Abstract
In plants, peroxisomes are the organelles involved in various metabolic processes and physiological functions including β-oxidation, mobilization of seed storage lipids, photorespiration, and hormone biosynthesis. We have recently shown that, in fungi and plants, peroxisomes play a vital role in biosynthesis of biotin, an essential cofactor required for various carboxylation and decarboxylation reactions. In fungi, the mutants defective in peroxisomal protein import exhibit biotin auxotrophy. The fungal BioF protein, a 7-keto-8-aminopelargonic acid (KAPA) synthase catalyzing the conversion of pimeloyl-CoA to KAPA in biotin biosynthesis, contains the peroxisomal targeting sequence 1 (PTS1), and its peroxisomal targeting is required for biotin biosynthesis. In plants, biotin biosynthesis is essential for embryo development. We have shown that the peroxisomal targeting sequences of the BioF proteins are conserved throughout the plant kingdom, and the Arabidopsis thaliana BioF protein is indeed localized in peroxisomes. Our findings suggest that peroxisomal localization of the BioF protein is evolutionarily conserved among eukaryotes, and required for biotin biosynthesis and plant growth and development.
Keywords: peroxisome, biotin, mitochondria, embryo development, fungi
The roles of Peroxisomes and the Molecular Mechanisms for Peroxisomal Protein Import in Plants
Peroxisomes are the eukaryotic organelles involved in various metabolic processes including fatty acid β-oxidation and secondary metabolisms. In plants, peroxisomes change their functions in response to developmental stages and environmental conditions. In germinating seedlings, peroxisomes act as glyoxysomes that execute glyoxylate cycle for storage lipid mobilization.1 During seedling transition, peroxisomes degrade enzymes required for the glyoxylate cycle and accumulate enzymes involved in photorespiration.2 In leaf, peroxisomes share metabolic processes with chloroplast and mitochondria to exhibit photorespiration.3 Peroxisomes are also required for biosynthesis of plant hormones including jasmonic acid and auxin.4,5 Loss of peroxisomal functions causes embryo lethality in plants, suggesting the importance of peroxisomes in plant development.6-10
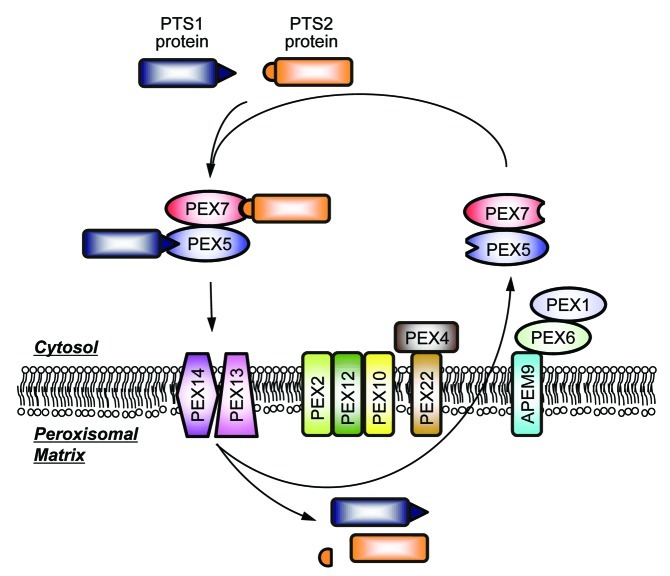
Figure 1. shows a schematic diagram of peroxisomal matrix protein import in plants. Peroxisomal matrix proteins are delivered from cytosol into peroxisomes by a series of the proteins called peroxins (PEX). There are two types of the peroxisomal targeting signals (PTS) for the import of peroxisomal matrix proteins: PTS1, a conserved tripeptide motif at the C-terminus, and PTS2, a nonapeptide with the motif R-L-X5-H-L typically located within 30 amino acids from the N-terminus. PTS1 and PTS2-containing proteins are recognized by the receptors PEX5 and PEX7, respectively, which interact with each other and import the proteins into peroxisomal matrix.11-14 The PEX5/7-PTS protein complexes are docked on the peroxisomal membrane via binding with the PEX13 and PEX14.11,15 In the peroxisomal matrix, PTS2 is removed by the activities of the proteases DEG1516,17 and LON2.18 After the translocation into peroxisomal matrix, the receptors appear to be relocated to