Abstract
Mediator is a conserved eukaryotic multiprotein complex required by RNA polymerase II for transcription of its target genes. Till date, there is no report explaining the signals that affect the overall concentration of individual Med subunits. In this report, we have analyzed the effect of different phytohormones and stresses on the transcript level of Med genes in Arabidopsis. Hormones like auxin and JA, and cold stress did not show significant effect. ABA moderately increased the transcript abundance of more than 70% of AtMed genes analyzed in this study. However, there was noticeable change in the transcript level of several AtMed genes in response to BR. Stresses like high light, dark and salt also caused significant change in the transcript abundance of many AtMed genes. These data reveal that different environmental cues can affect stoichiometric concentration of Med subunits by affecting the transcription of their respective genes. This may, in turn, affect the overall arrangement of functional Mediator complex. This also suggests that some subunits may have some specific functions to play in response different signals.
Keywords: Med subunit, Mediator, hormones, stress, transcription
Introduction
Mediator, a necessary component for RNA pol II mediated transcription of genes, was first discovered in yeast, and then subsequently was found in many fungi and metazoans.1,2 In 2007, the first plant Mediator complex was biochemically purified from Arabidopsis cell suspension culture.3 Following this, through bioinformatic analyses, subunits of Mediator complex were reported in many other plants including rice.2,4 Thus, Mediator complex is found in all the eukaryotes ranging from unicellular yeast to multicellular humans and plants suggesting its emergence before divergence of plants, animals and fungi. Consistent to this, in yeast and metazoans, Mediator has been found to play crucial roles in basic process of transcriptional regulation of eukaryotic genes. The genetic, biochemical and structural studies revealed the modular nature of the Mediator, wherein all the subunits are arranged in four modules; Head, Middle, Tail and Kinase.5,6 The ability of specific subunits of Mediator complex to interact with site-specific transcription factors and RNA Pol II make it suitable to play a role as a communication bridge between transcriptional regulators and transcriptional machinery. In yeast and animals, several nuclear receptors and nuclear receptor-like transcription factors have been found to target Med1, Med14 and Med15 for their function.7-12 Transcription factor VP16 interacts with Med17 and Med25 for its transcriptional activity, whereas E1A and ELK1 require Med23.13-16 Similarly, NANOG, β-catenin and REST target MED12.17-19 Subunits like Med5 and Med17 were shown to interact with RNA pol II and its associated GTFs.20-23 By chromatin immunoprecipitation experiments, Mediator was shown to co-localize with RNA pol II at many promoters, and additionally also at activator-bound regulatory elements.24-27 By virtue of its ability to interact directly with pol II, besides contributing a lot to the transcription initiation process, Mediator also plays important role during transcript elongation. A functional link between Mediator and elongation factor DSIF, physical interaction between Mediator and P-TEFb, and a functional commonality between MED31 and elongation factor SII suggest critical role of Mediator in the process of elongation of transcripts.28-30 There are many reports suggesting functional interactions of Mediator with chromatin co-factors. Acevedo et al. (2003) demonstrated synergistic function of Mediator and p300 during ERα-dependent transcription.31 Mediator can interact with both p300 and TFIID, and so might be coordinating the exchange of chromatin modifying co-factors and PIC components.32 This view is further strengthened by the observation of close association between Mediator and SAGA complex.33,34 There is a possibility that Mediator is also controlling epigenetic pathways as it was found in a complex of REST-Mediator-G9a.35 REST is a silencing factor and G9a is a methyltransferase, which methylates H3K9 at target genes.19
In plants, functions of some of the Mediator subunits have been established. In Arabidopsis, Med14, originally identified as SWP, affects cell number and shoot meristem development.36 AtMed14 coordinates with HDAC19 for its function.37 Moreover, AtMed14 might have a role in splicing as overexpression of its cDNA could complement the smp mutant phenotype.38 Other Mediator subunits like AtMed12, AtMed13 and AtCdk8 are also required for regulation of cell differentiation and development of plant.39-41 The multifaceted AtMed25/PFT1 is involved in regulation of flowering time in response to light quality, hormonal signaling, and biotic or abiotic responses, by the virtue of its ability to interact with several key transcriptional regulators.42-44 Three Arabidopsis Mediator subunits AtMed17, AtMed18 and AtMed20 were found to regulate levels of miRNAs.45 Authors suggested that in plants, Mediator might also be cooperating with other RNA polymerases. Consistent with this, AtMed36 subunit was co-purified with the largest subunit of RNA Pol V.46 Moreover, AtMed36 was predicted to be Fibrillarin which is involved in processing of rRNA.47 The molecular mechanism by which specific plant Mediator subunits regulate specific processes is not known. However, as the case of AtMed25 suggests, different subunits might be targeted by different activators and repressors for their respective transcriptional responses. Moreover, interaction of different specific subunits with the components of transcriptional machinery might be conveying the transcriptional signals to the RNA polymerase.
Thus, in all the eukaryotes, the main function of Mediator complex is to regulate the process of transcription. In such scenario it is logical to suggest that abundance of Med subunits can affect the overall composition of the complex, and hence the transcriptional response. We do not know if developmental or environmental signals affect the level of Med subunits and the resultant complex. There is no comprehensive report on transcriptional regulation of Med genes coding for Mediator subunits in any organism. In this report, we have tried to analyze the regulatory promoter elements of the Med genes, and studied the effect of different environmental cues on the transcript level of Med genes in Arabidopsis.
Results and Discussion
Gene structure and chromosomal distribution of Mediator subunit genes in Arabidopsis
Structure of all the genes coding for Mediator subunits in Arabidopsis were obtained from TAIR. The Mediator subunit genes contain variable number of introns ranging from 0 to 27. About 20% genes in Arabidopsis are intronless, but we found only one such Med subunit encoding gene, AtMed4.48 The AtMed2 and AtCdk8 genes have simple structure with one large intron, whereas AtMed35 has 27 introns. All the other Med subunit encoding genes in Arabidopsis were found to contain ≥ 2 introns (Fig. S1) suggesting their gradual evolution unlike rapid evolution of intronless genes by duplication or reverse transcription/integration.48 The intron/exon arrangement of all the Med genes in Arabidopsis are shown in Figure S1.
