Abstract
Penetration resistance against powdery mildews is one of the best-studied processes of plant innate immunity. One vital component is the plant syntaxin, PEN1, which is required for timely deposition of callose and extracellular membrane material, as well as PEN1 itself, at the attack sites. Recently, we reported that the ARF-GEF GNOM also is required for penetration resistance, mediating transport of recycled material, including PEN1, to the site of attack. The close relative of PEN1, SYP122, does not accumulate at the sites of attack nor does it affect penetration resistance. In support of this, we show here that in contrast to PEN1, SYP122 does not continuously recycle. Furthermore, by using a PEN1 transgene that is only transcribed in dividing cells, we show that papillary PEN1 accumulation is not dependent on de-novo protein synthesis. This emphasizes the involvement of recycling in penetration resistance, which possibly relates to the differences in function of the two syntaxins.
Keywords: fungal penetration, vesicle transport, defense, pathogen, syntaxin
Introduction
In plants, one type of innate immunity is represented by “penetration resistance,” where the plant executes a timely defense response to effectively hinder fungi from entering the plant cell. Penetration resistance is associated with a papilla response, where a dome-shaped cell wall apposition is deposited by the epidermal cell between the cell wall and plasma membrane (PM).1 For full protection against the non-host barley powdery mildew fungus, Blumeria graminis f.sp hordei (Bgh), Arabidopsis plants require the function of the PM localized syntaxin PEN1 (SYP121) and its interacting SNARE proteins, SNAP33 and VAMP721/2, all of which accumulate at the attack site.2-6 In Arabidopsis, the closest homolog of PEN1 is the PM localized SYP122. Interestingly, like PEN1, SYP122 interacts with SNAP33 and VAMP722, yet it does not accumulate at attacks sites or affect penetration resistance.7,8 SNARE proteins are required for membrane fusion,9 thereby establishing the need for vesicle trafficking in penetration resistance. Recently, we reported that inhibition of the ARF-GEF, GNOM, by application of brefeldin A (BFA), blocks papillary accumulation of GFP-PEN1 and impedes penetration resistance.8 GNOM is a BFA-sensitive ARF-GEF, mediating recycling of endocytosed proteins back to the PM, a function required for the correct localization of auxin-efflux carriers. GNOM is thus vital for root gravitropism and development.10,11
Based on our observations, we suggested that fully functional penetration resistance requires a GNOM-mediated trafficking pathway to re-cycle pre-existing material to the papillae.
In this report, we show that while the PEN1 population at the PM is highly dynamic and continuously recycles to endosomes and back to the PM, SYP122 is stably located at the PM. Furthermore, RFP-PEN1 was found to accumulate at attack sites in cells where no de-novo RFP-PEN1 protein synthesis occurs. Combined our observations underline the need for recycling during the deposition of material in papilla in order for them to confer efficient penetration resistance.
Results
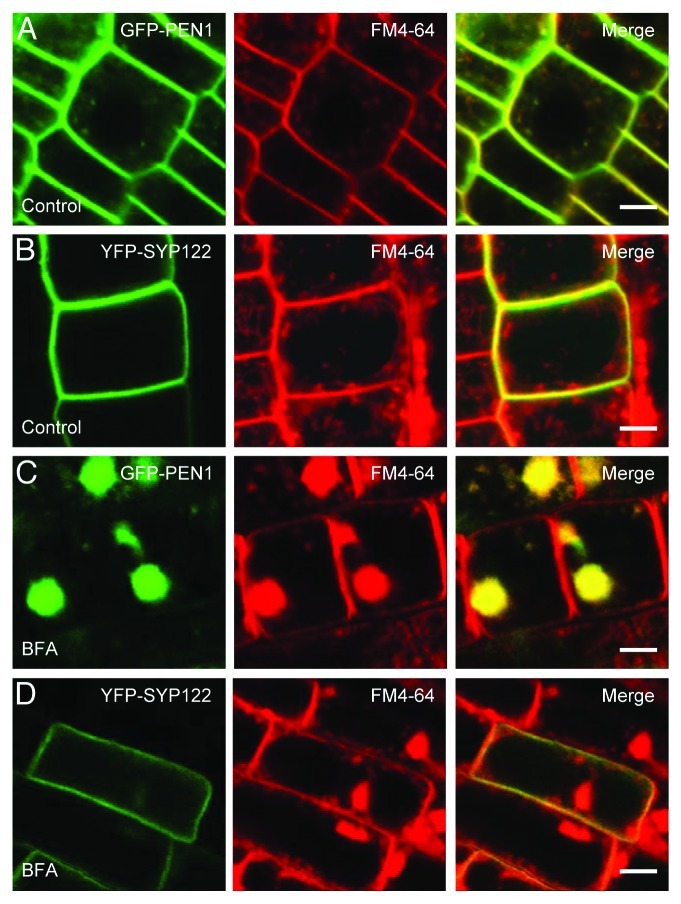
Previous investigations of syntaxin-mediated penetration resistance have shown that SYP122 does not accumulate at the site of attack by Bgh.8 Having found that BFA not only blocks papillary accumulation of PEN1, but also leads to accumulation of this protein in BFA bodies,8 we turned to look at the effects of BFA on SYP122 localization. As previously reported,12 GFP-PEN1 strongly labeled the PM, but also partly colocalized with the internalized FM4–64 dye in endosomes of untreated cells (Fig. 1). After 1 h. of BFA treatment, the GFP-PEN1 signal was almost exclusively found in BFA bodies together with FM4–64, indicating how dynamic this PM protein is. Interestingly, YFP-SYP122 only labeled the PM and did not respond to the application of BFA. This shows that PEN1 is likely cycling to and from the PM, although we cannot exclude that some PEN1 is stably located at endosomes. In contrast, SYP122 remains stably at the PM and does not undergo recycling. This observation provides a possible explanation for why SYP122 does not accumulate in the papilla although it is located to the PM together with PEN1 and interacts with the same SNARE proteins.
