Abstract
From a holistic perspective, the discovery of cellular plasticity, a very interesting property of totipotency, underlies many topical issues in biology with important medical applications, while transgenesis is a core research tool in biology. Partially known, some basic mechanisms involved in the regenerative property of cells and in their receptivity to transgenesis are common to plant and animal cells and highlight the principle of the unity of life. Transgenesis provides an important investigative instrument in plant physiology and is regarded as a valuable tool for crop improvement. The economic, social, cultural and scientific importance of cereals has led to a rich stream of research into their genetics, biology and evolution. Sustained efforts to achieve the results obtained in the fields of genetic engineering and applied biotechnology reflect this deep interest. Difficulties encountered in creating genetically modified cereals, especially wheat, highlighted the central notions of tissue culture regeneration and transformation competencies. From the perspective of combining or encountering these competencies in the same cell lineage, this reputedly recalcitrant species provides a stimulating biological system in which to explore the physiological and genetic complexity of both competencies. The former involves two phases, dedifferentiation and redifferentiation. Cells undergo development switches regulated by extrinsic and intrinsic factors. The re-entry into the cell division cycle progressively culminates in the development of organized structures. This is achieved by global chromatin reorganization associated with the reprogramming of the gene expression pattern. The latter is linked with surveillance mechanisms and DNA repair, aimed at maintaining genome integrity before cells move into mitosis, and with those mechanisms aimed at genome expression control and regulation. In order to clarify the biological basis of these two physiological properties and their interconnectedness, we look at both competencies at the core of defense/adaptive mechanisms and survival, between undifferentiated cell proliferation and organization, constituting a transition phase between two different dynamic regimes, a typical feature of critical dynamic systems. Opting for a candidate-gene strategy, several gene families could be proposed as relevant targets for investigating this hypothesis at the molecular level.
Keywords: adaptive and survival mechanisms, critical dynamic system, chromatin remodeling, developmental switch, gene expression reprogramming, gene transfer, regeneration and transformation competencies, totipotency
Introduction
What lies behind the ability of some organisms to rebuild themselves after injury? How does a cell give rise to a tissue, an organ or a whole individual? This question was selected by Science as one of 25 major puzzles that scientific research will face the next quarter century: a fundamental broad-ranging issue that will affect other scientific disciplines and society. This discovery of cellular plasticity has significance in cell therapy or regenerative medicine.1-4
Existing in only some animal cell types, the property of developmental plasticity is remarkable in plants: a complete organism can be constructed starting from a single somatic cell. This regenerative capacity of plant cells is similar to that of animal cells. At its core is the cellular dedifferentiation process, “the withdrawal of cells from a given differentiated state into a ‘stem cell’-like state that confers pluripotency”2,3,5.
The regenerative process has been widely applied in plants over the past 50 y, although the fundamentals are still largely unknown. The plant cell can provide an experimental model to help developmental biologists learn “how cell fate is fixed during the development process” and then “how the differentiated cell can preserve flexibility for development”2,5,6.
Somatic embryogenesis is a plant-specific phenomenon during which somatically differentiated plant cells regain totipotency and develop into embryos, representing the predominant means of plant regeneration in many species. Although somatic embryogenesis is widely used for in vitro plant propagation and has become the preferred method for regenerating most commercially cultivated biotech crops, little is known of the biological background of this process.7,8
Gene transfer technology is an important and additional tool in basic scientific research. The literature has numerous examples showing that transgenesis is a core research tool in biology, notably in allowing the direct testing of hypotheses in many aspects of plant physiology, plant development and basic functions, as well as the plant’s interactions with its environment, its adaptive behavior and its defense strategies. Transgenesis also provides a powerful tool for decoding the function of genes, and facilitating their incorporation in biotechnology program development. Genetically engineered crops represent an option with a range of possibilities for increasing productivity, diversification and the development of more sustainable agriculture.8-12
As stated by FAO, “Wheat remains the last big staple crop without approved genetically modified traits”13, “partly because its genetic engineering is technically harder than most other plants and partly because of political opposition”14. The acceptance of biotech wheat or genetically modified wheat has changed over the past years in the US and the political support for producing wheat in this way is growing globally13. The first biotech wheat is expected to be ready for commercialization by about 2017.15 Considerable progress has been achieved in the past decade, but the current capacity to transform Triticeae species lags behind that of other model plants, such as Arabidopsis thaliana or rice. The full potential of genetic transformation for advancing genetic understanding and driving crop improvement in the Triticeae has yet to be realized.16
For in vitro culture-based transformation methods, amenability to the culture stages required for gene transfer, selection and plant regeneration is the determinant of genetic transformation efficiency, which is controlled by intrinsic and external factors. This article describes the progress in understanding the regenerative property of plant cells and their receptivity to transgenesis. It explains how adaptive behaviors and cell survival mechanisms are involved, giving new insight into these competencies, as a response of a critical dynamic system. Based on these concepts, with wheat as the observed biological system, somatic embryogenesis as the regenerative pathway and particle bombardment as the gene transfer strategy, we propose a molecular approach to these processes, with special emphasis on a gene family historically related to developmental switch and now regarded as a key family in the life strategy in monocot.
Wheat as a Research Case Study
Whether conducting basic research, or focusing on the introduction of transgenes to produce improved genotypes with agronomic or quality traits, it is essential to be able to generate a large number of transgenic plants from almost any genotype. The production of large populations of transgenic plants is especially needed for high-throughput functional analysis, particularly for a species such as bread wheat, a plant with a huge genome (i.e., 5, 8 and 40 times the size of the genome, in Mb, of humans, maize and rice, respectively.17,18
As a plant of major economic and social importance, wheat has long been a central concern for many biotechnologists. But the genetic transformation protocol was difficult to define. Monocotyledonous species have long been described as recalcitrant in terms of in vitro regeneration. Compared with the two other major cereals, rice and maize, wheat appeared even more refractory.19
Considerable improvements in wheat transformation have been made in the past decade. But efficiency lags behind that of rice, which is ahead in terms of the development of reliable and robust transformation methods.20 The progress in obtaining very powerful methods for rice led to tools being developed likely to revolutionize plant basic research and applied biotechnology for this crop (Table 1).
Table 1. Comparison of transformation achievements in rice and wheat crops: Milestones in terms of efficiency and realizations.
| Efficiency of transformation technology from initial development to successful outcome | |||
|---|---|---|---|
| Rice | Wheat | ||
| Shortly after the development of transformation technology, efficiency (22%) was reported to be as high as that reported in dicots 21, 108-110. Although there are some varietal differences, yields increased by several hundred percent 24, 33, 111-113. |
Since the pioneering work in this area, there has been significant progress in the methodology, but it is still confined mainly to a few responsive varieties, with efficiency ranging from less that 1% to 10%; high throughput transformation channels, such those developed for rice, are not yet available 22-25, 114, 115. |
||
|
Functional genomics About 10,000 and 100,000 independent lines were generated for large-scale applications, such as T-DNA insertion mutagenesis and functional genomics, respectively 26, 27. RNAi silencing The first commercial improved cultivar produced by RNAi technology sought to reduce the level of undesirable metabolites to a value not yet achieved using conventional methods. The low glutenin content rice variety was useful for patients with kidney disease whose protein intake is restricted and who are unable to digest glutenin 28, 29. Gene targeting (GT) and genome editing A high-throughput Agrobacterium-mediated transformation model was essential for the development of GT in rice and has opened the door to the development of this technology for crop plants31-33 . Some 12,000 FOX (Full-length cDNA Over-eXpresser) lines were generated for analyzing gain-of-function phenotypes from large populations of transgenic plants overexpressing cDNAs of interest and others with unknown functions in rice30 . |
Functional genomics For crops where gene transfer efficiency and subsequent plant regeneration remain a limitation, a nontransgenic method for reverse genetics, called TILLING (Targeting Induced Local Lesions IN Genomes) has been developed. TILLING has been extended to the improvement of crops such as wheat and is seen as a useful general method for both functional genomics and the modulation of key traits116 . For functional genomics projects, it has been suggested that barley be used as a model for wheat due to its highly efficient transformation rates and smaller, less complex genome22 . RNAi silencing Several wheat studies aimed at improving human health through its resistant starch content or allergens have been conducted. High-amylose lines were generated using RNAi via downregulation of the two different isoforms of starch-branching enzyme, SBEIIa and SBEIIb. Improved indices of large-bowel health were demonstrated in rats28, 117. Reduced gliadin (gluten proteins associated with the development of celiac disease) was obtained in the ‘Bobwhite’ model cultivar118. Gene targeting (GT) and genome editing GT of a particular genomic location has been shown to be applicable to regenerable hexaploid wheat cells119 , but further advances are required before it can be routinely used22. |
||
1FOX: This gene-hunting system is a transgenic procedure that uses a normalized full-length cDNA collection under the control of the constitutive CaMV35S promoter; it offers a technique for identifying new gain-of-function in phenotypes. TILLING: This is a nontransgenic method for reverse genetics that uses molecular biology and genomics to identify point mutations in selected gene(s) amplified from a mutagenized population, using high-throughput detection platforms such as slab gel electrophoresis, capillary electrophoresis or dHPLC
Shortly after the development of DNA transfer technology for rice, efficiency was reported to be as high as that reported for dicots.21 As shown in recent reviews of cereal transformation work between the mid-1990s and the mid-2000s, rice transformation efficiency progressed from a few percent to several hundreds of percent, whereas it has progressed to a much lower extent in the case of wheat, reaching at best some 20%. In the past 5 y, work in this area has involved both biolistic particle and Agrobacterium-mediated wheat transformation methods, using immature embryos as explants as well as other types of explants, such as mature embryos, anther-derived calli, inflorescences, apical meristems and other floral organs. The results obtained and the ongoing work aimed at improving them further demonstrate the challenge that wheat represents. As a consequence of all these efforts, transformation efficiency continues to improve, but the need for an ideal transformation system remains (i.e., very effective, simple to perform, inexpensive, genotype-independent, and producing the required transgene expression) (see reviews in refs. 22–25, although even by the early 2000s generating a large number of rice T-DNA lines was no longer a limiting factor. Performances reached for rice transformation allowed multiple applications: generation of T-DNA insertion,26,27 RNA interferences28,29 and FOX libraries,30 as well as gene-targeting studies.31-33
Another obstacle to large-scale genetic manipulation is that there are significant differences among genotypes, with implications for efficiency and an accessible germplasm pool. Efforts are often needed to develop transformation protocols on a genotype-by-genotype basis. Genotypes that are recalcitrant in terms of tissue culture therefore have low transformation efficiencies. Despite comparable tissue culture performances, however, variable competencies for wheat transformation have been observed.34-36
Plant Biotechnology: In vitro Culture-Based Transformation Methods
Plant biotechnology is founded on the principles of cellular totipotency. With few exceptionsa, plant genetic engineering requires the use of tissue cultures (aseptic culture of cells, tissues or organs), the molecular biology tools along with the technology applied for gene transfer into the plant genomic DNA, and the ability to regenerate whole plants from the genetically modified cells, through ‘organogenesis’ (shoot induction followed by rhizogenesis) or ‘somatic embryogenesis’ (embryo formation from non-zygotic cell). In the latter case, somatic embryos present the basic body plan of the adult plant, possess a shoot and root tip and are vascularised. They form under specific conditions that differ from that required for shoot morphogenesis, followed by rooting.
