Abstract
Comparison of the expression of 13 genes involved in arbuscular mycorrhizal (AM) symbiosis was performed in a wild type tomato (Solanum lycopersicum cv 76R) and its reduced mycorrhizal colonization mutant rmc in response to colonization with Glomus fasiculatum. Four defense-related genes were induced to a similar extent in the mutant and wild type AM colonized plants, indicating a systemic response to AM colonization. Genes related to nutrient exchange between the symbiont partners showed higher expression in the AM roots of wild type plants than the mutant plants, which correlated with their arbuscular frequency. A symbiosis receptor kinase that is involved in both nodulation and AM symbiosis was not expressed in the rmc mutant. The fact that some colonization was observed in rmc was suggestive of the existence of an alternate colonization signaling pathway for AM symbiosis in this mutant.
Keywords: AM colonization, defense genes, nutrient exchange genes, reduced mycorrhizal colonization (rmc) tomato mutant, SYMRK
Arbscular mycorrhizal (AM) symbiosis is widely distributed in the plant kingdom, unlike rhizobial symbiosis that is observed only in four orders of the eurosid dicots.1 AM symbiosis involves provision of mineral nutrients to the plant by the fungus, which in turn derives carbon compounds from the plant.2 Other benefits of the symbiosis to plants include improved water relations and tolerance to some plant diseases.3 The fungal hyphae penetrate the root epidermis, spread inter-cellularly in the cortex and form arbuscules in the root cells of the inner cortex. Arbuscules are highly branched hyphal structures that are separated from the plant cytoplasm by a perifungal membrane.4 They are the main sites where nutrient exchange between the symbiotic partners occurs and their formation signifies the establishment of functional symbiosis.
Molecular events occurring during AM symbiosis have been studied in legumes, which also show an elaborate signaling pathway for establishing nodulation symbiosis.5 Two genes, NFR1 and NFR5 encoding receptor-like serine/threonine kinases with LysM domains, are involved in nodulation (Nod) factor perception in Lotus japonicus.6 Several downstream components of the Nod factor signaling cascade include the leucine-rich-repeat receptor kinase SYMRK, which is known to be involved in AM symbiosis besides nodulation symbiosis and is thought to act near the junction of fungal and rhizobial signaling cascades.7 Activation of SYMRK causes a transient increase in intracellular calcium levels. Downstream components of this signaling pathway include a calcium / calmodulin dependent protein kinase (CCAMK) and a protein CYCLOPS, whose function is not known. It is suggested that the evolutionarily more recent nodulation symbiosis has recruited this signaling pathway from the more ancient AM-symbiosis, since non-legumes like rice, tomato and Casuarina show orthologs of the legume genes involved in symbiosis signaling.8-10
A number of AM induced genes have been identified, which show expression only in AM colonized roots. Many of these are genes associated with defense responses of plants and it is reported that the initial stages of colonization by the fungal symbiont and biotrophic pathogens are similar.11 Some genes are associated with nutrient exchange that occurs between the two symbiont partners and include a low affinity phosphate transporter and genes involved in sugar and nitrogen metabolism.12 A few transcription factor genes are also induced by AM colonization, which are either related to regulation of expression of defense genes or genes involved in alteration of growth patterns in mycorrhizal roots.13
Symbiosis pathway mutants have been identified in leguminous (Lotus japonicus)14 as well as non-leguminous plants like Oryza sativa8 and Casuarina glauca.10 In these mutants, infection by AM fungi is either aborted before or after hyphal penetration of root cells. However, several AM induced genes were expressed in the SYMRK pathway mutants, suggesting that another pathway may be involved in AM signaling.15 A reduced mycorrhizal colonization mutant rmc has been identified in tomato,16 in which reduced symbiotic association was attributed to lack of penetration, inability to colonize the root cortex or a slower but successful colonization, depending on the species of the fungal interacting partner.17 The mycorrhizal phenotype of rmc mutants resembles that of dmi mutants of Medicago trunculata suggesting the possibility that RMC may be an ortholog of DMI genes, one of which codes for a symbiosis receptor kinase (DMI2).18 Though the RMC locus has been identified and is known to lie on chromosome 8, the function(s) encoded by this locus is not known.19 In this paper we have compared the expression of a few AM induced genes in the tomato rmc mutant and its wild type parent 76R. Our results indicate that reduced colonization in rmc could be attributed to the lack of SYMRK signaling pathway in this mutant.