cytosol for another round of the import process.19 It is hypothesized that PEX4, a cytosolic ubiquitin-conjugating enzyme, and the ubiquitin ligases PEX2, PEX10, and PEX12 cooperatively promote the receptor recycling by monoubiquitination of PEX5.20 PEX4 is anchored to the peroxisomal membrane by interaction with PEX22.21,22 Two AAA ATPases PEX1 and PEX6, which are tethered to peroxisome via interaction with ABERRANT PEROXISOME MORPHOLOGY9 (APEM9),23 are also likely to mediate relocation of the receptors to cytosol.24 PEX4, PEX5, PEX6, and PEX22 are also known to be involved in selective degradation of the glyoxylate cycle enzymes.25
Figure 1. Schematic diagrams of the peroxisomal matrix protein import.
Involvement of Peroxisomes in Biotin Biosynthesis in Fungi
In fungi, peroxisomes are essential for various metabolic processes and physiological functions. Their primary role is β-oxidation of fatty acids: the mutants lacking peroxisomes fail to grow on the minimal medium containing fatty acids as carbon source in the budding yeast Saccharomyces cerevisiae.26 In the filamentous fungus Aspergillus nidulans, similar observations were reported in the studies of the mutants lacking the peroxin genes.27 Peroxisomes are also required for methanol metabolism in the methylotrophic yeasts including Pichia pastoris.28 In filamentous fungi, peroxisomes are involved in secondary metabolism including penicillin biosynthesis,29 plant pathogenicity,30,31 sexual development,32,33 and formation of the Woronin body, an organelle for wound healing.34,35 We have reported that the Pex11 protein, which is required for peroxisomal division, is involved in the formation of Woronin body from peroxisomes in the filamentous fungus Aspergillus oryzae.35 To further clarify the role of peroxisomal proteins in the Woronin body formation, we generated Aspergillus mutants of which the Aopex5 and Aopex7 genes are disrupted. They are defective in protein import into the peroxisomal matrix due to the lack of the PTS1 and PTS2 receptors, respectively.36 These gene disruptants failed to grow on the minimal medium containing oleic acid as the sole carbon source due to the defects in peroxisomal β-oxidation. Unexpectedly, we found that they also exhibit growth deficiency on the minimal medium containing glucose as the sole carbon source, which is restored by addition of biotin and its biosynthetic precursors.36 It has been suggested that, in fungi, biotin is synthesized through the sequential activities of the Bio proteins: BioF, a 7-keto-8-aminopelargonic acid (KAPA) synthase catalyzing the conversion of pimeloyl-CoA to KAPA, BioD/A, a chimeric protein of a 7, 8-diaminopelargonic acid (DAPA) synthase and a desthiobiotin (DTB) synthase which converts KAPA to DTB, and BioB, a biotin synthase catalyzing the conversion of DTB to biotin.36-38 We have shown mitochondrial localization of the Aspergillus BioD/A protein, suggesting that mitochondria are the site for the conversion of KAPA to biotin.36 The Aspergillus BioF protein possesses the PTS1 sequence at the C terminus, and its peroxisomal targeting is required for biotin biosynthesis.36 Our study suggests that peroxisomes are functionally coupled with mitochondria and involved in biotin biosynthesis.