The key regeneration step is reached via the selection, proliferation and differentiation of transformed cells into organs or embryos. Whatever the developmental process, the ability to regenerate a large number of plants from in vitro transformed cells and tissues depends on the genetic background, the physiological status of the donor plant, the organ used as the explant and its physiological state/development, the culture medium and conditions, and the interactions among these factors.8,37
Another key point is the permanent character of the genetically transformed cells status. Genetic transformation can be defined as a process by which DNA is introduced into a cell. The process can be transient (i.e., the DNA does not integrate into the host genome, and transgene expression is eventually lost) or stable (i.e., the introduced DNA integrates into the host chromosomes and can, unless silenced, express from this integrated position). Agrobacterium-mediated and particle bombardment methods are the two most common techniques used for gene transfer into the nuclear DNA of crop plants.
Stable transformation efficiency for a given plant species is therefore determined by the efficiency of tissue culture and by the ability to stably transform target cells. Both these characteristics are regulated by genetic and physiological factors controlled by external factors.
The ability of somatic cells to undergo dedifferentiation and acquire a stem-cell-like state (i.e., the ability to self-renew and simultaneously give rise to new cells that eventually take specific differentiation patterns) is the major process underlying totipotency in multicellular organisms. After dedifferentiation, re-entering the cell cycle (progression between the G1 and S phases of the cycle) is the second requirement for pluripotentiality and for a culture-based transformation method.5
Dividing cells have also, however, turned out to be the most effective targets for transgene nuclear integration. In addition, the pivotal importance of cell cycle regulation at the time of transformation (i.e., S-phase, DNA replication) has been demonstrated for the indirect plant transformation procedure using Agrobacterium tumefaciens.38 For the methods based on the direct delivery of naked DNA, it is known from strategies used to achieve targeted gene repair reaction in a range of eukaryotic cells (e.g., oligonucleotide-directed gene repair, through the use of single-stranded DNA oligonucleotides in Saccharomyces and mammalian cells) that the cell cycle progression (S-phase) also affects the chances of nuclear integration. Notably, the models suggest that replicating genomes are more amenable to directed gene repair because the chromatin is in a more open configuration and the target sequence is more accessible.39
Transformation Process
Genome stability is ensured by mechanisms that coordinate DNA replication, DNA repair and chromosome segregation. DNA repair refers to a group of processes through which a cell senses and repairs damage to DNA molecules in order to ensure the transmission of the genome and to sustain organism viability40,41
Transgene Integration and Organization
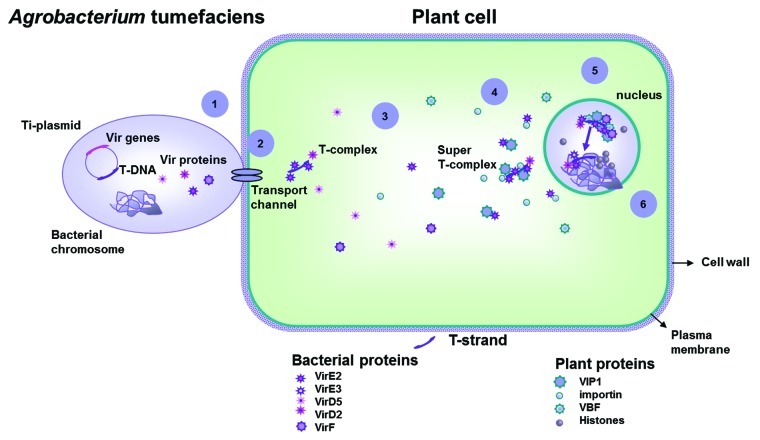
During Agrobacterium transformation, intracellular T-DNA transport and integration into the plant genome is a complex process requiring concerted action by bacterial and plant biological factors for each of the sequential steps (Fig. 1). Although much has been learned over the past two decades about the bacterial effectors involved and the molecular mechanisms by which Agrobacterium produces and transports its T-DNA into the host nucleus, the ultimate step of integration still needs to be elucidated, with so little known about the molecular basis for T-DNA integration.42
Figure 1. Agrobacterium transformation process depends on the concerted action of numerous bacterial and plant biological factors, since the intracellular T-DNA transport to its integration into the plant genome and expression. The final outcome of the interaction depends on the ability of the bacterium to hijack fundamental cellular processes and to by-pass or suppress the defenses of the host cell. 1. Agrobacterium attachment to the plant cell; 2. transfer of T-strands and virulence effector proteins through the plant plasma membrane into the plant cell; 3. T-complex and super-T-complex formation and subsequent cytoplasmic trafficking; 4. nuclear targeting; 5. Targeting of the super-Tcomplex to chromatin; 6. removal of proteins from the super-T-complex prior to T-DNA integration into the plant genome, transgene integration and expression (Fig. adapted from43).
Among the numerous plant genes and mechanisms that affect the overall transformation process, plant defense responses probably play an important role.43,44 During the Agrobacterium transformation process “plants perceive bacterium and the transferred transgenes as foreign invaders and use their defence systems to battle the infection process and expression of foreign genes”45. Under laboratory conditions, Agrobacterium can transform other eukaryotic non-plant species, including human cells.44 This remarkably wide host range for the production of genetically modified cells is attributed to the ability of Agrobacterium to hijack fundamental cellular processes shared by these evolutionarily distant organisms, while simultaneously repressing the defense response. Active at the integration step are the plant genes that contribute to chromatin structure and function, including nucleosome assembly factors, histones, histone deacetylases and acetyltransferases.43-45 Irrespective of the transformation method used (i.e., Agrobacterium or particle mediated), what characterizes the pattern of transgene integration in plant nuclear DNA is its unpredictable nature regarding location and configuration, being regularly accompanied by extensive sequence rearrangements (i.e., truncations, internal deletions, interspersions of genomic DNA and inversions).46 Site-specific transgene integration occurs at a very low frequency in plant cells compared with random integration, even when the introduced DNA contains large sequence stretches with homology to the host DNA.47 The random and often complex nature of integration sites and patterns have been attributed to the process of transgene integration into the plant nuclear genome, which is believed to be a random event localized at the position of naturally occurring chromosome breaks in the more accessible DNA areas.46
In eukaryotes, transgenes are thought to recruit enzymes from the DNA double-strand break (DSB) repair pathway for chromosomal integration. Yeasts, plants and vertebrates all have the ability to repair DSBs or integrate transgenes via two distinct mechanismsb—homologous recombination (HR) and non-homologous end-joining (NHEJ)—but they use these pathways to varying extents.48
Although scientists currently have an incomplete knowledge of the molecular basis of this process in plants, DNA integration is thought to mainly use an NHEJ-related mechanism, the low gene targeting efficiency reflecting the HR non-dominant pathway of DSB repair.48
Nonetheless, there are now strategies for efficient and precise genetic modification in a specific genomic target sequence. They are based on the activation of DNA damage signaling and repair. Single-stranded oligonucleotides (ssODNs) and zinc-finger nucleases (ZFNs) are two approaches used. This has been demonstrated in a number of animal and plant model organisms (such as Arabidopsis thaliana, maize, tobacco and soya)49,50.
Once inside the plant cell, whether this entry has been mediated by A. tumefaciens or direct DNA transfer, the transgene DNA integration into the nuclear genome is thought to depend largely on the host cell nucleic acid modifying enzyme activities (host proteins involved in DNA replication, repair and recombination), triggering the signal network for sensing DNA damage, the cell cycle control check point and DSB repair machinery. DSBs are highly toxic DNA injuries for living organisms; failure to repair such breaks can lead to genomic instability and death. Paradoxically, transgene integration is linked with the maintenance of genome integrity and survival mechanisms.
