Abstract
Recently, we have identified circadian clock genes as targets of Histone Monoubiquitination1 (HUB1) in Arabidopsis from a transcriptome comparison between the hub1–1 mutant and HUB1 overexpression lines. HUB1 affected the amplitudes of the circadian clock gene expression profiles in the hub1–1 mutant that coincided with reduced monoubiquitination of histone H2B at their coding regions. Here we showed that parameters for plant fitness are altered in HUB1 mutant and overexpression lines, suggesting that the histone H2B monoubiquitination status affects plant fitness.
Keywords: H2B monoubiquitination, germination, chloroplast development, seed parameters, yield, Arabidopsis thaliana
The circadian clock is an important upstream control mechanism of the basic metabolism, and small deviations in its regulation are detrimental to plant fitness.1,2 The effects on plant fitness can be measured by parameters, such as photosynthetic capacity, growth and vigor, flowering time and seed set. Recently, we have shown that HUB1 positively regulates expression levels of circadian clock regulators and input and downstream effector genes through histone H2B monoubiquitination at their coding regions,3 and that it acts in concert with transcription regulators FACT and ELONGATOR.3,4 In the hub1–1 mutant leaf growth was severely reduced by cell division and cell expansion defects,5 and an early flowering time phenotype of hub1 alleles is well documented.6,7 The requirement of HUB1 for circadian clock genes to reach maximal transcript levels prompted us to analyze here the impact of HUB1 perturbations on plant fitness through chloroplasts, growth, vigor and seed yield parameters.
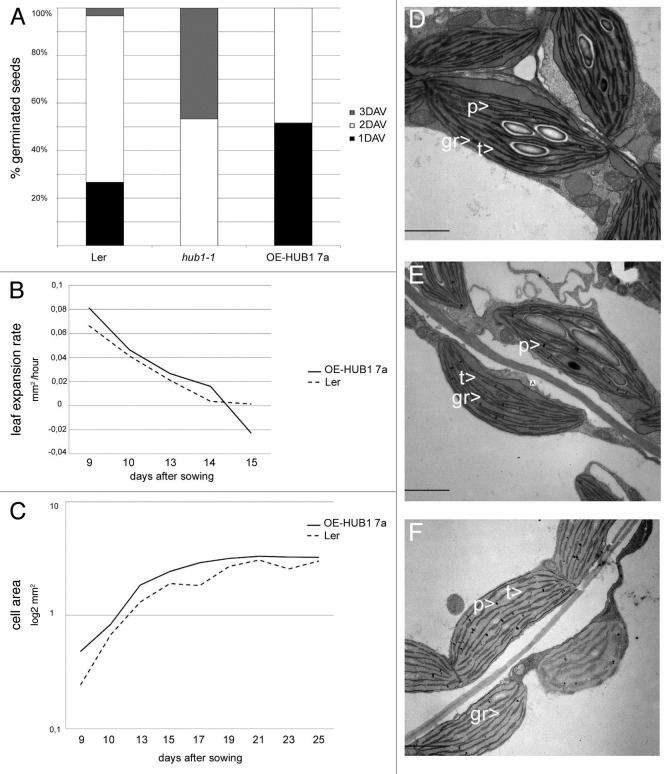
Germination vigor is a parameter that describes the synchronization of the germination rate and early growth performance that results in a competitive advantage in the field. We compared the germination vigor between vernalized low dormancy ecotype Ler and hub1–1 and OE-HUB1–7a 3 in the same background by following their germination time on MS media supplemented with 1% sucrose. The overexpression line was the first one to germinate on day 1 and 2, followed by wild-type Ler with the majority germinating on day 2, while for the hub1–1 mutant the germination mainly occurred on day 2 and 3 (Fig. 1A).
Figure 1. Fitness parameters in HUB1 perturbed lines. (A) % of germinated seeds 1, 2 or 3 d after vernalization (DAV). (B) Leaf 1 and 2 expansion rate in Ler and OE-HUB1–7a line during 9 to 15 d after sowing. (C) Cell area increase in Ler and OE-HUB1–7a line between 9 to 25 d after sowing. (D) Chloroplast ultrastructure in Ler grown under short-day conditions, (E) hub1–1 individual with weak phenotype and (F) hub1–1 individual with severe phenotype. gr, grana; p, plastoglobuli; t, thylakoid membrane. Bar = 2 µm (D, E and F).
