoplasia,24, 25 GATA6 also plays a key role in myogenesis and embryos without both GATA 4 and 6 do not develop heart tissue.26 Members of the myocyte enhancer factor 2 (MEF2) family of transcription factors also play key roles in CM differentiation by regulating cardiac muscle structural genes. Thus, multiple complex interactions between this highly conserved gene regulatory networks control the initial differentiation, proliferation and maturation of CMs. Apart from their functional role, many of these factors can be used as markers of emerging CMs in differentiating cultures of hPSC.
The sequential activation of the transcription factors that drive the formation of the nascent-precardiac mesoderm, and eventually determine cardiac cell fate is likely to be controlled in hESC and hiPSC in much the same way as it is in embryos. As evidence supporting this assumption, many of the successful protocols developed to induce cardiomyogenesis in pluripotent cells are based on activating or inhibiting these known signaling pathways. The same sequence of gene changes observed in vivo are also observed in differentiating hPSCs.27 Thus, a hierarchy of cardiac progenitors likely occurs during in vitro differentiation ultimately leading to the formation of CMs (see Figure 1).28
Figure 1. Model of Differentiation of Human PSC via Sequential Progenitors to Cardiomyocytes.
Based on available data, this simplified model shows the cardiomyocyte lineage hierarchy progressing through sequential progenitors identified by key transcription factors as well as known cell-surface markers. Some of the best characterized signaling pathways responsible for the sequential transitions in cell fate are shown.
Embryoid body-mediated differentiation
Cultured mouse pluripotent cells including embryonal carcinoma cells (mECCs) and embryonic stem cells (mESCs) provided the first opportunities to develop methods for in vitro differentiation of pluripotent cells to CMs. When mECC and mESC cultures were dissociated to single cells and aggregated, typically in hanging drops, they developed into spheroids with an inner layer of ectoderm-like cells and a single outer layer of endoderm. To reflect the similarity between these spheroids and early post implantation embryos they were termed embryoid bodies (EBs).29 Cells in EBs differentiated to derivatives of the three primary germ layers.30 Unlike normal embryonic development, these EBs were highly variable in structure and composition, but a fraction of the EBs exhibited spontaneously contracting regions containing CMs. These early studies identified critical parameters for optimizing in vitro cardiogenesis including: 1) the starting number of mESCs used to form aggregates; 2) the specific medium and serum; 3) the mESC line employed; 4) the time of EB plating to allow substrate attachment and outgrowth.31 More recent studies have attempted to improve the process further by testing a range of different growth factors and small molecules which are described elsewhere for the mouse system.32
The successful isolation of hESCs in 1998 led to efforts to differentiate these cells as EBs.3 However, it was quickly evident that unlike mESCs, hESCs did not readily tolerate enzymatic isolation to single cells, undergoing apoptosis and failing to form EBs. Therefore, the first successful EB-mediated differentiation of hESCs to CMs used collagenase IV to disperse hESC colonies (from the H9 line) into small clumps of 3–20 cells which were grown as suspension EBs in non-adherent plastic petri dishes.33 The EBs were transferred to 0.1% gelatin-coated culture or attachment, and beating areas were first observed 4 days after plating. Comparable 'spontaneous' EB differentiation protocols soon generated CMs from a variety of other hESC lines34, 35 and more recently for hiPSC lines.36 Furthermore, these early studies provided evidence that CMs were driving the contraction observed, based on gene expression patterns, immunolabeling for myofilament proteins and electron microscopy demonstrating sarcomeric organization typical of developing CMs. In addition, electrophysiological analysis showed that the cells generated spontaneous electrical field potentials and action potentials typical of early nodal-, atrial- and ventricular-like cells.33–35 However, these studies were unable to explicitly measure the number of CMs that were actually present in these EBs, and subsequent studies have suggested it was probably just a few percent. Significant line-to-line variability in the efficiency of CM formation was also observed.37 Reproducibility of these early protocols was problematic for a number of reasons including the inclusion of undefined components such as serum, the heterogeneous EB sizes and phenotypic 'drift' in the undifferentiated cell population dependent on basal culture conditions. Importantly, in the case of serum, as earlier investigators realized for mouse EB differentiation, testing of serum was essential to identify lots that promote cardiogenesis. Subsequent protocols, therefore, have focused on moving away from serum- and feeder-based culture conditions towards fully defined conditions and generating EBs of uniform and defined sizes (Figure 2).
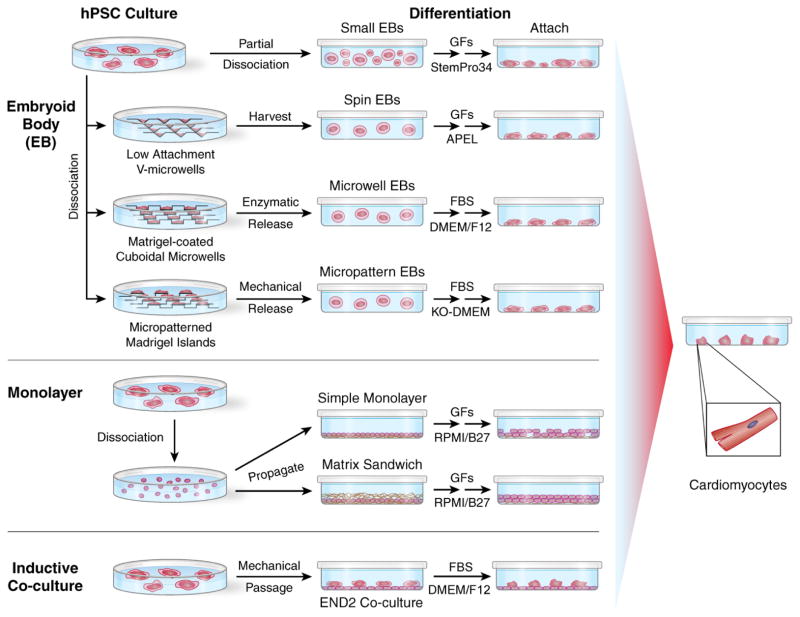
Figure 2. Current Methods for Cardiac Differentiation of Human PSCs.
The three major approaches for differentiation of human PSCs to CMs are summarized: embryoid body (EB), monolayer culture, and inductive co-culture. Methods for forming EBs range from a simple enzymatic partial dissociation of hPSC colonies to various methods to more precisely control EB cell number and EB size using microwells with forced aggregation (centrifugation), microwells to first expand hPSC colonies to a defined size, and micropatterned hPSC colonies of defined sizes. Alternatively, propagating hPSCs as monolayers on Matrigel with defined media can be used for cardiogenesis. For both EBs and monolayer approaches, stage-specific application of key growth factors (GFs) in defined media are required for optimal cardiogenesis, although some protocols use fetal bovine serum (FBS) or small molecules to induce cardiogenesis (see text for details of specific protocols). Co-culture of mechanically passaged hPSCs with visceral endodermal-like END2 cells takes advantage of cell signaling from END2 cells to promote cardiogenesis.
Media and Growth Factor Optimization for EB Differentiation
Protocols were developed using a variety of serum-free defined media for cardiac differentiation in EBs including commercially available media, APEL (StemCell Technologies) and StemPro34 (Invitrogen).38, 39 The APEL (Albumin, Polyvinylalcohol (PVA), Essential Lipids) medium is based on the chemically defined medium originally described by Johansson and Wiles,40 that permitted an objective assessment of the role of exogenously added growth factors in directing differentiation. APEL medium contains only recombinant human proteins (albumin, transferrin and insulin) and is free of animal or human products. StemPro-34 was developed to likewise be serum free without added growth factors and was first used to culture hematopoietic progenitors, but it has been utilized for EB cardiac differentiation more recently.38, 41
In the absence of serum to induce cardiogenesis, the newer protocols have tested a range of growth factors implicated in normal cardiac development including BMP4, ActivinA, FGF2, Wnt agonists and antagonists, and VEGF. Optimization of the growth factors for mesendoderm differentiation was assayed using a genetically tagged HES3 cell line (MIXL1GFP/w) in which the expression of GFP is linked to the expression of the primitive streak marker MIXL1.42 Using this cell line, over 90% of cells in day 3 spin EBs, differentiated in APEL medium containing BMP4, Activin A or combinations of the two factors, express MIXL1-GFP. Protocols in which cells are differentiated in BMP4 alone are biased towards posterior mesodermal derivatives such as primitive hematopoietic precursors42 whilst the use of high concentrations of Activin A result in endodermal differentiation.43
Cardiac lineages that differentiate from mesodermal progenitors are critically dependent on the concentration of growth factors including BMP4 and Activin A and the time of their addition and removal. The optimal concentration of these factors for any given hESC or hiPSC line may be determined by the cross titration of these factors.39, 41 Furthermore, for some growth factors such as Wnts that promote cardiomyogenesis at early stages, it may be important to inhibit signaling later on.44, 45 Cell line variability in growth factor requirements has been partially attributed to variations in endogenous growth factor signaling.41 Additional factors in protocols that can improve the efficiency or reproducibility of CM generation from EBs include vascular endothelial growth factor (VEGF),38 and the use of a lower concentration of insulin.46
Spin EBs
The heterogeneity of the EBs formed in the early studies adversely affected the reproducibility and synchronicity of differentiation. In order to improve the reliability and robustness of hESC differentiation, a protocol was developed in which defined numbers of enzymatically adapted hESCs47 were seeded into low attachment multiwell plates and aggregated by centrifugation.48 In this protocol, the centrifugation of undifferentiated cells into U- or V-bottomed wells leads to the formation of aggregates of identical size in each well, and by varying the number of cells in each well, the aggregate size can be precisely controlled.48, 49 A high throughput approach for the generation of EBs by forced aggregation has been described using a silicon wafer-based microfabrication technology to generate surfaces that permit the production of hundreds to thousands of EBs per cm2 which is commercially available as Aggrewells®.50 Using this approach, the optimal sized hEB for cardiogenesis was suggested to be in the range of 250–300 μm in diameter on day 3 of differentiation.49 The formation of these ‘spin EBs’ hinges upon the preadaption to enzymatic passage combined with the use APEL medium. The inclusion of PVA is essential for initial aggregation of the cells to form EBs. The use of spin EBs with optimized concentrations of growth factors is a preferred method for high efficiency differentiation which can be used without specialized apparatus.39
Microwell EBs
Engineered microwells provide a way to control hESC colony size as well as to generate EBs of uniform size. This technology involves the microfabrication of microwells of defined sizes.51, 52 The bottom of the microwells is coated with Matrigel extracellular matrix to promote cell adhesion, and the hESCs seeded into the microwells grow efficiently to fill the microwell and generate colonies of defined size. Growth of hESCs in microwells was found to be able to sustain self-renewal and lead to less background differentiation of hESCs than growth under standard 2-D culture conditions.52 By generating cuboidal microwells of different dimensions, different sized hESC colonies form. Removing the hESC colonies from the microwells resulted in cell aggregates of defined sizes that can be used for EB-mediated differentiation. With this approach, it was demonstrated that EBs of 100–300 μm X-Y dimension and 120 μm Z dimension were optimal for generating CMs.53 This microwell technology has the advantage of generating uniform-sized colonies of hESCs, which have minimal differentiation, and an appropriate substrate for subsequent differentiation. This provides a platform for the standardized input of hESCs for EB formation, similar to the spin EB method. Importantly, such technology also has the potential to be scaled up. However, the microwell technology is not currently commercially available at this time and requires access to the appropriate microengineering technology.