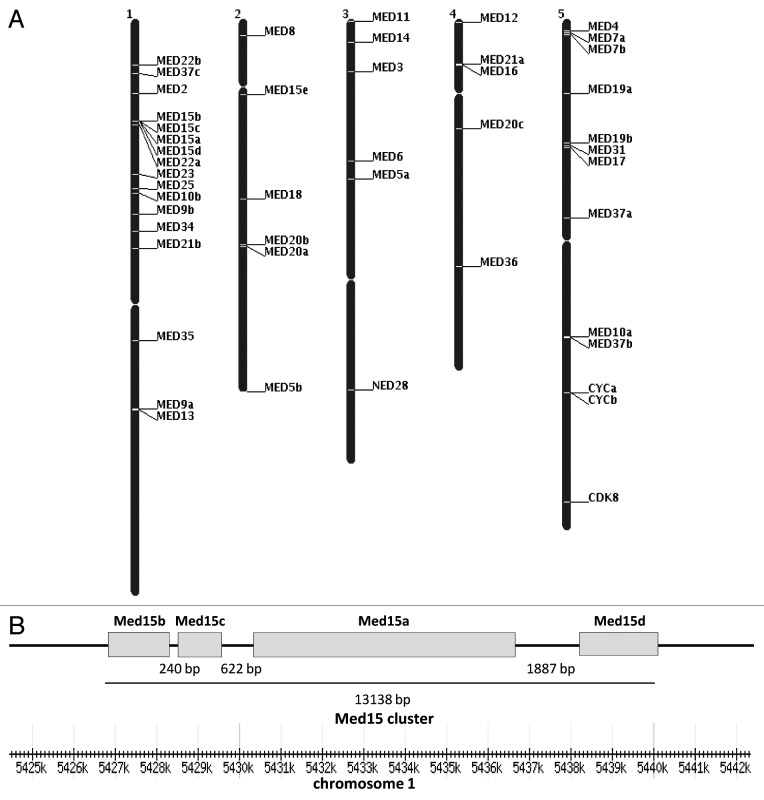
Location of Med subunit encoding genes mapped on Arabidopsis chromosome pseudomolecules available at TAIR is shown in Figure 1A. The exact positions and orientations of all the Med genes in Arabidopsis chromosome pseudomolecules are mentioned in Table S1. The AtMed genes were found to be scattered randomly on the chromosomes of Arabidopsis. Chromosome 1, which is the biggest one, code for 17 Med subunits, whereas the smallest chromosome (chromosome 5) harbour five Med genes. Thus, more than 36% of Med genes are located on chromosome 1. The interesting feature on chromosome 1 is presence of four Med15 paralogs (out of 5) in a span of just 13138 bases in the order of Med15b-Med15c-Med15a-Med15d (Fig. 1B). In this Med15 cluster, intergenic distance between Med15d and Med15a is 1887 bases, Med15a and Med15c is 622 bases, whereas Med15c is separated from Med15b by an intergenic region of just 240 bases. In Arabidopsis, among all the Med subunits, we found that Med15 has the maximum number of paralogs (Fig. 1A, Table S1). In fungi and metazoans, Med15 is required by several transcription factors for their transcriptional responses.9,11,12,49 Thus, in Arabidopsis, more number of Med15 paralogs might help in broadening the regulatory capability of Mediator complex by interacting with diverse transcription factors. In order to see if all these Med15 genes are expressed or not, we performed RT-PCR analysis and found that all the paralogs being expressed in one or the other tissues of Arabidopsis (Kumar and Thakur, in preparation). It will be interesting to study the functional relevance of these expressed Med15 genes.
Figure 1. (A) Genomic distribution of Med genes on Arabidopsis chromosomes. Chromosome numbers are indicated at the top of the black bars which represent chromosomes. (B) Arrangement of AtMed15 genes in the Med15 cluster on chromosome 1. Total length of Med15 cluster and intergenic distances between different AtMed15 genes are shown in bp. Scale at the bottom shows distance from the telomeric end.
In silico analysis of Med gene promoters
Since promoters dictate the expression pattern of a gene, we analyzed promoter regions of Med genes in Arabidopsis. The putative 2 kb AtMed promoter sequences were identified in silico. Bioinformatic analyses of these sequences revealed presence of highly represented regulatory elements known in plant gene promoters, including elements responsive to hormones and stresses. Most of these elements were present within 1 kb promoter. Figure 2 shows number of Arabidopsis Med genes carrying a particular responsive element in their promoter regions. Elements like ACGT, REalpha and I-box, responsible for response to light or darkness, were found to be present in more than 20 promoters of AtMed genes. Although in less number of AtMed genes, elements important for response to hormones like auxin (ARF, ASF1, AuxRE and SAUR), ABA (ABRE) or JA (T/G box and GCC core), and stresses like cold (LTRE) or salinity (SCAM) were also found. In order to understand the significance of presence of these elements in the upstream regulatory regions of AtMed genes, we studied the effect of hormones and stresses on the transcript abundance of 31 Med genes in Arabidopsis.
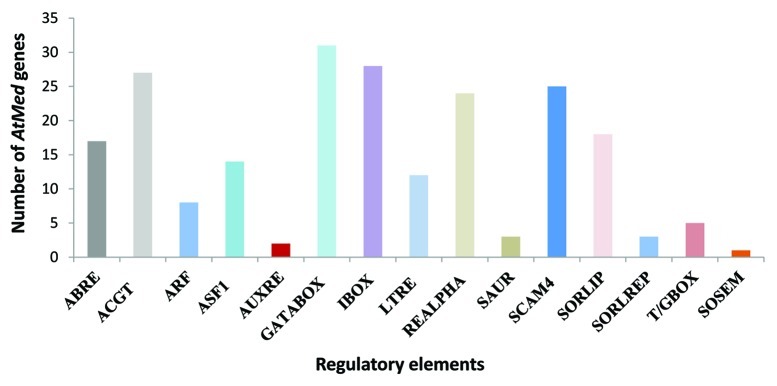
Figure 2. Total number of AtMed genes (y-axis) harboring a particular responsive element (shown on x-axis) in its promoter region. ABRE, ACGT, GATABOX (GATA box), IBOX (I box), REALPHA (REalpha), SORLIP and SORLREP are light/dark responsive elements. ARF, ASF1, AUXRE and SAUR are important for auxin response. ACGT and SOSEM represent ABA responsive elements. LTRE is responsive to cold and ABA. SCAM4 is important for response to salinity stress.