Seeds of Solanum lycopersicum cv 76R and its reduced mycorrhizal colonisation mutant (rmc) were procured from Dr. Susan Barker (University of Western Australia, Australia). A soil based AM fungal inoculum of Glomus fasciculatum consisting of spores, colonized root pieces and the surrounding soil was supplied by Dr. Joseph Bagyaraj, (Centre for Natural Biological Resources and Community Development, Bangalore, India). The seeds were treated with 1% HCl for 15 min, rinsed with water and sowed in autoclaved (121°C, 1.034 bars, for 1h) soil in seedling trays. Seedlings were carefully transplanted after 20 d to polythene bags containing about 2 kg autoclaved soil each, ensuring minimum root damage during transplantation. Inoculation with AM fungus was done at the time of transplantation by adding about 1g inoculum to the soil cavity in which the seedling was transplanted. Controls consisted of plants to which no AM inoculum was applied. Three replicates were used per treatment.
Roots were carefully harvested from 52 dpi plants by completely immersing the polythene bags in water. Roots were carefully washed to remove all particulates, cleared using 10% hot KOH solution and stained with 0.5% trypan blue (invam.wvu.edu/methods/mycorrhizae). Extent of colonization was measured from 30 root segments of 2 cm length per plant using the computer program ‘Mycocalc’.20 Student’s t test was applied to compare the colonization parameters in 76R and rmc.
Genes induced during mycorrhizal colonization were identified by searching for tomato orthologs of the rice AM-specific genes8 and from the microarray data available on AM colonized tomato plants9 (Table 1). RNA isolation was performed using root tissue (pooled from 3 plants) with Trizol reagent (Sigma Aldrich, cat. no. 93289) from 52 dpi plants and their respective controls. Extracted RNA (1 µg) was used as a template for reverse transcription with ImProm-II reverse transcriptase (Promega, cat. no. A3802). Gene specific primers were designed using primer Blast (blast.ncbi.nlm.nih.gov) and transcript abundance in the cDNA was studied at three dilutions by PCR amplification. Gene expression was normalized using a constitutively expressed Elongation factor1α (EF1α) gene. Cycling conditions used were: 94°C for 5 min, followed by 35 cycles of 94°C for 1 min, varying annealing temperatures (Table 1) for 0.45 min and 72°C for 1 min. Final extension was performed at 72°C for 10 min. Two genes AQP and GRAS4, showed saturation of PCR at 35 cycles and so 30 cycles of amplification were used. Similarly for EF1α, 25 cycles of PCR amplification were used so as to get transcript abundance-dependent differential expression. All amplified products were sequenced to confirm the transcripts amplified. Gel images were captured using a gel documentation system (Bio Rad, Gel Doc XR+, USA) and intensities of the bands were recorded.
Table 1. Genes used for studying AM-induced expression in tomato cv 76R and its reduced mycorrhizal colonization mutant rmc. Primer sequences, primer annealing temperatures and the expected product sizes are given.
| Gene Name | Accession number | Primer Sequence (5′-3′) | Annealing temperature(°C) | Product Size (bp) |
|---|---|---|---|---|
| Xyloglucan-specific fungal endoglucanase inhibitor |
AY155579.1 |
GTGTTCACTGGGCGGAGCGT TGACCTGATGAAAAGGCTGAGGC |
59 |
492 |
| Cysteine protease |
AF172856.1 |
GCTGGTGGCAGAGACTTCCAGC AGCAAGTGGTGCCGACAGCG |
59 |
343 |
| UDP-xylose phenolic glycosyltransferase |
AJ889012.1 |
TGGTGGTCGCGATCAAGCAGG ACCCCATGCCAATTCTTCCATTTGC |
59 |
682 |
| Indole acetic acid amido synthetase |
AC215447.2 |
AGGACCCGGCTAACCCACCC TCAACGTCGTCGTTCTGGAGTCC |
63 |
314 |
| Ethylene response element binding protein |
BT013241.1 |
GTTCGGAAGAGGCCATGGGGG TTGCTGACGTGGCGGTCTCG |
61 |
336 |
| GRAS family transcription factor |
DQ399826.1 |
TCCATGAGGGCTGGGGGACC GTCCTCGTCCTCGCGGGGAA |
59 |
450 |
| Low affinity phosphate LePT4 |
AY804012.2 |
ATGGTTTGACTTTCTTCTTTG TAGAAAGCACAAGGCGTAG |
57 |
418 |
| Aquaporin |
AB211518.1 |
AAGCGGCCTTGGCGGAGTTC ATCCCGTGCTCATGCCACCG |
59 |
330 |
| Sucrose synthase Sus 3 |
AJ011319.1 |
GCCATGAGCTGCGGTTTGCC GCTAGTGTCAACAGCCGGTCGG |
61 |
248 |
| Invertase (Vacuolar) |
Z12027.1 |
CCCCGAAAACTCCGCCTCTCG AAACACCTCTTGACGGCGGC |
63 |
200 |
| Invertase (Cell wall) Lin6 |
AB004558.1 |
AGGATGGGCCGGGGTTCACC GGCCCAAGACCACCTTGAACCG |
63 |
309 |
| Symbiosis receptor-like kinase |
AY940041.1 |
For semi-quantitative RT-PCR GCCGGCCAGACTTTCCATTGC ACCACTCTCCACAGCGCCTCA For Real-Time RT-PCR AGCTTGGTTGAATGGGCGAAACC ACCACTCTCCACAGCGCCTCA |
59 59 |
453 107 |
| Calcium dependent protein kinase | AB530160.1 | GCTCGGGTGCCGGATGACTC ACCGCATCCTCATAAGCCCCT |
59 | 491 |
Real-time RT PCR using the QuantiFast SYBR Green PCR kit (Qiagen, cat. no. 204054) was performed to confirm the differential expression of SYMRK using thermocycler (Eppendorf, RealPlex 2, Germany). Primers used were as shown in Table 1. Total amount of SYMRK cDNA in each reaction was normalized by co-amplification of the constitutively expressed EF1α gene.
