Abstract
Background
The genetic diversity and the clinical relevance of the drug-resistant Klebsiella pneumoniae isolates from hospital settings are largely unknown. We thus conducted this prospective study to analyze the molecular epidemiology of K. pneumoniae isolates from patients being treated in the 306 Hospital in Beijing, China for the period of November 1, 2010–October 31, 2011.
Methodology/Principal Findings
Antibiotic susceptibility testing, PCR amplification and sequencing of the drug resistance-associated genes, and multilocus sequence typing (MLST) were conducted. A total of 163 isolates were analyzed. The percentage of MDR, XDR and PDR isolates were 63.8% (104), 20.9 (34), and 1.8% (3), respectively. MLST results showed that 60 sequence types (STs) were identified, which were further separated by eBURST into 13 clonal complexes and 18 singletons. The most dominant ST was ST15 (10.4%). Seven new alleles and 24 new STs were first identified in this study. Multiple logistic regression analysis revealed that certain clinical characteristics were associated with those prevalent STs such as: from ICU, from medical ward, from community acquired infection, from patients without heart disease, from patients with treatment success, susceptible to extended spectrum cephalosporin, susceptible to cephamycins, susceptible to fluoroquinolones, and with MDR.
Conclusions/Significance
Our data indicate that certain drug-resistant K. pneumoniae clones are highly prevalent and are associated with certain clinical characteristics in hospital settings. Our study provides evidence demonstrating that intensive nosocomial infection control measures are urgently needed.
Introduction
Klebsiella pneumoniae is an important bacterial pathogen associated with community acquired (CA) and hospital acquired (HA) infections and has the potential to cause severe morbidity and mortality, particularly in immunocompromised patients [1]–[3]. Infections caused by drug-resistant K. pneumoniae isolates, especially those produce extended-spectrum beta-lactamases (ESBLs) and which are multidrug-resistant (MDR), extensively drug-resistant (XDR) or pandrug-resistant (PDR), are more difficult and expensive to treat with worse treatment outcome [4]–[8]. More recently, carbapenem-resistant K. pneumoniae have been reported worldwide as a consequence of acquisition of carbapenemase genes, and a large variety of carbapenemases have been identified in K. pneumoniae [9]–[14].
Rapid and discriminative genotyping methods are useful for determining the clonality of the isolates in nosocomial or household outbreaks [15], [16]. Multilocus sequencing typing (MLST) is a nucleotide sequence-based approach for characterizing bacterial isolates (http://www.mlst.net/), with the advantage over traditional pulsed-field gel electrophoresis (PFGE) of ease of manipulation and convenient comparison [17], [18]. In our previous study, we observed alarmingly high rates of MDR, XDR and PDR strains among K. pneumoniae isolates from a tertiary care hospital in Beijing, China [19]. In addition, data from that study indicate that many of the drug resistance genes were transmissible [19]. Since the genetic diversity, transmission patterns and the clinical relevance of the drug-resistant K. pneumoniae isolates from hospital settings are largely unknown, we thus further conducted this MLST genotyping analysis for K. pneumoniae isolates from the 306 Hospital, a tertiary care hospital in Beijing, China for the period of November 1, 2010–October 31, 2011 with an aim to assess the molecular epidemiology as well as clinical characteristics associated with prevalent K. pneumoniae clones.
Methods
Ethics statement
All of the investigation protocols in this study were approved by the institutional ethics committee of the 306 Hospital, Beijing, China. Written informed consent for K. pneumoniae isolates to be collected as well as for their information to be stored in the hospital database for research purposes was provided by participants. Written informed consent was obtained from the next of kin, caretakers, or guardians on the behalf of the minors/children participants involved in this study. Permission for using the information in the medical records of the patients for research purposes was obtained from the 306 Hospital. The Institute ethics committee of the 306 Hospital reviewed that relevant ethical issues in this study were considered.
Study population, bacterial isolate identification, and drug susceptibility testing
The 306 Hospital in Beijing, China is a tertiary care hospital, with 1,100 beds and approximately 25,000 hospital admissions per year. Consecutive non-repetitive K. pneumoniae isolates were collected from patients being treated in the 306 Hospital for the period of November 1, 2010–October 31, 2011. Isolates with ambiguous sequence data for one or more alleles were excluded from the analysis. All isolates were cultured in Luria-Bertani (LB) medium. A total of 175 isolates were confirmed as K. pneumoniae by 16S rDNA sequencing. Drug susceptibility testing (DST) for the K. pneumoniae isolates was performed using the bioMérieux VITEK2 system following manufacturer's instructions. The following 18 drugs were tested: ampicillin (AMP), piperacillin/tazobactam (TZP), ampicillin/sulbactam (SAM), cefazolin (CFZ), ceftriaxone (CRO), ceftazidime (CAZ), cefepime (FEP), cefotetan (CTT), ertapenem (ETP), imipenem (IM), aztreonam (ATM), ciprofloxacin (CIP), levofloxacin (LVX), gentamicin (GM), tobramycin (TOB), amikacin (AMK), trimethoprim-sulfamethoxazole (SXT), and nitrofurantoin (FD). The ESBLs were detected by the bioMérieux VITEK-2 AST-GN13 test. In some cases, the ESBL positivity was further confirmed by the double disk diffusion method according to standard protocols by the Clinical Laboratory Standard Institute (CLSI) [20]. Escherichia coli strains ATCC 25922 and ATCC 35218, Klebsiella pneumoniae strain ATCC 700603 and Pseudomonas aeruginosa strain ATCC 27853 were used as quality control strains for the DST. Clinical records of patients from whom the K. pneumoniae isolates were obtained were reviewed retrospectively.
PCR amplification and sequencing for drug resistance genes
Drug resistance-associated genes were detected by PCR and sequencing using 37 pairs of primers as described previously [19]. The drug resistance-associated genes examined include: bla CTX-M-1, bla CTX-M-2, bla CTX-M-3, bla CTX-M-8, bla CTX-M-9, bla CTX-M-10, bla CTX-M-14, bla CTX-M-25, bla SHV-group, bla TEM, bla KPC, bla NDM, bla IMP, bla VIM, bla OXA-48, bla CMY, bla DHA, bla FOX, dhfr, qnrA, qnrB, qnrC, qnrD, qnrS, aac(6′)-Ib-cr, qepA, gyrA, parC, aacA4, aacC1, aacC2, aadA1, aadB, aphA6, armA, rmtB, and Integron I. DNA sequences were annotated using the BLAST program at http://www.ncbi.nlm.nih.gov.
Genotyping of K. pneumoniae isolates by MLST analysis
Genotyping was determined by MLST analysis. MLST with seven genes (gapA, infB, mdh, pgi, phoE, rpoB and tonB) was performed on isolates according to the protocol described on the K. pneumoniae MLST website (www.pasteur.fr/mlst) [18]. Alleles and sequence types (STs) were assigned by using the MLST database (www.pasteur.fr/mlst/Kpneumoniae.html). Alleles and STs that had not been previously described were submitted to the curator of the database and were assigned new designations.
Assignment to clonal complexes
The program eBURST v 3.0 was used to identify the different clonal complexes [21]. Clonal complexes were defined as groups of two or more independent isolates that shared identical alleles at six or more loci; each complex was named after the putative founder ST. Data from additional 1,380 isolates of K. pneumoniae were obtained from the MLST isolate database deposited at the Pasteur Institute (http://www.pasteur.fr/cgi-bin/genopole/PF8/mlstdbnet.pl?file=klebs_isolates.xml) [18].