Involvement of Biotin Biosynthesis in Plant Embryo Development
Plants are the other eukaryotic organisms that synthesize biotin. Molecular genetic studies using the Arabidopsis mutants auxotrophic for biotin have revealed that biotin biosynthesis is vital for plant growth and development. A forward genetic screen isolated an embryo-lethal mutant, bio1–1, of which embryogenesis is arrested during globular to mature-cotyledon stages.39,40 The arrested embryo of the bio1–1 mutant plant accumulates virtually no detectable biotin,41 and is rescued by the exogenous addition of biotin as well as DTB.39 The bio1–1 mutant phenotype was complemented by a transgene containing the Escherichia coli bioA gene, indicating that the BIO1 gene encodes a DAPA synthase.42 In the Arabidopsis genome, the BIO1 gene is positioned immediately downstream of the BIO3 gene encoding a DTB synthase.43 Genetic complementation analysis revealed that the bio1 and bio3 mutants define a single locus which exhibits a chimeric gene transcript.43 A subsequent study demonstrated that this locus produces a chimeric protein of BIO3 and BIO1, which has a bifunctional enzyme activity catalyzing both of DTB synthase and DAPA synthase reactions.44 The BIO2 gene encodes a biotin synthase, which catalyzes the final step of biotin biosynthesis, and the bio2 mutation causes embryo lethality.40,45 While the bio1 mutant embryos abort at the torpedo or later stages, more than 50% of the bio2 mutant embryos abort at the early globular stage, implying that the bio2 mutation has more severe impact on embryo development than that of the bio1 mutation.40 These observations consistently suggest that biotin biosynthesis plays a vital role during embryo development in plants.
Recent studies have demonstrated that, in plants, the final three reactions of the biotin synthesis are likely to occur in mitochondria. The BIO2 protein contains a mitochondrial targeting sequence (MTS) at its N terminus.45-49 Biochemical studies showed that the recombinant BIO2 protein produced in E. coli is localized in the mitochondrial fraction from pea (Pisum sativum) leaf extracts,48 and its biotin synthase activity is detectable only in the mitochondrial fraction from pea leaf and potato (Solanum tuberosum) extracts.49 A molecular genetic study demonstrated that a truncated version of the BIO2 protein lacking MTS was not able to complement the bio2 mutant phenotype, regardless of exogenously added DTB, a substrate of the biotin synthase, suggesting that mitochondrial localization, rather than substrate availability, is required for the BIO2 activity.45 Recent studies showed that the BIO3-BIO1 fusion protein contains MTS at its N terminus,43,44 and biochemical analysis and fluorescent protein-based microscopic observations demonstrated its mitochondrial localization.44 These observations suggest that mitochondria are the main sites for a series of the reactions converting KAPA to biotin in plants.
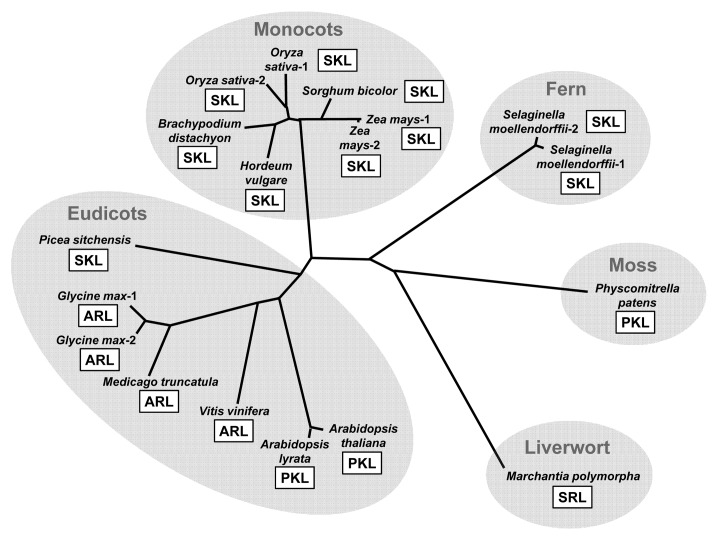