Colonization of tomato roots by Glomus faciculatum was seen in 52 dpi plants in the form of intercellular and extra-radical mycelia as well as arbuscule and vesicle formation. The wild type cultivar 76R and its reduced mycorrhizal colonization mutant (rmc) showed the presence of normal arbuscules and extra radical hyphae (Fig. 1), but the frequency of mycorrhiza in root system, intensity of mycorrhizal colonization and arbuscule abundance varied. All the colonization parameters were significantly lower in rmc as compared with 76R and arbuscule abundance was most severely reduced (Table 2). The colonization pattern in tomato roots (76R and its rmc mutant) has been reported to be similar for Glomus fasciculatum and Glomus intraradices interactions.17
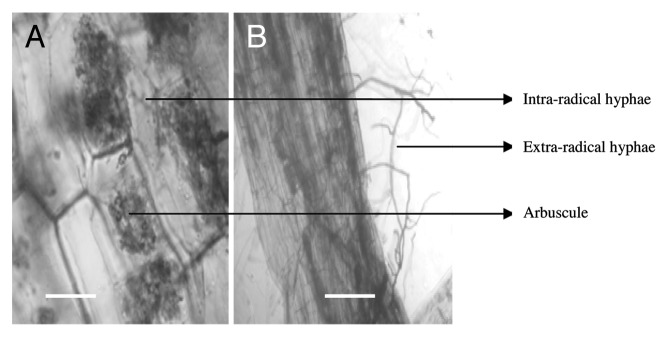
Figure 1.Solanum lycopersicum (76R) roots colonized with Glomus fasciculatum showing (A) arbuscules and (B) extraradical hyphae. Both these features were also observed in the colonization deficient mutant (rmc) but at a much lower frequency (see Table 2). The roots were cleared and stained with Trypan blue, 52 d after inoculum application. Bar = 25μm (A), 150μm (B).
Table 2. Measurement of the extent of Glomus fasciculatum colonization in roots of Solanum lycopersicum cv 76R (wt) and its colonization deficient mutant (rmc). The colonization parameters were calculated using Mycocalc software and represent mean % values of 30 root fragments per plant for 3 plants per treatment, along with standard deviations.
| Colonization parameter | Mean values % | |
|---|---|---|
| |
76R |
rmc |
| Frequency of mycorrhiza in root system |
91 ± 3.2 |
62 ± 6.2* |
| Intensity of mycorrhizal colonization in root system |
44 ± 3.1 |
19 ± 4.2* |
| Intensity of mycorrhizal colonization in root fragment |
52 ± 0.7 |
25 ± 2.9* |
| Arbuscule abundance in mycorrhizal part of root fragment |
37 ± 2.6 |
23 ± 2.4* |
| Arbuscule abundance in root system | 16 ± 2.9 | 2 ± 0.6* |
Means differ significantly between 76R and rmc (p < 0.05).