Hormonal regulation of transcription of AtMed genes
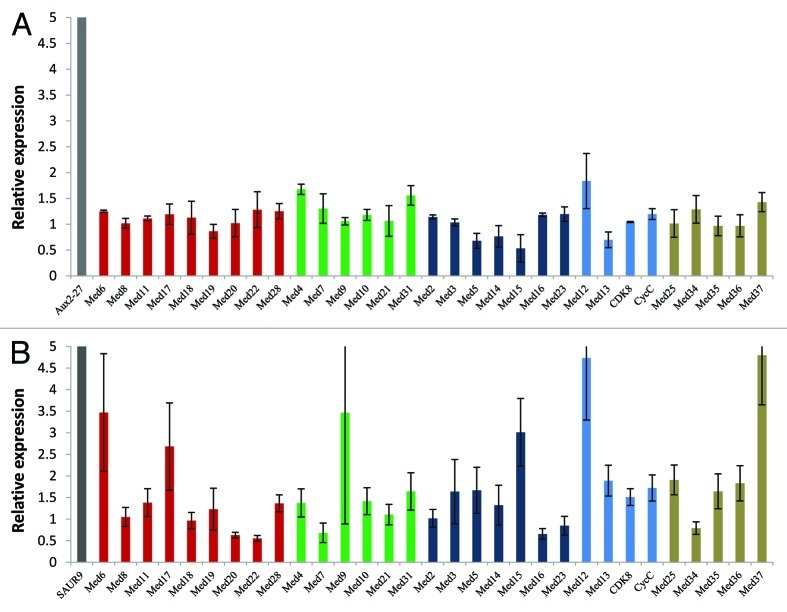
phytohormones play very important role in almost all the aspects of plant growth and development. They regulate transcription of a plethora of plant genes. In order to know if auxin affected abundance of Med subunits, we treated Arabidopsis seedlings with IAA and analyzed the change in transcript level of Med genes. Change in transcript level of Aux2-27 was used as positive control for auxin treatment.50 As evident from Figure 3A, very few AtMed genes were affected by IAA. Moreover, the effect on these few genes was not so robust. Three AtMed genes, AtMed12, AtMed4 and AtMed31, showed slight increase (> 1.5 fold) in their transcript levels, whereas AtMed15, AtMed5, AtMed13 and AtMed14 showed > 40% reduction. Interestingly, AtMed15 was severely reduced in the presence of auxin (Fig. 3A). It will be interesting to see how reduction in the level of this paralog of AtMed15 helps in auxin signaling. Just like auxin, brassinosteroid (BR) is another phytohormone that play very critical role in growth and differentiation of plants. A defect in BR signaling pathways can make plants dwarf and sterile. In order to understand the effect of BR on genes coding for AtMed subunits, change in their transcript levels in the Arabidopsis seedlings grown in the medium with BR were compared with that in the normally grown seedlings. Increase in transcript level of SAUR9 by BR was used as control.51 Surprisingly, unlike that in the case of auxin, several AtMed genes were significantly upregulated in the presence of BR (Fig. 3B). Transcription of six AtMed genes, AtMed6, AtMed17, AtMed9, AtMed15, AtMed12, and AtMed37, were increased by more than 2.5 fold. Some other AtMed genes were moderately upregulated (eight; more than 1.5 fold), although four (AtMed20, AtMed22, AtMed7 and AtMed16) were found to be slightly downregulated (less than 0.6 fold). Thus, although BR and auxin are known to act synergistically, expression of AtMed genes are affected differently by these two important plant hormones. This observation is significant because almost 40% auxin regulated genes are also affected by BRs, and many BR-regulated promoters have been found to harbour auxin responsive elements.52,53 There is a possibility that these hormones give signals for different arrangement of Med subunits for their transcriptional outputs.
Figure 3. Quantitative real-time RT-PCR analysis showing change in the transcript abundance of Med genes in Arabidopsis seedlings treated with (A) auxin or (B) brassinosteroid. Aux2–27 and SAUR9 were used as positive controls for respective treatments. The relative expression level represents fold change to control seedlings. Mean values from three independent experiments are shown. Error bars represent standard deviation. Red, green, dark blue and blue bars represent AtMed genes coding for subunits of Head, Middle, Tail and Kinase modules, respectively. Subunits, whose positions are not known, are shown in tan color.