Micropatterned EBs
An alternate engineering approach to generate EBs of defined sizes used micropatterning.54, 55 Using a microcontact printing approach, size-specified hESC colonies were plated from single-cell suspensions onto micropatterned extracellular matrix (Matrigel) islands. Such islands of matrix can be considered as localized niches for the hPSCs and manipulation of the matrix altered the behaviour of the cells. Varying the size of the Matrigel islands (200, 400, and 800 μm) varied the size of the hESCs colonies. By mechanically transferring entire colonies into suspension culture, size-controlled EBs were generated. These experiments likewise demonstrated an optimal size range for maximizing mesoderm formation and cardiac induction. It also produced more homogenously sized EBs than conventional methods. The approach is also amenable to scale-up, but it requires access to microcontact printing equipment.
Monolayer differentiation to CMs
An alternative approach for differentiation of hPSCs starts with cells grown as monolayers. Taking advantage of a relatively uniform monolayer of cells without the complex diffusional barriers present in EBs, application of growth factors and other interventions will theoretically be more readily controlled and reproducible. Furthermore, because hESCs maintained in some feeder-free culture systems such as mTESR do not readily form spin EBs (ESN, DAE and AGE unpublished results), directed differentiation of hESCs in monolayer format represents a convenient alternative. The monolayer protocols also do not require replating steps typical of EB protocols thus reducing the procedural steps as well as the use of tissue culture supplies.
Building on monolayer-based protocols for differentiation of endodermal derivatives,56 a differentiation protocol was described which plated H7 hESCs on Matrigel™ as a monolayer in the presence of mouse embryonic fibroblast conditioned media.57 To direct cardiac differentiation, cells are sequentially treated with Activin A for 24 hours followed by BMP4 in serum-free RPMI medium plus a B27 supplement. The protocol for H7 hESCs resulted in >30% CMs, and with a Percoll gradient centrifugation step the CMs were enriched to >70%, which was a dramatic improvement over the EB method for this cell line. However, this initial protocol has not been successfully applied to many other hESC or hiPSC lines, and so a number of modifications have been tested such as addition of Wnt3a at the induction of differentiation.58
An alternative approach employs APEL medium59 instead of RPMI/B27 when cytokines are added for induction. In this protocol, enzymatically adapted hESCs grown on MEFs can be dissociated into single cells and replated onto Matrigel™ in knockout serum replacer (KOSR, Invitrogen) hESC culture medium. Cells attach and form large numbers of small colonies after 24–48 hr, at which time the medium is replaced with APEL medium containing cytokines that induce cardiac mesoderm, specifically BMP4, Activin A and WNT3A. This differentiation process is again critically dependent upon the ratio of BMP4 to Activin A, the time of addition and the time of removal of growth factors since each step in the process of mesoderm formation, specification and differentiation requires its own growth factor combination and concentration. Furthermore, as for EB-based methods, optimization of the concentrations of BMP4 and Activin A for individual specific cell lines has proven important. Although it has been implied that differentiation of CMs in monolayer culture is more efficient than in EBs,60 results are influenced by the differentiation media used to generate the EBs and the monolayers. For example, when APEL medium is used with both protocols, preparations containing about ~30% CMs are obtained.39
A recent innovation for monolayer protocols utilizes application of extracellular matrix in combination with cytokines, since extracellular matrix proteins play fundamental roles in development and can complement responses to soluble cytokines.61 For this matrix sandwich approach, hPSC are seeded as single cells on Matrigel (Growth Factor Reduced) coated plates and cultured in mTeSR1 media. When cells reach 90% confluence after 2–4 days, a matrix overlay is applied on the monolayer of cells by mixing the Matrigel with mTeSR1 media. Cells grow for another 1–2 days in mTeSR1 media to reach 100% confluence, then Activin A with Matrigel (Growth Factor Reduced) is added in basal media of RPMI/B27 minus insulin supplement to treat the cells for 24 hours, followed by BMP4 and bFGF for 4 days in the same basal medium. Flow cytometry analysis by cTnT labelling has demonstrated high purity (40–90%) of CMs and high yield (4–11CMs / input hESC/hiPSC) for a range of human ESC and hiPSC lines.61 Thus, using extracellular matrix to supplement soluble growth factor signalling can enable reproducible and robust cardiac differentiation across multiple hESC and hiPSC lines. This matrix sandwich monolayer protocol is a preferred approach to generate relatively high purity and yields of CMs.
Inductive co-culture
Derivation of CMs from hPSC by co-culture with visceral endodermal-like cells is particularly effective for hESC and hiPSC being passaged mechanically and was first described by Mummery et al.,62 with later refinements described by Passier et al.63 Visceral endoderm plays an important inductive role in the differentiation of cardiogenic precursor cells in the adjacent mesoderm in vivo in developing embryos. Beating areas were observed in hESC colonies during co-culture with a mouse visceral endoderm-like cell-line (END-2). In the first experiments, END-2 cells were seeded on a 12-well plate, mitotically inactivated with mitomycin C and co-cultured with the hESC line, HES2. Beating areas formed in approximately 35% of the wells after 12 days in co-culture. However, only about 2–3% of the cells were CMs. It was later shown that both serum supplementation of the culture medium and mTeSR®, a serum free culture medium designed for expansion of undifferentiated cells in the absence of feeder cells, reduced the efficiency of differentiation. But if the medium was completely serum- and insulin-free, the number of beating colonies increased 10-fold, and each beating colony contained ~25% CMs based on staining with sarcomeric α-actinin antibodies. Insulin was the most important component of the medium that inhibited mesoderm formation in favor of ectoderm.46, 64 The protocol is effective not only in hESC but also in hiPSC65 provided they are derived from mechanically passaged colonies of undifferentiated cells on mouse embryonic fibroblast feeders cells (MEFs). This protocol is particularly useful for testing the ability of both hESC and hiPSC to form cardiac mesoderm cells at an early stage of their derivation, since the cells are generally passaged mechanically and the primary goal of the experiment is simply to demonstrate ability to form functional CMs. Under these circumstances, co-culture is convenient since (i) it requires very few cells, (ii) it is simple and rapid and (iii) it yields CMs in sufficient numbers and quality to detect visible beating and identify sarcomere structures by immunofluorescent staining. The protocol is described in detail elsewhere.66 Skeletal myoblasts do not form under these conditions. Another stromal cell line, OP9, has also been reported to induce mesoderm differentiation and cardiogenesis illustrating the general concept of co-culture with another cell line as supporting directed (cardiac) differentiation.67
Cardiovascular progenitors derived from human pluripotent stem cells
In the previous section, we presented various methods which result in hPSCs forming cultures containing beating cardiomyocytes. This process of hPSC differentiation is thought to proceed through a similar hierarchy of cardiac progenitors as described for cardiac development (Figure 1). This concept is supported by the sequential pattern of gene expression suggesting the ordered formation of distinct progenitor populations.27 However, these progenitor populations are only beginning to be isolated and defined in detail. The earliest defined mesodermal progenitors differentiating from human ESCs were identified by the expression of neuronal cell adhesion molecule (NCAM/CD56) and loss of expression of epithelial cell adhesion molecule (EpCAM/CD326).68 These mesodermal progenitors are multipotent and can give rise to all mesodermal lineages including CMs. Later in differentiation, a population of cells expressing low levels of KDR and no cell surface C-KIT (KDRlow/C-KIT-) was isolated at day 6 of EB-mediated differentiation.38 Based on colony forming assays, the KDRlow/C-KIT- cell population contained cardiovascular colony forming cells which gave rise to CMs, smooth muscle cells and endothelial cells. If the KDRlow/C-KIT- cells were plated as monolayers, 50% of the differentiating cells were cardiac troponinT positive CMs. More recently the Keller group shows that that KDR and Platelet Derived Growth Factor Receptorα (PDGFR α) are co-expressed in the emerging cardiac mesoderm.41 Further, when the KDR/PDGFRα double positive population is cultured over 80% of cells differentiate to CMs. However, a disadvantage of using KDR as a cell surface marker to identify cardiac progenitors is that it is also expressed in undifferentiated hESCs.
Another marker which has been proposed to isolate cardiac progenitors from human and nonhuman primate ESCs is the stage-specific embryonic antigen-1 (SSEA-1).69 Unlike mouse ESCs, primate and human ESCs do not express SSEA-1 in the undifferentiated state; however, upon the initiation of differentiation, SSEA-1 is quickly upregulated. In a protocol in which enzymatically passaged hESCs or hiPSCs are cultured on MEFs, a 4 or 6 day treatment of cultures with BMP2 (10 ng/ml) in the presence of the FGF receptor inhibitor SU5402 (1 μM) produced a culture containing ~50% SSEA-1+ cells. The SSEA-1+ cells were isolated by antibody-conjugated magnetic beads and expressed a variety of markers of cardiac progenitors including MESP1, MEF2C, NKX2-5, GATA4, TBX5, TBX18 and ISL1. Single cell clones expanded from these SSEA-1+ cells appeared to include multipotent cardiac progenitors that could differentiate into CMs, smooth muscle cells, and endothelial cells. In contrast to undifferentiated ESCs and hiPSCs, the purified SSEA-1+ cells were unable form teratomas following transplantation to immunocompromised animals.69 However, SSEA1 marks all early differentiated derivatives of hPSC and is only appropriate for selection of cells of the cardiac lineage when hPSC have been exposed to cardiomyogenic growth factors and/or induction conditions.
An alternative approach for isolating cardiac progenitors involves the use of genetically engineered reporter ESC lines. Typically these reporter lines express fluorescent proteins such as GFP, either inserted by homologous recombination into the locus of a cardiac specific gene, or randomly inserted as a transgene regulated by cardiac-specific promoters /enhancers to enable the identification and isolation of the cells of interest. This has been a particularly powerful strategy used to generate a variety reporter mESC lines based on the expression of reporters for Nkx2-5, Isl1, and Mesp1,20, 70–72 but this approach is only starting to be employed for hPSCs. By targeting the ISL1 locus in hESCs, a population of Isl1+ cardiovascular progenitors was isolated that was derived from the secondary heart field.73 These progenitors were capable of limited proliferation in culture in the presence of Wnt3A and gave rise to clones of smooth muscle cells with some clones of CM and endothelial cell lineages. The Isl1+ progenitors can be isolated prior to the expression of KDR and subsequently an Isl1+/KDR+ progenitor population was identified which may overlap with the KDR+/c-kit− population described above.
An NKX2-5eGFP/w targeted hESC line has been described recently which enabled the identification of cardiac-specific progenitors based on eGFP expression.39, 74 Characterization of the first cells to express eGFP during cardiac differentiation using both antibody arrays and gene expression arrays identified the cell surface marker signal regulatory protein alpha (SIRPA/CD172a) as specifically present on the eGFP positive progenitors relative to the eGFP negative cells. SIRPA expression was not detected in the KDR/PDGFRα double positive population of cardiac mesoderm cells present in earlier in differentiation, but its expression starts in the cardiac committed progenitors. SIRPA expression persists specifically in the CMs. Interestingly, SIRPA is not expressed in the developing mouse heart, in contrast to the human heart, which highlights the need for defining markers in the human system. Furthermore, none of the described cell surface markers are exclusively expressed on cardiac cell lineages, and so they need to be used in conjunction with other markers and cellular characteristics to identify various progenitors specifically. Both new engineered hPSC lines and cell surface markers are anticipated that will provide further definition and as well as isolation of the hierarchy of cardiac progenitors.