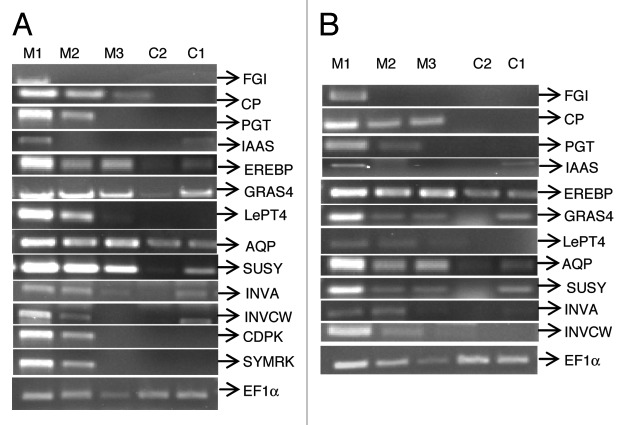
Expression pattern of 13 AM induced genes in 76R and rmc were analyzed from the band intensities in (a) AM roots/control roots for 76R and rmc respectively and (b) 76R AM roots/rmc AM roots (Fig. 2; Table 3). Comparative analysis of AM induced gene expression in 76R and rmc mutant revealed two patterns of gene expression. Some genes were AM-specific and were not expressed in non-colonized roots, while others though induced in response to AM colonization, also showed some basal level expression in non-colonized roots. Functionally these genes could be broadly categorized into (a) defense related genes, which included a fungal endoglucanase inhibitor (FGI), a cysteine protease (CP), a phenolic glycosyltransferase (PGT) and an IAA amidosynthetase (IAAS) (b) signal transduction and transcription regulation genes, which included a calcium dependent protein kinase (CDPK), an ethylene response element binding protein (EREBP) and a GRAS domain transcription factor (GRAS4) (c) transporters and primary metabolism genes, which included a low affinity phosphate transporter (LePT4), an aquaporin protein (AQP), sucrose synthase (SUSY), vacuolar invertase (INVA) and a cell wall invertase (INVCW) (d) nodulation symbiosis pathway gene, symbiosis receptor kinase (SymRK).
Figure 2. Expression of AM induced genes in Solanum lycopersicum cv 76R (A) and its colonization deficient mutant (rmc) (B), 52 dpi with Glomus fasciculatum. Transcript abundance was analyzed using semiquantitative RT-PCR. M1, M2 and M3 represent 0, 5 and 10-fold dilutions of the cDNA prepared from AM roots and C1 and C2 represent 0 and 5-fold dilution of the cDNA prepared from non-colonized roots. Constitutively expressed EF 1α was used as a loading control for comparing expression levels. The genes studied were: FGI (Fungal endoglucanase inhibitor); CP (Cysteine Protease); PGT (Phenolic glycosyltransferase); IAAS (IAA amido synthetase); EREBP (Ethylene response element binding protein); GRAS4 (GRAS family transcription factor); LePT4 (Low affinity phosphate Transporter); AQP (Aquaporin); SUSY (Sucrose synthase); INVA (Vacuolar Invertase); INVCW (Cell wall Invertase Lin6); CDPK (Calcium dependent protein kinase); SYMRK (Symbiosis receptor-like kinase).
Table 3. Fold expression of 13 a.m.-induced genes in Solanum lycopersicum cv 76R and its reduced mycorrhizal colonization mutant (rmc) analyzed using semiquantitative RT-PCR. The band intensities (see Figure 2) were normalized using constitutively expressed EF1α. Fold-expression was determined as the ratio of normalized intensities in (i) AM colonized roots (M1 in Figure 2) and non-colonized roots (C1 in Figure 2) for 76R and rmc respectively and (ii) AM colonized roots of 76R (M1 in Figure 2A) and AM colonized roots of rmc (M1 in Figure 2B).