Transgene Fate and Genome Stability
Epigenetic silencing (of transgenes and endogenous genes) triggered by DNA or RNA sequence interactions can occur at the transcriptional level (transcriptional gene silencing, TGS: inhibition of transcription that involves DNA gene-specific methylation or histone methylation ‘genome modification’) or at the post-transcriptional level (post-transcriptional gene silencing, PTGS: RNA transcription, but no mRNA accumulation due to specific degradation).51
Gene silencing in transgenic plants is the consequence of triggering plant’s diverse defense mechanisms that usually act on natural foreign, parasitic or ‘invasive’ nucleic acids (e.g., transposable elements, viroids, RNA and DNA viruses, and bacterial DNA), with a number of parallels with the immune system of mammals.52,53 Transgenes or their transcripts can resemble these cellular invaders in several ways, making them targets of host protective reactions.52
Regeneration Process
Since the in vitro regeneration step is a basic requirement of current plant biotechnologies, whatever the experimental system used (protoplasts, callus, or tissue fragments), the final constraint remains – how can the frequency and the speed of plant regeneration be increased?
Developmental State: Cellular Redox State
Compared with animals, somatic plant cells are not terminally differentiated and can regain totipotency. The ‘G0’ or ‘quiescent’ state is therefore far more flexible. The same terminology, however, is used in plant biology, and marks plant cells as being out of the cell cycle with differentiated functions. Cell cycle re-entry of differentiated cells can occur in plants in many circumstances during normal development, in response to environmental stimuli, or it can be induced under in vitro conditions. “Cell cycle entry (G0-to-G1) transition may represent a regulatory node where the cellular redox status has a central role”54.
Many biological processes involve a series of redox reactions in which electrons are transferred from a donor molecule to an acceptor molecule. The redox state of a cell indirectly indicates energy availability, since many biologically important redox components must be in a reduced form in order to carry out relevant metabolic reactions. Redox changes influence many aspects of cell activities and functions in the cytoplasm and in the nucleus.55 The redox potential has been implicated in the regulation of a variety of biological activities, such as proliferation and growth arrest, and it appears to correlate with the biological status of an animal cell (i.e., exponential growth phase, differentiation, and apoptosis).56 Similarly, the influence of the intracellular redox state on developmental processes within the plant system has also been recognized57,58 (Fig. 2).
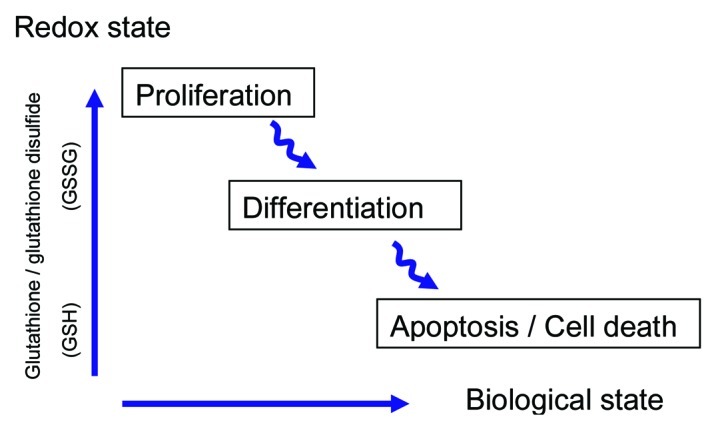
Figure 2. Redox state in the life cycle of cells The intracellular redox state is the resulting balance between the reduced and oxidized forms of several key redox pairs. The tripeptide glutathione (GSH; γ-glu–cys–gly) is a major thiol antioxidant found ubiquitously in tissues of higher organisms. The glutathione/glutathione disulfide (2GSH /GSSG) couple is the most abundant redox couple in a cell, and can serve as an important indicator of redox environment (i.e., based on the GSH/GSSG ratio with GSH and GSSG being the reduced and the oxidized forms, respectively). The oxidized form becomes progressively more abundant in the life cycle of animal cells, being correlated with the functional state, from division to apoptosis, through differentiation (figure adapted from56). In plants, the glutathione redox pair also acts as a modulator of development and morphogenesis. Elevated concentrations of cellular GSH, and ultimately a higher GSH/GSSG ratio, are often associated with rapidly growing tissues, including meristematic regions, whereas increased GSSG contents often correlate with dormancy and cell death. Exogenous additions of reduced or oxidized glutathione strongly influence in vitro developmental events: GSH promotes cell proliferation, while GSSG promotes organized development, notably somatic embryogenesis development.106
Plant Developmental Plasticity: Environment, ROS and Antioxidant System
Reactive oxygen species (ROS) or active oxygen species (AOS) are a highly reactive group of oxygen-containing molecules, characterized by the presence of unpaired valence electron. ROS are generated as common by-products of aerobic metabolism, as well as in response to environmental inducers. ROS accumulation is controlled by the rate of production as well as the rate of elimination by the antioxidative system (i.e., living cells have both enzymatic and non-enzymatic mechanisms). Numerous redox regulatory mechanisms in the membrane-bound and soluble phases operate, and they are intimately linked at many levels to ensure homeostasis.58
Under severe environmental conditions, ROS production might exceed the antioxidant capacity of cells; excess concentrations of ROS result in cumulative oxidative damage to biological molecules within the cell (DNA, RNA, proteins and lipids) and might trigger cell death, whereas ‘mild’ stress activates adaptive responses to environmental challenges. In this process, ROS at low levels are known to act as important signaling molecules, in both animals and plants. This dual role of ROS depends mainly on the concentration, pulse duration, site of action and physiological state of the cells.59,60
Plant development involves a high level of plasticity in response to a wide variety of environmental constraints, with an important role being played by a range of protective responses in these developmental ’switches’. Different stresses/environmental cues can induce similar conserved morphogenic responses in different plant species, but this stress-induced modulation of growth (stress-induced morphogenic response, SIMR) is mediated by many interchangeable molecular pathways. From a thermodynamic point of view, “not the specific pathway, but solely its beginning and end points have any biological relevance”59.
From the physiological point of view, the ROS/antioxidant system is recognized as an important element of the morphological plasticity, one of the centerpieces of the redirection of morphogenesis in response to environmental alterations. At the plant scale, cellular redox homeostasis is considered to be an ‘integrator’ of information from the metabolism and the environment, acting at the interface of the organismal and cellular response levels, integrating the stress signals that the plant perceives with the control of local redox poise.59
In wheat, the abiotic stresses induced somatic embryogenesis from the leaf segments, and the implication of genes in defense and anti-oxidation was evident in the expression of this developmental pathway.61,62 Among the proteins associated with the metabolism of ROS in cereals, the apoplastic oxalate oxidases (OXOs) are particularly noteworthy, given the diversity of information in the literature supporting their implication in developmental reprogramming and differentiation,63-67 including via somatic embryogenesis,65,68,69 and in stress and defense responses triggered by biotic and abiotic stresses (for review, see ref. 70).
Developmental Switch in Plant Cells: Crosstalk Between Stress, ROS and Hormonal Signaling Pathways
At the cell scale, SIMRs are characterized by elongation inhibition, redistribution of cell cycle activity through the plant (i.e., cessation of meristematic activity in some parts of the plant, but localized stimulation of cell division in a distal zone) and alterations in cell differentiation status.59 In many other cases, stress exposure induces dedifferentiation.71
In relation, in particular, to cell division and growth in planta, ROS/redox signaling/antioxidant interaction is seen as a switching node at the core of plant cell physiology. This convergence hub between stress signaling and development allows specific responses to environmental challenges to be orchestrated via crosstalk with phytohormones (mainly auxine and ethylene) and sugars.58
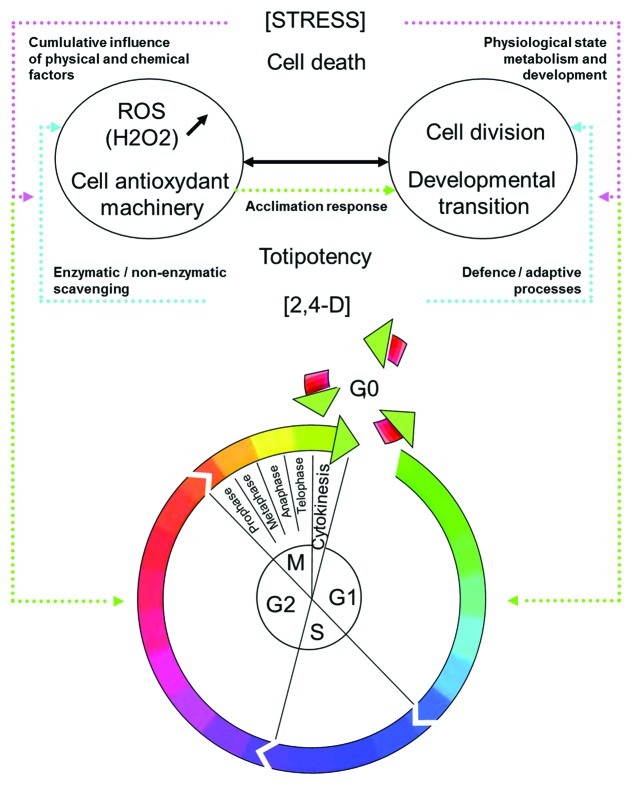
The developmental switching of somatic cells toward the regenerative pathway in vitro involves significant adjustments in response to the environmental conditions (i.e., a complete reorganization of their physiology, metabolism and morphology).7 The physiological response is the result of the integration of many events transduced into a network of signaling pathways.72Figure 3 illustrates how ROS, cellular antioxidant machinery activity, defense responses and plant hormones occupy a central place in this transduction network.54,58
Figure 3. Developmental switch, cell cycle activation and developmental transition: the 2,4-D induced developmental switch during somatic embryogenesis. The fate of individual cells (to enter the cell cycle, maintain proliferation competence, become quiescent, expand, differentiate or die) depends on the perception of various signals. Complementary interactions between stress, ROS/redox signaling and auxins control the plant cell responses during the regenerative pathway. The outcome depends on two main parameters: the level of stress and the physiological state of the cells (depending on the genotype, cell identity, developmental state). If the stress level exceeds cellular tolerance, the cells die. Both the higher level of intracellular H2O2 and the activity of cellular antioxidant machinery have been shown to be crucial for cell division activation and the expression of totipotency. The level of intracellular H2O2, cellular antioxidant machinery activity, defense responses and the rate of cell division activation are interdependent, and together are subordinated to the concentration of exogenous 2,4-D supplied54,58,72,107).