Selection of CMs from mixed differentiated cultures
Strategies to isolate CMs from mixed differentiating cultures have also been developed which include physical methods, genetic methods, and nongenetic selection. The ability to isolate relatively pure populations of CMs is essential to multiple applications using the cells. For example, studies of global gene expression are more readily interpretable if a purified population of cells is studied. Cardiac drug-screening assays also require relatively pure populations of CMs so that the observed signals can be attributed to effects on CMs. Finally, clinical applications proposing to use CMs derived from hPSCs will need to be free of contaminating pluripotent stem cells and any other undesired cell type to minimize the risk of tumor formation and other adverse outcomes.
The first described methods for CM selection from hESC EBs relied on physical methods. Following manual dissection and dissociation of contracting areas from EBs, between 2–70% of the cells stained for cardiac markers, depending on the particular aggregate dissected.34, 62 Alternatively, by taking cells isolated from contracting aggregates and using Percoll gradient purification, a further 4-fold enrichment could be obtained (up to 70% of cells stained positively for cardiac markers) in a particular cell fraction compared with the initial differentiated cell suspension.35 Although this method provided enrichment for CMs, it was labor intensive and critically dependent on the successful formation of EBs with well-defined larger contracting outgrowths.
A second method for selection of CMs involves genetically engineering stem cell lines to express a drug resistance gene or reporter protein gene under the control of a cardiac-specific promoter. This strategy is based on the original study by Field and colleagues who engineered mESCs with the cardiac α-myosin heavy chain (αMHC) promoter driving the expression of an antibiotic resistance gene (aminoglycoside phosphotransferase) which enabled selection of relatively pure cultures of CMs in the presence of neomycin.75 Subsequently, a similar approach was employed using a variety of cardiac promoters with different selection or reporter proteins in mESCs.76–80 Selection for CMs was achieved either using drug selection to eliminate cells not expressing the drug resistance gene or fluorescence activated cell sorting (FACS) for cells expressing fluorescent reporter proteins. In the case of hESCs, the αMHC promoter has also been expressed ectopically and used to drive fluorescent protein expression (GFP or mCherry) or puromycin resistance to allow selection of hESC-CMs.81–83 Likewise, an engineered NKX2-5eGFP/w knock-in hESC line has been described that enables isolation of GFP+ CMs. However, because αMHC and NKX2-5 are expressed in all types of CMs, the resulting population of selected CMs represents a mixture of nodal-, atrial- and ventricular-like CMs. In an effort to select specifically for ventricular-like CMs, Gepstein and colleagues linked the myosin light chain 2v (MLC2v) promoter to GFP expression, enabling a highly enriched population of ventricular-like cells to be purified by flow cytometry.84 Conversely, the proximal promoter-enhancer element from the cGATA6 gene was used to identify nodal-like hESC-derived CMs.85 Using this approach, it was found that neuregulin-1β increased the relative abundance of nodal-like CMs, demonstrating the utility of such markers for lineage enrichment strategies.85
Future studies will likely test a variety of additional cardiac-relevant promoters as well as various selection strategies to enhance or refine selection. The subtleties of the promoter/enhancer chosen will be of critical importance, and issues such as species differences between well-characterized mouse regulatory elements and often less well characterized human elements will need to be considered. While certain proteins exhibit cardiac-specific expression and knock-in to the native locus gives similar results to expressing a characterized promoter sequence, in other cases where genes are more broadly expressed, selected promoter/enhancer elements which give a greater degree of tissue specificity may be more appropriately chosen. The choice of using drug-selection or reporter protein cassettes or a combination will depend on the planned application. For enrichment of CMs during culture, drug selection may be the preferred approach because it does not require cell isolation and sorting, and highly pure populations of CMs have been obtained by this route. The timing of drug selection during differentiation is critical and may affect the ability of CMs to mature in culture.86 Alternatively, if the goal is to track the time course of differentiation and localize CMs in differentiating cultures, then fluorescent reporter proteins are optimal. Likewise, CMs with fluorescent or other reporters such as luciferase can be tracked in vivo following transplantation in animal models.87 Finally, it is essential to consider the approach for genetically modifying the cells lines. There are multiple techniques to introduce new reporter constructs including lentiviral transduction, homologous recombination, random integration, integrase-mediated recombination, zinc finger nuclease assisted recombination and others.86, 88, 89 Each genetic engineering approach has advantages and disadvantages as described elsewhere.88
Nongenetic methods for isolating CMs hold appeal which might be more readily applied across various cell lines with specificity and efficiency. Cell surface marker proteins have been utilized with great success in the hematopoietic system to isolate and define various cell lineages, but cell surface markers on CMs are only just starting to be defined. The cell surface protein SIRPA present on cardiac progenitors is also expressed on differentiated CMs and can be used for selection.39, 74 VCAM1 is a cell-surface protein also identified on differentiated CMs which can be used for purification.39, 90 Alternatively, an approach taking advantage of the high density of mitochondria present in mature CMs compared to other cell types has recently been described.91 Using a fluorescent vital dye which labels mitochondria, tetramethylrhodamine methyl ester perchlorate (TMRM), CMs derived from hESCs could be isolated at greater than 99% purity. The dye rapidly washes out suggesting limited long-term impact or toxicity on the selected population. The limitation with this method is that it is only effective on relatively late or mature hESC-derived CMs, maintained in culture 8 weeks or more.
In summary, techniques for the separation and purification of CMs from mixed cultures are still being refined. Highly enriched populations of CMs have been obtained using various techniques, but the relative efficiency of these techniques and practical limitations will drive further improvements. In addition, strategies to isolate different types of CMs and different maturities of CMs are required and, therefore, will be the focus of continued development. Approaches such as different reporters in the same line, as described for mESCs, may be a way forward to improve cell type specificity.92
Scalable cardiac differentiation of hPSC
For some applications, large-scale production of CMs from hPSCs is of critical importance. Clinical therapies envisioning the use of hPSC derived-CMs may require 108 to 109 cells per patient dose, which reflects the amount of working myocardium lost in a large myocardial infarction. Additionally, high throughput screening of libraries of small molecules using hPSC-derived CMs for safety or efficacy signals will require large numbers of CMs, although the testing of selected lead compounds is already possible with lesser cell numbers available with current technologies.8 To enable such applications, both the starting PSC and the differentiation process will need to be scalable. Progress has been made on the scalable culture of human PSCs using defined, humanized culture media and reagents using a variety of bioreactor formats.93, 94 Current bioreactor technology allows continuous perfusion and automated control of pH, oxygen tension, temperature, and other relevant parameters. This eliminates the large swings in media pH and in the concentrations of metabolites and nutrients which typically occur with medium changes in standard static culture formats. Although we will focus only on the scalable generation of hPSC-derived CMs, it is important to acknowledge that limiting background differentiation and maintaining karyotype stability remain critical issues for the large scale propagation of hPSC.
Large-scale differentiation of mESCs to CMs was first described using suspension culture of EBs.95–98 Suspension cultures allow a greater cell number:culture volume ratio and hence are more efficient. However, cultures need to be dynamic to prevent the rafting of EBs and to allow adequate exchange of metabolites, nutrients, and growth factors as well as maintaining homeostasis of physicochemical parameters such as pH and oxygen levels. A variety of dynamic culture systems such as spinner flasks, stirred bioreactors and rotary culture have been used. In one study, to avoid agglomeration, the EBs were encapsulated in hydrogel which did not impair cellular viability or differentiation.95 In another study, single cell suspensions of mESCs were added to a bioreactor and conditions were optimized so aggregates of appropriate size formed and generated EBs.74 By combining genetic selection using the αMHC promoter with the suspension EB differentiation, as described above, relatively pure populations of mESC-derived CMs could be generated with up to 109 CMs from a 2 L bioreactor.96 Other refinements of the bioprocess such as differentiation under conditions of low oxygen (4% oxygen tension) or continuous perfusion of the bioreactors have been suggested to further improve CM yield.95, 99–101 To translate this to hESC cells, micropatterning was used to generate size-specified aggregates of hESCs which, when loaded into a bioreactor, formed EBs that differentiated into CMs.102 Likewise, other dynamic culture systems for human EBs that generate CMs have been described.100, 101 However, these initial studies using hESCs did not clearly describe yields of CMs and did not use selection strategies to further purify CMs. Nevertheless, large-scale production of hESC and hiPSC derived-CMs has been demonstrated in commercial products now available.103
Basic characterization of hPSC-derived CMs
Determining the success of differentiation protocols requires detailed characterization of the resulting CMs. Typically, the first sign of successful differentiation to CMs is the appearance of contractile foci in the culture. Although early studies using EB-based differentiation protocols usually reported the percentage of contracting EBs as a measure for cardiac differentiation, more recent studies have described the proportion of cardiomyocytes, estimated by intracellular flow cytometry for a cardiac-specific protein such as cardiac troponin T. This more accurately reflects the efficacy of the protocol. In order to better characterize and quantify the resulting cardiomyocytes, a variety of approaches have been used, ranging from analyses of gene expression to functional studies. These complementary assays may be considered to span a range of stringencies. The lowest stringency includes RT-PCR evaluation for expression of a variety of cardiac genes, from transcription factors to myofilament proteins. An assay slightly higher in the hierarchy is provided by immunolabeling of the CMs with antibodies to a variety of myofilament proteins (e.g. αMHC, βMHC, MLC2a, MLC2v, cardiac troponin T, cardiac troponin I, α-actinin) in order to demonstrate organized myofilaments typical of early cardiomyocytes (Figure 3). Electron microscopy can also be used to demonstrate the ultrastructure typical of CMs including bundles of myofilaments and abundant mitochondria.
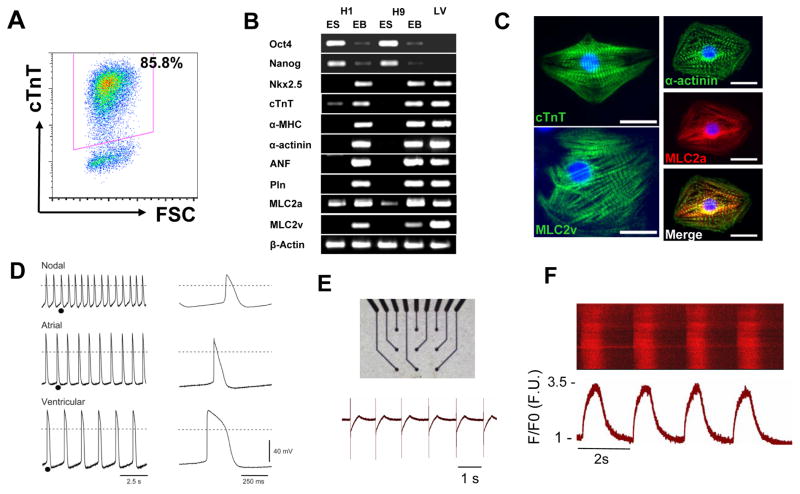
Figure 3. Basic Characterization of Human PSC-derived Cardiomyocytes.