Sequence analysis
The proportions of nucleotide alterations that led to a change in the amino acid sequence (non-synonymous substitution, dn) and the proportions of nucleotide alterations that did not lead to a change in the amino acid sequence (synonymous substitution, ds) were calculated with START2 [22]. Phylogenetic analysis was performed using ClonalFrame algorithm with the software package ClonalFrame version 1.1 [23], using 50,000 burn-in cycles and 100,000 further iterations. Maximum likelihood tree was constructed using PhyML 3.0 under a GTR+I+G model based on the alignment of concatenated sequence from the seven MLST gene loci with 1000 bootstrap replicates [24].
Measurement of clonality
The standardized index of association (IAS) for the seven loci was calculated using START2 software and 1,000 iterations [22]. IAS was also estimated using 764 unique STs including the new STs in this study and existing STs in the entire MLST isolate database as a whole.
Definitions
Every patient was counted only once during his hospital stay regardless of the number of positive cultures. Each case was differentiated between CA infection and HA infection based on a temporal definition. A CA case was defined as a case with known carriage of K. pneumoniae on admission or with the first culture positive for these bacteria within 48 h of admission [6], [25]. A HA case was defined by the first culture positive obtained more than 48 h after admission. MDR was defined as acquired non-susceptibility to at least one agent in three or more antimicrobial categories, XDR was defined as non-susceptibility to at least one agent in all but two or fewer antimicrobial categories (i.e. bacterial isolates remain susceptible to only one or two categories) and PDR was defined as non-susceptibility to all agents in all antimicrobial categories [7].
Statistical analyses
Data were entered and analysed using the statistical package SPSS for windows (version 15). Two people were independently cross-checked each entry to ensure the quality of data entered into the computer. For categorical data, different groups were compared using the Chi-square test. Univariate and multivariate analyses were used to determine the factors associated with prevalent K. pnuemoniae clones. The factors examined were shown in Table S5. All Potentially associated factors were included in a logistic regression model for multivariate analysis, and they were eliminated using a backward stepwise selection method using a P value threshold of 0.1 for the variables to be remained in the model. Mantel-Haenszel odds ratios (ORs), 95% confidence intervals (CIs) and corresponding P values were reported. P value of <0.05 was considered to be statistically significant.
Results
Demographic and clinical characteristics of the patients
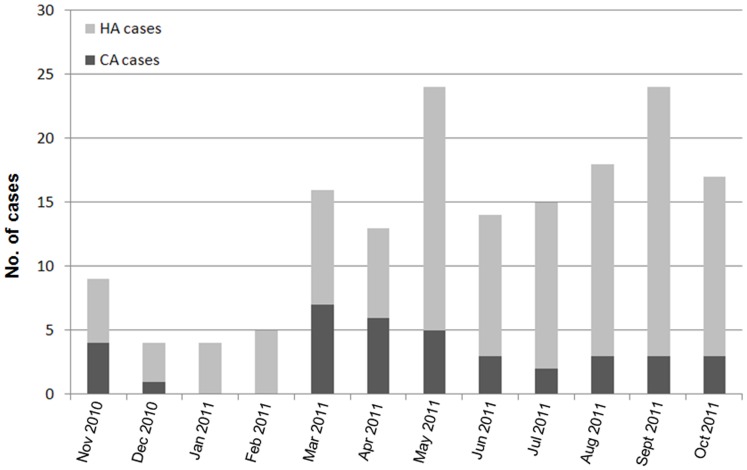
From November 1, 2010 to October 31, 2011, a total of 175 non-repetitive hospitalized patients who had K. pneumoniae isolates available were subjected to DST at the 306 Hospital. Twelve (6.9%) patients were excluded from the study as a result of low-quality sequencing results for one or more of the 7 house-keeping genes of their isolates. Among the remaining 163 patients, 126 (77.3%) were HA cases and 37 (22.7%) were CA cases. Eighty (49.1%) isolates were ESBL positive and 83 (50.9%) isolates were ESBL negative. The proportion of the male and female patients were 73.6% (120/163) and 26.4% (43/163), respectively. Fifty-four (33.1%) of the patients were Beijing residents and the rest were from other provinces of China (non-Beijing residents). The median (±SD) age of the patients was 74.50±19.16 years (range 1–98 years). The majority of the patients were from medical ward (75/163, 46.0%) and intensive care unit (ICU) (49/163, 30.1%). The main source of the specimens was sputum (121/163, 74.2%). The proportion of MDR, XDR, PDR, and other types of K. pneumoniae isolates were 63.8% (104/163), 20.9% (34/163), 1.8% (3/163) and 13.5% (22/163) respectively. Notably, the rates of resistance to most drugs were much higher among ESBL positive isolates than ESBL negative isolates. The epidemiology of CA and HA cases based on the admission date is shown in Fig. 1. More detailed information on relevant demographic and clinical characteristics of the study population is summarized in Table 1.
Figure 1. Epidemiology of hospitalized patients that were colonized or infected with K. pneumoniae isolates in the 306 Hospital during November 1, 2010–October 31, 2011.
Note: HA: hospital acquired; CA: community acquired.
Table 1. Demographic and clinical characteristics of the patients.
| Characteristics | Total n = 163 (%) | ESBL positive cases n = 80 (%) | ESBL negative cases n = 83 (%) | P value |
| Gender | 0.166 | |||
| Male | 120 (73.6) | 55 (68.8) | 65 (78.3) | |
| Female | 43 (26.4) | 25 (31.3) | 18 (21.7) | |
| Age group, years | ||||
| <15 | 6 (3.7) | 3 (3.8) | 3 (3.6) | 0.963 |