In contrast to the other three enzymes, the KAPA synthase has been less characterized in plants. Pinon et al. (2005) showed that the AtBioF gene encodes an ortholog of the bacterial KAPA synthase, which is able to complement an E. coli mutant deficient in KAPA synthase activity.50 They also demonstrated that the recombinant AtBioF protein purified from E. coli extracts has the catalytic properties of KAPA synthase. Using the AtBioF protein of which C terminus is fused to GFP, they concluded AtBioF as a cytosolic enzyme.50 We found that the AtBioF protein contains a PTS1 sequence at its C terminus.36 While the C-terminal fusion of GFP allowed AtBioF to be cytosolic as shown in the previous study, the N-terminal fusion of GFP showed co-localization of AtBioF with a fluorescent peroxisomal marker, and the PTS1-deleted version of AtBioF was diffused throughout the cytoplasm, suggesting that AtBioF is localized to peroxisome by the activity of its PTS1 sequence.36 In addition, our phylogenetic analysis revealed that the orthologs of AtBioF from various plant species possess the PTS1 sequences at their C termini (Fig. 2), suggesting that the peroxisomal localization of the BioF protein is conserved throughout the plant kingdom. Our findings suggest that the plant BioF protein is an evolutionally conserved peroxisomal enzyme. The reaction product KAPA is likely to be transported from peroxisome to mitochondria as a substrate for the subsequent series of the biotin biosynthesis reactions. Thus, metabolic interaction between peroxisomes and mitochondria is required for biotin biosynthesis in plants, which in turn may play a vital role during plant embryo development.
Figure 2. Phylogenetic relationship of the plant KAPA synthases. The amino acid residues of their C-terminal peroxisomal targeting signals (PTS1) are also shown with open squares. The full-length amino acid sequences of the plant KAPA synthases were aligned using Clustal W program (version 2.1) and the unrooted phylogenetic tree was constructed using the neighbor-joining method. The GenBank accession numbers for the sequences retrieved are as follows: Arabidopsis thaliana, NP_974731.1; Arabidopsis lyrata, XP_002871105.1; Oryza sativa-1, BAD87813.1; Oryza sativa-2, NP_001065381.1; Hordeum vulgare, BAK03504.1; Brachypodium distachyon, XP_003574335.1; Sorgham bicolor, XP_002467492.1; Zea mays-1, ACG35792.1; Zea mays-2, ACG35881.1; Selaginella moellendorffii-1, XP_002969752.1; Selaginella moellendorffii-2, XP_002981364.1; Physcomitrella patens, XP_001769874.1; Picea sitchensis, ABR18106.1; Vinis vinifera, XP_002268950.1; Medicago truncatula, XP_003598166.1; Glycine max-1, XP_003527547.1; Glycine max-2, XP_003522881.1. The peptide sequence of the putative Marchantia polymorpha KAPA synthase was confirmed by PCR amplification and sequencing of its cDNA based on the information from the Marchantia expression sequence tag database (S. Yamaoka, unpublished data).
Conclusion and Perspective
We proposed a new model for the biotin biosynthesis in plants (Fig. 3). In peroxisomes, KAPA is synthesized from pimeloyl-CoA by the BioF protein, and transported to mitochondria. Then, KAPA is converted to biotin by the BIO3-BIO1 and BIO2 proteins in mitochondria. By analogy with the fungal system,36 the production of pimeloyl-CoA may involve the proteins containing PTS1 and PTS2. A recent fungal study also implies that peroxisomal β-oxidation is involved in biotin biosynthesis.38
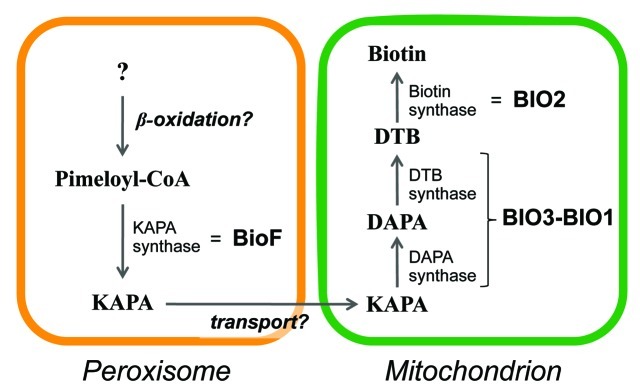
Figure 3. A model of the biotin biosynthetic pathway in plants. KAPA, 7-keto-8-aminopelargonic acid; DAPA, 7, 8-diaminopelargonic acid; DTB, desthiobiotin.