A) Flow cytometry provides a quantitative method to evaluate the relative yield and purity of CMs resulting from differentiation protocols by measuring the number of cells expressing cardiac-specific proteins such as cardiac troponin T (cTnT). B) RT-PCR can be used as a first assessment for changes in gene expression typical of cardiogenesis by assaying for the expression of cardiac transcription factors, myofilament proteins, and Ca2+-cycling proteins. C) Immunofluorescence microscopy using antibodies specific for myofilament proteins determine if cells exhibit organized sarcomeres typical of CMs. D) Functional assessment of CMs can be provided by cellular electrophysiology measurements to determine if cardiac action potentials typical of different types of CMs are present. E) Extracellular field potential measurements by multielectrode arrays provide another method to detect spontaneous electrical activity in the CMs preparations. F) Detection of Ca2+ transients typical of CMs provides another assessment of the functional integrity of the differentiating CMs. Cells loaded with the Ca2+ indicator Fluo-3 were imaged using laser scanning confocal microscopy in the line scan mode with Ca2+ transients displayed and time versus normalized Ca2+ transient intensity (F/F0) shown below. (Panels B and D modified from Zhang et al., 2009)
The highest stringency assays are primarily functional assays which demonstrate the integrated cellular phenotype. A key property of CMs is generation of action potentials (APs) necessary for propagation of the electrical signal in the myocardium and for triggering intracellular Ca2+ release and ensuing mechanical contraction. Demonstrating cardiac APs using microelectrode recordings can provide a high level of evidence that functional CMs are present. Furthermore, properties of the AP such as the AP amplitude, spontaneous phase 4 depolarization, maximum diastolic potential, AD duration and rate of rise of the action potential can provide information regarding the type and maturity of CM.34, 36, 62 The majority of CMs display ventricular-like APs, but atrial-like and nodal-like APs are also detected (Figure 3). The properties of the APs suggest that the hPSC-derived CMs are relatively immature in phenotype compared to adult CMs which is consistent with other characteristics of the cells (see Table). Furthermore, the CMs typically show evidence for a range of maturities, and for CMs with more depolarized maximum diastolic potentials, discrimination between APs representative of different types of CMs is difficult given overlap between the properties of more mature nodal CMs and immature CMs of all types. The multiple underlying ionic currents responsible for generating the cardiac APs can also be studied in detail using single cell patch clamp methodology,103, 104 but this is typically beyond the scope of basic characterization studies. However, such detailed electrophysiology is of particular importance in investigations evaluating basic mechanisms of arrhythmia such as recent studies using patient-specific hiPSCs to study inherited long QT syndrome.7, 105, 106 Multielectrode arrays (MEAs) can be used to demonstrate spontaneous electrical signals (field potentials) and monitor the electrophysiology of multicellular preparations providing information on parameters such as conduction velocity among CMs; however, these assays do not have single cell resolution like microelectrode recordings.107 Nevertheless, MEAs are more rapid and less technically demanding, and patterns typical of CMs preparations are becoming evident. Intracellular Ca2+ can also be monitored using fluorescent Ca2+ indicators to determine if cardiac typical cardiac Ca2+ transients are present. Overall, it is important to note that molecular assays alone are not sufficient to confirm CM identity; functional assays in the highest stringency category are also essential.
Table.
Markers and characteristics associated with immature versus mature human ventricular cardiomyocytes
| Characteristic/marker | Immature CMs* | Mature CMs** | |
|---|---|---|---|
| Electrical | Spontaneous contracting | + | − |
| Maximum diastolic potential | ~−40mV | ~−85mV | |
| AP upstroke velocity | 5–50V/s | 150–300V/s | |
| AP amplitude | 70–90 mV | 110–120 mV | |
| Ionic currents | IK1 (inward rectifier) | − | + |
| IKr (hERG channel) | low | high | |
| If (funny current) | high | low | |
| Structural proteins | Sarcomeres (striations) | Disorganized | Organized |
| Myosin light chain (MLC)2v | +/− | +++ | |
| MLC2a | +++ | + | |
| Myosin heavy chain (MHC) | αMHC≫βMHC | βMHC≫αMHC | |
| Transverse(T-) tubules | − | + | |
| Metabolism | Mitochondria: | low | high |
| number | low | high | |
| membrane potential cristae density | low | high | |
| Glucose metabolism | high | low | |
| Fatty acid metabolism | low | high | |
| Atrial Natriuretic Factor (ANF) | low | high | |
| Morphology | Cell volume | small | large |
| Shape | circular/irregular | rectangular | |
| Cell cycle | Proliferation | + | − |
| Cyclin D1 | high | low | |
| p21 | low | high |
human fetal and stem cell-derived ventricular cardiomyocytes
human adult ventricular cardiomyocytes
Emerging strategies for improving and troubleshooting CM differentiation from hPSCs
Although protocols for cardiac differentiation have advanced dramatically over the past decade, significant limitations still remain regarding line-to-line variability in the yield and purity of CMs, reproducibility of protocols, cost of reagents, and in the complexity of the protocols. For example, differences in individual hESC and hiPSC lines with respect to their yield of cardiomyocytes even when using identical protocols have been described.37, 49 Various reasons have been given for this interline variability including differences in the initial state of pluripotency,21, 22 conditions used to maintain lines, epigenetic status determined by the tissue of origin in the case of hiPSC,24, 108 and intrinsic differences in endogenous growth factor production between individual lines.41, 58, 109 More robust differentiation protocols that are broadly successful across cell lines are needed.
Recent protocols have largely replaced undefined serum with the sequential application of growth factors and more recently small molecules to provide more reproducible results. Many of the growth factors are costly to obtain, degrade rapidly, and do not readily diffuse into complex multicellular aggregates. In addition, they exhibit lot-to-lot variation in their bioactivity. Thus, efforts at identifying chemically synthesized small molecules to promote the differentiation process are being actively pursued. A variety of small molecules that promote cardiogenesis of mouse ESCs have been identified using strategies including high throughput screens with various engineered reporter lines.110–112 These mouse ESC studies clearly determined that the timing of application is of critical importance; nevertheless, only a few of these results have been extrapolated to human PSCs at this time. Inhibitors of Wnt signalling, IWR-1, IWP-3, and XAV939 have been shown to promote cardiogenesis when added later in differentiation following mesoderm formation,113 and recently activation of canonical Wnt signalling by glycogen synthase kinase inhibition (CHIR99021 or 6-bromoindirubin-30-oxime) followed by inhibition of Wnt signalling (IWP2 or IWP2) was found to be sufficient in a monolayer protocol in the absence of exogenous growth factors to produce robust cardiogenesis from multiple hPSC lines.114 In addition, inhibition of the Activin/Nodal/TGF-β pathway using SB-431542 promoted cardiogenesis when added following mesoderm specification at least in the HES2 line where continued expression of low levels of endogenous nodal has been suggested.41 Inhibition of the p38 MAPK pathway with SB203580 has also been found to enhance cardiogenesis of hESCs.115 Future studies will undoubtedly identify additional small molecules that can be utilized to promote hPSC cardiogenesis in a more cost effective and reproducible fashion.
The redox status of ESCs which has also been implicated in the differentiation process may be amenable to optimization. Investigation of the metabolome of ESCs has demonstrated an abundance of unsaturated metabolites and differentiation is associated with the formation of downstream oxidized metabolites.116 Intracellular oxidation has been linked to the balance of stem cell self-renewal and differentiation117 and culture of undifferentiated hESCs in low O2 reduces background differentiation.118, 119 Interestingly, generation of reactive oxygen species (ROS) by oxidative enzymes such as NADPH oxidase has been implicated in the cardiac differentiation of mESCs.120, 121 Performing the initial stage of differentiation in a 5% O2 environment has been found to increase the effectiveness of hPSC cardiogenesis by some investigators.38, 41, 109 The role of redox state and ROS in cardiogenesis merits further study and may be a useful tool to modulate differentiation.
Refinements in the composition and mechanical properties of extracellular matrix (ECM) preparations used in cardiac differentiation protocols may also provide important advances. Major changes in ECM occur during normal development, and so it seems likely that ECM will be influence in vitro differentiation.122 The interaction of cells with ECM ligands in combination with soluble growth factors impacts on cell fate decisions including proliferation and differentiation. Not only do ECM proteins interact directly with cell surface receptors to activate signalling pathways, but the ECM also transmits an important mechanical component. Additionally, cyclic strain in developing heart tissue derived from hPSCs is thought to result in maturation of cardiomyocytes.123 Indeed, failure to undergo complete maturation is still one of the major limitations to using hPSC derived CMs in multiple applications such as disease modelling and drug discovery. Dynamic remodelling of ECM is also likely to involve activation of matrix proteinases. It will be valuable to define the specific ECM composition optimal for promoting cardiogenesis as many of the previous studies have used Matrigel, which is an undefined product secreted from a rodent tumour cell line. Progress in identifying novel synthetic substrates to grow undifferentiated hESCs suggests that similar advances may empower CM differentiation protocols in the future.124–127
Given the lack of harmonization of current differentiation protocols and the increasing number of new investigators using these protocols, some simple approaches for troubleshooting can be followed. First, it is critical that the input undifferentiated hPSCs be maintained optimally with minimal background differentiation.21 Furthermore, the culture methods for maintaining the hPSCs (e.g. feeder free in specific medium, mechanical passage) must be matched precisely to the published differentiation protocol as these impact the cells’ initial responses and the final outcome. If the hPSCs used for differentiation are recovered from cryopreservation, they should initially be passaged at least two times under the indicated conditions before differentiation. Investigators without experience in hPSC culture need training first in this methodology before success can be anticipated with differentiation protocols. Secondly, high quality growth factors and small molecules should be obtained ideally matching the protocols regarding source. In some cases, it is necessary to test a range of growth factor concentrations to optimize each individual line.41 For new investigators, it is also essential to have a validated control hPSC line that has been shown to differentiate well with a given protocol. Ideally, such hPSCs should be obtained from quality controlled cell banks. Future harmonization of protocols is an important goal, and control transgenic cell lines, in which reporter genes are driven by cardiac specific promoters,39, 128 may be useful in benchmarking protocols between laboratories and provide ways of standardizing operator and reagent variability.
Summarizing remarks
The demonstration that human ESCs can give rise to functional cardiomyocytes has promoted much discussion about the potential utility of hPSC-CMs. At the forefront of popular interest has been the promise of cardiomyocyte replacement therapies. However, as experimental data continue to reveal the challenges for clinical application using hPSCs, the goal of hPSC-CMs mediated cardiac repair appears elusive.129 Nevertheless, for drug safety pharmacology and disease modelling following the advent of hiPSC, it may transpire that human PSCs can have a much greater impact on cardiovascular health than simply through transplantation to replace CMs lost in disease. Several recent studies generating hiPSCs from patients with cardiac disease have demonstrated that the phenotype is retained.5, 7, 105, 106 More importantly, these studies showed that the cardiomyocytes were suitable for testing the effects of drugs on the disease. Once further explored, this will provide unprecedented opportunities for developing novel treatment strategies particularly, since robust and efficient methods for the differentiation and maturation of cardiomyocytes from hPSC are well underway.
Methodological Reviews Text Box.
Methodological Reviews discuss methods that are of broad interest to the community of cardiovascular investigators and that enable a better understanding of cardiovascular biology, particularly recent technologies in which the methods are still in flux and/or not widely known. It is hoped that these articles, written by recognized experts, will be useful to all investigators, but especially to early-career investigators.
Acknowledgments
We apologize to those authors whose excellent work we have not cited due to space restrictions. We gratefully acknowledge Randall Loaiza Montoya for providing Ca2+ data shown in Figure 3.