| Product | Fold-expression in AM roots / non-AM roots | Fold-expression in 76R AM roots / rmc AM roots | |
|---|---|---|---|
| |
76R |
rmc |
|
| Xyloglucan-specific fungal endoglucanase inhibitor |
Only expressed in AM colonized roots |
Only expressed in AM colonized roots |
1.1 |
| Cysteine protease |
Only expressed in AM colonized roots |
Only expressed in AM colonized roots |
1.2 |
| UDP-xylose phenolic glycosyltransferase |
Only expressed in AM colonized roots |
Only expressed in AM colonized roots |
3.0 |
| IAA amido synthetase |
2.6 |
2.5 |
1.3 |
| EREBP |
5.5 |
3.4 |
1.2 |
| GRAS4 transcription factor |
3.6 |
3.5 |
1.1 |
| Phosphate Transporter |
Only expressed in AM colonized roots |
Only expressed in AM colonized roots |
4.0 |
| Aquaporin |
3.7 |
6.6 |
0.7 |
| Sucrose synthase |
5.2 |
3.0 |
2.0 |
| Vacuolar invertase |
1.5 |
1.5 |
2.6 |
| Cell wall invertase |
1.7 |
1.3 |
1.5 |
| Calcium dependent protein kinase |
Only expressed in AM colonized roots |
Not expressed in AM colonized roots |
— |
| Symbiosis receptor-like kinase | Only expressed in AM colonized roots | Not expressed in AM colonized roots |
— |
Of the defense genes studied FGI, CP and PGT showed expression only in the mycorrhizal roots while IAAS was expressed in both the mycorrhizal as well as control roots. The defense related genes IAAS, CP and FGI showed similar levels of expression in the AM roots of 76R and rmc (Table 3). The products of these genes could be classified as pathogenesis-related (PR) proteins, since they did not show any expression in non-AM roots of 76R or rmc. Plants are known to develop similar defense responses to biotrophic pathogenic and symbiotic fungi.11 For example, rice roots colonized by AM fungi were known to produce PR proteins like chitinases, which can hydrolyse fungal cell walls.21 PGT has been reported to be induced in response to systemic pathogen infection in tomato.22 An IAA amido synthetase that has been reported to play a role in expression of basal immunity in rice23 was seen to have about 2-fold expression in response to AM colonization indicating its probable role in attaining IAA homeostasis after the establishment of symbiosis.
The AM fungi have to evade host defense mechanisms in order to establish successful mycorrhizal colonization. Defense genes like extracellular acidic β-1,3-glucanase, PR-1 and chitinase, though induced during the early stages of colonization showed lower expression than mock-inoculated controls or rmc at 42 d post-colonization.24 However an induction of the defense genes studied by us was observed in both, the wild type cultivar 76R and its rmc mutant. Expression of the defense genes appeared to depend not on the extent of colonization, but on the colonization event itself, which was indicative of a systemic response. Split-root experiments with tomato colonized by Glomus mosseae revealed a systemic bio-protective effect in roots where the non-mycorrhizal portion of the mycorrhizal root system also exhibited resistance.25
The transcription factors EREBP and GRAS4 showed higher expression in AM roots of both 76R and rmc, while CDPK, a signaling intermediate was expressed only in AM roots of 76R. Ethylene responsive transcription factors are widely known to play a role in regulating gene expression in response to biotrophic and necrotrophic pathogens.26 CDPKs have also been reported to play a role in AM signaling in Medicago trunculata.27
The LePT4 and sugar metabolism genes, SUSY, INVA and INVCW were induced in response to AM colonization, in both 76R and rmc, but the level of expression was at least 2-fold higher in 76R. The fungal symbiont is known to provide phosphate to the AM roots and acquire sugars from the roots in exchange. Expression of symbiosis-specific Pi transporters in AM roots has been reported.28 An increase in the activities and expression of sucrose metabolizing enzymes, invertase and sucrose synthase has also been reported in the AM roots of Trifolium repens and tomato, which was independent of the improved phosphate nutrition of colonized roots.29,30 That arbuscules are the sites of nutrient exchange is known from various studies. For example, a symbiosis-specific phosphate transporter (MtPT4) was shown to localize to the periarbuscular membrane.31 Likewise a sucrose synthase (MtSucS1) knockout mutant showed defects in arbuscule development and maintenance.32 Hence unlike the defense genes that were induced in response to systemic signals provided by the fungal symbiont, expression of the sugar metabolism and phosphate transport genes correlated with the extent of arbuscular development in AM roots. The water channel protein AQP, which is also known to transport other small molecules like NH4+ ions, showed higher expression in AM roots, but in this case, AM roots of rmc showed higher levels of expression than those of 76R.
The nodulation symbiosis pathway gene homolog in tomato (SYMRK) was expressed in AM roots of 76R but not in the poorly colonized rmc roots (Fig. 2). Transcript abundance of SYMRK was about 200-fold higher in AM colonized roots of 76R as compared with AM colonized roots of rmc (Fig. 3). SYMRK is a key component of the symbiosis signaling pathway, common to nodulation, actinorrhizal and mycorrhizal symbiosis.10 During nodulation, Nod factors are known to be perceived by specific receptor-like serine / threonine kinases with LysM domains, which bring about the activation of SYMRK. However receptors for Myc factors, which have been identified as small, lipophilic molecules with chitin backbone, are not known.33 Activation of SYMRK brings about calcium spiking, which is important for the expression of downstream genes involved in the establishment of symbiosis.34 In tomato, the CDPK gene was expressed only in AM colonized 76R but not in rmc roots suggesting that its expression may be dependent on calcium spiking caused by SYMRK.