| 15–60 | 31 (19.0) | 14 (17.5) | 17 (20.5) | 0.628 |
| >60 | 126 (77.3) | 63 (78.8) | 63 (75.9) | 0.665 |
| Residence situation | 0.406 | |||
| Beijing resident | 54 (33.1) | 29 (36.3) | 25 (30.1) | |
| Non-Beijing resident | 109 (66.9) | 51 (63.8) | 58 (69.9) | |
| Hospital location | ||||
| Emergency room | 8 (4.9) | 3 (3.8) | 5 (6.0) | 0.502 |
| Intensive care unit | 49 (30.1) | 27 (33.8) | 22 (26.5) | 0.313 |
| Medical ward | 75 (46.0) | 37 (46.3) | 38 (45.8) | 0.952 |
| Surgical ward | 31 (19.0) | 13 (16.3) | 18 (21.7) | 0.377 |
| Sources of specimens | ||||
| Sputum | 121 (74.2) | 60 (75.0) | 61 (73.5) | 0.826 |
| Urine | 12 (7.4) | 9 (11.3) | 3 (3.6) | 0.062 |
| Throat or nose swabs | 13 (8.0) | 3 (3.8) | 10 (12.0) | 0.051 |
| Blood | 8 (4.9) | 5 (6.3) | 3 (3.6) | 0.436 |
| Others | 9 (5.5) | 3 (3.8) | 6 (7.2) | 0.331 |
| Underlying diseases | ||||
| Pneumonia | 14 (8.6) | 6 (7.5) | 8 (9.6) | 0.610 |
| Diabetes mellitus | 40 (24.5) | 19 (23.8) | 21 (25.3) | 0.786 |
| Chronic bronchitis | 11 (6.7) | 6 (7.5) | 5 (6.0) | 0.721 |
| Chronic obstructive pulmonary disease | 14 (8.6) | 7 (8.8) | 7 (8.4) | 0.959 |
| Abnormal liver function | 36 (22.1) | 13 (16.3) | 23 (27.7) | 0.070 |
| Renal dysfunction | 35 (21.5) | 21 (26.3) | 14 (16.9) | 0.152 |
| Hypertension | 84 (51.5) | 39 (48.8) | 45 (54.2) | 0.427 |
| Heart disease | 55 (33.7) | 30 (37.5) | 25 (30.1) | 0.337 |
| Cerebral infarction | 41 (25.2) | 22 (27.5) | 19 (22.9) | 0.520 |
| Pulmonary infection | 81 (49.7) | 42 (52.5) | 39 (47.0) | 0.513 |
| Urinary tract infection | 11 (6.7) | 4 (5.0) | 7 (8.4) | 0.383 |
| Infection acquired model | 0.665 | |||
| Community acquired | 37 (22.7) | 17 (21.3) | 20 (24.1) | |
| Hospital acquired | 126 (77.3) | 63 (78.8) | 63 (75.9) | |
| Drug resistance profiles | ||||
| Penicillins | 162 (99.4) | 80 (100.0) | 82 (98.8) | 0.325 |
| 1st and 2nd generation cephalosporins | 86 (52.8) | 78 (97.5) | 8 (9.6) | <0.001 |
| 3rd and 4th generation cephalosporins | 86 (52.8) | 77 (96.3) | 9 (10.8) | <0.001 |
| Cephamycins | 108 (66.3) | 37 (46.3) | 71 (85.5) | <0.001 |
| Carbapenems | 12 (7.4) | 8 (10.0) | 4 (4.8) | 0.205 |
| Monobactams | 83 (50.9) | 77 (96.3) | 6 (7.2) | <0.001 |
| Fluoroquinolones | 72 (44.2) | 53 (66.3) | 19 (22.9) | <0.001 |
| Aminoglycosides | 90 (55.2) | 71 (88.8) | 19 (22.9) | <0.001 |
| Folate pathway inhibitors | 94 (57.7) | 76 (95.0) | 18 (21.7) | <0.001 |
| Nitrofurantoin | 145 (89.0) | 77 (96.3) | 68 (81.9) | 0.004 |
| Drug resistance types | ||||
| MDR-KP | 104 (63.8) | 49 (61.3) | 55 (66.3) | 0.505 |
| XDR-KP | 34 (20.9) | 22 (27.5) | 12 (14.5) | 0.040 |
| PDR-KP | 3 (1.8) | 3 (3.8) | 0 | 0.075 |
| Other types of KP | 22 (13.5) | 6 (7.5) | 16 (19.3) | 0.028 |
| Treatment outcome | 0.751 | |||
| Treatment success | 138 (84.7) | 67 (83.8) | 71 (85.5) | |
| Died | 25 (15.3) | 13 (16.3) | 12 (14.5) |
MLST analysis of the K. pneumoniae isolates and identification of clonal complexes
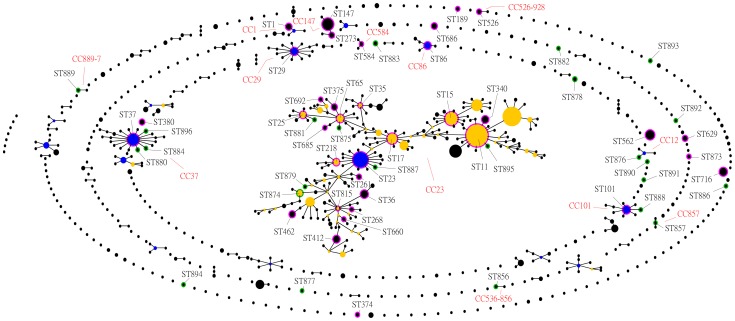
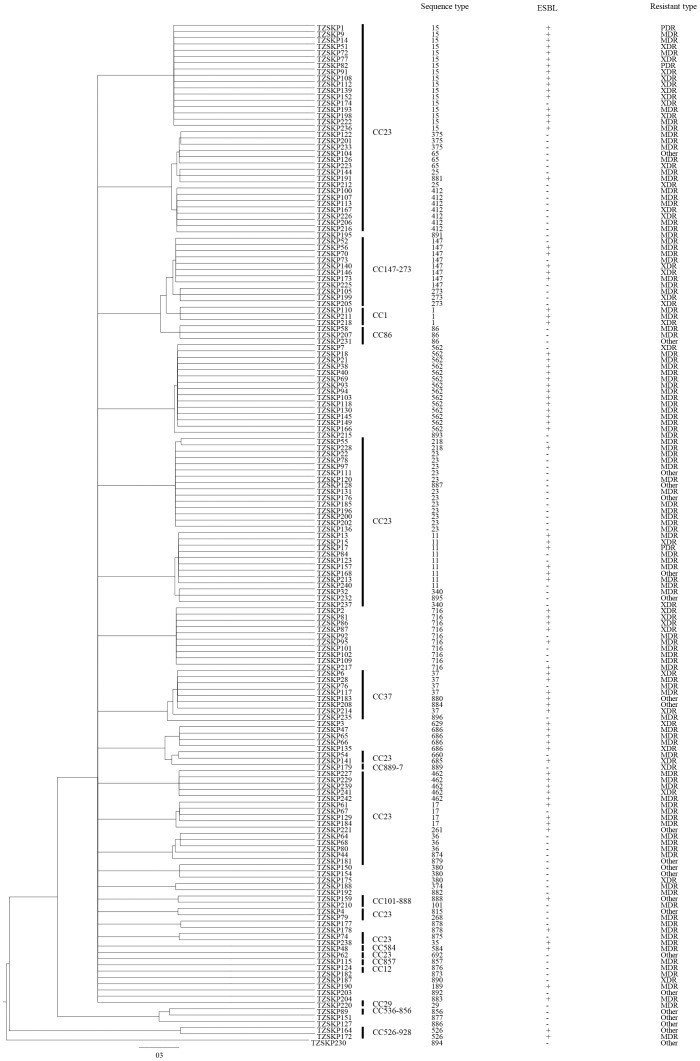
MLST was conducted to determine the extent of genotypic diversity among the K. pneumoniae isolates. Sixty different STs were identified by MLST. Thirty-seven of the STs contained single isolates, while 23 STs included between 2 and 17 isolates. 24 (40%) of the 60 STs had not been previously identified, 16 of these differed from recognized STs at only a single locus, 5 of these (ST877, ST882, ST883, ST890 and ST891) differed from recognized STs at two loci, and 3 of these (ST886, ST893 and ST894) differed from recognized STs at three loci (Table S1). The 60 STs generated in this data set were separated by eBURST into 13 clonal complexes and 18 singletons with the default stringent definition of the groups by sharing alleles at 6 of 7 loci (Fig. 2 and 3, Table 2, and Table S1). The most dominant ST was ST15 (10.4%, 17/163), followed by ST562 (8.6%, 14/163), ST23 (7.4%, 12/163), ST716 (6.1%, 10/163), ST11 (5.5%, 9/163), ST147 (4.9%, 8/163). Those 6 STs accounted for 42.9% (70/163) of the total isolates, and those 70 isolates were thus designated prevalent clones in this study. Two of the three PDR isolates in this study belonged to ST15.
Figure 2. Comparative eBURST analysis showing the clonal assignment of the STs present in this study compared to that of the STs in the entire K. pneumonia MLST isolate database.
Only STs in this study are given, and lines connect single locus variants. The names of the CCs are based on the ST assigned as the founder genotype of the complex shown in blue. Subgroup founders are shown in yellow. The relative size of the circles indicates their prevalence. New STs identified in this study are highlighted by a green halo; STs present in the database and this study are highlighted by a pink halo.
Figure 3. 50% majority-rule consensus phylogenetic tree derived from ClonalFrame for the 7 housekeeping loci in K. pneumoniae, displaying the clonal relationship between the STs and drug resistance of the K. pneumoniae population.