Figure 1. Syntaxins PEN1 and SYP122 differ in sensitivity to BFA. Roots expressing functional GFP-PEN1 (A and C) or YFP-SYP122 (B and D), were stained with FM4- 64 for 1 h without (A-B) or with 50 μM BFA (C-D). Scale bars are equivalent to 5 μm.
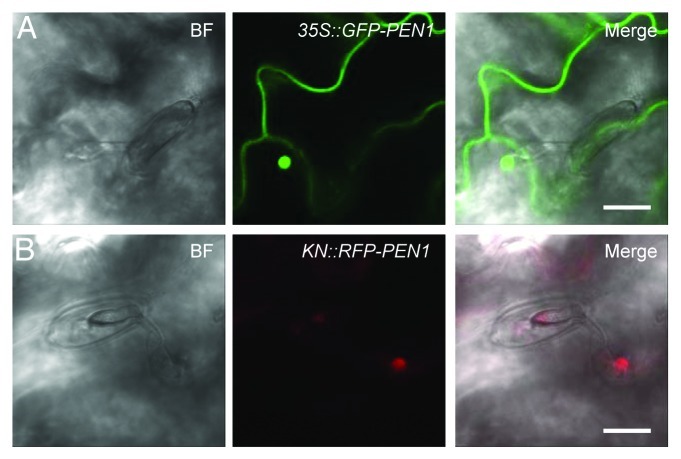
Reichardt et al.12 found that PEN1 expressed under the KNOLLE promoter (pKN), the activity of which is confined to late G2 and M phases of cell cycle,13,14 rescues penetration resistance in pen1–1. This means that in these plants, PEN1 is stable and functional in fully expanded leaves, despite absence of transcription from this gene. Our finding that GNOM is required for normal papilla formation and penetration resistance, strongly suggests that plants rely on fast recycling of papilla material in order to block the powdery mildew fungus from entering. Accordingly, premade PEN1, which confers penetration resistance,12 should also accumulate in the papillae. We therefore turned to analyze whether premade RFP-PEN1 would also accumulate in papillae upon Bgh attack. Although the pKN-driven RFP-PEN1 signal was somewhat weaker than the signal from GFP-PEN1, driven by the 35S-promoter in our control line, we found a clear accumulation of RFP-PEN1 at the Bgh attack sites (Fig. 2). This finding supports the notion that papillary PEN1 accumulation does not require de-novo protein synthesis and a functional secretory pathway, but instead relies on recycling of premade protein.
Figure 2. Papillary accumulation of premade PEN1 in response to Bgh. Leaves of plants expressing GFP-PEN1 (A) and RFP-PEN1 (B), under 35S and KNOLLE (KN) promoter regulation, respectively, monitored 24 h after inoculation with Bgh. Scale bars are equivalent to 10 μm.
Combined, our investigations underline the involvement of recycling in papilla formation and explain why PEN1, but not SYP122, accumulates in these cell wall appositions. It is tempting to speculate that the functional difference in penetration resistance between these two closely related syntaxins is connected to their difference in recycling. However, we find it unlikely that the PEN1 papilla localization per se plays a role in penetration resistance, as PEN1 is considered to be transported there on multivesicular-body derived exosomes,8 in which syntaxins should have no function. Therefore, the difference in recycling taking place in unstressed cells (Fig. 1) is attracting our attention in relation to how PEN1 mediates penetration resistance.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank Gerd Jürgens for providing transgenic lines and The Carlsberg Foundation, Denmark, for financial support to M.E.N.