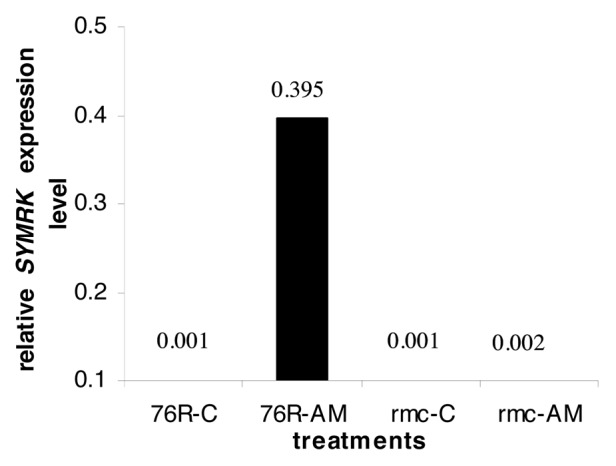
Figure 3. Real-time PCR analysis of transcript abundance of SYMRK in 76R and rmc. Values represent mRNA levels of SYMRK normalized to transcript abundance of the constitutively expressed gene EF1α.
The rmc mutant of tomato, which lacked SYMRK expression, did however show some extent of colonization, which indicated the possibility of an alternate, SYMRK-independent pathway being employed for establishing symbiosis in the mutant. The sym15 and castor mutants of Lotus japonicus14 and the dmi mutants of Medicago trunculata,18 which showed mycorrhizal colonization in spite of a non-functional SYMRK pathway also suggested the presence of an alternate pathway for symbiosis signaling. Lack of SYMRK expression could be a probable cause for the fewer arbuscules detected in rmc, since the kinase is known to play an important role during penetration of the inner cortical cells by the fungal symbiont partner as seen in Casuarina.10 This is a first report offering a functional explanation for the mycorrhizal colonization deficiency observed in rmc mutants. It is possible however that up stream components of the SYMRK pathway, like NFP in Medicago trunculata35 or NFR1 and NFR5 in Lotus japonicus6 could be responsible for the lack of expression of SYMRK in the tomato reduced mycorrhizal colonization (rmc) mutant.
Acknowledgments
The authors thank Dr. Barker for providing tomato seeds of 76R and rmc, Dr. J. Bagyaraj for providing Glomus fasciculatum soil inoculum and Department of Science and Technology, Government of India, for financial assistance.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/psb/article/20156
References
- 1.Soltis DE, Soltis PS, Morgan DR, Swensen SM, Mullin BC, Dowd JM, et al. Chloroplast gene sequence data suggest a single origin of the predisposition for symbiotic nitrogen fixation in angiosperms. Proc Natl Acad Sci U S A. 1995;92:2647–51. doi: 10.1073/pnas.92.7.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sawers RJH, Yang S-Y, Gutjahr C, Paszkowski U. The molecular components of nutrient exchange in arbuscular mycorrhizal interactions. In: Siddiqui ZA, Akthar MS, Futai K. Eds. Mycorrhizae: Sustainable Agriculture and Forestry. Netherlands: Springer, 2008: 37-59. [Google Scholar]
- 3.Newsham KK, Fitter AH, Watkinson AR. Arbuscular mycorrhiza protect an annual grass from root pathogenic fungi in the field. J Ecol. 1995;83:991–1000. doi: 10.2307/2261180. [DOI] [Google Scholar]
- 4.Paszkowski U. A journey through signaling in arbuscular mycorrhizal symbioses 2006. New Phytol. 2006;172:35–46. doi: 10.1111/j.1469-8137.2006.01840.x. [DOI] [PubMed] [Google Scholar]
- 5.Parniske M. Arbuscular mycorrhiza: the mother of plant root endosymbioses. Nat Rev Microbiol. 2008;6:763–75. doi: 10.1038/nrmicro1987. [DOI] [PubMed] [Google Scholar]
- 6.Radutoiu S, Madsen LH, Madsen EB, Felle HH, Umehara Y, Grønlund M, et al. Plant recognition of symbiotic bacteria requires two LysM receptor-like kinases. Nature. 2003;425:585–92. doi: 10.1038/nature02039. [DOI] [PubMed] [Google Scholar]
- 7.Stracke S, Kistner C, Yoshida S, Mulder L, Sato S, Kaneko T, et al. A plant receptor-like kinase required for both bacterial and fungal symbiosis. Nature. 2002;417:959–62. doi: 10.1038/nature00841. [DOI] [PubMed] [Google Scholar]
- 8.Gutjahr C, Banba M, Croset V, An K, Miyao A, An G, et al. Arbuscular mycorrhiza-specific signaling in rice transcends the common symbiosis signaling pathway. Plant Cell. 2008;20:2989–3005. doi: 10.1105/tpc.108.062414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fiorilli V, Catoni M, Miozzi L, Novero M, Accotto GP, Lanfranco L. Global and cell-type gene expression profiles in tomato plants colonized by an arbuscular mycorrhizal fungus. New Phytol. 2009;184:975–87. doi: 10.1111/j.1469-8137.2009.03031.x. [DOI] [PubMed] [Google Scholar]