Table 2. Distribution of sequence types in K. pneumoniae clonal complexes.
| Clonal complex | Sequence type | Allelic profile | No. of isolates | No. of isolates | |||
| ESBL+ | ESBL - | HA | CA | ||||
| bla CTX-M | bla TEM/bla SHV | ||||||
| CC23 | 23 | 2-1-1-1-9-4-12 | 0 | 0 | 12 | 7 | 5 |
| 11 | 3-3-1-1-1-1-4 | 5 | 4 | 3 | 5 | 4 | |
| 15 | 1-1-1-1-1-1-1 | 14 | 11 | 1 | 10 | 7 | |
| 17 | 2-1-1-1-4-4-4 | 3 | 2 | 1 | 3 | 1 | |
| 65 | 2-1-2-1-10-4-13 | 0 | 0 | 3 | 3 | 0 | |
| 340 | 3-3-1-1-1-1-18 | 0 | 0 | 2 | 2 | 0 | |
| 895 | 3-3-1-1-1-1-42 | 0 | 0 | 1 | 1 | 0 | |
| 815 | 2-1-2-1-7-1-12 | 0 | 0 | 1 | 1 | 0 | |
| 35 | 2-1-2-1-10-1-19 | 1 | 1 | 0 | 1 | 0 | |
| 268 | 2-1-2-1-7-1-81 | 0 | 0 | 1 | 0 | 1 | |
| 875 | 2-1-2-1-10-4-19 | 0 | 0 | 1 | 0 | 1 | |
| 25 | 2-1-1-1-10-4-13 | 0 | 0 | 2 | 1 | 1 | |
| 36 | 2-1-2-1-7-1-7 | 0 | 0 | 3 | 3 | 0 | |
| 218 | 2-3-1-1-9-4-12 | 1 | 0 | 1 | 1 | 1 | |
| 375 | 43-1-2-1-10-4-13 | 0 | 0 | 3 | 3 | 0 | |
| 412 | 2-1-2-1-9-1-112 | 0 | 0 | 7 | 6 | 1 | |
| 660 | 2-1-2-1-4-1-25 | 0 | 0 | 1 | 0 | 1 | |
| 685 | 2-1-2-1-3-4-25 | 1 | 1 | 0 | 1 | 0 | |
| 874 | 4-1-1-1-7-1-22 | 1 | 0 | 1 | 1 | 0 | |
| 887 | 2-1-1-1-21-4-12 | 0 | 0 | 1 | 1 | 0 | |
| 692 | 2-1-2-1-1-4-42 | 0 | 0 | 1 | 0 | 1 | |
| 879 | 2-1-1-1-145-1-9 | 0 | 0 | 1 | 1 | 0 | |
| 261 | 2-1-1-1-4-27-12 | 0 | 0 | 0 | 1 | 0 | |
| 462 | 2-1-1-6-1-4-12 | 4 | 2 | 0 | 3 | 2 | |
| 881 | 2-68-1-1-10-4-13 | 1 | 1 | 0 | 1 | 0 | |
| CC37 | 37 | 2-9-2-1-13-1-16 | 3 | 4 | 1 | 3 | 2 |
| 896 | 2-9-2-1-13-1-38 | 0 | 0 | 1 | 1 | 0 | |
| 884 | 77-9-2-1-13-1-16 | 1 | 1 | 0 | 1 | 0 | |
| 880 | 2-9-2-1-1-1-16 | 1 | 1 | 0 | 1 | 0 | |
| CC29 | 29 | 2-3-2-2-6-4-4 | 0 | 0 | 1 | 1 | 0 |
| CC101 | 101 | 2-6-1-5-4-1-6 | 0 | 0 | 1 | 1 | 0 |
| 888 | 2-6-1-5-4-1-4 | 1 | 1 | 0 | 1 | 0 | |
| CC86 | 86 | 9-4-2-1-1-1-27 | 0 | 0 | 3 | 3 | 0 |
| CC1 | 1 | 4-4-1-1-7-4-10 | 3 | 2 | 0 | 2 | 1 |
| CC12 | 876 | 6-3-1-1-12-1-110 | 0 | 0 | 1 | 1 | 0 |
| CC147 | 147 | 3-4-6-1-7-4-38 | 5 | 3 | 3 | 5 | 3 |
| 273 | 3-4-6-1-7-4-4 | 0 | 0 | 3 | 2 | 1 | |
| CC857 | 857 | 2-35-2-35-56-24-19 | 0 | 0 | 1 | 1 | 0 |
| CC584 | 584 | 4-1-2-1-1-7-4 | 1 | 1 | 0 | 1 | 0 |
| CC526-928 | 526 | 38-19-53-58-73-21-130 | 2 | 2 | 0 | 2 | 0 |
| CC536-856 | 856 | 16-18-21-27-39-22-105 | 0 | 0 | 1 | 1 | 0 |
| CC889-7 | 889 | 3-1-1-4-3-1-19 | 0 | 0 | 1 | 0 | 1 |
| Singleton | 894 | 18-15-18-61-93-37-99 | 0 | 0 | 1 | 1 | 0 |
| Singleton | 893 | 25-1-101-1-10-1-100 | 0 | 0 | 1 | 1 | 0 |
| Singleton | 892 | 2-1-65-2-5-4-36 | 0 | 0 | 1 | 1 | 0 |
| Singleton | 891 | 3-1-2-1-9-1-184 | 0 | 0 | 1 | 1 | 0 |
| Singleton | 890 | 2-1-1-8-10-4-61 | 0 | 0 | 1 | 1 | 0 |
| Singleton | 886 | 16-1-21-27-29-17-183 | 0 | 0 | 1 | 1 | 0 |
| Singleton | 883 | 2-5-1-1-9-4-9 | 1 | 1 | 0 | 0 | 1 |
| Singleton | 882 | 2-1-1-37-1-27-19 | 0 | 0 | 1 | 1 | 0 |
| Singleton | 878 | 2-5-1-1-144-1-4 | 1 | 1 | 1 | 2 | 0 |
| Singleton | 877 | 59-24-21-78-54-22-67 | 0 | 0 | 1 | 0 | 1 |
| Singleton | 189 | 2-3-41-1-17-4-46 | 1 | 0 | 0 | 1 | 0 |
| Singleton | 374 | 2-3-58-37-10-27-9 | 0 | 0 | 1 | 1 | 0 |
| Singleton | 380 | 2-1-1-1-1-4-19 | 0 | 0 | 3 | 3 | 0 |
| Singleton | 562 | 2-5-1-1-10-1-139 | 9 | 8 | 1 | 13 | 1 |
| Singleton | 629 | 2-3-87-1-12-1-26 | 1 | 1 | 0 | 1 | 0 |
| Singleton | 686 | 4-1-1-3-3-5-54 | 4 | 3 | 0 | 4 | 0 |
| Singleton | 716 | 71-1-1-2-16-4-164 | 5 | 5 | 4 | 10 | 0 |
| Singleton | 873 | 14-1-2-1-7-4-182 | 0 | 0 | 1 | 1 | 0 |
Measurement of clonality and selection pressure
Analysis of the data set of 163 isolates from patients yielded an IAS value of 0.1251. This was decreased to 0.0841 when only one representative of each sequence type was included. Significant linkage disequilibrium was detected in both analyses. It remained significant when only one representative isolate for each ST in the entire isolate database were considered (IAS = 0.1133), thus the observed linkage disequilibrium is not due to sampling bias. The values of IAS in these analyses were low suggesting the weakly clonal population. The dn/ds ratios for all loci were significantly less than 1 (Table S2), indicating there was no strong positive selective pressure on the genes.