Somatic Embryogenesis: Determination, Initiation and Expression
It is not known why certain genotypes, explants or cells are more amenable to somatic embryogenesis than others. The answer probably lies in the complicated interaction of genetic and physiological factors. Embryogenic development is determined mainly by the physiological state of the cell, which is subordinated to genetics, developmental (cell identity and developmental state) and environmental signals.7
Embryogenic capability is viewed as a general feature, whose expression depends on many circumstances, and many conditions can be used to initiate this developmental pathway.7 We consider this concept of somatic embryogenesis to resemble the SIMR concept, thought to be part of a general acclimation strategy whereby plant growth is redirected to reduce exposure to mild stress.59 A close connection, or overlapping, between in vitro embryogenesis and stress response pathways has often been highlighted. In practice, somatic embryogenesis induction is usually achieved by a stress and/or hormone treatment of somatic cells. At the physiological scale, the activation of key regulators is preceded by the stress-induced reprogramming of cellular metabolism.7
At the molecular level, such a developmental alternative pre-exists in the plant genome expression potential, but is repressed in somatic cells. The developmental switching in somatic cells toward the embryogenic pathway occurs under the cumulative influence of physical and chemical inductive conditions and requires profound and dynamic gene expression reprogramming.73,74 The developmental switching indicates that the prevailing program needs to be erased or greatly altered, and thereafter involves the activation of various signal cascades and differential gene expression giving the somatic cells the ability to manifest the embryogenic pathway.7,74 The induction of somatic embryogenesis is, instead, a release from suppression, controlled at the chromatin level, through a global and dynamic reorganization of chromatin structure.7,75
Chromatin Remodelling: At the Crossroads of the Two Physiological Processes
All cells in a multicellular organism share the same genomic sequence, but their morphology and function can differ greatly among cell types and in their differentiation state. This high cellular diversity stems from differences in the nuclear program. These changes in gene activity are orchestrated in a development- and tissue-specific manner, as well as in response to diverse stimuli. Central to this plasticity is the dynamic organization of genetic information.76 In addition to the DNA sequence itself, an extra layer of information, termed 'epigenetic code’ or 'epigenome', determines which genes are expressed in a particular cell type at a given time. Cellular DNA is never naked; it always forms a complex with various proteins. This complex is the core structure underlying the dynamics of the transcriptional output of the genome and replication.77
Dynamic Organization of Genetic Information
Chromatin is the complex formed by DNA and protein partners (histone and non-histone proteins) whose physical function is to package the enormous amount of linear genome information into the limited volume of a cell (i.e., up to about one-million-fold compaction relative to an extended double helix of DNA). The structural unit of chromatin is the nucleosome, which consists of a hundred base pairs of DNA double helix wrapped around an octamer of core histones. The multiple levels of nucleosome stacking up and the folding of nucleosomes into large-scale configurations eventually form the characteristic chromosome configuration, microscopically visible in mitotic nuclei. Apart from providing a scaffold for the compaction of nuclear DNA in the nucleus, this DNA-protein assembly is also important for the regulation of nuclear functions.76,77
Nuclear organization as multiple levels of chromatin packaging might appear to be static at a cytological level, but chromatin is actually a dynamic assemblage in which proteins are constantly associating and dissociating. The regulation of the chromatin structure to expose or conceal a particular DNA segment is controlled by the dynamic interplay between sequence-specific DNA-binding proteins, histone variants, histone-modifying enzymes, nucleosome remodelers and other chromatin-associated factors.78
Chromatin Hallmarks of Pluripotency
During differentiation, lineage- and developmental-specific transcription factors activate the expression of specific sets of genes. Contrary to the assumption that stem cells selectively express specific ‘stem cell genes’, widespread transcription in coding and non-coding regions has been found in animal embryogenic stem cells. This global transcription was accompanied by a disproportionate expression of general transcription factors and chromatin-remodelling proteins (including histone acetyltransferases, histone deacetylases and histone methyltransferases). This transcriptional hyperactivity marks a competent chromatin state encompassing the widespread expression of most signatures of differentiated cells, but at a very low level, whereas the transcriptional landscape becomes more discrete and the genome undergoes large-scale silencing as cells differentiate.
In animal pluripotent embryogenic cells, chromatin is morphologically distinct in that heterochromatin is organized in larger and fewer domains, which become smaller, more abundant and hypercondensed as differentiation proceeds.71,79
In planta large-scale chromatin decompaction is a transient state that accompanies changes in the nuclear program in response to a developmental transition (seedling development and the floral transition) or (a)biotic changing conditions. Such large-scale decondensation is exemplified in the cells of mesophyll undergoing conversion to protoplasts and dedifferentiation in vitro.76 Although less pronounced, heterochromatin decondensation was also found in mutants that affect chromatin.76,80 The dynamics of the chromatin structure in the context of somatic embryogenesis regeneration has not been specifically documented for wheat. The existence, however, of typical chromatin remodelling activity, similar to that of other eukaryotes, has been demonstrated for this species.81
This distinguishing and widespread chromatin decondensation of heterochromatin is a common feature between animal stem cells and dedifferentiated plant cells. In a plant cell undergoing a regenerative process, dedifferentiation followed by cell cycle activation are two successive processes marked by this characteristic change in chromatin morphology.71 On the other hand, the genetic and epigenetic instability that can be observed during cellular dedifferentiation could pose a significant challenge, notably in regenerative medicine.3,80,82
Chromatin Architecture and Genetic Transformability
Chromatin fibers are naturally highly condensed under physiological conditions. The role of the epigenetic code in transcriptional regulation is long established.83 More recently, these mechanisms have been implicated in DNA damage detection and repair. DNA repair requires a high degree of coordination between the DNA-repair machinery and the chromatin modifying/remodelling complexes, which regulates the accessibility of DNA in chromatin.41
The reversible condensed/relaxed chromatin status alternates with cell cycle progression. In proliferating cells, most of the chromatin is assembled during DNA replication.80,84 In different eukaryotic cells and for various gene transfer strategies, the positive incidence of the S-phase on DNA transfer has been at least partly attributed to DNA accessibility while chromosomal DNA is replicating.39
During Agrobacterium- and bombardment plant cell transformation, however, transgenes integrate randomly into the genome without regard to DNA sequence, transcriptional activity or DNA methylation status. Since the DSB in the eukaryote host genome represents the primary target site of transgene integration, integration per se relies largely on the host DSB repair machinery (DSB sensing and/or repair proteins).46,48,51,85
The chromatin architecture surrounding the DSB has a critical impact on the ability of cells to mount an effective DNA damage response (i.e., in the detection and repair of these lesions).41 Among the numerous genes that contribute to chromatin structure and function, those that affect Agrobacterium-mediated transformation include histone, histone deacetylases and acetyltransferases.43,44 Histone acetylation has long been known to induce the structural relaxation of chromatin that makes DNA more accessible. The observations in yeast and Arabidopsis suggest that histone acetylation/deacetylation balance is important for facilitating T-DNA integration. In addition, the overexpression of several histones increases Agrobacterium-mediated transformation and the transient expression of electroporated transgene by protecting incoming DNA from nuclease degradation. Histones and their modifications might also play a role in T-DNA integration itself by recruiting DNA repair or replication enzymes.42-44
In wheat, although there is no direct evidence of the implication of DNA repair mechanisms in genetic transformation, the existence of histone acetylation and phosphorylation activities involved in DNA damage repair has recently been demonstrated.86
New Insights into the Regenerative Property of Plant Cells and their Receptivity to Transgenesis
Given the above considerations, we put forward the hypothesis that both competencies are at the core of defense/adaptive and survival mechanisms, between chaos and organization, as a response of critical dynamic systems.
Notion of Competence
Both the embryogenic potential and the ability to accept transgenes, generally formulated as competencies, are anthropocentric views of the ability of living cells to make the adjustments necessary for them to survive environmental fluctuations.
Land plants are sessile and constantly experience periodic environmental fluctuations. In contrast to animals, which change their behavior, because of immovability plants change their body plan. Unlike animal immune systems, each individual plant cell must orchestrate its own defense, as well as respond to cues from its neighbors.