Sources of Funding
Funding: C.L.M receives lab funding from the Netherlands Heart Foundation, EU FP7 (“InduStem” PIAP-GA-2008-230675), ZonMW (114000101), The Netherlands Institute of Regenerative Medicine and the Netherlands Proteomics Consortium (050-040-250). AGE is a Senior Research Fellow of the NHMRC and receives lab funding from the NHMRC, ARC, the Australian Stem Cell Centre, JDRF and QNRF. DAE is funded by the ASCC, NHRMC and QNRF. T.J.K receives funding from the National Institutes of Health, R01EB007534, R01HL08416, and U01HL099773.
Non-standard Abbreviations and Acronyms
- AP
action potential
- APEL
Albumin, Polyvinylalcohol (PVA), Essential Lipids
- BMPs
bone morphogenetic proteins
- CMs
cardiomyocytes
- EB
embryoid bodies
- ECM
extracellular matrix
- EpCAM/CD326
epithelial cell adhesion molecule
- FACS
fluorescence activated cell sorting
- FGFs
fibroblast growth factors
- hESCs
human embryonic stem cells
- hiPSCs
human induced pluripotent stem cells
- hPSCs
human pluripotent stem cells
- MLC2a
myosin light chain 2a
- MLC2v
myosin light chain 2v
- MHC
myosin heavy chain
- MEAs
Multielectrode arrays
- mECCs
mouse embryonal carcinoma cells
- MEF2
myocyte enhancer factor 2
- MEFs
mouse embryonic fibroblasts
- mESCs
mouse embryonic stem cells
- NCAM/CD56
neuronal cell adhesion molecule
- ROS
reactive oxygen species
- SIRPA/CD172a
signal regulatory protein alpha
- SRF
serum response factor
- SSEA-1
stage-specific embryonic antigen-1
- TMRM
tetramethylrhodamine methyl ester perchlorate
- VEGF
vascular endothelial growth factor
- WNTs
Wingless/INT proteins
Footnotes
Disclosures
T.J.K. is a founding shareholder and consultant for Cellular Dynamics International which produces hPSC-derived CMs for drug discovery, toxicity testing, and research applications.
In May 2012, the average time from submission to first decision for all original research papers submitted to Circulation Research was 12.0 days.
References
- 1.Park IH, Lerou PH, Zhao R, Huo H, Daley GQ. Generation of human-induced pluripotent stem cells. Nat Protoc. 2008;3:1180–1186. doi: 10.1038/nprot.2008.92. [DOI] [PubMed] [Google Scholar]
- 2.Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 3.Thomson JA, Itskovitz-Eldor J, Shapiro SS, Waknitz MA, Swiergiel JJ, Marshall VS, Jones JM. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- 4.Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA, Ruotti V, Stewart R, Slukvin II, Thomson JA. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 5.Carvajal-Vergara X, Sevilla A, D'Souza SL, Ang YS, Schaniel C, Lee DF, Yang L, Kaplan AD, Adler ED, Rozov R, Ge Y, Cohen N, Edelmann LJ, Chang B, Waghray A, Su J, Pardo S, Lichtenbelt KD, Tartaglia M, Gelb BD, Lemischka IR. Patient-specific induced pluripotent stem-cell-derived models of leopard syndrome. Nature. 2010;465:808–812. doi: 10.1038/nature09005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Itzhaki I, Maizels L, Huber I, Zwi-Dantsis L, Caspi O, Winterstern A, Feldman O, Gepstein A, Arbel G, Hammerman H, Boulos M, Gepstein L. Modelling the long qt syndrome with induced pluripotent stem cells. Nature. 2011 doi: 10.1038/nature09747. [DOI] [PubMed] [Google Scholar]
- 7.Moretti A, Bellin M, Welling A, Jung CB, Lam JT, Bott-Flugel L, Dorn T, Goedel A, Hohnke C, Hofmann F, Seyfarth M, Sinnecker D, Schomig A, Laugwitz KL. Patient-specific induced pluripotent stem-cell models for long-qt syndrome. N Engl J Med. 2010;363:1397–1409. doi: 10.1056/NEJMoa0908679. [DOI] [PubMed] [Google Scholar]
- 8.Braam SR, Tertoolen L, van de Stolpe A, Meyer T, Passier R, Mummery CL. Prediction of drug-induced cardiotoxicity using human embryonic stem cell-derived cardiomyocytes. Stem Cell Res. 2010;4:107–116. doi: 10.1016/j.scr.2009.11.004. [DOI] [PubMed] [Google Scholar]
- 9.Anson BD, Kolaja KL, Kamp TJ. Opportunities for use of human ips cells in predictive toxicology. Clinical pharmacology and therapeutics. 2011;89:754–758. doi: 10.1038/clpt.2011.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van Laake LW, Passier R, Doevendans PA, Mummery CL. Human embryonic stem cell-derived cardiomyocytes and cardiac repair in rodents. Circ Res. 2008;102:1008–1010. doi: 10.1161/CIRCRESAHA.108.175505. [DOI] [PubMed] [Google Scholar]
- 11.van Laake LW, Passier R, Monshouwer-Kloots J, Verkleij AJ, Lips DJ, Freund C, den Ouden K, Ward-van Oostwaard D, Korving J, Tertoolen LG, van Echteld CJ, Doevendans PA, Mummery CL. Human embryonic stem cell-derived cardiomyocytes survive and mature in the mouse heart and transiently improve function after myocardial infarction. Stem Cell Res. 2007;1:9–24. doi: 10.1016/j.scr.2007.06.001. [DOI] [PubMed] [Google Scholar]
- 12.Abu-Issa R, Kirby ML. Heart field: From mesoderm to heart tube. Annu Rev Cell Dev Biol. 2007;23:45–68. doi: 10.1146/annurev.cellbio.23.090506.123331. [DOI] [PubMed] [Google Scholar]
- 13.Olson EN. Gene regulatory networks in the evolution and development of the heart. Science. 2006;313:1922–1927. doi: 10.1126/science.1132292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Olson EN, Schneider MD. Sizing up the heart: Development redux in disease. Genes Dev. 2003;17:1937–1956. doi: 10.1101/gad.1110103. [DOI] [PubMed] [Google Scholar]
- 15.Brand T. Heart development: Molecular insights into cardiac specification and early morphogenesis. Dev Biol. 2003;258:1–19. doi: 10.1016/s0012-1606(03)00112-x. [DOI] [PubMed] [Google Scholar]
- 16.Cohen ED, Tian Y, Morrisey EE. Wnt signaling: An essential regulator of cardiovascular differentiation, morphogenesis and progenitor self-renewal. Development. 2008;135:789–798. doi: 10.1242/dev.016865. [DOI] [PubMed] [Google Scholar]
- 17.Park EJ, Watanabe Y, Smyth G, Miyagawa-Tomita S, Meyers E, Klingensmith J, Camenisch T, Buckingham M, Moon AM. An fgf autocrine loop initiated in second heart field mesoderm regulates morphogenesis at the arterial pole of the heart. Development. 2008;135:3599–3610. doi: 10.1242/dev.025437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ilagan R, Abu-Issa R, Brown D, Yang YP, Jiao K, Schwartz RJ, Klingensmith J, Meyers EN. Fgf8 is required for anterior heart field development. Development. 2006;133:2435–2445. doi: 10.1242/dev.02408. [DOI] [PubMed] [Google Scholar]
- 19.Wu SM. Mesp1 at the heart of mesoderm lineage specification. Cell Stem Cell. 2008;3:1–2. doi: 10.1016/j.stem.2008.06.017. [DOI] [PubMed] [Google Scholar]
- 20.Bondue A, Tannler S, Chiapparo G, Chabab S, Ramialison M, Paulissen C, Beck B, Harvey R, Blanpain C. Defining the earliest step of cardiovascular progenitor specification during embryonic stem cell differentiation. J Cell Biol. 2011;192:751–765. doi: 10.1083/jcb.201007063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kuo CT, Morrisey EE, Anandappa R, Sigrist K, Lu MM, Parmacek MS, Soudais C, Leiden JM. Gata4 transcription factor is required for ventral morphogenesis and heart tube formation. Genes Dev. 1997;11:1048–1060. doi: 10.1101/gad.11.8.1048. [DOI] [PubMed] [Google Scholar]
- 22.Molkentin JD, Lin Q, Duncan SA, Olson EN. Requirement of the transcription factor gata4 for heart tube formation and ventral morphogenesis. Genes Dev. 1997;11:1061–1072. doi: 10.1101/gad.11.8.1061. [DOI] [PubMed] [Google Scholar]
- 23.Watt AJ, Battle MA, Li J, Duncan SA. Gata4 is essential for formation of the proepicardium and regulates cardiogenesis. Proc Natl Acad Sci U S A. 2004;101:12573–12578. doi: 10.1073/pnas.0400752101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pu WT, Ishiwata T, Juraszek AL, Ma Q, Izumo S. Gata4 is a dosage-sensitive regulator of cardiac morphogenesis. Dev Biol. 2004;275:235–244. doi: 10.1016/j.ydbio.2004.08.008. [DOI] [PubMed] [Google Scholar]
- 25.Zeisberg EM, Ma Q, Juraszek AL, Moses K, Schwartz RJ, Izumo S, Pu WT. Morphogenesis of the right ventricle requires myocardial expression of gata4. J Clin Invest. 2005;115:1522–1531. doi: 10.1172/JCI23769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhao R, Watt AJ, Battle MA, Li J, Bondow BJ, Duncan SA. Loss of both gata4 and gata6 blocks cardiac myocyte differentiation and results in acardia in mice. Dev Biol. 2008;317:614–619. doi: 10.1016/j.ydbio.2008.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Beqqali A, Kloots J, Ward-van Oostwaard D, Mummery C, Passier R. Genome-wide transcriptional profiling of human embryonic stem cells differentiating to cardiomyocytes. Stem Cells. 2006;24:1956–1967. doi: 10.1634/stemcells.2006-0054. [DOI] [PubMed] [Google Scholar]
- 28.Martin-Puig S, Wang Z, Chien KR. Lives of a heart cell: Tracing the origins of cardiac progenitors. Cell Stem Cell. 2008;2:320–331. doi: 10.1016/j.stem.2008.03.010. [DOI] [PubMed] [Google Scholar]
- 29.Stevens LC. Embryonic potency of embryoid bodies derived from a transplantable testicular teratoma of the mouse. Dev Biol. 1960;2:285–297. doi: 10.1016/0012-1606(60)90010-5. [DOI] [PubMed] [Google Scholar]
- 30.Doetschman TC, Eistetter H, Katz M, Schmidt W, Kemler R. The in vitro development of blastocyst-derived embryonic stem cell lines: Formation of visceral yolk sac, blood islands and myocardium. J EmbryolExp Morphol. 1985;87:27–45. [PubMed] [Google Scholar]
- 31.Boheler KR, Czyz J, Tweedie D, Yang HT, Anisimov SV, Wobus AM. Differentiation of pluripotent embryonic stem cells into cardiomyocytes. Circ Res. 2002;91:189–201. doi: 10.1161/01.res.0000027865.61704.32. [DOI] [PubMed] [Google Scholar]
- 32.Keller G. Embryonic stem cell differentiation: Emergence of a new era in biology and medicine. Genes Dev. 2005;19:1129–1155. doi: 10.1101/gad.1303605. [DOI] [PubMed] [Google Scholar]