Phylogenetic analysis
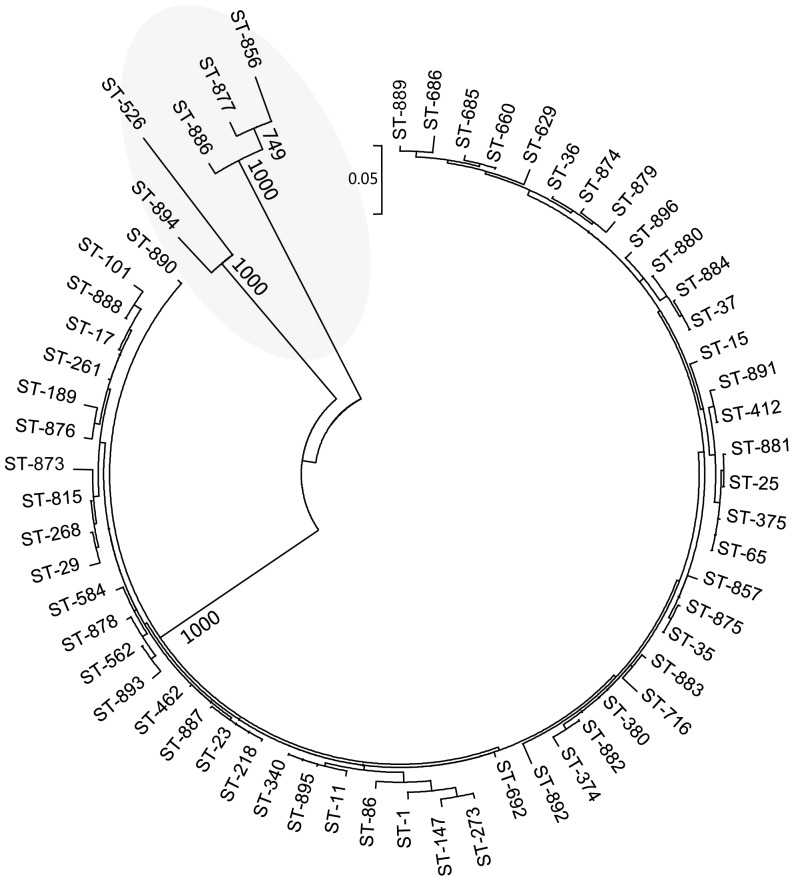
The maximum likelihood tree show a phylogenetically distinct cluster of related STs (ST877, ST886, ST894, ST856 and ST526) is formed with strong bootstrap support. Four of them (ST877, ST886, ST894 and ST856) are new STs in this study, and 2 of these (ST886 and ST894) differed from recognized STs at three loci (Fig. 4). The drug resistance profiles and epidemiological information of those clones belonging to phylogenetically distinct cluster of related STs are shown in Table S3.
Figure 4. Phylogenetic tree of K. pneumoniae isolates with one representative in each ST as derived from concatenated sequences of the 7 gene loci used in MLST.
The tree was constructed using the maximum likelihood method. The phylogenetically distinct cluster is shaded.
Drug resistance profiles of K. pneumoniae isolates grouped by CCs and STs
The detailed information on drug resistance profiles of K. pneumoniae isolates grouped by CCs is shown in Table 3. All CCs showed high proportion of resistance to penicillin. We did not observe significant differences between ESBL blaCTX-M and blaTEM/SHV groups (Table 2 and Table 3). We also compared the drug resistance profiles, the corresponding drug resistance-associated genes, as well as the clinical characteristics of the prevalent K. pneumoniae clones (Table S4). We observed that some isolates with the same STs were from patients who were hospitalized in the same period and who shared the same wards, especially in the first ward of the department of respiration, neuro-intensive care unit (NICU), and ICU. In addition, we detected a large variety of ESBL genes (such as bla SHV and bla CTX-M, and bla TEM), as well as genes associated with resistance to fluoroquinolones (such as qnrA, qnrB, qnrC, qnrD, qnrS, aac(6′)-Ib-cr, and qepA) and aminoglycosides (such as aacC2, addA1, and aacA4) in the prevalent clones. We detected the bla OXA-48 gene in one of the PDR K. pneumoniae isolate (TZSKP-82).
Table 3. Drug resistance profiles of K. pneumoniae isolates grouped by clonal complex.
| Clonal complex (No. of isolates) | Drug resistance types, No. (%) of isolates | Drug resistance, No. (%) of isolates | ||||||||||||
| MDR | XDR | PDR | Penicillin | Non-extended spectrum cephalosporins | Extended spectrum cephalosporins(bla CTX-M) | Extended spectrum cephalosporins(bla TEM/bla SHV) | Cephamycins | Carbapenems | Monobactams | Fluoroquinolones | Aminoglycosides | Folate pathway inhibitors | Nitrofurantoin | |
| CC1 (3) | 2(66.7) | 1(33.3) | 0 | 3 (100.0) | 3 (100.0) | 3(100.0) | 2 (66.7) | 3 (100.0) | 1(33.3) | 3(100.0) | 3(100.0) | 2 (66.7) | 2 (66.7) | 3(100.0) |
| CC12 (1) | 1(100.0) | 0 | 0 | 1(100.0) | 0 | 0 | 0 | 1(100.0) | 0 | 0 | 0 | 0 | 0 | 1(100.0) |
| CC23 (82) | 52(63.4) | 17(20.7) | 3(3.7) | 82 (100.0) | 39 (47.6) | 33 (40.2) | 26 (31.7) | 61(74.4) | 6(7.3) | 39(47.6) | 34(41.5) | 41(50.0) | 42(51.2) | 73(89.0) |
| CC29 (1) | 1(100.0) | 0 | 0 | 1(100.0) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1(100.0) |
| CC37 (8) | 4 (50.0) | 2 (25.0) | 0 | 8(100.0) | 6 (75.0) | 5 (62.5) | 6 (75.0) | 4(50.0) | 0 | 6(75.0) | 3(37.5) | 5(62.5) | 4(50.0) | 7(87.5) |
| CC86 (3) | 2(66.7) | 0 | 0 | 3(100.0) | 0 | 0 | 0 | 2(66.7) | 0 | 0 | 0 | 0 | 0 | 1(33.3) |
| CC101 (2) | 1(50.0) | 0 | 0 | 2(100.0) | 1(50.0) | 1(50.0) | 1(50.0) | 1(50.0) | 0 | 1(50.0) | 0 | 1(50.0) | 1(50.0) | 2(100.0) |
| CC147 (11) | 7 (63.6) | 4 (36.4) | 0 | 11(100.0) | 8 (72.7) | 4 (36.4) | 5 (45.5) | 7 (63.6) | 1(9.1) | 5 (45.5) | 11(100.0) | 10(90.9) | 11(100.0) | 11(100.0) |
| CC526-928 (2) | 1 (50.0) | 0 | 0 | 2(100.0) | 2(100.0) | 2(100.0) | 2(100.0) | 0 | 0 | 2(100.0) | 2(100.0) | 2(100.0) | 2(100.0) | 2(100.0) |
| CC536-856 (1) | 0 | 0 | 0 | 1(100.0) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| CC584 (1) | 1(100.0) | 0 | 0 | 1(100.0) | 1(100.0) | 1(100.0) | 1(100.0) | 0 | 0 | 1(100.0) | 0 | 1(100.0) | 1(100.0) | 1(100.0) |
| CC857 (1) | 1(100.0) | 0 | 0 | 1(100.0) | 0 | 0 | 0 | 1(100.0) | 0 | 0 | 0 | 0 | 0 | 1(100.0) |
| CC889-7 (1) | 0 | 1(100.0) | 0 | 1(100.0) | 0 | 0 | 0 | 1(100.0) | 0 | 0 | 0 | 0 | 0 | 1(100.0) |
| Singletons(46) | 31(67.4) | 9(19.6) | 0 | 45(97.8) | 26 (56.5) | 21(45.7) | 19(41.3) | 27(58.7) | 3(6.5) | 26(56.5) | 19(41.3) | 28(60.9) | 31(67.4) | 41(89.1) |
| Total(163) | 104(63.8) | 34(20.9) | 3(1.8) | 162(99.4) | 86(52.8) | 70(42.9) | 62(38.0) | 108(66.3) | 11(6.7) | 83(50.9) | 72(44.2) | 90(55.2) | 94(57.7) | 145(89.0) |
Factors associated with prevalent K. pneumoniae clones
Factors associated with prevalent STs compared with non-prevalent STs upon univariate and multivariate analysis are shown in Table S5 and Table 4, respectively. Multiple logistic regression analysis revealed that isolates from ICU (OR, 13.802), from medical ward (OR, 5.154), from community acquired infection (OR, 3.106), from patients without heart disease (OR, 3.446), from patients with treatment success (OR, 6.691), susceptible to extended spectrum cephalosporins (OR, 8.633), susceptible to cephamycins (OR, 3.430), susceptible to fluoroquinolones (OR, 6.247), with MDR (OR, 3.111) were significantly associated with the prevalent STs.