In plants, embryo development requires biotin biosynthesis9,40,43 as well as peroxisomal functions.6-10 Our study has recently revealed that both fungi and plants use the evolutionally conserved pathway for biotin biosynthesis which is shared with peroxisomes and mitochondria.36 These observations may imply that biotin biosynthesis is one of the major functions of peroxisomes during plant embryo development. Future studies based on the analogy between fungi and plants may give further insights on the importance of biotin biosynthesis and peroxisomal functions during plant growth and development.
Acknowledgments
We thank Drs. Kimitsune Ishizaki and Takayuki Kohchi (Kyoto University, Japan) for providing cDNA and sequence informations of Marchantia. This study was supported by Grant-in-Aid for Young Scientist from the Japan Society for the Promotion of Science (to J.M. and S.Y.).
Footnotes
Previously published online: www.landesbioscience.com/journals/psb/article/22405
References
- 1.Mano S, Nishimura M. Plant peroxisomes. Vitam Horm. 2005;72:111–54. doi: 10.1016/S0083-6729(05)72004-5. [DOI] [PubMed] [Google Scholar]
- 2.Nishimura M, Hayashi M, Kato A, Yamaguchi K, Mano S. Functional transformation of microbodies in higher plant cells. Cell Struct Funct. 1996;21:387–93. doi: 10.1247/csf.21.387. [DOI] [PubMed] [Google Scholar]
- 3.Reumann S, Weber AP. Plant peroxisomes respire in the light: some gaps of the photorespiratory C2 cycle have become filled–others remain. Biochim Biophys Acta 2006; 1763:1496-510. [DOI] [PubMed]
- 4.Weber H. Fatty acid-derived signals in plants. Trends Plant Sci. 2002;7:217–24. doi: 10.1016/S1360-1385(02)02250-1. [DOI] [PubMed] [Google Scholar]
- 5.Woodward AW, Bartel B. Auxin: regulation, action, and interaction. Ann Bot. 2005;95:707–35. doi: 10.1093/aob/mci083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hu J, Aguirre M, Peto C, Alonso J, Ecker J, Chory J. A role for peroxisomes in photomorphogenesis and development of Arabidopsis. Science. 2002;297:405–9. doi: 10.1126/science.1073633. [DOI] [PubMed] [Google Scholar]
- 7.Schumann U, Wanner G, Veenhuis M, Schmid M, Gietl C. AthPEX10, a nuclear gene essential for peroxisome and storage organelle formation during Arabidopsis embryogenesis. Proc Natl Acad Sci U S A. 2003;100:9626–31. doi: 10.1073/pnas.1633697100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sparkes IA, Brandizzi F, Slocombe SP, El-Shami M, Hawes C, Baker A. An Arabidopsis pex10 null mutant is embryo lethal, implicating peroxisomes in an essential role during plant embryogenesis. Plant Physiol. 2003;133:1809–19. doi: 10.1104/pp.103.031252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tzafrir I, Pena-Muralla R, Dickerman A, Berg M, Rogers R, Hutchens S, et al. Identification of genes required for embryo development in Arabidopsis. Plant Physiol. 2004;135:1206–20. doi: 10.1104/pp.104.045179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fan J, Quan S, Orth T, Awai C, Chory J, Hu J. The Arabidopsis PEX12 gene is required for peroxisome biogenesis and is essential for development. Plant Physiol. 2005;139:231–9. doi: 10.1104/pp.105.066811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nito K, Hayashi M, Nishimura M. Direct interaction and determination of binding domains among peroxisomal import factors in Arabidopsis thaliana. Plant Cell Physiol. 2002;43:355–66. doi: 10.1093/pcp/pcf057. [DOI] [PubMed] [Google Scholar]