- 33.Kehat I, Kenyagin-Karsenti D, Snir M, Segev H, Amit M, Gepstein A, Livne E, Binah O, Itskovitz-Eldor J, Gepstein L. Human embryonic stem cells can differentiate into myocytes with structural and functional properties of cardiomyocytes. J Clin Invest. 2001;108:407–414. doi: 10.1172/JCI12131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.He JQ, Ma Y, Lee Y, Thomson JA, Kamp TJ. Human embryonic stem cells develop into multiple types of cardiac myocytes: Action potential characterization. Circ Res. 2003;93:32–39. doi: 10.1161/01.RES.0000080317.92718.99. [DOI] [PubMed] [Google Scholar]
- 35.Xu C, Police S, Rao N, Carpenter MK. Characterization and enrichment of cardiomyocytes derived from human embryonic stem cells. Circ Res. 2002;91:501–508. doi: 10.1161/01.res.0000035254.80718.91. [DOI] [PubMed] [Google Scholar]
- 36.Zhang J, Wilson GF, Soerens AG, Koonce CH, Yu J, Palecek SP, Thomson JA, Kamp TJ. Functional cardiomyocytes derived from human induced pluripotent stem cells. Circ Res. 2009;104:e30–41. doi: 10.1161/CIRCRESAHA.108.192237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Osafune K, Caron L, Borowiak M, Martinez RJ, Fitz-Gerald CS, Sato Y, Cowan CA, Chien KR, Melton DA. Marked differences in differentiation propensity among human embryonic stem cell lines. Nat Biotechnol. 2008;26:313–315. doi: 10.1038/nbt1383. [DOI] [PubMed] [Google Scholar]
- 38.Yang L, Soonpaa MH, Adler ED, Roepke TK, Kattman SJ, Kennedy M, Henckaerts E, Bonham K, Abbott GW, Linden RM, Field LJ, Keller GM. Human cardiovascular progenitor cells develop from a kdr+ embryonic-stem-cell-derived population. Nature. 2008;453:524–528. doi: 10.1038/nature06894. [DOI] [PubMed] [Google Scholar]
- 39.Elliott DA, Braam SR, Koutsis K, Ng ES, Jenny R, Lagerqvist EL, Biben C, Hatzistavrou T, Hirst CE, Yu QC, Skelton RJ, Ward-van Oostwaard D, Lim SM, Khammy O, Li X, Hawes SM, Davis RP, Goulburn AL, Passier R, Prall OW, Haynes JM, Pouton CW, Kaye DM, Mummery CL, Elefanty AG, Stanley EG. Nkx2-5(egfp/w) hescs for isolation of human cardiac progenitors and cardiomyocytes. Nat Methods. 2011;8:1037–1040. doi: 10.1038/nmeth.1740. [DOI] [PubMed] [Google Scholar]
- 40.Johansson BM, Wiles MV. Evidence for involvement of activin a and bone morphogenetic protein 4 in mammalian mesoderm and hematopoietic development. Mol Cell Biol. 1995;15:141–151. doi: 10.1128/mcb.15.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kattman SJ, Witty AD, Gagliardi M, Dubois NC, Niapour M, Hotta A, Ellis J, Keller G. Stage-specific optimization of activin/nodal and bmp signaling promotes cardiac differentiation of mouse and human pluripotent stem cell lines. Cell Stem Cell. 2011;8:228–240. doi: 10.1016/j.stem.2010.12.008. [DOI] [PubMed] [Google Scholar]
- 42.Davis RP, Ng ES, Costa M, Mossman AK, Sourris K, Elefanty AG, Stanley EG. Targeting a gfp reporter gene to the mixl1 locus of human embryonic stem cells identifies human primitive streak-like cells and enables isolation of primitive hematopoietic precursors. Blood. 2008;111:1876–1884. doi: 10.1182/blood-2007-06-093609. [DOI] [PubMed] [Google Scholar]
- 43.Kubo A, Shinozaki K, Shannon JM, Kouskoff V, Kennedy M, Woo S, Fehling HJ, Keller G. Development of definitive endoderm from embryonic stem cells in culture. Development. 2004;131:1651–1662. doi: 10.1242/dev.01044. [DOI] [PubMed] [Google Scholar]
- 44.Ueno S, Weidinger G, Osugi T, Kohn AD, Golob JL, Pabon L, Reinecke H, Moon RT, Murry CE. Biphasic role for wnt/beta-catenin signaling in cardiac specification in zebrafish and embryonic stem cells. Proc Natl Acad Sci U S A. 2007;104:9685–9690. doi: 10.1073/pnas.0702859104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ren Y, Lee MY, Schliffke S, Paavola J, Amos PJ, Ge X, Ye M, Zhu S, Senyei G, Lum L, Ehrlich BE, Qyang Y. Small molecule wnt inhibitors enhance the efficiency of bmp-4-directed cardiac differentiation of human pluripotent stem cells. J Mol Cell Cardiol. 2011;51:280–287. doi: 10.1016/j.yjmcc.2011.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Freund C, Ward-van Oostwaard D, Monshouwer-Kloots J, van den Brink S, van Rooijen M, Xu X, Zweigerdt R, Mummery C, Passier R. Insulin redirects differentiation from cardiogenic mesoderm and endoderm to neuroectoderm in differentiating human embryonic stem cells. Stem Cells. 2008;26:724–733. doi: 10.1634/stemcells.2007-0617. [DOI] [PubMed] [Google Scholar]
- 47.Costa M, Dottori M, Sourris K, Jamshidi P, Hatzistavrou T, Davis R, Azzola L, Jackson S, Lim SM, Pera M, Elefanty AG, Stanley EG. A method for genetic modification of human embryonic stem cells using electroporation. Nat Protoc. 2007;2:792–796. doi: 10.1038/nprot.2007.105. [DOI] [PubMed] [Google Scholar]
- 48.Ng ES, Davis RP, Azzola L, Stanley EG, Elefanty AG. Forced aggregation of defined numbers of human embryonic stem cells into embryoid bodies fosters robust, reproducible hematopoietic differentiation. Blood. 2005;106:1601–1603. doi: 10.1182/blood-2005-03-0987. [DOI] [PubMed] [Google Scholar]
- 49.Burridge PW, Anderson D, Priddle H, Barbadillo MMD, Chamberlain S, Allegrucci C, Young LE, Denning C. Improved human embryonic stem cell embryoid body homogeneity and cardiomyocyte differentiation from a novel v-96 plate aggregation system highlights interline variability. Stem Cells. 2007;25:929–938. doi: 10.1634/stemcells.2006-0598. [DOI] [PubMed] [Google Scholar]
- 50.Ungrin MD, Joshi C, Nica A, Bauwens C, Zandstra PW. Reproducible, ultra high-throughput formation of multicellular organization from single cell suspension-derived human embryonic stem cell aggregates. PLoS One. 2008;3:e1565. doi: 10.1371/journal.pone.0001565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Khademhosseini A, Ferreira L, Blumling J, III, Yeh J, Karp JM, Fukuda J, Langer R. Co-culture of human embryonic stem cells with murine embryonic fibroblasts on microwell-patterned substrates. Biomaterials. 2006;27:5968–5977. doi: 10.1016/j.biomaterials.2006.06.035. [DOI] [PubMed] [Google Scholar]
- 52.Mohr JC, de Pablo JJ, Palecek SP. 3-d microwell culture of human embryonic stem cells. Biomaterials. 2006;27:6032–6042. doi: 10.1016/j.biomaterials.2006.07.012. [DOI] [PubMed] [Google Scholar]
- 53.Mohr JC, Zhang J, Azarin SM, Soerens AG, de Pablo JJ, Thomson JA, Lyons GE, Palecek SP, Kamp TJ. The microwell control of embryoid body size in order to regulate cardiac differentiation of human embryonic stem cells. Biomaterials. 2010;31:1885–1893. doi: 10.1016/j.biomaterials.2009.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bauwens CL, Peerani R, Niebruegge S, Woodhouse KA, Kumacheva E, Husain M, Zandstra PW. Control of human embryonic stem cell colony and aggregate size heterogeneity influences differentiation trajectories. Stem Cells. 2008;26:2300–2310. doi: 10.1634/stemcells.2008-0183. [DOI] [PubMed] [Google Scholar]
- 55.Peerani R, Rao BM, Bauwens C, Yin T, Wood GA, Nagy A, Kumacheva E, Zandstra PW. Niche-mediated control of human embryonic stem cell self-renewal and differentiation. EMBO J. 2007;26:4744–4755. doi: 10.1038/sj.emboj.7601896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.D'Amour KA, Agulnick AD, Eliazer S, Kelly OG, Kroon E, Baetge EE. Efficient differentiation of human embryonic stem cells to definitive endoderm. Nat Biotechnol. 2005;23:1534–1541. doi: 10.1038/nbt1163. [DOI] [PubMed] [Google Scholar]
- 57.Laflamme MA, Chen KY, Naumova AV, Muskheli V, Fugate JA, Dupras SK, Reinecke H, Xu C, Hassanipour M, Police S, O'Sullivan C, Collins L, Chen Y, Minami E, Gill EA, Ueno S, Yuan C, Gold J, Murry CE. Cardiomyocytes derived from human embryonic stem cells in pro-survival factors enhance function of infarcted rat hearts. Nat Biotechnol. 2007;25:1015–1024. doi: 10.1038/nbt1327. [DOI] [PubMed] [Google Scholar]
- 58.Paige SL, Osugi T, Afanasiev OK, Pabon L, Reinecke H, Murry CE. Endogenous wnt/beta-catenin signaling is required for cardiac differentiation in human embryonic stem cells. PLoS One. 2010;5:e11134. doi: 10.1371/journal.pone.0011134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ng ES, Davis R, Stanley EG, Elefanty AG. A protocol describing the use of a recombinant protein-based, animal product-free medium (apel) for human embryonic stem cell differentiation as spin embryoid bodies. Nat Protoc. 2008;3:768–776. doi: 10.1038/nprot.2008.42. [DOI] [PubMed] [Google Scholar]
- 60.Mignone JL, Kreutziger KL, Paige SL, Murry CE. Cardiogenesis from human embryonic stem cells. Circ J. 2010;74:2517–2526. doi: 10.1253/circj.cj-10-0958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang j, Raval KK, Lian X, Herman AM, Wilson GF, Barron MR, Yu J, Palecek SP, Thompson JA, Kamp TJ. Matrix-promoted efficient cardiac differentiation of human ips and es cells. Circulation. 2010;122:A20724. [Google Scholar]
- 62.Mummery C, Ward-van Oostwaard D, Doevendans P, Spijker R, van den BS, Hassink R, van der HM, Opthof T, Pera M, de la Riviere AB, Passier R, Tertoolen L. Differentiation of human embryonic stem cells to cardiomyocytes: Role of coculture with visceral endoderm-like cells. Circulation. 2003;107:2733–2740. doi: 10.1161/01.CIR.0000068356.38592.68. [DOI] [PubMed] [Google Scholar]
- 63.Passier R, Oostwaard DW, Snapper J, Kloots J, Hassink RJ, Kuijk E, Roelen B, de la Riviere AB, Mummery C. Increased cardiomyocyte differentiation from human embryonic stem cells in serum-free cultures. Stem Cells. 2005;23:772–780. doi: 10.1634/stemcells.2004-0184. [DOI] [PubMed] [Google Scholar]