Table 4. Multivariate logistic regression analysis for factors independently associated with prevalent K. pneumoniae clonesa.
| Variablesb | Univariate analysis | Multivariate analysis Univariate analysis | ||
| Odds Ratio (95% CI) | P value | Odds Ratio (95% CI) | P value | |
| From intensive care unit | 1.597 (0.814–3.135) | 0.174 | 13.802 (3.180–59.891) | <0.001 |
| From medical ward | 1.198 (0.643–2.232) | 0.570 | 5.154 (1.452–18.296) | 0.011 |
| Community acquired infection | 1.788 (0.854–3.742) | 0.123 | 3.106 (1.005–9.602) | 0.049 |
| With diabetes mellitus | 1.244 (0.603–2.565) | 0.554 | 2.776 (0.981–7.854) | 0.054 |
| With abnormal liver function | 1.630 (0.770–3.452) | 0.202 | 2.551 (0.924–7.042) | 0.071 |
| With cerebral infarction | 1.529 (0.746–3.136) | 0.246 | 2.528 (0.926–6.900) | 0.070 |
| Without heart disease | 1.250 (0.640–2.440) | 0.513 | 3.446 (1.298–9.150) | 0.013 |
| Treatment success | 1.408 (0.582–3.406) | 0.447 | 6.691 (1.884–23.770) | 0.003 |
| Susceptible to 3rd and 4th generation cephalosporins | 4.448 (2.268–8.722) | <0.001 | 8.633 (2.515–29.640) | 0.001 |
| Susceptible to cephamycins | 1.072 (0.556–2.068) | 0.836 | 3.430 (1.078–10.921) | 0.037 |
| Susceptible to fluoroquinolones | 5.558 (2.823–10.943) | <0.001 | 6.247 (2.117–18.439) | 0.001 |
| MDR | 1.291 (0.674–2.473) | 0.442 | 3.111 (1.113–8.693) | 0.030 |
Prevalent clones include a total of 70 isolates with the following STs: ST15 (17), ST562 (14), ST23 (12), ST716 (10), ST11 (9), ST147 (8).
All variables included in the univariate analysis (shown in Table S5) were included in the logistic regression model for multivariate analysis, and they were eliminated using a backward stepwise selection method using a P value threshold of 0.1 for the variables to remain in the model. P values<0.05 were considered to be statistically significant.
Discussion
The present study describes the genetic diversity of drug-resistant K. pneumoniae isolates in a tertiary hospital in Beijing. Twenty-four new STs were detected, demonstrating that the MLST database is still novel and continuously growing. The isolates originated from both CA and HA infections. It is noteworthy that in contrast to the observations from a study from Germany [1], the levels of resistance were equally high among both HA and CA K. pneumoniae isolates in our study, indicating a very large reservoir of resistance in the community around Beijing. We further analyzed the drug resistance profiles, the corresponding drug resistance-associated genes, as well as the clinical characteristics of those isolates with the same STs. We observed that the rates of resistance to most drugs were much higher among ESBL positive isolates than ESBL negative isolates. But we did not observe significant differences between ESBL blaCTX-M and blaTEM/blaSHV groups in their distribution of STs or CCs. We noticed that some prevalent isolates with the same STs were from patients who were hospitalized in the same period and who shared the same wards, especially in the first ward of the department of respiration, NICU, and ICU, this observation suggests that those clonal isolates were transmitted in the hospital, causing infections among immunocompromised patients in those wards. In addition, many patients with this ST shared the same wards such as the first ward of the department of respiration and cardiac care unit, suggesting there is currently an ongoing transmission of isolates of this ST in those wards of the hospital.
Notably, among the three isolates of PDR K. pneumoniae, two isolates belonged to ST15, which is the most prevalent ST in this study. The clinical and molecular epidemiological data suggest that the majority of those ST15 isolates were from patients with severe underlying diseases such as pulmonary infection, renal dysfunction, heart failure, and chronic obstructive pulmonary disease, etc. A study from Spain reported that VIM-1 producing K. pneumoniae ST15 clone has a high capacity to spread among ICU patients with severe underlying conditions [2]. ST15 is also widespread in other countries such as Denmark, Hungary, Korea, Malaysia, Singapore and Taiwan [26]–[28]. Interestingly, one of the PDR K. pneumoniae isolate (TZSKP-82) possesses the bla OXA-48 gene. OXA-48 carbapenemases were first isolated from K. pneumoniae in Turkey in 2008 [10], [29]. To the best of our knowledge, this is the first documented case of OXA-48-producing K. pneumoniae in China.
ST23 was another prevalent ST in this study. ST23 was the primary founder of CC23, and isolates sharing this ST were found in other countries. Previous studies showed that ST23 isolates were closely related to liver abscess [30], [31]. However, the ST23 isolates in this study were diagnosed with different kinds of illnesses including cerebral infarction, renal dysfunction, and liver abscess, etc.
Another frequently identified ST is ST11, which is a single locus variant of ST258. ST258 is a well known lineage of K. pneumoniae which plays an important role in the global spread of carbapenemases. ST258 was not found in our study. There are only a few nucleotide differences between ST11 and ST258 in their tonB alleles. ST258 was proposed to be probably arisen from ST11 by acquisition of the tonB-79 allele, followed by acquisition of carbapenem-resistance genes on mobile elements [32]. A recent study was conducted to analyze carbapenem-resistant K. pneumoniae isolates from 13 hospitals in nine cities covering five provinces in China, and they found that ST11 was the most dominant clone among the 95 carbapenem-resistant K. pneumoniae isolates in China [33]. Although ST11 is not the most dominant one in our study, it is among one of the prevalent clones and all those ST11 isolates harbored ESBL genes. In addition, the genetic relatedness of ST11 with ST258 is of great concern.
From the maxiumum likelihood tree, a phylogenetically distinct cluster of related STs (ST877, ST886, ST894, ST856 and ST526) is formed. Four of them (ST877, ST886, ST894 and ST856) are new STs in this study, and 2 of these (ST886 and ST894) differed from recognized STs at three loci. These results suggest a recent clone is emerging locally. Further identification of the drug resistance profiles and epidemiological information of the 4 isolates with new STs imply that they are relatively susceptible (with resistance to only a few drugs and no resistance genes detected).