Plants have evolved complex sensing-signaling pathways in response to various stimuli and have acquired plasticity in metabolic functions and developmental switches to cope with changing environmental conditions.58,59 Their whole physiology, metabolism and developmental processes reflect their accommodation and adjustment to different environmental factors stemming from biotic as well as abiotic stresses that never act alone.87,88
In order to survive, they must respond in a rapid and adequate way via the nuclear program. Dynamic chromatin is an integral part of the mechanism that facilitates such a response. Interplay between modifications of histones, and the relationship among histone modifications, DNA methylation and RNA interference, facilitate proper expression of target genes in response to light, temperature, abiotic and biotic stresses, and polyploidy.76,84
Compared with plants, cells in mammals are less responsive to environmental changes. Environmentally induced reprogramming of cells during adaptation indicates that developmental plasticity is a typical characteristic of plants.76,87 Somatic embryogenesis, which is a notable illustration of plant cell totipotency, is an example of developmental plasticity that can require profound reprogramming of gene expression based on chromatin remodelling.74,87 Chromatin remodelling regulates the balance between pluripotency and differentiation.71
There are also emerging examples of factors known to play roles in plant immune responses, which also play roles in plant morphology. The intersections between immune responses and morphological regulation in plants might be epigenetic.89,90 As demonstrated in mammals, cells might use the same basic mechanism to regulate distinct cellular processes, such as transcription and DNA repair.91
Various external agents and cell metabolism are known to be responsible for DNA modifications. Due to the immobility of plants, shoot apical and marginal meristems are continuously exposed to putative DNA-damaging conditions (e.g., UV-B light). When DNA is damaged, complex mechanisms that recognize and repair aberrant nucleotides are activated. They lead to cell cycle arrest and allow the cell to repair the damaged DNA. In contrast to mammals, however, the action of the DNA damage checkpoint process rarely results in apoptotic cell death in the case of failure of repair. Rather, plant cells start differentiating. “Pushing malicious cells into the differentiation pathway might be sufficient to avoid transmission of damaged genomes to the next generation”92.
Characteristic of Critical Dynamic Systems
Chromatin behaves as a highly dynamic cellular component, but also exhibits a large combinatorial complexity beyond the DNA sequence that conforms to the epigenetic landscape.80
Inferred from microarray experiments (i.e., hundreds of microarray data from different living organisms), the regulatory interactions among genes, the global dynamics of the genetic network, have recently been recognized to operate close to criticality.93 Criticality is a typical feature of dynamic systems characterized by the compromise between robustness and adaptability: (i) the system is robust enough as to guarantee stability; (ii) the system is flexible enough to recognize and integrate specific external signals under a broad range of external conditions.93,94
This compromise characterizes dynamic systems that operate close to a phase of transition between order and chaos. Many examples occur in nature (e.g., avalanches, financial markets, earthquakes, the brain). These systems can integrate process, transfer information and respond faster and more reliably than non-critical systems. This precise, measurable and well-characterized property of criticality gives the system the ability to collectively respond and adapt to an often rapidly changing environment. These properties at the genetic level might constitute a fundamental evolutionary mechanism that generates the great diversity of dynamically robust living forms.93
Critical phases are often observed during the regulation of plant development. Competence windows in development (also termed ‘developmental windows’) are characterized by an enhanced sensitivity to specific endogenous or exogenous factors. Competence windows have been described not only for normal development, but also for the capacity of the organism to adjust to a constraint.95
We consider that, within the explant tissue that is dismantled, some cells are in the ‘in vitro competence window’, a critical period/window of high physiological responsiveness during which there might be a reorganization of the regulation networks. These cells are able to develop effective mechanisms to sense and respond to the peculiar and severe in vitro conditions (e.g., through the embryogenic developmental pathway alternative). The response is communicated and orchestrated at each level of organization. The entire process helps to ensure that the biological system endures life-threatening conditions and ultimately survives.
Interestingly, cells capable of undergoing rejuvenation share common features with cells undergoing senescence (i.e., including widespread chromatin decondensation and similar expression profiles of chromatin modifying genes that favor the acquisition of a decondensed chromatin configuration, shrinkage/disruption of the nucleolus, decondensation of rRNA).71 However, some, but not all, dedifferentiated cells become competent for embryogenesis. Depending on the type of stimulus and the complex interacting pathways (as presented in section "Regeneration process" and the subsections within), dedifferentiated cells can be induced to redifferentiate, re-enter the cell cycle or die. Further, the active process of cell death (programmed cell death, PCD) has long been considered to be an integral part of plant development or responses to pathogens.96 The observed correlation between the manifestation of PCD during the development of somatic embryos is not uncommon, and has been extensively studied in gymnosperms.97
During the ongoing process of the embryogenesis pathway, within the subpopulation that ultimately participates in establishing the polar embryo, cells undergo development switches several times, which are associated with re-entry to the cell cycle, coordinated changes and a balance between euchromatin and heterochromatin, and the reprogramming of gene expression.
From the transgenesis perspective, each cell is a complex and dynamic system that goes through a sequence of physiological stages over time. On the premise that these physiological events might interfere with their transformation receptiveness, from the biologist’s point of view, the critical window to gene transfer has to be found during a transition period where cells are actively dividing before they redifferentiate. The competence window coincides with physical and physiological accessibility, very early in the process of cell proliferation toward organization, while cells are reachable and it is possible to take advantage of the native DNA repair mechanisms and probably reduced immune defense.
Concluding Remarks
The regeneration from in vitro-grown cultured tissues is a key point for creating genetically transformed plants. During the process, the re-entry into the cell cycle plays a crucial role in the expression of interconnected cellular properties, physiological totipotency and the ability to accept a transgene stably integrated into the nuclear genome.
A dynamic change in chromatin structure and gene activity is a fundamental theme in the basic mechanisms involved in the regenerative property of cells and their receptivity to transgenesis. For both properties, the adaptive dimension of the critical period refers to a series of transition/competence windows from dedifferentiation to reorganization.
Experimentally, transcriptional profiling has shown that a large number of genes are differentially expressed during somatic embryogenesis induction or differentiation. The genome-wide shift in the transcriptome involves multiple cellular pathways interconnecting within an intricate functional network, including signal transduction cascades, defense, anti-oxidation, programmed cell death/senescence, hormone response/metabolism and cell division.62,98 Transcription studies of chosen genes would enable the diverse cellular and physiological responses involved in this important reprogramming process to be investigated. In view of the importance of ROS, a metabolomic study could also be conducted to determine the biochemical events associated with these responses.
Opting for a candidate-gene strategy, some genes might deserve special attention (e.g., those for which expression data might provide spatio-temporal indicators relative to the establishment of plant defense mechanisms, the antioxidant apparatus dynamics, cell adjustment to changing environmental conditions, DNA-repair machinery and chromatin modifying/remodelling complexes, reprogramming of genome expression during somatic embryogenesis induction, proliferative state competent for gene transfer and transgene integration). Where cereals, and wheat in particular, are the focus of a case study, the germin oxalate oxidases (OXOs) could be worth considering. The involvement of OXOs in plant stress-response, global defense strategy in monocots, ROS (H2O2) and calcium release, cell wall remodelling and reinforcement, developmental reprogramming and differentiation (including through somatic embryogenesis) make this gene family an appropriate target for tracing the successive stages of manifestation of both competencies,68-70,99-105 either by looking at the contribution of individual members of this family or by monitoring the overall response.
Finally, bearing in mind the criticality concept, a global approach based on transcriptomics, proteomics and metabolomics in relation to chromatin dynamics could be developed in order to better understand differential responses among wheat genotypes to in vitro culture regeneration and transgenesis.
Notes
aExcept for the floral dip procedure used in Arabidopsis or other in planta procedures developed for some other species. In the case of 'the floral dip procedure', transformed plantlets are directly obtained after inflorescence agroinfiltration and seedling selection.
bIn animals and plants, although HR is an essential process for maintaining genome integrity, the NHEJ alternative repair pathway, which is relatively inaccurate (i.e., random integration frequently accompanied by insertion or deletion of DNA sequences), is predominant.
Footnotes
Previously published online: www.landesbioscience.com/journals/psb/article/22424
References
- 1.Kennedy D, Norman C. What don’t we know? Science. 2005;309:75. doi: 10.1126/science.309.5731.75. [DOI] [PubMed] [Google Scholar]
- 2.Vogel G. How does a single somatic cell become a whole plant? Science. 2005;309:86. doi: 10.1126/science.309.5731.86. [DOI] [PubMed] [Google Scholar]
- 3.Grafi G. The complexity of cellular dedifferentiation: implications for regenerative medicine. Trends Biotechnol. 2009;27:329–32. doi: 10.1016/j.tibtech.2009.02.007. [DOI] [PubMed] [Google Scholar]
- 4.Yamanaka S, Blau HM. Nuclear reprogramming to a pluripotent state by three approaches. Nature. 2010;465:704–12. doi: 10.1038/nature09229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Grafi G. How cells dedifferentiate: a lesson from plants. Dev Biol. 2004;268:1–6. doi: 10.1016/j.ydbio.2003.12.027. [DOI] [PubMed] [Google Scholar]
- 6.Costa S, Shaw P. ‘Open minded’ cells: how cells can change fate. Trends Cell Biol. 2007;17:101–6. doi: 10.1016/j.tcb.2006.12.005. [DOI] [PubMed] [Google Scholar]
- 7.Fehér A. The initiation phase of somatic embryogenesis: what we know and what we don't. Acta Biol Szeged. 2008;52:53–6. [Google Scholar]
- 8.Vasil IK. A history of plant biotechnology: from the Cell Theory of Schleiden and Schwann to biotech crops. Plant Cell Rep. 2008;27:1423–40. doi: 10.1007/s00299-008-0571-4. [DOI] [PubMed] [Google Scholar]
- 9.Hilson P. Cloned sequence repertoires for small- and large-scale biology. Trends Plant Sci. 2006;11:133–41. doi: 10.1016/j.tplants.2006.01.006. [DOI] [PubMed] [Google Scholar]
- 10.Dixon RA, Bouton JH, Narasimhamoorthy B, Saha M, Wang Z-Y, May GD. Beyond Structural Genomics for Plant Science Advances in Agronomy. Adv Agron. 2007;95:77–161. doi: 10.1016/S0065-2113(07)95002-6. [DOI] [Google Scholar]
- 11.Fedoroff NV. The past, present and future of crop genetic modification. N Biotechnol. 2010;27:461–5. doi: 10.1016/j.nbt.2009.12.004. [DOI] [PubMed] [Google Scholar]
- 12.Raymond Park J, McFarlane I, Hartley Phipps R, Ceddia G. The role of transgenic crops in sustainable development. Plant Biotechnol J. 2011;9:2–21. doi: 10.1111/j.1467-7652.2010.00565.x. [DOI] [PubMed] [Google Scholar]
- 13.James C. Global Status of Commercialized Biotech/GM Crops: 2009. ISAAA Brief No 41 ISAAA: Ithaca, NY 2009. [Google Scholar]
- 14.FAO. Monthly News Report on Grains Issue 59 – February 2010. http://wwwfaoorg/fileadmin/templates/est/COMM_MARKETS_MONITORING/Grains/Documents/MNR0210pdf, 2010.