To further investigate the role of HUB1 in growth, we analyzed how HUB1 overexpression would enhance the juvenile leaf growth rate. Arabidopsis leaf growth occurs in three phases, namely proliferation, expansion and maturation, and these transitions coincide with 9, 12 and 18 d post germination, respectively.8 The leaf area, length and width of OE-HUB1- 7a 3 were analyzed every 24 or 48 h from day 7 to 25. The analysis of the leaf expansion rate showed an enhanced rate for overexpression lines (Fig. 1B) and the mature leaf areas were statistically significantly larger than those of the wild type (p < 0.001) (Table 1). To obtain similar insights into the underlying cellular responses in the overexpression lines as were shown for the mutant,5 the epidermal cell sizes were analyzed every 48 h between days 9 and 25 (Fig. 1C). Indeed, the calculated size of adaxial epidermal leaf cells in the HUB1 overexpression line was significantly larger than that in the Ler control during all growth stages (Fig. 1C). Accordingly, in the hub1–1 mutant, reduced cell areas were observed earlier despite increased endoreduplication levels that usually drive cell expansion.5
Table 1. Comparison of the mean area, length and width of juvenile leaf lamina (1st and 2nd) between OE-HUB1–7a and Ler (n = 15, mean ± SD).
| Genotype | Area (cm) | Length (cm) | Width (cm) | |||
|---|---|---|---|---|---|---|
|
OE-HUB1–7a |
27.333 ± 6.667 |
** |
0.633 ± 0.097 |
* |
0.547 ± 0.078 |
NS |
| Ler | 20.033 ± 6.156 | 0.56 ± 0.097 | 0.505 ± 0.091 |
Significant (p < 0.05); **highly significant (p < 0.01); NS non-significant
The leaf growth effects in HUB1 misexpression lines could be influenced by developmental and cell cycle genes,5 as well as by reduced metabolic and photosynthetic activities as indicated by phenotypic and transcriptomic analysis.3,9 To verify the state of the photosynthetic apparatus in the lines, we analyzed the chloroplast ultrastructure in young leaves of wild type, and weak (rather normal leaves) and severe (small and pale-colored leaves) hub1–1 mutants (Fig. 1D-F). Reduced thylakoid membrane and grana structures were observed in the hub1–1 mutants in both short and long day conditions. In the most severe hub1–1 mutant individuals, the thylakoid membranes were totally misoriented (Fig. 1F), while in the weak mutant individuals they remained poorly connected and poorly stacked (Fig. 1E). The number of plastoglobuli, which are spherical, lipid-rich bodies with a putative function in lipid and sugar metabolism and abiotic stress response, had increased in the stroma of the hub1–1 mutants (Fig. 1E and F). The overexpression line was visually comparable to the wild type (data not shown).
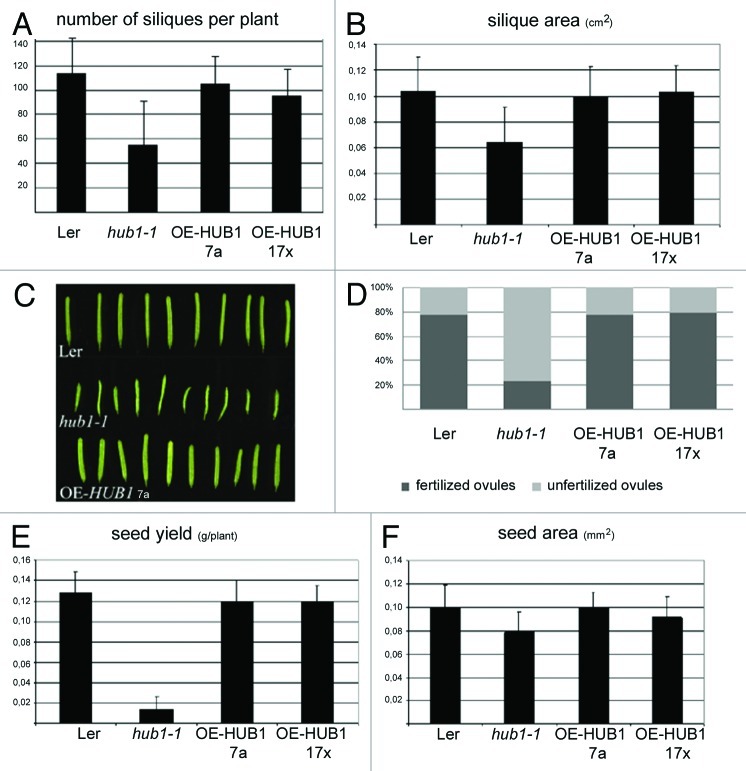
All hub1 mutant alleles flower earlier than the wild type, predicting a reduced reproductive fitness. We analyzed the effects of the HUB1 misexpression on silique and seed phenotypes such as silique number and shape, and size and number of seeds. The average number of siliques per plant was 115 siliques in the wild-type and overexpression lines, OE-HUB1–7a 3and OE-HUB1–17 ×3 and 55 siliques in the hub1–1 mutant (Fig. 2A). The area and length of hub1–1 siliques were severely reduced (Fig. 2B); whereas the silique thickness of the HUB1 overexpression lines had increased (Fig. 2C). In hub1–1, many seeds were missing due to 50% to 80% of unfertilized ovules (Fig. 2D). The total seed yield from dried siliques of the hub1–1 plants was dramatically lower than that of the Ler wild type (Fig. 2E) and the seed size was reduced (Fig. 2F). While the silique and seed parameters were severely affected in the hub1–1 mutant, no significant effects on the seed parameters were observed in the HUB1 overexpression lines, probably because the Arabidopsis seed development does not depend on cell expansion. Taken together, important aspects of plant fitness were found to be affected by HUB1 misexpression, namely HUB1 overexpression enhanced germination vigor and green biomass production, while the hub1–1 mutation impaired seed yield.