One of the challenges for infection control is to discern the prevalent clones as well as their clinical relevance, especially the treatment outcome, of those isolates, so as to provide information for better management measures. We thus further examined the association between certain prevalent K. pneumoniae isolates (based on the frequency of the STs) and the demographic and clinical features as well as mortality of the patients from whom the isolates were obtained. We noticed that some results (For example, the results for the association with the intensive care unit) from univariant and multivariant analysis differ greatly. We think that the results from the multivariant analysis should be more reliable and those variables with significant P values in the multivariant analysis were identified to be independently associated with prevalent K. pneumoniae clones after excluding some less significant variables and taking into consideration of the confounding factors during the multivariate analysis. Data from multiple logistic regression analysis revealed that isolates from ICU, from medical ward, from community acquired infection, and with MDR were significantly associated with those prevalent clones. In addition, we noticed that those prevalent clones were more frequently associated with patients without heart disease, who were susceptible to extended spectrum cephalosporins, cephamycins and fluoroquinolones, and who had better treatment outcome. A recent study from Taiwan reported that the ESBL positive E. coli ST131, which has emerged in bloodstream infections in Taiwan, is not related to more health-care-associated risk factors, and the E. coli bacteremia caused by this clone did not exhibited a higher mortality rate [34]. Thus the prevalent clones are not always the most virulent ones or those associated with more severe clinical features or outcome.
Since the prevalent clones have a great potential of transmission among patients, the observation that those clones were significantly associated with MDR, HA infection, as well as nosocomial infections in the crowded ICU, together with the identification of a large variety of drug resistance-associated genes, particularly those ESBL genes, as well as genes associated with resistance to fluoroquinolones and aminoglycosides [35]–[41], in those prevalent clones suggest that although those isolates are associated with less severe clinical features and outcome, they could be a dangerous reservoir for transmission of drug resistance genes, thus warrant a high degree of awareness and monitoring of those drug resistance determinants in clinical isolates. In addition, these isolates from China had different ESBL genotypes, implying multiple acquisition events and the presence of multiple circulating variants of the clone.
In conclusion, the diversity of the genotypes and the complexity of the resistance phenotypes and determinants found, as well as the potential for widespread dissemination of those prevalent isolates detected in our study suggest that certain possibly less virulent (based on the clinical manifestations of the patients) but highly transmissible drug-resistant clones of K. pneumoniae isolates are currently prevalent among patients in hospital settings in Beijing, emphasizing the continuous hospital-wide surveillance of phenotypic and genotypic drug resistance data, as well clinical characteristics and treatment outcome for the prevalent K. pneumoniae clones is necessary to understand the spread of those successful clones, so as to make better infection control measure against nosocomial infection caused by K. pneumoniae and Enterobacteriaceae, which are closely related to K. pneumoniae and interchange resistance determinants frequently with them. Further in-depth investigation of other important population genetic forces, such as gene flow, natural selection, etc., with more extensive sampling, would validate the interesting observation of an inverse relationship between prevalence and virulence in a statistically robust fashion, as well as to provide more insights into the spatial and temporal population dynamics of drug-resistant K. pneumoniae isolates.
Supporting Information
ST and CC for all K. pneumoniae isolates in this study.
(DOC)
Variation in loci used in the present K. pneumoniae MLST scheme.
(DOC)
Drug resistance profiles and epidemiological information of the K. pneumoniae clones belong to phylogenetically distinct cluster of related STs.
(DOC)
Drug resistance profiles and epidemiological information of the prevalent K. pneumoniae clones.
(DOC)
Univariate logistic regression analysis for factors associated with prevalent K. pneumoniae clones.
(DOC)
Acknowledgments
We thank platform Genotyping of Pathogens and Public Health (Institut Pasteur, Paris, France) for coding MLST alleles and profiles available at www.pasteur.fr/mlst.
Funding Statement
This study was supported by a grant from the National Basic Research Program of China (grant number 2012CB518700); a grant from National Natural Science Foundation of China (NSFC) (grant No. 30700975); a grant from the Knowledge Innovation Program of the Chinese Academy of Sciences (KSCX2-EW-J-6); and the Merieux Research Grant program. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Kohlenberg A, Schwab F, Rüden H (2012) Wide dissemination of extended-spectrum beta-lactamase (ESBL)-producing Escherichia coli and Klebsiella spp. in acute care and rehabilitation hospitals. Epidemiol Infect 140: 528–534. [DOI] [PubMed] [Google Scholar]
- 2. Sánchez-Romero I, Asensio A, Oteo J, Muñoz-Algarra M, Isidoro B, et al. (2012) Nosocomial Outbreak of VIM-1-producing Klebsiella pneumoniae of multilocus sequence type 15: Molecular basis, clinical risk factors, and outcome. Antimicrob Agents Chemother 56: 420–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Zhang R, Wang XD, Cai JC, Zhou HW, Lv HX, et al. (2011) Outbreak of Klebsiella pneumoniae carbapenemase 2-producing K. pneumoniae with high qnr prevalence in a Chinese hospital. J Med Microbiol 60: 977–982. [DOI] [PubMed] [Google Scholar]
- 4. Ben-David D, Kordevani R, Keller N, Tal I, Marzel A, et al. (2012) Outcome of carbapenem resistant Klebsiella pneumoniae bloodstream infections. Clin Microbiol Infect 18: 54–60. [DOI] [PubMed] [Google Scholar]
- 5. Daikos GL, Petrikkos P, Psichogiou M, Kosmidis C, Vryonis E, et al. (2009) Prospective observational study of the impact of VIM-1 metallo-beta-lactamase on the outcome of patients with Klebsiella pneumoniae bloodstream infections. Antimicrob Agents Chemother 53: 1868–1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lautenbach E, Patel JB, Bilker WB, Edelstein PH, Fishman NO (2001) Extended-spectrumβ-lactamase-producing Escherichia coli and Klebsiella pneumoniae: risk factors for infection and impact of resistance on outcomes. Clin Infect Dis 32: 1162–1171. [DOI] [PubMed] [Google Scholar]
- 7. Magiorakos AP, Srinivasan A, Carey RB, Carmeli Y, Falagas ME, et al. (2012) Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect 18: 268–281. [DOI] [PubMed] [Google Scholar]
- 8. Schlesinger J, Navon-Venezia S, Chmelnitsky I, Hammer-Münz O, Leavitt A, et al. (2005) Extended-spectrum beta-lactamases among Enterobacter isolates obtained in Tel Aviv, Israel. Antimicrob Agents Chemother 49: 1150–1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chudácková E, Bergerová T, Fajfrlík K, Cervená D, Urbásková P, et al. (2010) Carbapenem-nonsusceptible strains of Klebsiella pneumoniae producing SHV-5 and/or DHA-1 beta-lactamases in a Czech hospital. FEMS Microbiol Lett 309: 62–70. [DOI] [PubMed] [Google Scholar]
- 10. Cuzon G, Ouanich J, Gondret R, Naas T, Nordmann P (2011) Outbreak of OXA-48-positive carbapenem-resistant Klebsiella pneumoniae isolates in France. Antimicrob Agents Chemother 55: 2420–2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hammerum AM, Toleman MA, Hansen F, Kristensen B, Lester CH, et al. (2010) Global spread of New Delhi metallo-beta-lactamase 1. Lancet Infect Dis 10: 829–830. [DOI] [PubMed] [Google Scholar]
- 12. Nordmann P, Naas T, Poirel L (2011) Global Spread of Carbapenemase-producing Enterobacteriaceae . Emerg Infect Dis 17: 1791–1798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Nordmann P, Poirel L, Carrër A, Toleman MA, Walsh TR (2010) How To Detect NDM-1 Producers. J Clin Microbiol 49: 718–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Thomson KS (2010) Extended-Spectrum-β-Lactamase, AmpC, and Carbapenemase issues. J Clin Microbiol 48: 1019–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Nagano N, Nagano Y, Toyama M, Kimura K, Tamura T, et al. (2012) Nosocomial spread of multidrug-resistant group B streptococci with reduced penicillin susceptibility belonging to clonal complex 1. J Antimicrob Chemother 67: 849–856. [DOI] [PubMed] [Google Scholar]
- 16. Rotariu O, Smith-Palmer A, Cowden J, Bessell PR, Innocent GT, et al. (2010) Putative household outbreaks of campylobacteriosis typically comprise single MLST genotypes. Epidemiol Infect 138: 1744–1747. [DOI] [PubMed] [Google Scholar]
- 17. Aanensen DM, Spratt BG (2005) The multilocus sequence typing network: mlst.net. Nucleic Acids Res 33: W728–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Laure D, Virginie P, Jan V, Grimont PA, Brisse S (2005) Multilocus Sequence Typing of Klebsiella pneumoniae Nosocomial Isolates. J Clin Microbiol 43: 4178–4182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Li B, Yi Y, Wang Q, Woo PCY, Tan L, et al. (2012) Analysis of drug resistance determinants in Klebsiella pneumoniae isolates from a tertiary-care hospital in Beijing, China. PLoS One 7: e42280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Performance Standards for antimicrobial Susceptibility Testing; Twenty-first Informational Supplement. Clinical and Laboratory Standards Institute (CLSI); 2011.