- 64.Xu XQ, Graichen R, Soo SY, Balakrishnan T, Rahmat SN, Sieh S, Tham SC, Freund C, Moore J, Mummery C, Colman A, Zweigerdt R, Davidson BP. Chemically defined medium supporting cardiomyocyte differentiation of human embryonic stem cells. Differentiation. 2008;76:958–970. doi: 10.1111/j.1432-0436.2008.00284.x. [DOI] [PubMed] [Google Scholar]
- 65.Freund C, Davis RP, Gkatzis K, Ward-van Oostwaard D, Mummery CL. The first reported generation of human induced pluripotent stem cells (ips cells) and ips cell-derived cardiomyocytes in the netherlands. Neth Heart J. 2010;18:51–54. [PMC free article] [PubMed] [Google Scholar]
- 66.Mummery C, Ward D, Passier R. Differentiation of human embryonic stem cells to cardiomyocytes by coculture with endoderm in serum free medium. Curr Protoc Stem Cell Biol. 2007 doi: 10.1002/9780470151808.sc01f02s2. 2:1:F.2.1–1F.2.14. [DOI] [PubMed] [Google Scholar]
- 67.Narazaki G, Uosaki H, Teranishi M, Okita K, Kim B, Matsuoka S, Yamanaka S, Yamashita JK. Directed and systematic differentiation of cardiovascular cells from mouse induced pluripotent stem cells. Circulation. 2008;118:498–506. doi: 10.1161/CIRCULATIONAHA.108.769562. [DOI] [PubMed] [Google Scholar]
- 68.Evseenko D, Zhu Y, Schenke-Layland K, Kuo J, Latour B, Ge S, Scholes J, Dravid G, Li X, MacLellan WR, Crooks GM. Mapping the first stages of mesoderm commitment during differentiation of human embryonic stem cells. Proc Natl Acad Sci U S A. 2010;107:13742–13747. doi: 10.1073/pnas.1002077107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Blin G, Nury D, Stefanovic S, Neri T, Guillevic O, Brinon B, Bellamy V, Rucker-Martin C, Barbry P, Bel A, Bruneval P, Cowan C, Pouly J, Mitalipov S, Gouadon E, Binder P, Hagege A, Desnos M, Renaud JF, Menasche P, Puceat M. A purified population of multipotent cardiovascular progenitors derived from primate pluripotent stem cells engrafts in postmyocardial infarcted nonhuman primates. J Clin Invest. 2010;120:1125–1139. doi: 10.1172/JCI40120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kattman SJ, Huber TL, Keller GM. Multipotent flk-1+ cardiovascular progenitor cells give rise to the cardiomyocyte, endothelial, and vascular smooth muscle lineages. Dev Cell. 2006;11:723–732. doi: 10.1016/j.devcel.2006.10.002. [DOI] [PubMed] [Google Scholar]
- 71.Moretti A, Caron L, Nakano A, Lam JT, Bernshausen A, Chen Y, Qyang Y, Bu L, Sasaki M, Martin-Puig S, Sun Y, Evans SM, Laugwitz KL, Chien KR. Multipotent embryonic isl1+ progenitor cells lead to cardiac, smooth muscle, and endothelial cell diversification. Cell. 2006;127:1151–1165. doi: 10.1016/j.cell.2006.10.029. [DOI] [PubMed] [Google Scholar]
- 72.Wu SM, Fujiwara Y, Cibulsky SM, Clapham DE, Lien CL, Schultheiss TM, Orkin SH. Developmental origin of a bipotential myocardial and smooth muscle cell precursor in the mammalian heart. Cell. 2006;127:1137–1150. doi: 10.1016/j.cell.2006.10.028. [DOI] [PubMed] [Google Scholar]
- 73.Bu L, Jiang X, Martin-Puig S, Caron L, Zhu S, Shao Y, Roberts DJ, Huang PL, Domian IJ, Chien KR. Human isl1 heart progenitors generate diverse multipotent cardiovascular cell lineages. Nature. 2009;460:113–117. doi: 10.1038/nature08191. [DOI] [PubMed] [Google Scholar]
- 74.Dubois NC, Craft AM, Sharma P, Elliott DA, Stanley EG, Elefanty AG, Gramolini A, Keller G. Sirpa is a specific cell-surface marker for isolating cardiomyocytes derived from human pluripotent stem cells. Nat Biotechnol. 2011;29:1011–1018. doi: 10.1038/nbt.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Klug MG, Soonpaa MH, Koh GY, Field LJ. Genetically selected cardiomyocytes from differentiating embronic stem cells form stable intracardiac grafts. J Clin Invest. 1996;98:216–224. doi: 10.1172/JCI118769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Fijnvandraat AC, van Ginneken AC, Schumacher CA, Boheler KR, Lekanne Deprez RH, Christoffels VM, Moorman AF. Cardiomyocytes purified from differentiated embryonic stem cells exhibit characteristics of early chamber myocardium. J Mol Cell Cardiol. 2003;35:1461–1472. doi: 10.1016/j.yjmcc.2003.09.011. [DOI] [PubMed] [Google Scholar]
- 77.Gassanov N, Er F, Zagidullin N, Hoppe UC. Endothelin induces differentiation of anp-egfp expressing embryonic stem cells towards a pacemaker phenotype. Faseb J. 2004;18:1710–1712. doi: 10.1096/fj.04-1619fje. [DOI] [PubMed] [Google Scholar]
- 78.Metzger JM, Lin WI, Samuelson LC. Vital staining of cardiac myocytes during embryonic stem cell cardiogenesis in vitro. Circ Res. 1996;78:547–552. doi: 10.1161/01.res.78.4.547. [DOI] [PubMed] [Google Scholar]
- 79.Meyer N, Jaconi M, Landopoulou A, Fort P, Puceat M. A fluorescent reporter gene as a marker for ventricular specification in es-derived cardiac cells. FEBS Letters. 2000;478:151–158. doi: 10.1016/s0014-5793(00)01839-1. [DOI] [PubMed] [Google Scholar]
- 80.Muller M, Fleischmann BK, Selbert S, Ji GJ, Endl E, Middeler G, Muller OJ, Schlenke P, Frese S, Wobus AM, Hescheler J, Katus HA, Franz WM. Selection of ventricular-like cardiomyocytes from es cells in vitro. Faseb J. 2000;14:2540–2548. doi: 10.1096/fj.00-0002com. [DOI] [PubMed] [Google Scholar]
- 81.Anderson D, Self T, Mellor IR, Goh G, Hill SJ, Denning C. Transgenic enrichment of cardiomyocytes from human embryonic stem cells. Mol Ther. 2007;15:2027–2036. doi: 10.1038/sj.mt.6300303. [DOI] [PubMed] [Google Scholar]
- 82.Kita-Matsuo H, Barcova M, Prigozhina N, Salomonis N, Wei K, Jacot JG, Nelson B, Spiering S, Haverslag R, Kim C, Talantova M, Bajpai R, Calzolari D, Terskikh A, McCulloch AD, Price JH, Conklin BR, Chen HS, Mercola M. Lentiviral vectors and protocols for creation of stable hesc lines for fluorescent tracking and drug resistance selection of cardiomyocytes. PLoS One. 2009;4:e5046. doi: 10.1371/journal.pone.0005046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Xu XQ, Zweigerdt R, Soo SY, Ngoh ZX, Tham SC, Wang ST, Graichen R, Davidson B, Colman A, Sun W. Highly enriched cardiomyocytes from human embryonic stem cells. Cytotherapy. 2008;10:376–389. doi: 10.1080/14653240802105307. [DOI] [PubMed] [Google Scholar]
- 84.Huber I, Itzhaki I, Caspi O, Arbel G, Tzukerman M, Gepstein A, Habib M, Yankelson L, Kehat I, Gepstein L. Identification and selection of cardiomyocytes during human embryonic stem cell differentiation. Faseb J. 2007;21:2551–2563. doi: 10.1096/fj.05-5711com. [DOI] [PubMed] [Google Scholar]
- 85.Zhu WZ, Xie Y, Moyes KW, Gold JD, Askari B, Laflamme MA. Neuregulin/erbb signaling regulates cardiac subtype specification in differentiating human embryonic stem cells. Circ Res. 2010;107:776–786. doi: 10.1161/CIRCRESAHA.110.223917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kim C, Majdi M, Xia P, Wei KA, Talantova M, Spiering S, Nelson B, Mercola M, Chen HS. Non-cardiomyocytes influence the electrophysiological maturation of human embryonic stem cell-derived cardiomyocytes during differentiation. Stem Cells Dev. 2010;19:783–795. doi: 10.1089/scd.2009.0349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Cao F, Lin S, Xie X, Ray P, Patel M, Zhang X, Drukker M, Dylla SJ, Connolly AJ, Chen X, Weissman IL, Gambhir SS, Wu JC. In vivo visualization of embryonic stem cell survival, proliferation, and migration after cardiac delivery. Circulation. 2006;113:1005–1014. doi: 10.1161/CIRCULATIONAHA.105.588954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Giudice A, Trounson A. Genetic modification of human embryonic stem cells for derivation of target cells. Cell Stem Cell. 2008;2:422–433. doi: 10.1016/j.stem.2008.04.003. [DOI] [PubMed] [Google Scholar]
- 89.Elefanty AG, Stanley EG. Reshaping pluripotent stem cells. Nat Biotechnol. 2009;27:823–824. doi: 10.1038/nbt0909-823. [DOI] [PubMed] [Google Scholar]
- 90.Uosaki H, Fukushima H, Takeuchi A, Matsuoka S, Nakatsuji N, Yamanaka S, Yamashita JK. Efficient and scalable purification of cardiomyocytes from human embryonic and induced pluripotent stem cells by vcam1 surface expression. PLoS One. 2011;6:e23657. doi: 10.1371/journal.pone.0023657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hattori F, Chen H, Yamashita H, Tohyama S, Satoh YS, Yuasa S, Li W, Yamakawa H, Tanaka T, Onitsuka T, Shimoji K, Ohno Y, Egashira T, Kaneda R, Murata M, Hidaka K, Morisaki T, Sasaki E, Suzuki T, Sano M, Makino S, Oikawa S, Fukuda K. Nongenetic method for purifying stem cell-derived cardiomyocytes. Nat Methods. 2010;7:61–66. doi: 10.1038/nmeth.1403. [DOI] [PubMed] [Google Scholar]
- 92.Domian IJ, Chiravuri M, van der Meer P, Feinberg AW, Shi X, Shao Y, Wu SM, Parker KK, Chien KR. Generation of functional ventricular heart muscle from mouse ventricular progenitor cells. Science. 2009;326:426–429. doi: 10.1126/science.1177350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Azarin SM, Palecek SP. Development of scalable culture systems for human embryonic stem cells. Biochem Eng J. 2010;48:378. doi: 10.1016/j.bej.2009.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Krawetz R, Taiani JT, Liu S, Meng G, Li X, Kallos MS, Rancourt DE. Large-scale expansion of pluripotent human embryonic stem cells in stirred-suspension bioreactors. Tissue Eng Part C Methods. 2010;16:573–582. doi: 10.1089/ten.TEC.2009.0228. [DOI] [PubMed] [Google Scholar]
- 95.Bauwens C, Yin T, Dang S, Peerani R, Zandstra PW. Development of a perfusion fed bioreactor for embryonic stem cell-derived cardiomyocyte generation: Oxygen-mediated enhancement of cardiomyocyte output. Biotechnol Bioeng. 2005;90:452–461. doi: 10.1002/bit.20445. [DOI] [PubMed] [Google Scholar]
- 96.Schroeder M, Niebruegge S, Werner A, Willbold E, Burg M, Ruediger M, Field LJ, Lehmann J, Zweigerdt R. Differentiation and lineage selection of mouse embryonic stem cells in a stirred bench scale bioreactor with automated process control. Biotechnol Bioeng. 2005;92:920–933. doi: 10.1002/bit.20668. [DOI] [PubMed] [Google Scholar]