- 12.Hayashi M, Yagi M, Nito K, Kamada T, Nishimura M. Differential contribution of two peroxisomal protein receptors to the maintenance of peroxisomal functions in Arabidopsis. J Biol Chem. 2005;280:14829–35. doi: 10.1074/jbc.M411005200. [DOI] [PubMed] [Google Scholar]
- 13.Woodward AW, Bartel B. The Arabidopsis peroxisomal targeting signal type 2 receptor PEX7 is necessary for peroxisome function and dependent on PEX5. Mol Biol Cell. 2005;16:573–83. doi: 10.1091/mbc.E04-05-0422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Khan BR, Zolman BK. pex5 Mutants that differentially disrupt PTS1 and PTS2 peroxisomal matrix protein import in Arabidopsis. Plant Physiol. 2010;154:1602–15. doi: 10.1104/pp.110.162479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mano S, Nakamori C, Nito K, Kondo M, Nishimura M. The Arabidopsis pex12 and pex13 mutants are defective in both PTS1- and PTS2-dependent protein transport to peroxisomes. Plant J. 2006;47:604–18. doi: 10.1111/j.1365-313X.2006.02809.x. [DOI] [PubMed] [Google Scholar]
- 16.Helm M, Lück C, Prestele J, Hierl G, Huesgen PF, Fröhlich T, et al. Dual specificities of the glyoxysomal/peroxisomal processing protease Deg15 in higher plants. Proc Natl Acad Sci U S A. 2007;104:11501–6. doi: 10.1073/pnas.0704733104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schuhmann H, Huesgen PF, Gietl C, Adamska I. The DEG15 serine protease cleaves peroxisomal targeting signal 2-containing proteins in Arabidopsis. Plant Physiol. 2008;148:1847–56. doi: 10.1104/pp.108.125377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lingard MJ, Bartel B. Arabidopsis LON2 is necessary for peroxisomal function and sustained matrix protein import. Plant Physiol. 2009;151:1354–65. doi: 10.1104/pp.109.142505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ratzel SE, Lingard MJ, Woodward AW, Bartel B. Reducing PEX13 expression ameliorates physiological defects of late-acting peroxin mutants. Traffic. 2011;12:121–34. doi: 10.1111/j.1600-0854.2010.01136.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hu J, Baker A, Bartel B, Linka N, Mullen RT, Reumann S, et al. Plant peroxisomes: biogenesis and function. Plant Cell. 2012;24:2279–303. doi: 10.1105/tpc.112.096586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zolman BK, Monroe-Augustus M, Silva ID, Bartel B. Identification and functional characterization of Arabidopsis PEROXIN4 and the interacting protein PEROXIN22. Plant Cell. 2005;17:3422–35. doi: 10.1105/tpc.105.035691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nito K, Kamigaki A, Kondo M, Hayashi M, Nishimura M. Functional classification of Arabidopsis peroxisome biogenesis factors proposed from analyses of knockdown mutants. Plant Cell Physiol. 2007;48:763–74. doi: 10.1093/pcp/pcm053. [DOI] [PubMed] [Google Scholar]
- 23.Goto S, Mano S, Nakamori C, Nishimura M. Arabidopsis ABERRANT PEROXISOME MORPHOLOGY9 is a peroxin that recruits the PEX1-PEX6 complex to peroxisomes. Plant Cell. 2011;23:1573–87. doi: 10.1105/tpc.110.080770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zolman BK, Bartel B. An Arabidopsis indole-3-butyric acid-response mutant defective in PEROXIN6, an apparent ATPase implicated in peroxisomal function. Proc Natl Acad Sci U S A. 2004;101:1786–91. doi: 10.1073/pnas.0304368101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lingard