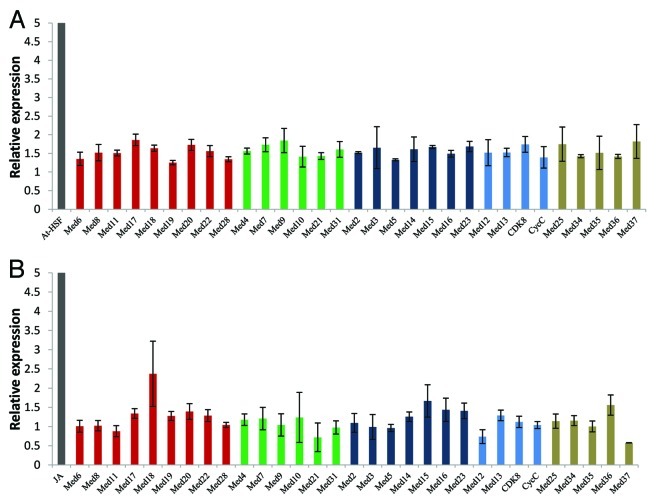
Phytohormone, abscisic acid (ABA), in addition to many growth and development related processes, is important for plants in response to several abiotic stresses. When Arabidopsis seedlings were grown in media with ABA, not much of the significant effect was observed in transcript level of Med genes, though transcript levels of many were increased by more than just 1.5 fold (Fig. 4A). These moderately upregulated Med genes were AtMed17, 9, 37, 25, 7, 20, 23, 15, 3, 18, 14, 31, 4, 22, and AtCdk8. In the case of ABA, not a single AtMed gene was downregulated. Relative expression of AtHsf was used as a positive control for effect of ABA on gene expression. Jasmonic acid (JA) is a jasmonate class of plant hormone that promotes plant defense responses against pathogen attack and physical wound. In order to know if JA changes the level of any Med subunit, we analyzed the transcript levels of 31 Med genes in the Arabidopsis seedlings grown in the agar media with JA (Fig. 4B). Upregulation in the transcript level of JAZ in comparison to the seedlings grown in normal agar medium without JA was used as control.54 AtMed15 and AtMed36 showed little more than 1.5 fold increase in the transcript level, whereas AtMed18 showed more than 2 fold increase (Fig. 4B). On the other hand, transcript levels of AtMed12, AtMed21 and AtMed37 decreased in the seedlings grown in JA. All other AtMed genes did not show any difference in their transcript abundance in response to JA treatment. Thus, except AtMed18 as in the case of JA, no other AtMed gene showed significant upregulation in response to two main stress response regulating hormones, JA and ABA. In Arabidopsis, partial loss of function of AtMed18 causes developmental defects like shorter petioles, abnormal phyllotaxy, late flowering, abnormal flowers, and reduced fertility.45 These phenotypes are regulated by signaling pathways evoked by diverse plant hormones. It will be interesting to see if hormones, like JA, require AtMed18 for these functions.
Figure 4. Quantitative real-time RT-PCR analysis showing change in the transcript abundance of Med genes in Arabidopsis seedlings treated with (A) abcisic acid or (B) jasmonic acid. AtHSF and JA were used as positive controls for respective treatments. The relative expression level represents fold change to control seedlings. Mean values from three independent experiments are shown. Error bars represent standard deviation. Red, green, dark blue and blue bars represent AtMed genes coding for subunits of Head, Middle, Tail and Kinase modules, respectively. Subunits, whose positions are not known, are shown in tan color.
Effect of light on the transcription of AtMed genes
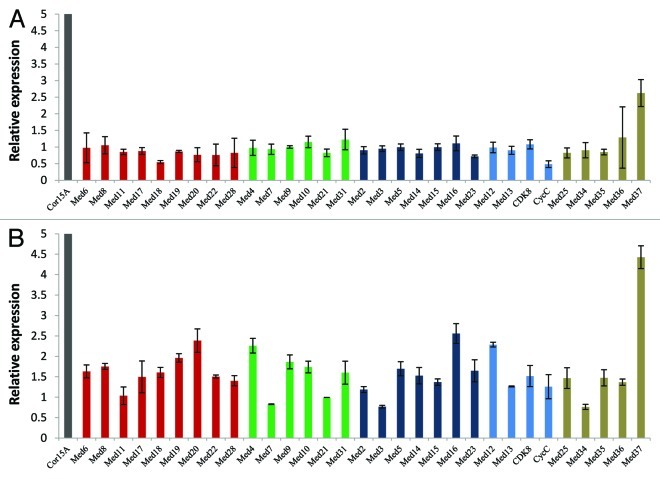
Being sessile organisms, plants possess a well organized system to assess the quality of light and utilize it for their growth and survival, and to modulate different developmental programmes including flowering. In order to see if light affected stoichiometric concentration of Med subunits in plants, we analyzed their transcript abundance in Arabidopsis seedlings either treated with more than normal range of light intensity or grown in complete darkness. In etiolated seedlings, transcription of AtMed15 and AtMed37 was significantly upregulated, more than 2-fold, in comparison to normally grown seedlings under 14 h light and 10 h dark cycle (Fig. 5A). Additionally, there was slight increase (> 1.5 fold) in the transcript levels of AtMed14 and AtMed19 in these seedlings. Interestingly, more number of AtMed genes were downregulated in dark. Transcript levels of AtMed22, AtCycC, AtMed36, AtMed17, AtMed3, AtMed20, AtMed25, AtMed11, AtMed21 and AtMed18 were reduced to half or less in etiolated seedlings in comparison to that in normally grown seedlings (Fig. 5A). Arabidopsis belongs to the category of long day plants, so it is sensitive to complete darkness. This might be the reason that it downregulates its transcriptional machinery when grown in complete darkness. Consistent with the observation in etiolated seedlings, many AtMed genes were upregulated when normally grown Arabidopsis seedlings were subjected to high light intensity (150 W m−2) for 3 h. Transcript level of five genes, AtMed12, AtMed25, AtMed17, AtMed23 and AtMed15, increased more than 2-fold in response to high light, though nine more AtMed genes were moderately upregulated, more than 1.5 fold and less than 2 fold (Fig. 5B). Thus, presence and absence of light, which is an essential requirement for plant growth and development, affect expression of most of the AtMed genes. It is noticeable that, except AtMed15 and AtMed37, all other Med genes were either upregulated by high light or downregulated by dark. Surprisingly, AtMed15 and AtMed37 were found to be positively affected by both the high light and complete darkness. There is a possibility that transcription factors required for transcriptional response of light/dark might be targeting them for their response. In fungi and animals, Med15 subunit has been found to interact with several transcription factors. According to bioinformatic analyses, Med37 seems to be a plant specific subunit, suggesting its importance in light/dark mediated plant specific process. As mentioned earlier, we used high intensity of white light for this study. Since different photoreceptors perceive different wavelengths of light to regulate complex photomorphogenic events in plants, it will be interesting to study the role of Med subunits in UV, red, far-red and blue light responses.