- 10.Gherbi H, Markmann K, Svistoonoff S, Estevan J, Autran D, Giczey G, et al. SymRK defines a common genetic basis for plant root endosymbioses with arbuscular mycorrhiza fungi, rhizobia, and Frankiabacteria. Proc Natl Acad Sci U S A. 2008;105:4928–32. doi: 10.1073/pnas.0710618105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Heupel S, Roser B, Kuhn H, Lebrun MH, Villalba F, Requena N. Erl1, a novel era-like GTPase from Magnaporthe oryzae, is required for full root virulence and is conserved in the mutualistic symbiont Glomus intraradices. Mol Plant Microbe Interact. 2010;23:67–81. doi: 10.1094/MPMI-23-1-0067. [DOI] [PubMed] [Google Scholar]
- 12.Délano-Frier JP, Tejeda-Sartorius M. Unraveling the network: Novel developments in the understanding of signaling and nutrient exchange mechanisms in the arbuscular mycorrhizal symbiosis. Plant Signal Behav. 2008;3:936–44. doi: 10.4161/psb.6789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu J, Maldonado-Mendoza I, Lopez-Meyer M, Cheung F, Town CD, Harrison MJ. Arbuscular mycorrhizal symbiosis is accompanied by local and systemic alterations in gene expression and an increase in disease resistance in the shoots. Plant J. 2007;50:529–44. doi: 10.1111/j.1365-313X.2007.03069.x. [DOI] [PubMed] [Google Scholar]
- 14.Kistner C, Winzer T, Pitzschke A, Mulder L, Sato S, Kaneko T, et al. Seven Lotus japonicus genes required for transcriptional reprogramming of the root during fungal and bacterial symbiosis. Plant Cell. 2005;17:2217–29. doi: 10.1105/tpc.105.032714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bonfante P, Requena N. Dating in the dark: how roots respond to fungal signals to establish arbuscular mycorrhizal symbiosis. Curr Opin Plant Biol. 2011;14:451–7. doi: 10.1016/j.pbi.2011.03.014. [DOI] [PubMed] [Google Scholar]
- 16.Barker S, Stummer B, Gao L, Dispain I, O’Connor P, Smith S. A mutant in Lycopersicon esculentum Mill. with highly reduced VA mycorrhizal colonization: isolation and preliminary characterization. Plant J. 1998;15:791–7. doi: 10.1046/j.1365-313X.1998.00252.x. [DOI] [PubMed] [Google Scholar]
- 17.Gao LL, Delp G, Smith SE. Colonization patterns in a mycorrhiza-defective mutant tomato vary with different arbuscular-mycorrhizal fungi. New Phytol. 2001;151:477–91. doi: 10.1046/j.0028-646x.2001.00193.x. [DOI] [Google Scholar]
- 18.Harrison MJ. Signaling in the arbuscular mycorrhizal symbiosis. Annu Rev Microbiol. 2005;59:19–42. doi: 10.1146/annurev.micro.58.030603.123749. [DOI] [PubMed] [Google Scholar]
- 19.Larkan NJ, Smith SE, Barker SJ. Position of the reduced mycorrhizal colonisation (Rmc) locus on the tomato genome map. Mycorrhiza. 2007;17:311–8. doi: 10.1007/s00572-007-0106-9. [DOI] [PubMed] [Google Scholar]
- 20.Trouvelot A, Kough JL, Gianinazzi-Pearson V. Mesure du taux de mycorhization VAd‘un system radiculaire. Recherche de methods d’estimation ayant une signification fonctionnele. In: Gianinazzi PV, Gianinazzi S Eds. Mycorrhizae: Physiology and Genetics. Paris: INRA, 1986: 217–22. [Google Scholar]
- 21.Campos-Soriano L, García-Garrido JM, San Segundo B. Activation of basal defense mechanisms of rice plants by Glomus intraradices does not affect the arbuscular mycorrhizal symbiosis. New Phytol. 2010;188:597–614. doi: 10.1111/j.1469-8137.2010.03386.x. [DOI] [PubMed] [Google Scholar]