MJ, Monroe-Augustus M, Bartel B. Peroxisome-associated matrix protein degradation in Arabidopsis. Proc Natl Acad Sci U S A. 2009;106:4561–6. doi: 10.1073/pnas.0811329106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Erdmann R, Veenhuis M, Mertens D, Kunau WH. Isolation of peroxisome-deficient mutants of Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1989;86:5419–23. doi: 10.1073/pnas.86.14.5419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hynes MJ, Murray SL, Khew GS, Davis MA. Genetic analysis of the role of peroxisomes in the utilization of acetate and fatty acids in Aspergillus nidulans. Genetics. 2008;178:1355–69. doi: 10.1534/genetics.107.085795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van der Klei IJ, Yurimoto H, Sakai Y, Veenhuis M. The significance of peroxisomes in methanol metabolism in methylotrophic yeast. Biochim Biophys Acta 2006; 1763:1453-62. [DOI] [PubMed]
- 29.Bartoszewska M, Opaliński L, Veenhuis M, van der Klei IJ. The significance of peroxisomes in secondary metabolite biosynthesis in filamentous fungi. Biotechnol Lett. 2011;33:1921–31. doi: 10.1007/s10529-011-0664-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kimura A, Takano Y, Furusawa I, Okuno T. Peroxisomal metabolic function is required for appressorium-mediated plant infection by Colletotrichum lagenarium. Plant Cell. 2001;13:1945–57. doi: 10.1105/TPC.010084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Asakura M, Okuno T, Takano Y. Multiple contributions of peroxisomal metabolic function to fungal pathogenicity in Colletotrichum lagenarium. Appl Environ Microbiol. 2006;72:6345–54. doi: 10.1128/AEM.00988-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bonnet C, Espagne E, Zickler D, Boisnard S, Bourdais A, Berteaux-Lecellier V. The peroxisomal import proteins PEX2, PEX5 and PEX7 are differently involved in Podospora anserina sexual cycle. Mol Microbiol. 2006;62:157–69. doi: 10.1111/j.1365-2958.2006.05353.x. [DOI] [PubMed] [Google Scholar]
- 33.Peraza-Reyes L, Zickler D, Berteaux-Lecellier V. The peroxisome RING-finger complex is required for meiocyte formation in the fungus Podospora anserina. Traffic. 2008;9:1998–2009. doi: 10.1111/j.1600-0854.2008.00812.x. [DOI] [PubMed] [Google Scholar]
- 34.Liu F, Ng SK, Lu Y, Low W, Lai J, Jedd G. Making two organelles from one: Woronin body biogenesis by peroxisomal protein sorting. J Cell Biol. 2008;180:325–39. doi: 10.1083/jcb.200705049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Escaño CS, Juvvadi PR, Jin FJ, Takahashi T, Koyama Y, Yamashita S, et al. Disruption of the Aopex11-1 gene involved in peroxisome proliferation leads to impaired Woronin body formation in Aspergillus oryzae. Eukaryot Cell. 2009;8:296–305. doi: 10.1128/EC.00197-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tanabe Y, Maruyama J, Yamaoka S, Yahagi D, Matsuo I, Tsutsumi N, et al. Peroxisomes are involved in biotin biosynthesis in Aspergillus and Arabidopsis. J Biol Chem. 2011;286:30455–61. doi: 10.1074/jbc.M111.247338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Magliano P, Flipphi M, Sanglard D, Poirier Y. Characterization of the Aspergillus nidulans biotin biosynthetic gene cluster and use of the bioDA gene as a new transformation marker. Fungal Genet Biol. 2011;48:208–15. doi: 10.1016/j.fgb.2010.08.004. [DOI] [PubMed] [Google Scholar]