Figure 5. Quantitative real-time RT-PCR analysis showing change in the transcript abundance of Med genes in Arabidopsis seedlings (A) grown in complete darkness or (B) treated with high light. Cor15A were used as positive control for high light stress treatment. The relative expression level represents fold change to control seedlings. Mean values from three independent experiments are shown. Error bars represent standard deviation. Red, green, dark blue and blue bars represent AtMed genes coding for subunits of Head, Middle, Tail and Kinase modules, respectively. Subunits, whose positions are not known, are shown in tan color.
Stress mediated changes in the transcription of AtMed genes
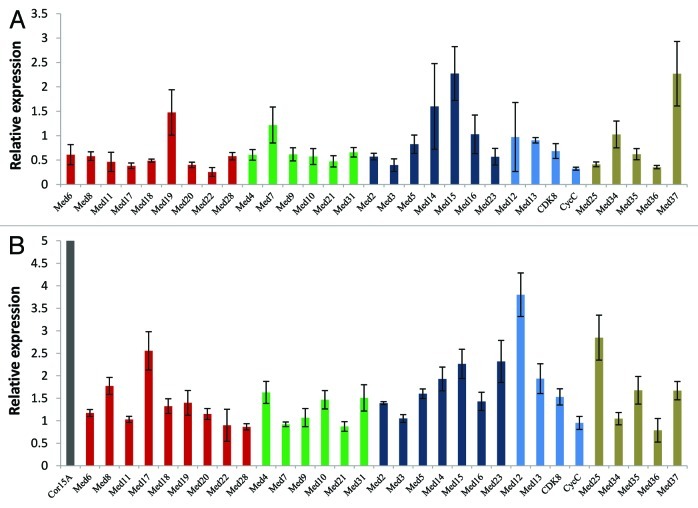
Plants face so many stresses in their lifetime. In order to cope up with these stresses, they have different network of gene expression. We analyzed effect of cold or salt on the transcription of AtMed genes. In the Arabidopsis seedlings kept in cold for 24 h, transcript level of AtMed37 increased more than 2.5 folds whereas that of AtMed18 and AtCycC decreased 2-fold (Fig. 6A). Three AtMed genes, AtMed20, AtMed22 and AtMed23, were slightly downregulated. There was not much effect on any other AtMed gene. Even, transcription of AtMed16, which was characterized as SENSITIVE TO FREEZING6 (SFR6) having prominent role in cold acclimation, was not affected by cold.55 Increase in Cor15A transcript was used as a positive control to study the effect of cold.56 A number of literatures exist which explains transcriptome profile of different plants including Arabidopsis, in response to NaCl treatment which is used as a laboratory method to mimic stress caused by salinity in the soil or water used for irrigation.57 In our analysis, five AtMed genes (AtMed37, AtMed16, AtMed20, AtMed12 and AtMed4) showed more than 2-fold increase in their transcript abundance in response to presence of NaCl in the media, whereas 13 other AtMed genes were moderately upregulated by 1.5 fold or more (Fig. 6B). Expression of Cor15A was assessed as the control for salinity stress.58 A couple of AtMed genes, AtMed3 and AtMed34, showed slight reduction in their transcript level under salt stress. Thus, just like ABA response, a big number of AtMed genes were found to be upregulated by NaCl in Arabidopsis. This supports the fact that ABA plays important role in response to salt stress. Significant overlap in the Med genes showing increased transcription in the presence of ABA or salt indicate a possibility, that during salt or other stress, ABA maintains the increased concentration of functional Mediator complex by increasing the abundance of most of the subunits.
Figure 6. Quantitative real-time RT-PCR analysis showing change in the transcript abundance of Med genes in Arabidopsis seedlings subjected to (A) cold stress or (B) salinity stress treatment. Cor15A was used as positive control for both the stresses. The relative expression level represents fold change to control seedlings. Mean values from three independent experiments are shown. Error bars represent standard deviation. Red, green, dark blue and blue bars represent AtMed genes coding for subunits of Head, Middle, Tail and Kinase modules, respectively. Subunits, whose positions are not known, are shown in tan color.
Materials and Methods
Plant materials and treatments
Arabidopsis thaliana wild-type seeds (ecotype Columbia Col 0) were sterilized and then sown on MS-agar (Murashige and Skoog medium supplemented with 0.8% agar and 2% sucrose) plates. Seeds were stratified in dark for 48 h at 4°C, and then transferred to growth room. The seedlings were grown at 22°C for 10 d under 16 h light and 8 h dark cycle. According to the experimental requirements, 10 d old seedlings were transferred to MS-agar with 25 µM ABA or 30 µM JA or 200 mM NaCL, and grown for another 3 d. For cold stress treatment, 10 d old seedlings on MS-agar plates were incubated at 4°C for 24 h. IAA and BR treatments were given in liquid medium. For this, 10 d old seedlings were transferred to a flask containing liquid MS medium supplemented with 10 µM IAA or 1 µM Epibrassinolide, and then were shaken at 100 rpm for 3 h. For light stress treatment, 10 d old seedlings were incubated under high light (800µE m−2 s−2) in phytotrone (PGV36) for 3 h. After required growth and treatments, seedlings were harvested, immediately frozen in liquid nitrogen and then stored in -80°C till further use.
Isolation of RNA and reverse transcription
Total RNA was isolated from Arabidopsis seedlings using Spectrum Plant Total RNA Kit (SIGMA Life Sciences) following the manufacturer’s instructions. The amount and quality of RNA was checked by Nano Nanodrop ND-1000 Spectrophotometer (Thermo SCIENTIFIC) and further confirmed by electrophoresis in 1.2% formamide agarose gel. For each sample, 5μg of total RNA was reverse transcribed to cDNA in 33 μl reaction using First- Strand cDNA Synthesis Kit (GE Healthcare). Reverse transcription were performed at 37°C for 1 h. 50 times diluted cDNA mix was used for qPCR. qPCR reaction included SYBR Green Master Mix (Applied Biosystem), a gene specific primers and 2µl of the 50 times diluted cDNA. List of primers used in this study are given in Table S2.