- 22.Tárraga S, Lisón P, López-Gresa MP, Torres C, Rodrigo I, Bellés JM, et al. Molecular cloning and characterization of a novel tomato xylosyltransferase specific for gentisic acid. J Exp Bot. 2010;61:4325–38. doi: 10.1093/jxb/erq234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ding X, Cao Y, Huang L, Zhao J, Xu C, Li X, et al. Activation of the indole-3-acetic acid-amido synthetase GH3-8 suppresses expansin expression and promotes salicylate- and jasmonate-independent basal immunity in rice. Plant Cell. 2008;20:228–40. doi: 10.1105/tpc.107.055657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gao LL, Knogge W, Delp G, Smith FA, Smith SE. Expression patterns of defense-related genes in different types of arbuscular mycorrhizal development in wild-type and mycorrhiza-defective mutant tomato. Mol Plant Microbe Interact. 2004;17:1103–13. doi: 10.1094/MPMI.2004.17.10.1103. [DOI] [PubMed] [Google Scholar]
- 25.Pozo MJ, Cordier C, Dumas-Gaudot E, Gianinazzi S, Barea JM, Azcón-Aguilar C. Localized versus systemic effect of arbuscular mycorrhizal fungi on defence responses to Phytophthora infection in tomato plants. J Exp Bot. 2002;53:525–34. doi: 10.1093/jexbot/53.368.525. [DOI] [PubMed] [Google Scholar]
- 26.McGrath KC, Dombrecht B, Manners JM, Schenk PM, Edgar CI, Maclean DJ, et al. Repressor- and activator-type ethylene response factors functioning in jasmonate signaling and disease resistance identified via a genome-wide screen of Arabidopsis transcription factor gene expression. Plant Physiol. 2005;139:949–59. doi: 10.1104/pp.105.068544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ivashuta S, Liu J, Liu J, Lohar DP, Haridas S, Bucciarelli B, et al. RNA interference identifies a calcium-dependent protein kinase involved in Medicago truncatula root development. Plant Cell. 2005;17:2911–21. doi: 10.1105/tpc.105.035394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bucher M. Functional biology of plant phosphate uptake at root and mycorrhiza interfaces. New Phytol. 2007;173:11–26. doi: 10.1111/j.1469-8137.2006.01935.x. [DOI] [PubMed] [Google Scholar]
- 29.Wright DP, Read DJ, Scholes JP. Mycorrhizal sink strength influences whole plant carbon balance of Trifolium repens L. Plant Cell Environ. 1998;21:881–91. doi: 10.1046/j.1365-3040.1998.00351.x. [DOI] [Google Scholar]
- 30.Garcia-Rodriguez S, Azcon-Aguilera C, Ferrol N. Transcriptional regulation of host enzymes involved in the cleavage of sucrose during arbuscular mycorrhizal symbiosis. Physiol Plant. 2007;129:737–46. doi: 10.1111/j.1399-3054.2007.00873.x. [DOI] [Google Scholar]
- 31.Harrison MJ, Dewbre GR, Liu JY. A phosphate transporter from Medicago truncatula involved in the acquisition of phosphate released by arbuscular mycorrhizal fungi. Plant Cell. 2002;14:2413–29. doi: 10.1105/tpc.004861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Baier MC, Keck M, Gödde V, Niehaus K, Küster H, Hohnjec N. Knockdown of the symbiotic sucrose synthase MtSucS1 affects arbuscule maturation and maintenance in mycorrhizal roots of Medicago truncatula. Plant Physiol. 2010;152:1000–14. doi: 10.1104/pp.109.149898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bonfante P, Genre A. Mechanisms underlying beneficial plant-fungus interactions in mycorrhizal symbiosis. Nat Commun. 2010;1:48. doi: 10.1038/ncomms1046. [DOI] [PubMed] [Google Scholar]
- 34.Ercolin F, Reinhardt D. Successful joint ventures of plants: arbuscular mycorrhiza and beyond. Trends Plant Sci. 2011;16:356–62. doi: 10.1016/j.tplants.2011.03.006. [DOI] [PubMed] [Google Scholar]
- 35.Arrighi JF, Barre A, Ben Amor B, Bersoult A, Soriano LC, Mirabella R, et al. The Medicago truncatula lysin [corrected] motif-receptor-like kinase gene family includes NFP and new nodule-expressed genes. Plant Physiol. 2006;142:265–79. doi: 10.1104/pp.106.084657. [DOI] [PMC free article] [PubMed] [Google Scholar]