- 38.Magliano P, Flipphi M, Arpat BA, Delessert S, Poirier Y. Contributions of the peroxisome and β-oxidation cycle to biotin synthesis in fungi. J Biol Chem. 2011;286:42133–40. doi: 10.1074/jbc.M111.279687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schneider T, Dinkins R, Robinson K, Shellhammer J, Meinke DW. An embryo-lethal mutant of Arabidopsis thaliana is a biotin auxotroph. Dev Biol. 1989;131:161–7. doi: 10.1016/S0012-1606(89)80047-8. [DOI] [PubMed] [Google Scholar]
- 40.Patton DA, Schetter AL, Franzmann LH, Nelson K, Ward ER, Meinke DW. An embryo-defective mutant of arabidopsis disrupted in the final step of biotin synthesis. Plant Physiol. 1998;116:935–46. doi: 10.1104/pp.116.3.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shellhammer J, Meinke DW. Arrested embryos from the bio1 auxotroph of Arabidopsis contain reduced levels of biotin. Plant Physiol. 1990;93:1162–7. doi: 10.1104/pp.93.3.1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Patton DA, Volrath S, Ward ER. Complementation of an Arabidopsis thaliana biotin auxotroph with an Escherichia coli biotin biosynthetic gene. Mol Gen Genet. 1996;251:261–6. doi: 10.1007/BF02172516. [DOI] [PubMed] [Google Scholar]
- 43.Muralla R, Chen E, Sweeney C, Gray JA, Dickerman A, Nikolau BJ, et al. A bifunctional locus (BIO3-BIO1) required for biotin biosynthesis in Arabidopsis. Plant Physiol. 2008;146:60–73. doi: 10.1104/pp.107.107409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cobessi D, Dumas R, Pautre V, Meinguet C, Ferrer JL, Alban C. Biochemical and structural characterization of the Arabidopsis bifunctional enzyme dethiobiotin synthetase-diaminopelargonic acid aminotransferase: evidence for substrate channeling in biotin synthesis. Plant Cell. 2012;24:1608–25. doi: 10.1105/tpc.112.097675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Arnal N, Alban C, Quadrado M, Grandjean O, Mireau H. The Arabidopsis Bio2 protein requires mitochondrial targeting for activity. Plant Mol Biol. 2006;62:471–9. doi: 10.1007/s11103-006-9034-x. [DOI] [PubMed] [Google Scholar]
- 46.Patton DA, Johnson M, Ward ER. Biotin synthase from Arabidopsis thaliana. cDNA isolation and characterization of gene expression. Plant Physiol. 1996;112:371–8. doi: 10.1104/pp.112.1.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Weaver LM, Yu F, Wurtele ES, Nikolau BJ. Characterization of the cDNA and gene coding for the biotin synthase of Arabidopsis thaliana. Plant Physiol. 1996;110:1021–8. doi: 10.1104/pp.110.3.1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Baldet P, Alban C, Douce R. Biotin synthesis in higher plants: purification of bioB gene product equivalent from Arabidopsis thaliana overexpressed in Escherichia coli and its subcellular localization in pea leaf cells. FEBS Lett. 1997;419:206–10. doi: 10.1016/S0014-5793(97)01458-0. [DOI] [PubMed] [Google Scholar]
- 49.Picciocchi A, Douce R, Alban C. Biochemical characterization of the Arabidopsis biotin synthase reaction. The importance of mitochondria in biotin synthesis. Plant Physiol. 2001;127:1224–33. doi: 10.1104/pp.010346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pinon V, Ravanel S, Douce R, Alban C. Biotin synthesis in plants. The first committed step of the pathway is catalyzed by a cytosolic 7-keto-8-aminopelargonic acid synthase. Plant Physiol. 2005;139:1666–76. doi: 10.1104/pp.105.070144. [DOI] [PMC free article] [PubMed] [Google Scholar]