Real time PCR analysis
β-Actin was used as an internal control. Primers for all the genes were designed by using coding sequences of Arabidopsis genes available on TAIR database http://www.arabidopsis.org/ by using the online available software Integrated DNA Technologies Oligo Analyzer3.1 for calculating the Tm and GC content of primers. Details of primers used in this study are given in Table S2. The reaction conditions were: (95°C for 5 min, followed by 40 cycles at 95°C for 30 sec and 60°C for 60 sec, and then a final extension of 72°C for 5 min). qPCR was performed and analyzed in 7900 HT Real-Time PCR Detection System (Applied Biosystem). At least three biological replicates have been analyzed for every gene. Three technical replicates have been employed for each biological replicate. The threshold cycles (Ct) of each tested genes were averaged for triplicate reactions and the values were normalized to endogenous control of β-Actin gene (Ct Target gene – Ct Endogenous control) then normalized to reference sample for calculating the fold change.
Prediction of AtMed gene promoters and mapping of genes on Arabidopsis chromosomes
The promoter sequences (2000bp upstream 5′UTR region) were obtained from the sequence of BAC clones carrying the AtMed genes. The BAC sequences were obtained after BLASTX search on NCBI database (http://blast.ncbi.nlm.nih.gov) using cds of AtMed genes as the queries. The PLACE Database available on www.dna.affrc.go.jp was used to get different abiotic stress- and hormone-responsive elements. The TAIR based chromosome map tool www.arabidopsis.org/ was used to find the location of all the Mediator genes.
Conclusion
In plants, since the initial purification and characterization of Mediator in 2007, a number of reports have emerged revealing the role of the subunits of this complex in diverse processes like hormonal signaling, plant growth and development, flowering, and response to different biotic and abiotic stresses.42 Thus, just like in fungi and metazoans, in plants too, several pathways converge on Mediator. There is no information on the direct effect of these pathways on the transcriptional regulation of Med gene expression. As summarized in Figure 7, in this study, some hormones like BR and ABA, and stresses like high light, darkness and salinity, affected transcript abundance of many AtMed genes. On the other hand, hormones like auxin and JA, and cold stress did not affect so many AtMed genes. To our knowledge, this is the first such study on Med genes in any eukaryotic organism. We found that BR signaling had significant impact (increase) on the transcript level of at least six AtMed genes; two coding for subunits of Head module, one for each of the other modules. Product of AtMed37, which is highly upregulated by BR, has not yet been assigned to any module. It will be interesting to find out if these subunits can make a functional core structure or they provide the peripheral variation to the interface, to play some role in transcriptional response of BRs. We can however, not rule out the specific role of these subunits in BR signaling. A similar observation was made in the case of salinity stress. In another scenario, ABA moderately upregulated more than 70% of tested Med genes in young Arabidopsis seedlings (Fig. 4A and Fig. 7). Many reports have suggested that among all the hormones, ABA affects maximum number of plant genes (> 10% of protein coding genes in Arabidopsis) at the transcript level suggesting its pivotal role in plants.59,60 Thus, there is requirement of more transcriptional machineries to meet the demand. Our results suggest that ABA increases the overall concentration of Med subunits to form more Mediator complexes to cover all the target promoters. Consistent with this, we made similar observation in the case of Arabidopsis seedlings growing in the presence of NaCl mimicking salinity stress that use ABA pathway for its transcriptional responses. One other striking observation was downregulation of almost 68% of tested Med genes in etiolated Arabidopsis seedlings (Fig. 7). Seedlings grown in complete darkness adopt a skotomorphogenic program of development by actively repressing transcription of most of the genes to conserve the reserved energy. Thus, it seems that since transcriptional activities are down, expression level of Med genes coding for Mediator complex, an important component of transcriptional machinery, are also down in the dark grown seedlings. On the other hand, exposure of etiolated seedlings to light is known to induce massive reprogramming of the transcriptome.61 Transcript abundance of more than 20% of Arabidopsis genes increased when etiolated seedlings were irradiated with light for just about one hour.62,63 Thus, it is not surprising that more than 45% of AtMed genes are upregulated in response to high light (Fig. 7). Arabidopsis and other land plant cells require light for growth and differentiation. We hypothesize that in complete darkness, Arabidopsis downregulates its Mediator and transcriptional machinery. When etiolated seedlings are exposed to light, components of transcriptional machinery including the Med subunits, are increased to fulfil the demand of increased transcriptional activity to support the photomorphogenic development. Earlier, in mammalian study, a drastic reduction and nearly complete loss of Mediator subunits was reported in differentiated myotube cells.64 This suggests that the function of Mediator in differentiation is conserved in plants and animals. As analyzed in the Venn diagramme, many AtMed genes were upregulated by more than one treatment (Fig. 8A). This suggests that different signaling pathways merge on one or more Med subunits. Contrarily, very less number of AtMed genes was downregulated by two or more individual treatments (Fig. 8B). Thus, different signaling pathways seem to merge on Med subunits preferably by increasing its stoichiometric level in the complex, confirming the role of Med complex as an integrative hub for diverse signaling cascades.
Figure 7. Summary of changes in the transcript abundance of AtMed genes in response to different hormones or stresses. Color legends explaining the range of fold changes are shown at the bottom.