- 15.James C. Global Status of Commercialized Biotech/GM Crops: 2010. ISAAA Brief No 42 ISAAA: Ithaca, NY 2010. [Google Scholar]
- 16.Kumlehn J, Hensel G. Genetic transformation technology in the Triticeae. Breed Sci. 2009;59:553–60. doi: 10.1270/jsbbs.59.553. [DOI] [Google Scholar]
- 17.Paterson AH, Freeling M, Sasaki T. Grains of knowledge: genomics of model cereals. Genome Res. 2005;15:1643–50. doi: 10.1101/gr.3725905. [DOI] [PubMed] [Google Scholar]
- 18.Eckardt NA. Grass Genome Evolution. Plant Cell. 2008;20:3–4. doi: 10.1105/tpc.108.058586. [DOI] [Google Scholar]
- 19.Vasil IK. The story of transgenic cereals: the challenge, the debate, and the solution - A historical perspective. In Vitro Cell Dev Biol Plant. 2005;41:577–83. doi: 10.1079/IVP2005654. [DOI] [Google Scholar]
- 20.Chen H, Lin Y, Zhang Q. Rice. In: Kempken F, Jung C, eds. Genetic Modification of Plants: Springer Berlin Heidelberg, 2010:423-51. [Google Scholar]
- 21.Rashid H, Yokoi S, Toriyama K, Hinata K. Transgenic plant production mediated by Agrobacterium in Indica rice. Plant Cell Rep. 1996;15:727–30. doi: 10.1007/BF00232216. [DOI] [PubMed] [Google Scholar]
- 22.Harwood WA. Advances and remaining challenges in the transformation of barley and wheat. J Exp Bot. 2012;63:1791–8. doi: 10.1093/jxb/err380. [DOI] [PubMed] [Google Scholar]
- 23.Pérez-Piñeiro P, Gago J, Landín M, Galleg PP. http://www.intechopen.com/books/transgenic-plants-advances-and-limitations/agrobacterium-mediated-transformation-of Agrobacterium-Mediated Transformation of Wheat: General Overview and New Approaches to Model and Identify the Key Factors involved. In: Çiftçi YÖ, ed. Transgenic Plants - Advances and Limitations, 2012.
- 24.Shrawat AK, Lörz H. Agrobacterium-mediated transformation of cereals: a promising approach crossing barriers. Plant Biotechnol J. 2006;4:575–603. doi: 10.1111/j.1467-7652.2006.00209.x. [DOI] [PubMed] [Google Scholar]
- 25.Xia L, Ma Y, He Y, Jones HD. GM wheat development in China: current status and challenges to commercialization. J Exp Bot. 2012;63:1785–90. doi: 10.1093/jxb/err342. [DOI] [PubMed] [Google Scholar]
- 26.Sallaud C, Gay C, Larmande P, Bès M, Piffanelli P, Piégu B, et al. High throughput T-DNA insertion mutagenesis in rice: a first step towards in silico reverse genetics. Plant J. 2004;39:450–64. doi: 10.1111/j.1365-313X.2004.02145.x. [DOI] [PubMed] [Google Scholar]
- 27.Yang Y, Peng H, Huang H, Wu J, Jia S, Huang D, et al. Large-scale production of enhancer trapping lines for rice functional genomics. Plant Sci. 2004;167:281–8. doi: 10.1016/j.plantsci.2004.03.026. [DOI] [Google Scholar]
- 28.Jagtap UB, Gurav RG, Bapat VA. Role of RNA interference in plant improvement. Naturwissenschaften. 2011;98:473–92. doi: 10.1007/s00114-011-0798-8. [DOI] [PubMed] [Google Scholar]
- 29.Kusaba M, Miyahara K, Iida S, Fukuoka H, Takano T, Sassa H, et al. Low glutelin content1: a dominant mutation that suppresses the glutelin multigene family via RNA silencing in rice. Plant Cell. 2003;15:1455–67. doi: 10.1105/tpc.011452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nakamura H, Hakata M, Amano K, Miyao A, Toki N, Kajikawa M, et al. A genome-wide gain-of-function analysis of rice genes using the FOX-hunting system. Plant molecular biology: Springer Netherlands, 2007:357-71. [DOI] [PubMed]
- 31.Saika H, Oikawa A, Matsuda F, Onodera H, Saito K, Toki S. Application of gene targeting to designed mutation breeding of high-tryptophan rice. Plant Physiol. 2011;156:1269–77. doi: 10.1104/pp.111.175778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Saika H, Toki S. Towards a Highly Efficient Gene Targeting System in Higher Plants JARQ. 2009;43:81–5. [Google Scholar]
- 33.Terada R, Asao H, Iida S. A large-scale Agrobacterium-mediated transformation procedure with a strong positive-negative selection for gene targeting in rice (Oryza sativa L.) Plant Cell Rep. 2004;22:653–9. doi: 10.1007/s00299-003-0752-0. [DOI] [PubMed] [Google Scholar]
- 34.Lazzeri PA, Jones HD. Transgenic Wheat, Barley and Oats: Production and Characterization. Springer, 2009. [DOI] [PubMed] [Google Scholar]
- 35.Greer MS, Kovalchuk I, Eudes F. Ammonium nitrate improves direct somatic embryogenesis and biolistic transformation of Triticum aestivum. New Biotechnology - Special issue on Biocatalysis and Agricultural Biotechnology: number 1 2009; 26:44-52. [DOI] [PubMed]
- 36.Pellegrineschi A, Noguera LM, Skovmand B, Brito RM, Velazquez L, Salgado MM, et al. Identification of highly transformable wheat genotypes for mass production of fertile transgenic plants. Genome. 2002;45:421–30. doi: 10.1139/g01-154. [DOI] [PubMed] [Google Scholar]
- 37.Finer JJ. Plant Nuclear Transformation. In: Kempken F, Jung C, eds. Genetic Modification of Plants: Springer Berlin Heidelberg, 2010:3-21. [Google Scholar]
- 38.Arias RS, Filichkin SA, Strauss SH. Divide and conquer: development and cell cycle genes in plant transformation. Trends Biotechnol. 2006;24:267–73. doi: 10.1016/j.tibtech.2006.04.007. [DOI] [PubMed] [Google Scholar]
- 39.Engstrom JU, Suzuki T, Kmiec EB. Regulation of targeted gene repair by intrinsic cellular processes. Bioessays. 2009;31:159–68. doi: 10.1002/bies.200800119. [DOI] [PubMed] [Google Scholar]
- 40.Allen C, Ashley AK, Hromas R, Nickoloff JA. More forks on the road to replication stress recovery. J Mol Cell Biol. 2011;3:4–12. doi: 10.1093/jmcb/mjq049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xu Y, Price BD. Chromatin dynamics and the repair of DNA double strand breaks. Cell Cycle. 2011;10:261–7. doi: 10.4161/cc.10.2.14543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Magori S, Citovsky V. Epigenetic control of Agrobacterium T-DNA integration. Biochimica et Biophysica Acta (BBA) - Gene Regulatory Mechanisms. 2011 doi: 10.1016/j.bbagrm.2011.01.007. [Corrected Proof.] In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gelvin SB. Plant proteins involved in Agrobacterium-mediated genetic transformation. Annu Rev Phytopathol. 2010;48:45–68. doi: 10.1146/annurev-phyto-080508-081852. [DOI] [PubMed] [Google Scholar]
- 44.Pitzschke A, Hirt H. New insights into an old story: Agrobacterium-induced tumour formation in plants by plant transformation. 2010; 29:1021-32. [DOI] [PMC free article] [PubMed]
- 45.Citovsky V, Kozlovsky SV, Lacroix B, Zaltsman A, Dafny-Yelin M, Vyas S, et al. Biological systems of the host cell involved in Agrobacterium infection. Cell Microbiol. 2007;9:9–20. doi: 10.1111/j.1462-5822.2006.00830.x. [DOI] [PubMed] [Google Scholar]
- 46.Kohli A, Melendi PG, Abranches R, Capell T, Stoger E, Christou P. The Quest to Understand the Basis and Mechanisms that Control Expression of Introduced Transgenes in Crop Plants. Plant Signal Behav. 2006;1:185–95. doi: 10.4161/psb.1.4.3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schaefer DG. Gene targeting in Physcomitrella patens. Curr Opin Plant Biol. 2001;4:143–50. doi: 10.1016/S1369-5266(00)00150-3. [DOI] [PubMed] [Google Scholar]
- 48.Osakabe K, Abe K, Endo M, Toki S. Regulatory Mechanisms of Homologous Recombination in Higher Plants. In: Pua EC, Davey MR, eds. Plant Developmental Biology - Biotechnological Perspectives: Springer Berlin Heidelberg, 2010:371-91. [Google Scholar]
- 49.Olsen PA, Solhaug A, Booth JA, Gelazauskaite M, Krauss S. Cellular responses to targeted genomic sequence modification using single-stranded oligonucleotides and zinc-finger nucleases. DNA Repair (Amst) 2009;8:298–308. doi: 10.1016/j.dnarep.2008.11.011. [DOI] [PubMed] [Google Scholar]
- 50.Urnov FD, Rebar EJ, Holmes MC, Zhang HS, Gregory PD. Genome editing with engineered zinc finger nucleases. 2010; 11:636-46. [DOI] [PubMed]