Footnotes
Previously published online: www.landesbioscience.com/journals/psb/article/22304
References
- 1.Hückelhoven R, Panstruga R. Cell biology of the plant-powdery mildew interaction. Curr Opin Plant Biol. 2011;14:738–46. doi: 10.1016/j.pbi.2011.08.002. [DOI] [PubMed] [Google Scholar]
- 2.Collins NC, Thordal-Christensen H, Lipka V, Bau S, Kombrink E, Qiu JL, et al. SNARE-protein-mediated disease resistance at the plant cell wall. Nature. 2003;425:973–7. doi: 10.1038/nature02076. [DOI] [PubMed] [Google Scholar]
- 3.Assaad FF, Qiu JL, Youngs H, Ehrhardt D, Zimmerli L, Kalde M, et al. The PEN1 syntaxin defines a novel cellular compartment upon fungal attack and is required for the timely assembly of papillae. Mol Biol Cell. 2004;15:5118–29. doi: 10.1091/mbc.E04-02-0140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kwon C, Neu C, Pajonk S, Yun HS, Lipka U, Humphry M, et al. Co-option of a default secretory pathway for plant immune responses. Nature. 2008;451:835–40. doi: 10.1038/nature06545. [DOI] [PubMed] [Google Scholar]
- 5.Bhat RA, Miklis M, Schmelzer E, Schulze-Lefert P, Panstruga R. Recruitment and interaction dynamics of plant penetration resistance components in a plasma membrane microdomain. Proc Natl Acad Sci U S A. 2005;102:3135–40. doi: 10.1073/pnas.0500012102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kwaaitaal M, Keinath NF, Pajonk S, Biskup C, Panstruga R. Combined bimolecular fluorescence complementation and Forster resonance energy transfer reveals ternary SNARE complex formation in living plant cells. Plant Physiol. 2010;152:1135–47. doi: 10.1104/pp.109.151142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pajonk S, Kwon C, Clemens N, Panstruga R, Schulze-Lefert P. Activity determinants and functional specialization of Arabidopsis PEN1 syntaxin in innate immunity. J Biol Chem. 2008;283:26974–84. doi: 10.1074/jbc.M805236200. [DOI] [PubMed] [Google Scholar]
- 8.Nielsen ME, Feechan A, Böhlenius H, Ueda T, Thordal-Christensen H. Arabidopsis ARF-GTP exchange factor, GNOM, mediates transport required for innate immunity and focal accumulation of syntaxin PEN1. Proc Natl Acad Sci U S A. 2012;109:11443–8. doi: 10.1073/pnas.1117596109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jahn R, Scheller RH. SNAREs--engines for membrane fusion. Nat Rev Mol Cell Biol. 2006;7:631–43. doi: 10.1038/nrm2002. [DOI] [PubMed] [Google Scholar]
- 10.Richter S, Anders N, Wolters H, Beckmann H, Thomann A, Heinrich R, et al. Role of the GNOM gene in Arabidopsis apical-basal patterning--From mutant phenotype to cellular mechanism of protein action. Eur J Cell Biol. 2010;89:138–44. doi: 10.1016/j.ejcb.2009.11.020. [DOI] [PubMed] [Google Scholar]
- 11.Geldner N, Anders N, Wolters H, Keicher J, Kornberger W, Muller P, et al. The Arabidopsis GNOM ARF-GEF mediates endosomal recycling, auxin transport, and auxin-dependent plant growth. Cell. 2003;112:219–30. doi: 10.1016/S0092-8674(03)00003-5. [DOI] [PubMed] [Google Scholar]
- 12.Reichardt I, Slane D, El Kasmi F, Knöll C, Fuchs R, Mayer U, et al. Mechanisms of functional specificity among plasma-membrane syntaxins in Arabidopsis. Traffic. 2011;12:1269–80. doi: 10.1111/j.1600-0854.2011.01222.x. [DOI] [PubMed] [Google Scholar]
- 13.Lukowitz W, Mayer U, Jürgens G. Cytokinesis in the Arabidopsis embryo involves the syntaxin-related KNOLLE gene product. Cell. 1996;84:61–71. doi: 10.1016/S0092-8674(00)80993-9. [DOI] [PubMed] [Google Scholar]
- 14.Haga N, Kato K, Murase M, Araki S, Kubo M, Demura T, et al. R1R2R3-Myb proteins positively regulate cytokinesis through activation of KNOLLE transcription in Arabidopsis thaliana. Development. 2007;134:1101–10. doi: 10.1242/dev.02801. [DOI] [PubMed] [Google Scholar]