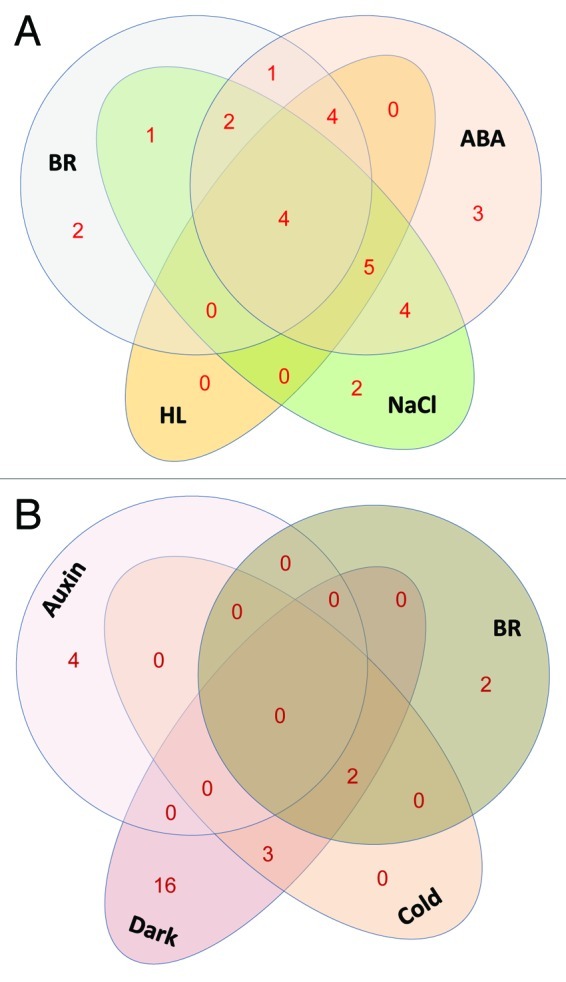
Figure 8. Schematic representation of number of AtMed genes commonly upregulat ed (A) or downregulated (B) by different factors (hormones or stresses) mentioned in bold black letters. BR, brassinosteroid; ABA, abscic acid; HL, high white light. Numerals denote number of AtMed genes.
Considering intrinsic modularity of the complex, variation may arise in the function of the plant Mediator complex. One level of heterogeneity may arise due to preferential incorporation of one of the subunit paralogs, into the functional complex. For example, replacement of one AtMed15 paralog with any other four remaining paralogs. Additional variation may arise from overexpression or substoichiometric expression of a given subunit or a set of subunits relative to other Med subunits. The factors, including hormones or stresses, affecting the expression level of Med subunits, may thus, be affecting the overall composition of functional Mediator complex, making composition and arrangement of the subunits a context specific response.
Supplementary Material
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgment
This work was supported by IYBA grant (BT/BI/12/045/2008) from Department of Biotechnology, Ministry of Science and Technology, Government of India. We acknowledge the Central Instrumentation Facility of NIPGR for real-time PCR reactions.
Glossary
Abbreviations:
- Med
Mediator
- AtMed
Arabidopsis thaliana Mediator
- IAA
indole acetic acid
- JA
jasmonic acid
- ABA
abscic acid
- BR
brassinosteroid
Footnotes
Previously published online: www.landesbioscience.com/journals/psb/article/22438
References
- 1.Boube M, Joulia L, Cribbs DL, Bourbon HM. Evidence for a mediator of RNA polymerase II transcriptional regulation conserved from yeast to man. Cell. 2002;110:143–51. doi: 10.1016/S0092-8674(02)00830-9. [DOI] [PubMed] [Google Scholar]
- 2.Bourbon HM. Comparative genomics supports a deep evolutionary origin for the large, four-module transcriptional mediator complex. Nucleic Acids Res. 2008;36:3993–4008. doi: 10.1093/nar/gkn349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bäckström S, Elfving N, Nilsson R, Wingsle G, Björklund S. Purification of a plant mediator from Arabidopsis thaliana identifies PFT1 as the Med25 subunit. Mol Cell. 2007;26:717–29. doi: 10.1016/j.molcel.2007.05.007. [DOI] [PubMed] [Google Scholar]
- 4.Mathur S, Vyas S, Kapoor S, Tyagi AK. The Mediator complex in plants: structure, phylogeny, and expression profiling of representative genes in a dicot (Arabidopsis) and a monocot (rice) during reproduction and abiotic stress. Plant Physiol. 2011;157:1609–27. doi: 10.1104/pp.111.188300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Malik S, Roeder RG. The metazoan Mediator co-activator complex as an integrative hub for transcriptional regulation. Nat Rev Genet. 2010;11:761–72. doi: 10.1038/nrg2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Conaway RC, Conaway JW. Origins and activity of the Mediator complex. Semin Cell Dev Biol. 2011;22:729–34. doi: 10.1016/j.semcdb.2011.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ge K, Guermah M, Yuan CX, Ito M, Wallberg AE, Spiegelman BM, et al. Transcription coactivator TRAP220 is required for PPAR γ 2-stimulated adipogenesis. Nature. 2002;417:563–7. doi: 10.1038/417563a. [DOI] [PubMed] [Google Scholar]
- 8.Kang YK, Guermah M, Yuan CX, Roeder RG. The TRAP/Mediator coactivator complex interacts directly with estrogen receptors α and β through the TRAP220 subunit and directly enhances estrogen receptor function in vitro. Proc Natl Acad Sci U S A. 2002;99:2642–7. doi: 10.1073/pnas.261715899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang F, Vought BW, Satterlee JS, Walker AK, Jim Sun ZY, Watts JL, et al. An ARC/Mediator subunit required for SREBP control of cholesterol and lipid homeostasis. Nature. 2006;442:700–4. doi: 10.1038/nature04942. [DOI] [PubMed] [Google Scholar]
- 10.Taubert S, Van Gilst MR, Hansen M, Yamamoto KR. A Mediator subunit, MDT-15, integrates regulation of fatty acid metabolism by NHR-49-dependent and -independent pathways in C. elegans. Genes Dev. 2006;20:1137–49. doi: 10.1101/gad.1395406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thakur JK, Arthanari H, Yang F, Pan SJ, Fan X, Breger J, et al. A nuclear receptor-like pathway regulating multidrug resistance in fungi. Nature. 2008;452:604–9. doi: 10.1038/nature06836. [DOI] [PubMed] [Google Scholar]
- 12.Thakur JK, Arthanari H, Yang F, Chau KH, Wagner G, Näär AM. Mediator subunit Gal11p/MED15 is required for fatty acid-dependent gene activation by yeast transcription factor Oaf1p. J Biol Chem. 2009;284:4422–8. doi: 10.1074/jbc.M808263200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Boyer TG, Martin ME, Lees E, Ricciardi RP, Berk AJ. Mammalian Srb/Mediator complex is targeted by adenovirus E1A protein. Nature. 1999;399:276–9. doi: 10.1038/20466. [DOI] [PubMed] [Google Scholar]