- 51.Matzke MA, Matzke AJM. Planting the seeds of a new paradigm. PLoS Biol. 2004;2:E133. doi: 10.1371/journal.pbio.0020133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Matzke MA, Mette MF, Matzke AJM. Transgene silencing by the host genome defense: implications for the evolution of epigenetic control mechanisms in plants and vertebrates. Plant Mol Biol. 2000;43:401–15. doi: 10.1023/A:1006484806925. [DOI] [PubMed] [Google Scholar]
- 53.Waterhouse PM, Wang MB, Lough T. Gene silencing as an adaptive defence against viruses. Nature. 2001;411:834–42. doi: 10.1038/35081168. [DOI] [PubMed] [Google Scholar]
- 54.Fehér A, Otvös K, Pasternak TP, Szandtner AP. The involvement of reactive oxygen species (ROS) in the cell cycle activation (G(0)-to-G(1) transition) of plant cells. Plant Signal Behav. 2008;3:823–6. doi: 10.4161/psb.3.10.5908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Go YM, Jones DP. Redox control systems in the nucleus: mechanisms and functions. Antioxid Redox Signal. 2010;13:489–509. doi: 10.1089/ars.2009.3021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jones DP. Extracellular redox state: refining the definition of oxidative stress in aging. Rejuvenation Res. 2006;9:169–81. doi: 10.1089/rej.2006.9.169. [DOI] [PubMed] [Google Scholar]
- 57.Stasolla C. Glutathione redox regulation of in vitro embryogenesis. Plant Physiol Biochem. 2010;48:319–27. doi: 10.1016/j.plaphy.2009.10.007. [DOI] [PubMed] [Google Scholar]
- 58.Foyer CH, Noctor G. Redox regulation in photosynthetic organisms: signaling, acclimation, and practical implications. Antioxid Redox Signal. 2009;11:861–905. doi: 10.1089/ars.2008.2177. [DOI] [PubMed] [Google Scholar]
- 59.Potters G, Pasternak TP, Guisez Y, Jansen MAK. Different stresses, similar morphogenic responses: integrating a plethora of pathways. Plant Cell Environ. 2009;32:158–69. doi: 10.1111/j.1365-3040.2008.01908.x. [DOI] [PubMed] [Google Scholar]
- 60.Dowling DK, Simmons LW. Reactive oxygen species as universal constraints in life-history evolution. Proc Biol Sci. 2009;276:1737–45. doi: 10.1098/rspb.2008.1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Patnaik D, Mahalakshmi A, Khurana P. Effect of water stress and heavy metals on induction of somatic embryogenesis in wheat leaf base cultures. Indian J Exp Biol. 2005;43:740–5. [PubMed] [Google Scholar]
- 62.Singla B, Tyagi AK, Khurana JP, Khurana P. Analysis of expression profile of selected genes expressed during auxin-induced somatic embryogenesis in leaf base system of wheat (Triticum aestivum) and their possible interactions. Plant Mol Biol. 2007;65:677–92. doi: 10.1007/s11103-007-9234-z. [DOI] [PubMed] [Google Scholar]
- 63.Lane BG, Dunwell JM, Ray JA, Schmitt MR, Cuming AC. Germin, a protein marker of early plant development, is an oxalate oxidase. J Biol Chem. 1993;268:12239–42. [PubMed] [Google Scholar]
- 64.Caliskan M, Cuming AC. Spatial specificity of H2O2-generating oxalate oxidase gene expression during wheat embryo germination. Plant J. 1998;15:165–71. doi: 10.1046/j.1365-313X.1998.00191.x. [DOI] [PubMed] [Google Scholar]
- 65.Çaliskan M. Germin, an oxalate oxidase, has a function in many aspects of plant life. Turk J Biol. 2000;24:717–24. [Google Scholar]
- 66.Caliskan M, Cuming AC. Temporal and spatial determination of germin biosynthesis in wheat tissues. Turk J Biol. 2000;24:775–82. [Google Scholar]
- 67.Lane BG. Oxalate oxidases and differentiating surface structure in wheat: germins. Biochem J. 2000;349:309–21. doi: 10.1042/0264-6021:3490309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Caliskan M, Turet M, Cuming AC. Formation of wheat ( Triticum aestivum L.) embryogenic callus involves peroxide-generating germin-like oxalate oxidase. Planta. 2004;219:132–40. doi: 10.1007/s00425-003-1199-9. [DOI] [PubMed] [Google Scholar]
- 69.Jacquard C, Mazeyrat-Gourbeyre F, Devaux P, Boutilier K, Baillieul F, Clément C. Microspore embryogenesis in barley: anther pre-treatment stimulates plant defence gene expression. Planta. 2009;229:393–402. doi: 10.1007/s00425-008-0838-6. [DOI] [PubMed] [Google Scholar]
- 70.Dunwell JM, Gibbins JG, Mahmood T, Naqvi SMS. Germin and germin-like proteins: evolution, structure, and function. Crit Rev Plant Sci. 2008;27:342–75. doi: 10.1080/07352680802333938. [DOI] [Google Scholar]
- 71.Grafi G, Chalifa-Caspi V, Nagar T, Plaschkes I, Barak S, Ransbotyn V. Plant response to stress meets dedifferentiation. Planta: Springer Berlin / Heidelberg, 2011:433-8. [DOI] [PubMed]
- 72.Wu G, Shao HB, Chu LY, Cai JW. Insights into molecular mechanisms of mutual effect between plants and the environment. A review. Agron Sustain Dev. 2007;27:69–78. doi: 10.1051/agro:2006031. [DOI] [Google Scholar]
- 73.von Arnold S, Sabala I, Bozhkov P, Dyachok J, Filonova L. Developmental pathways of somatic embryogenesis. Plant Cell Tissue Organ Cult. 2002;69:233–49. doi: 10.1023/A:1015673200621. [DOI] [Google Scholar]
- 74.Fehér A. Why Somatic Plant Cells Start to form Embryos? In: Mujib A, Å amaj J, eds. Somatic Embryogenesis: Springer Berlin / Heidelberg, 2006:85-101. [Google Scholar]
- 75.Zhao J, Morozova N, Williams L, Libs L, Avivi Y, Grafi G. Two phases of chromatin decondensation during dedifferentiation of plant cells: distinction between competence for cell fate switch and a commitment for S phase. J Biol Chem. 2001;276:22772–8. doi: 10.1074/jbc.M101756200. [DOI] [PubMed] [Google Scholar]
- 76.Fransz P, de Jong H. From nucleosome to chromosome: a dynamic organization of genetic information. Plant J. 2011;66:4–17. doi: 10.1111/j.1365-313X.2011.04526.x. [DOI] [PubMed] [Google Scholar]
- 77.Deal RB, Henikoff S. Capturing the dynamic epigenome. Genome Biol. 2010;11:218. doi: 10.1186/gb-2010-11-10-218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Deal RB, Henikoff S. Gene regulation: A chromatin thermostat. Nature. 2010;463:887–8. doi: 10.1038/463887a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Efroni S, Duttagupta R, Cheng J, Dehghani H, Hoeppner DJ, Dash C, et al. Global transcription in pluripotent embryonic stem cells. Cell Stem Cell. 2008;2:437–47. doi: 10.1016/j.stem.2008.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Desvoyes B, Sanchez MP, Ramirez-Parra E, Gutierrez C. Impact of nucleosome dynamics and histone modifications on cell proliferation during Arabidopsis development. Heredity (Edinb) 2010;105:80–91. doi: 10.1038/hdy.2010.50. [DOI] [PubMed] [Google Scholar]
- 81.Raut VV, Pandey SM, Sainis JK. Histone octamer trans-transfer: a signature mechanism of ATP-dependent chromatin remodelling unravelled in wheat nuclear extract. Ann Bot. 2011;108:1235–46. doi: 10.1093/aob/mcr232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Tanurdzic M, Vaughn MW, Jiang H, Lee T-J, Slotkin RK, Sosinski B, et al. Epigenomic consequences of immortalized plant cell suspension culture. PLoS Biol. 2008;6:2880–95. doi: 10.1371/journal.pbio.0060302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Cockerill PN. Structure and function of active chromatin and DNase I Hypersensitive Sites. FEBS Journal 2011:no. [DOI] [PubMed]
- 84.Chen ZJ, Tian L. Roles of dynamic and reversible histone acetylation in plant development and polyploidy. Biochim Biophys Acta 2007; 1769:295-307. [DOI] [PMC free article] [PubMed]
- 85.Schaefer DG, Delacote F, Charlot F, Vrielynck N, Guyon-Debast A, Le Guin S, et al. RAD51 loss of function abolishes gene targeting and de-represses illegitimate integration in the moss Physcomitrella patens. DNA Repair (Amst) 2010;9:526–33. doi: 10.1016/j.dnarep.2010.02.001. [DOI] [PubMed] [Google Scholar]
- 86.Raut VV, Sainis JK. 60Co-γ radiation induces differential acetylation and phosphorylation of histones H3 and H4 in wheat. Plant Biol (Stuttg) 2012;14:110–7. doi: 10.1111/j.1438-8677.2011.00463.x. [DOI] [PubMed] [Google Scholar]