- 97.Zweigerdt R, Burg M, Willbold E, Abts H, Ruediger M. Generation of confluent cardiomyocyte monolayers derived from embryonic stem cells in suspension: A cell source for new therapies and screening strategies. Cytotherapy. 2003;5:399–413. doi: 10.1080/14653240310003062. [DOI] [PubMed] [Google Scholar]
- 98.Zandstra PW, Bauwens C, Yin T, Liu Q, Schiller H, Zweigerdt R, Pasumarthi KB, Field LJ. Scalable production of embryonic stem cell-derived cardiomyocytes. Tissue Eng. 2003;9:767–778. doi: 10.1089/107632703768247449. [DOI] [PubMed] [Google Scholar]
- 99.Niebruegge S, Nehring A, Bar H, Schroeder M, Zweigerdt R, Lehmann J. Cardiomyocyte production in mass suspension culture: Embryonic stem cells as a source for great amounts of functional cardiomyocytes. Tissue Eng Part A. 2008;14:1591–1601. doi: 10.1089/ten.tea.2007.0247. [DOI] [PubMed] [Google Scholar]
- 100.Jing D, Parikh A, Tzanakakis ES. Cardiac cell generation from encapsulated embryonic stem cells in static and scalable culture systems. Cell Transplant. 2010;19:1397–1412. doi: 10.3727/096368910X513955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Yirme G, Amit M, Laevsky I, Osenberg S, Itskovitz-Eldor J. Establishing a dynamic process for the formation, propagation, and differentiation of human embryoid bodies. Stem Cells Dev. 2008;17:1227–1241. doi: 10.1089/scd.2007.0272. [DOI] [PubMed] [Google Scholar]
- 102.Niebruegge S, Bauwens CL, Peerani R, Thavandiran N, Masse S, Sevaptisidis E, Nanthakumar K, Woodhouse K, Husain M, Kumacheva E, Zandstra PW. Generation of human embryonic stem cell-derived mesoderm and cardiac cells using size-specified aggregates in an oxygen-controlled bioreactor. Biotechnol Bioeng. 2009;102:493–507. doi: 10.1002/bit.22065. [DOI] [PubMed] [Google Scholar]
- 103.Ma J, Guo L, Fiene SJ, Anson BD, Thomson JA, Kamp TJ, Kolaja KL, Swanson BJ, January CT. High purity human-induced pluripotent stem cell-derived cardiomyocytes: Electrophysiological properties of action potentials and ionic currents. Am J Physiol Heart Circ Physiol. 2011;301:H2006–2017. doi: 10.1152/ajpheart.00694.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Sartiani L, Bettiol E, Stillitano F, Mugelli A, Cerbai E, Jaconi ME. Developmental changes in cardiomyocytes differentiated from human embryonic stem cells: A molecular and electrophysiological approach. Stem Cells. 2007;25:1136–1144. doi: 10.1634/stemcells.2006-0466. [DOI] [PubMed] [Google Scholar]
- 105.Yazawa M, Hsueh B, Jia X, Pasca AM, Bernstein JA, Hallmayer J, Dolmetsch RE. Using induced pluripotent stem cells to investigate cardiac phenotypes in timothy syndrome. Nature. 2011;471:230–234. doi: 10.1038/nature09855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Itzhaki I, Maizels L, Huber I, Zwi-Dantsis L, Caspi O, Winterstern A, Feldman O, Gepstein A, Arbel G, Hammerman H, Boulos M, Gepstein L. Modelling the long qt syndrome with induced pluripotent stem cells. Nature. 2011;471:225–229. doi: 10.1038/nature09747. [DOI] [PubMed] [Google Scholar]
- 107.Kehat I, Gepstein A, Spira A, Itskovitz-Eldor J, Gepstein L. High-resolution electrophysiological assessment of human embryonic stem cell-derived cardiomyocytes: A novel in vitro model for the study of conduction. Circ Res. 2002;91:659–661. doi: 10.1161/01.res.0000039084.30342.9b. [DOI] [PubMed] [Google Scholar]
- 108.Bock C, Kiskinis E, Verstappen G, Gu H, Boulting G, Smith ZD, Ziller M, Croft GF, Amoroso MW, Oakley DH, Gnirke A, Eggan K, Meissner A. Reference maps of human es and ips cell variation enable high-throughput characterization of pluripotent cell lines. Cell. 2011;144:439–452. doi: 10.1016/j.cell.2010.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Burridge PW, Thompson S, Millrod MA, Weinberg S, Yuan X, Peters A, Mahairaki V, Koliatsos VE, Tung L, Zambidis ET. A universal system for highly efficient cardiac differentiation of human induced pluripotent stem cells that eliminates interline variability. PLoS One. 2011;6:e18293. doi: 10.1371/journal.pone.0018293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Takahashi T, Lord B, Schulze PC, Fryer RM, Sarang SS, Gullans SR, Lee RT. Ascorbic acid enhances differentiation of embryonic stem cells into cardiac myocytes. Circulation. 2003;107:1912–1916. doi: 10.1161/01.CIR.0000064899.53876.A3. [DOI] [PubMed] [Google Scholar]
- 111.Ding S, Schultz PG. A role for chemistry in stem cell biology. Nat Biotechnol. 2004;22:833–840. doi: 10.1038/nbt987. [DOI] [PubMed] [Google Scholar]
- 112.Sadek H, Hannack B, Choe E, Wang J, Latif S, Garry MG, Garry DJ, Longgood J, Frantz DE, Olson EN, Hsieh J, Schneider JW. Cardiogenic small molecules that enhance myocardial repair by stem cells. Proc Natl Acad Sci U S A. 2008;105:6063–6068. doi: 10.1073/pnas.0711507105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Willems E, Spiering S, Davidovics H, Lanier M, Xia Z, Dawson M, Cashman J, Mercola M. Small-molecule inhibitors of the wnt pathway potently promote cardiomyocytes from human embryonic stem cell-derived mesoderm. Circ Res. 2011;109:360–364. doi: 10.1161/CIRCRESAHA.111.249540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Lian X, Hsiao S, Wilson G, Zhu K, Hazeltine LB, Azarin SM, Raval K, Zhang J, Kamp TJ, Palecek SP. Robust cardiomyocyte differentiation from human pluripotent stem cells via temporal modulation of canonical wnt signaling. Proc Natl Acad Sci U S A. 2012;109 doi: 10.1073/pnas.1200250109. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Graichen R, Xu X, Braam SR, Balakrishnan T, Norfiza S, Sieh S, Soo SY, Tham SC, Mummery C, Colman A, Zweigerdt R, Davidson BP. Enhanced cardiomyogenesis of human embryonic stem cells by a small molecular inhibitor of p38 mapk. Differentiation. 2008;76:357–370. doi: 10.1111/j.1432-0436.2007.00236.x. [DOI] [PubMed] [Google Scholar]
- 116.Yanes O, Clark J, Wong DM, Patti GJ, Sanchez-Ruiz A, Benton HP, Trauger SA, Desponts C, Ding S, Siuzdak G. Metabolic oxidation regulates embryonic stem cell differentiation. Nat Chem Biol. 2010;6:411–417. doi: 10.1038/nchembio.364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Smith J, Ladi E, Mayer-Proschel M, Noble M. Redox state is a central modulator of the balance between self-renewal and differentiation in a dividing glial precursor cell. Proc Natl Acad Sci U S A. 2000;97:10032–10037. doi: 10.1073/pnas.170209797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Ezashi T, Das P, Roberts RM. Low o2 tensions and the prevention of differentiation of hes cells. Proc Natl Acad Sci U S A. 2005;102:4783–4788. doi: 10.1073/pnas.0501283102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Lin Q, Lee YJ, Yun Z. Differentiation arrest by hypoxia. J Biol Chem. 2006;281:30678–30683. doi: 10.1074/jbc.C600120200. [DOI] [PubMed] [Google Scholar]
- 120.Buggisch M, Ateghang B, Ruhe C, Strobel C, Lange S, Wartenberg M, Sauer H. Stimulation of es-cell-derived cardiomyogenesis and neonatal cardiac cell proliferation by reactive oxygen species and nadph oxidase. J Cell Sci. 2007;120:885–894. doi: 10.1242/jcs.03386. [DOI] [PubMed] [Google Scholar]
- 121.Li J, Stouffs M, Serrander L, Banfi B, Bettiol E, Charnay Y, Steger K, Krause KH, Jaconi ME. The nadph oxidase nox4 drives cardiac differentiation: Role in regulating cardiac transcription factors and map kinase activation. Mol Biol Cell. 2006;17:3978–3988. doi: 10.1091/mbc.E05-06-0532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Rozario T, DeSimone DW. The extracellular matrix in development and morphogenesis: A dynamic view. Dev Biol. 2010;341:126–140. doi: 10.1016/j.ydbio.2009.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Chien KR, Domian IJ, Parker KK. Cardiogenesis and the complex biology of regenerative cardiovascular medicine. Science. 2008;322:1494–1497. doi: 10.1126/science.1163267. [DOI] [PubMed] [Google Scholar]
- 124.Melkoumian Z, Weber JL, Weber DM, Fadeev AG, Zhou Y, Dolley-Sonneville P, Yang J, Qiu L, Priest CA, Shogbon C, Martin AW, Nelson J, West P, Beltzer JP, Pal S, Brandenberger R. Synthetic peptide-acrylate surfaces for long-term self-renewal and cardiomyocyte differentiation of human embryonic stem cells. Nat Biotechnol. 2010;28:606–610. doi: 10.1038/nbt.1629. [DOI] [PubMed] [Google Scholar]
- 125.Rodin S, Domogatskaya A, Strom S, Hansson EM, Chien KR, Inzunza J, Hovatta O, Tryggvason K. Long-term self-renewal of human pluripotent stem cells on human recombinant laminin-511. Nat Biotechnol. 2010;28:611–615. doi: 10.1038/nbt.1620. [DOI] [PubMed] [Google Scholar]
- 126.Villa-Diaz LG, Nandivada H, Ding J, Nogueira-de-Souza NC, Krebsbach PH, O'Shea KS, Lahann J, Smith GD. Synthetic polymer coatings for long-term growth of human embryonic stem cells. Nat Biotechnol. 2010;28:581–583. doi: 10.1038/nbt.1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Klim JR, Li L, Wrighton PJ, Piekarczyk MS, Kiessling LL. A defined glycosaminoglycan-binding substratum for human pluripotent stem cells. Nat Methods. 2010;7:989–994. doi: 10.1038/nmeth.1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Dixon JE, Dick E, Rajamohan D, Shakesheff KM, Denning C. Directed differentiation of human embryonic stem cells to interrogate the cardiac gene regulatory network. Mol Ther. 2011;19:1695–1703. doi: 10.1038/mt.2011.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Passier R, van Laake LW, Mummery CL. Stem-cell-based therapy and lessons from the heart. Nature. 2008;453:322–329. doi: 10.1038/nature07040. [DOI] [PubMed] [Google Scholar]