Figure 2. Mean silique parameters and seed yield ± SD of the Ler, hub1–1 and OE-HUB1–7a and OE-HUB117 × lines with resp. 20- and 10-fold overexpression levels.3 (A) Number of siliques per plant. (B) Silique areas. (C) Siliques of Ler, hub1–1 and OE-HUB1. (D) Fertilized vs. unfertilized ovules in siliques. (E) Seed yield. (F) Seed areas.
The circadian clock has a well documented role in the temporal control of metabolic activities, and key metabolic processes are driven by genes that are expressed in a circadian manner.10 The phenotypes observed in hub1–1, such as reduced growth and seed yield, are characteristic for reduced fitness observed in mutants with defects in the circadian clock and/or photosynthetic capacity.1,2,11 On the other hand, the HUB1 overexpression lines showed enhanced germination vigor as well as growth of leaves. HUB1-mediated activities in transcription elongation might account for reduced and enhanced germination vigor in the mutant and overexpression lines because a new transcription activity during germination assists in the rapidity and uniformity of the process.12 Taken together, our data indicate that HUB1 might be a component of fitness and, through its role in the regulation of the transcription of several biological processes,3,5 could indeed be a hub for plant fitness.
Kinematic Leaf Growth Analysis
The kinematic leaf growth analysis was done as described.5,8 For statistical analysis, the data were compared pair wise and the leaf length, width and area and cell area measurements were subjected to t-tests.
Transmission Electron Microscopy of Chloroplasts
Leaves 1 and 2 from 3-week-old wild-type (Ler) and hub1–1 seedlings, grown under short-day conditions, were cut into small pieces and immersed in a fixative solution of 2% paraformaldehyde and 2.5% glutaraldehyde, and postfixed in 1% OsO4 with 1.5% K3Fe(CN)6 in 0.1 M Na-Cacodylate buffer, pH 7.2. Samples were dehydrated through a graded ethanol series, including a bulk staining with 2% uranyl acetate at the 50% ethanol step, followed by embedding in Spurr’s resin. Ultrathin sections were made with an ultramicrotome (Leica) EM UC6 and post-stained in EM AC20 (Leica) for 40 min in uranyl acetate at 20°C and for 10 min in lead stain at 20°C. Grids were viewed with a 1010 transmission electron microscope (JEOL; http://www.jeol.com/) operating at 80 kV.
Silique and Seed Analyses
The silique numbers and shapes were recorded by scanning and photography. For silique analysis, the green and mature siliques were harvested and the length and width parameters were measured using the ImageJ software program. The number of unfertilized ovules was determined using microscopy. Seeds were harvested from the dried siliques and seed weight was determined by counting 200 seeds for weighing. The ImageJ software program was used to determine the area of the seeds. These yield parameters were subjected to t-tests.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank Annick Bleys for help in preparing the manuscript. K.H. is indebted to the Agency for Innovation by Science and Technology in Flanders (IWT) for a research fellowship, T.M.B. to the European Union-Human Resources and Mobility for an Early Stage Training grant (MEST-CT-2004–514632) and O.O. to the Vlaamse Interuniversitaire Raad (VLIR) for a predoctoral fellowship.