- 21. Feil EJ, Li BC, Aanensen DM, Hanage WP, Spratt BG (2004) eBURST: inferring patterns of evolutionary descent among clusters of related bacterial genotypes from multilocus sequence typing data. J Bacteriol 186: 1518–1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Jolley KA, Feil EJ, Chan MS, Maiden MC (2001) Sequence type analysis and recombinational tests (START). Bioinformatics 17: 1230–1231. [DOI] [PubMed] [Google Scholar]
- 23. Didelot X, Falush D (2006) Inference of bacterial microevolution using multilocus sequence data. Genetics 175: 1251–1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Guindon S, Lethiec F, Duroux P, Gascuel O (2005) PHYML Online–a web server for fast maximum likelihood-based phylogenetic inference. Nucleic Acids Res 33: W557–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Orsi GB, Scorzolini L, Franchi C, Mondillo V, Rosa G, et al. (2006) Hospital-acquired infection surveillance in a neurosurgical intensive care unit. J Hospital Infect 64: 23–29. [DOI] [PubMed] [Google Scholar]
- 26. Damjanova I, Tóth A, Pászti J, Hajbel-Vékony G, Jakab M, et al. (2008) Expansion and countrywide dissemination of ST11, ST15 and ST147 ciprofloxacin-resistant CTX-M-15-type beta-lactamase-producing Klebsiella pneumoniae epidemic clones in Hungary in 2005–the new ‘MRSAs’? J Antimicrob Chemother 62: 978–985. [DOI] [PubMed] [Google Scholar]
- 27. Lee MY, Ko KS, Kang CI, Chung DR, Peck KR, et al. (2011) High prevalence of CTX-M-15-producing Klebsiella pneumoniae isolates in Asian countries: diverse clones and clonal dissemination. Int J Antimicrob Agents 38: 160–163. [DOI] [PubMed] [Google Scholar]
- 28. Nielsen JB, Skov MN, Jørgensen RL, Heltberg O, Hansen DS, et al. (2011) Identification of CTX-M15-, SHV-28-producing Klebsiella pneumoniae ST15 as an epidemic clone in the Copenhagen area using a semi-automated Rep-PCR typing assay. Eur. J Clin Microbiol Infect Dis 30: 773–778. [DOI] [PubMed] [Google Scholar]
- 29. Carrër A, Poirel L, Eraksoy H, Cagatay AA, Badur S, et al. (2008) Spread of OXA-48-Positive Carbapenem-Resistant Klebsiella pneumoniae Isolates in Istanbul, Turkey. Antimicrob Agents Chemother 52: 2950–2954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chung DR, Lee H, Park MH, Jung SI, Chang HH, et al. (2012) Fecal carriage of serotype K1 Klebsiella pneumoniae ST23 strains closely related to liver abscess isolates in Koreans living in Korea. Eur. J Clin Microbiol Infect Dis 31: 481–486. [DOI] [PubMed] [Google Scholar]
- 31. Siu LK, Fung CP, Chang FY, Lee N, Yeh KM, et al. (2011) Molecular typing and virulence analysis of serotype K1 Klebsiella pneumoniae strains isolated from liver abscess patients and stool samples from noninfectious subjects in Hong Kong, Singapore, and Taiwan. J Clin Microbiol 49: 3761–3765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Breurec S, Guessennd N, Timinouni M, Le TA, Cao V, et al. (2012) Klebsiella pneumoniae resistant to third-generation cephalosporins in five African and two Vietnamese major towns: multiclonal population structure with two major international clonal groups, CG15 and CG258. Clin Microbiol Infect doi:10.1111/j.1469-0691.2012.03805.x. [DOI] [PubMed] [Google Scholar]
- 33. Qi Y, Wei Z, Ji S, Du X, Shen P, et al. (2011) ST11, the dominant clone of KPC-producing Klebsiella pneumoniae in China. J Antimicrob Chemother 66: 307–312. [DOI] [PubMed] [Google Scholar]
- 34. Chung HC, Lai CH, Lin JN, Huang CK, Liang SH, et al. (2012) Bacteremia caused by extended-spectrum-β-lactamase-producing Escherichia coli sequence type ST131 and non-ST131 clones: comparison of demographic data, clinical features, and mortality. Antimicrob Agents Chemother 56: 618–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Chmelnitsky I, Hermesh O, Navon-Venezia S, Strahilevitz J, Carmeli Y (2009) Detection of aac(6′)-Ib-cr in KPC-producing Klebsiella pneumoniae isolates from Tel Aviv, Israel. J Antimicro Chemother 64: 718–722. [DOI] [PubMed] [Google Scholar]
- 36. Hujer KM, Hujer AM, Hulten EA, Bajaksouzian S, Adams JM, et al. (2006) Analysis of antibiotic resistance genes in multidrug-resistant Acinetobacter sp. isolates from military and civilian patients treated at the Walter Reed Army Medical Center. Antimicrob Agents Chemother 50: 4114–4123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Jemima SA, Verghese S (2008) Molecular characterization of nosocomial CTX-M type β-lactamase producing Enterobacteriaceae from a tertiary care hospital in south India. Indian J Med Microbiol 26: 365–368. [DOI] [PubMed] [Google Scholar]
- 38. Kaczmarek FM, Dib-Hajj F, Shang W, Gootz TD (2006) High-Level Carbapenem Resistance in a Klebsiella pneumoniae Clinical Isolate Is Due to the Combination of bla ACT-1 β-Lactamase Production, Porin OmpK35/36 Insertional Inactivation, and Down-Regulation of the Phosphate Transport Porin PhoE. Antimicrob Agents Chemother 50: 3396–3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ramirez MS, Tolmasky ME (2010) Aminoglycoside modifying enzymes. Drug Resist Update 13: 151–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Yamane K, Wachino J, Suzuki S, Kimura K, Shibata N, et al. (2007) New plasmid-mediated fluoroquinolone efflux pump, QepA, found in an Escherichia coli clinical isolate. Antimicrob Agents Chemother 51: 3354–3360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Zimhony O, Chmelnitsky I, Bardenstein R, Goland S, Hammer Muntz O, et al. (2006) Endocarditis caused by extended-spectrum-β-lactamase-producing Klebsiella pneumoniae: emergence of resistance to ciprofloxacin and piperacillin-tazobactam during treatment despite initial susceptibility. Antimicrob Agents Chemother 50: 3179–3182. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
ST and CC for all K. pneumoniae isolates in this study.
(DOC)
Variation in loci used in the present K. pneumoniae MLST scheme.
(DOC)
Drug resistance profiles and epidemiological information of the K. pneumoniae clones belong to phylogenetically distinct cluster of related STs.
(DOC)
Drug resistance profiles and epidemiological information of the prevalent K. pneumoniae clones.
(DOC)
Univariate logistic regression analysis for factors associated with prevalent K. pneumoniae clones.
(DOC)