- 87.Arnholdt-Schmitt B. Stress-induced cell reprogramming. A role for global genome regulation? Plant Physiol. 2004;136:2579–86. doi: 10.1104/pp.104.042531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Meyer AJ, Hell R. Glutathione homeostasis and redox-regulation by sulfhydryl groups. Photosynth Res. 2005;86:435–57. doi: 10.1007/s11120-005-8425-1. [DOI] [PubMed] [Google Scholar]
- 89.Alvarez ME, Nota F, Cambiagno DA. Epigenetic control of plant immunity. Mol Plant Pathol. 2010;11:563–76. doi: 10.1111/j.1364-3703.2010.00621.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Uchida N, Tasaka M. Intersections between immune responses and morphological regulation in plants. J Exp Bot. 2010;61:2539–47. doi: 10.1093/jxb/erq126. [DOI] [PubMed] [Google Scholar]
- 91.Murr R, Loizou JI, Yang Y-G, Cuenin C, Li H, Wang Z-Q, et al. Histone acetylation by Trrap-Tip60 modulates loading of repair proteins and repair of DNA double-strand breaks. Nat Cell Biol. 2006;8:91–9. doi: 10.1038/ncb1343. [DOI] [PubMed] [Google Scholar]
- 92.De Veylder L, Beeckman T, Inze D. The ins and outs of the plant cell cycle. 2007; 8:655-65. [DOI] [PubMed]
- 93.Balleza E, Alvarez-Buylla ER, Chaos A, Kauffman S, Shmulevich I, Aldana M. Critical dynamics in genetic regulatory networks: examples from four kingdoms. PLoS One. 2008;3:e2456. doi: 10.1371/journal.pone.0002456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Lehar J, Krueger A, Zimmermann G, Borisy A. High-order combination effects and biological robustness. 2008; 4. [DOI] [PMC free article] [PubMed]
- 95.Amzallag GN. Maturation of integrated functions during development. I. Modifications of the regulatory network during transition periods in Sorghum bicolor. Plant Cell Environ. 2001;24:337–45. doi: 10.1046/j.1365-3040.2001.00675.x. [DOI] [Google Scholar]
- 96.Greenberg JT. Programmed cell death: a way of life for plants. Proc Natl Acad Sci U S A. 1996;93:12094–7. doi: 10.1073/pnas.93.22.12094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Filonova LH, Bozhkov PV, Brukhin VB, Daniel G, Zhivotovsky B, von Arnold S. Two waves of programmed cell death occur during formation and development of somatic embryos in the gymnosperm, Norway spruce. J Cell Sci. 2000;113:4399–411. doi: 10.1242/jcs.113.24.4399. [DOI] [PubMed] [Google Scholar]
- 98.Chakrabarty D, Trivedi PK, Shri M, Misra P, Asif MH, Dubey S, et al. Differential transcriptional expression following thidiazuron-induced callus differentiation developmental shifts in rice. Plant Biol (Stuttg) 2010;12:46–59. doi: 10.1111/j.1438-8677.2009.00213.x. [DOI] [PubMed] [Google Scholar]
- 99.Lane BG. Oxalate, germin, and the extracellular matrix of higher plants. FASEB J. 1994;8:294–301. doi: 10.1096/fasebj.8.3.8143935. [DOI] [PubMed] [Google Scholar]
- 100.Davoine C, Le Deunff E, Ledger N, Avice J-C, Billard J-P, Dumas B, et al. Specific and constitutive expression of oxalate oxidase during the ageing of leaf sheaths of ryegrass stubble. Plant Cell Environ. 2001;24:1033–43. doi: 10.1046/j.1365-3040.2001.00757.x. [DOI] [Google Scholar]
- 101.Lane BG. Oxalate, germins, and higher-plant pathogens. IUBMB Life. 2002;53:67–75. doi: 10.1080/15216540211474. [DOI] [PubMed] [Google Scholar]
- 102.Christensen AB, Thordal-Christensen H, Zimmermann G, Gjetting T, Lyngkjaer MF, Dudler R, et al. The germinlike protein GLP4 exhibits superoxide dismutase activity and is an important component of quantitative resistance in wheat and barley. Mol Plant Microbe Interact. 2004;17:109–17. doi: 10.1094/MPMI.2004.17.1.109. [DOI] [PubMed] [Google Scholar]
- 103.Le Deunff E, Davoine C, Le Dantec C, Billard J-P, Huault C. Oxidative burst and expression of germin/oxo genes during wounding of ryegrass leaf blades: comparison with senescence of leaf sheaths. Plant J. 2004;38:421–31. doi: 10.1111/j.1365-313X.2004.02056.x. [DOI] [PubMed] [Google Scholar]
- 104.Zimmermann G, Bäumlein H, Mock H-P, Himmelbach A, Schweizer P. The multigene family encoding germin-like proteins of barley. Regulation and function in Basal host resistance. Plant Physiol. 2006;142:181–92. doi: 10.1104/pp.106.083824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Manosalva PM, Davidson RM, Liu B, Zhu X, Hulbert SH, Leung H, et al. A germin-like protein gene family functions as a complex quantitative trait locus conferring broad-spectrum disease resistance in rice. Plant Physiol. 2009;149:286–96. doi: 10.1104/pp.108.128348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Yeung EC, Belmonte MF, Tu LTT, Stasolla C. Glutathione modulation of in vitro development (2005) In Vitro Cell Dev Biol Plant. 2005;41:584–90. doi: 10.1079/IVP2005683. [DOI] [Google Scholar]
- 107.Pasternak T, Ötvös K, Domoki M, Fehér A. Linked activation of cell division and oxidative stress defense in alfalfa leaf protoplast-derived cells is dependent on exogenous auxin. Plant Growth Regul. 2007;51:109–17. doi: 10.1007/s10725-006-9152-0. [DOI] [Google Scholar]
- 108.Datta SK, Peterhans A, Datta K, Potrykus I. Genetically Engineered Fertile Indica-Rice Recovered from Protoplasts. Nat Biotechnol. 1990;8:736–40. doi: 10.1038/nbt0890-736. [DOI] [Google Scholar]
- 109.Hiei Y, Ohta S, Komari T, Kumashiro T. Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J. 1994;6:271–82. doi: 10.1046/j.1365-313X.1994.6020271.x. [DOI] [PubMed] [Google Scholar]
- 110.Tada Y, Sakamoto M, Fujimura T. Efficient gene introduction into rice by electroporation and analysis of transgenic plants: use of electroporation buffer lacking chloride ions. Theor Appl Genet. 1990;80:475–80. doi: 10.1007/BF00226748. [DOI] [PubMed] [Google Scholar]
- 111.Karthikeyan A, Shilpha J, Karutha Pandian S, Ramesh M. Agrobacterium-mediated transformation of indica rice cv. ADT 43. Plant Cell Tissue Organ Cult. 2012;109:153–65. doi: 10.1007/s11240-011-0083-8. [DOI] [Google Scholar]
- 112.Ozawa K. Establishment of a high efficiency Agrobacterium-mediated transformation system of rice (Oryza sativa L.) Plant Sci. 2009;176:522–7. doi: 10.1016/j.plantsci.2009.01.013. [DOI] [PubMed] [Google Scholar]
- 113.Ozawa K. A high-efficiency Agrobacterium-mediated transformation system of rice (Oryza sativa L.). In: Dunwell JM, Wetten AC, eds.: Humana Press, 2012:51-7. [DOI] [PubMed] [Google Scholar]
- 114.Patnaik D, Khurana P. Wheat biotechnology: A mini review. Electronic J. Biotechnology Electronic J Biotechnol. http://www.ejbiotechnology.info/index.php/ejbiotechnology/article/viewFile/v4n2-4/903, 2001:74-102.
- 115.Vasil V, Castillo A, Fromm M, Vasil I. Herbicide resistant fertile transgenic wheat plants obtained by microprojectile bombardment of regenerable embryogenic callus. Bio-Technology. 1992;10:667–74. [Google Scholar]
- 116.Slade AJ, Knauf VC. TILLING moves beyond functional genomics into crop improvement. Transgenic Res. 2005;14:109–15. doi: 10.1007/s11248-005-2770-x. [DOI] [PubMed] [Google Scholar]
- 117.Regina A, Bird A, Topping D, Bowden S, Freeman J, Barsby T, et al. High-amylose wheat generated by RNA interference improves indices of large-bowel health in rats. Proc Natl Acad Sci U S A. 2006;103:3546–51. doi: 10.1073/pnas.0510737103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Gil-Humanes J, Pistón F, Hernando A, Alvarez JB, Shewry PR, Barro F. Silencing of [gamma]-gliadins by RNA interference (RNAi) in bread wheat. J Cereal Sci. 2008;48:565–8. doi: 10.1016/j.jcs.2008.03.005. [DOI] [Google Scholar]
- 119.Dong C, Beetham P, Vincent K, Sharp P. Oligonucleotide-directed gene repair in wheat using a transient plasmid gene repair assay system. Plant Cell Rep. 2006;25:457–65. doi: 10.1007/s00299-005-0098-x. [DOI] [PubMed] [Google Scholar]