Glossary
Abbreviations:
- HUB1
histone H2B monoubiquitination 1
- FACT
“Facilitates Chromatin Transcription”
- OE-HUB1
overexpression of HUB1
Footnotes
Current affiliation: Department of Agricultural Sciences; University of Helsinki; Helsinki, Finland
Current affiliation: Department of Biochemistry and Biotechnology; Kenyatta University; Nairobi, Kenya
Previously published online: www.landesbioscience.com/journals/psb/article/22326
References
- 1.Dodd AN, Salathia N, Hall A, Kévei E, Tóth R, Nagy F, et al. Plant circadian clocks increase photosynthesis, growth, survival, and competitive advantage. Science. 2005;309:630–3. doi: 10.1126/science.1115581. [DOI] [PubMed] [Google Scholar]
- 2.Ni Z, Kim E-D, Ha M, Lackey E, Liu J, Zhang Y, et al. Altered circadian rhythms regulate growth vigour in hybrids and allopolyploids. Nature. 2009;457:327–31. doi: 10.1038/nature07523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Himanen K, Woloszynska M, Boccardi TM, De Groeve S, Nelissen H, Bruno L, et al. Histone H2B monoubiquitination is required to reach maximal transcript levels of circadian clock genes in Arabidopsis. Plant J 2012; July 5. doi: 10.1111/j.1365-313X.2012.05071.x. [Epub ahead of print] PMID:22762858. [DOI] [PubMed]
- 4.Lolas IB, Himanen K, Grønlund JT, Lynggaard C, Houben A, Melzer M, et al. The transcript elongation factor FACT affects Arabidopsis vegetative and reproductive development and genetically interacts with HUB1/2. Plant J. 2010;61:686–97. doi: 10.1111/j.1365-313X.2009.04096.x. [DOI] [PubMed] [Google Scholar]
- 5.Fleury D, Himanen K, Cnops G, Nelissen H, Boccardi TM, Maere S, et al. The Arabidopsis thaliana homolog of yeast BRE1 has a function in cell cycle regulation during early leaf and root growth. Plant Cell. 2007;19:417–32. doi: 10.1105/tpc.106.041319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cao Y, Dai Y, Cui S, Ma L. Histone H2B monoubiquitination in the chromatin of FLOWERING LOCUS C regulates flowering time in Arabidopsis. Plant Cell. 2008;20:2586–602. doi: 10.1105/tpc.108.062760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xu L, Ménard R, Berr A, Fuchs J, Cognat V, Meyer D, et al. The E2 ubiquitin-conjugating enzymes, AtUBC1 and AtUBC2, play redundant roles and are involved in activation of FLC expression and repression of flowering in Arabidopsis thaliana. Plant J. 2009;57:279–88. doi: 10.1111/j.1365-313X.2008.03684.x. [DOI] [PubMed] [Google Scholar]
- 8.Fiorani F, Beemster GTS. Quantitative analyses of cell division in plants. Plant Mol Biol. 2006;60:963–79. doi: 10.1007/s11103-005-4065-2. [DOI] [PubMed] [Google Scholar]
- 9.Liu Y, Koornneef M, Soppe WJJ. The absence of histone H2B monoubiquitination in the Arabidopsis hub1 (rdo4) mutant reveals a role for chromatin remodeling in seed dormancy. Plant Cell. 2007;19:433–44. doi: 10.1105/tpc.106.049221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Harmer SL. The circadian system in higher plants. Annu Rev Plant Biol. 2009;60:357–77. doi: 10.1146/annurev.arplant.043008.092054. [DOI] [PubMed] [Google Scholar]
- 11.Athanasiou K, Dyson BC, Webster RE, Johnson GN. Dynamic acclimation of photosynthesis increases plant fitness in changing environments. Plant Physiol. 2010;152:366–73. doi: 10.1104/pp.109.149351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Holdsworth MJ, Finch-Savage WE, Grappin P, Job D. Post-genomics dissection of seed dormancy and germination. Trends Plant Sci. 2008;13:7–13. doi: 10.1016/j.tplants.2007.11.002. [DOI] [PubMed] [Google Scholar]