- 14.Ito M, Yuan CX, Malik S, Gu W, Fondell JD, Yamamura S, et al. Identity between TRAP and SMCC complexes indicates novel pathways for the function of nuclear receptors and diverse mammalian activators. Mol Cell. 1999;3:361–70. doi: 10.1016/S1097-2765(00)80463-3. [DOI] [PubMed] [Google Scholar]
- 15.Yang F, DeBeaumont R, Zhou S, Näär AM. The activator-recruited cofactor/Mediator coactivator subunit ARC92 is a functionally important target of the VP16 transcriptional activator. Proc Natl Acad Sci U S A. 2004;101:2339–44. doi: 10.1073/pnas.0308676100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang G, Balamotis MA, Stevens JL, Yamaguchi Y, Handa H, Berk AJ. Mediator requirement for both recruitment and postrecruitment steps in transcription initiation. Mol Cell. 2005;17:683–94. doi: 10.1016/j.molcel.2005.02.010. [DOI] [PubMed] [Google Scholar]
- 17.Kim S, Xu X, Hecht A, Boyer TG. Mediator is a transducer of Wnt/β-catenin signaling. J Biol Chem. 2006;281:14066–75. doi: 10.1074/jbc.M602696200. [DOI] [PubMed] [Google Scholar]
- 18.Wang X, Yang N, Uno E, Roeder RG, Guo S. A subunit of the mediator complex regulates vertebrate neuronal development. Proc Natl Acad Sci U S A. 2006;103:17284–9. doi: 10.1073/pnas.0605414103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ding N, Zhou H, Esteve PO, Chin HG, Kim S, Xu X, et al. Mediator links epigenetic silencing of neuronal gene expression with x-linked mental retardation. Mol Cell. 2008;31:347–59. doi: 10.1016/j.molcel.2008.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim YJ, Björklund S, Li Y, Sayre MH, Kornberg RD. A multiprotein mediator of transcriptional activation and its interaction with the C-terminal repeat domain of RNA polymerase II. Cell. 1994;77:599–608. doi: 10.1016/0092-8674(94)90221-6. [DOI] [PubMed] [Google Scholar]
- 21.Benoff B, Yang H, Lawson CL, Parkinson G, Liu J, Blatter E, et al. Structural basis of transcription activation: the CAP-alpha CTD-DNA complex. Science. 2002;297:1562–6. doi: 10.1126/science.1076376. [DOI] [PubMed] [Google Scholar]
- 22.Esnault C, Ghavi-Helm Y, Brun S, Soutourina J, Van Berkum N, Boschiero C, et al. Mediator-dependent recruitment of TFIIH modules in preinitiation complex. Mol Cell. 2008;31:337–46. doi: 10.1016/j.molcel.2008.06.021. [DOI] [PubMed] [Google Scholar]
- 23.Soutourina J, Wydau S, Ambroise Y, Boschiero C, Werner M. Direct interaction of RNA polymerase II and mediator required for transcription in vivo. Science. 2011;331:1451–4. doi: 10.1126/science.1200188. [DOI] [PubMed] [Google Scholar]
- 24.Andrau JC, van de Pasch L, Lijnzaad P, Bijma T, Koerkamp MG, van de Peppel J, et al. Genome-wide location of the coactivator mediator: Binding without activation and transient Cdk8 interaction on DNA. Mol Cell. 2006;22:179–92. doi: 10.1016/j.molcel.2006.03.023. [DOI] [PubMed] [Google Scholar]
- 25.Zhu X, Wirén M, Sinha I, Rasmussen NN, Linder T, Holmberg S, et al. Genome-wide occupancy profile of mediator and the Srb8-11 module reveals interactions with coding regions. Mol Cell. 2006;22:169–78. doi: 10.1016/j.molcel.2006.03.032. [DOI] [PubMed] [Google Scholar]
- 26.Ansari SA, He Q, Morse RH. Mediator complex association with constitutively transcribed genes in yeast. Proc Natl Acad Sci U S A. 2009;106:16734–9. doi: 10.1073/pnas.0905103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kagey MH, Newman JJ, Bilodeau S, Zhan Y, Orlando DA, van Berkum NL, et al. Mediator and cohesin connect gene expression and chromatin architecture. Nature. 2010;467:430–5. doi: 10.1038/nature09380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang Z, Yik JH, Chen R, He N, Jang MK, Ozato K, et al. Recruitment of P-TEFb for stimulation of transcriptional elongation by the bromodomain protein Brd4. Mol Cell. 2005;19:535–45. doi: 10.1016/j.molcel.2005.06.029. [DOI] [PubMed] [Google Scholar]
- 29.Malik S, Barrero MJ, Jones T. Identification of a regulator of transcription elongation as an accessory factor for the human Mediator coactivator. Proc Natl Acad Sci U S A. 2007;104:6182–7. doi: 10.1073/pnas.0608717104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guglielmi B, Soutourina J, Esnault C, Werner M. TFIIS elongation factor and Mediator act in conjunction during transcription initiation in vivo. Proc Natl Acad Sci U S A. 2007;104:16062–7. doi: 10.1073/pnas.0704534104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Acevedo ML, Kraus WL. Mediator and p300/CBP-steroid receptor coactivator complexes have distinct roles, but function synergistically, during estrogen receptor α-dependent transcription with chromatin templates. Mol Cell Biol. 2003;23:335–48. doi: 10.1128/MCB.23.1.335-348.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Black JC, Choi JE, Lombardo SR, Carey M. A mechanism for coordinating chromatin modification and preinitiation complex assembly. Mol Cell. 2006;23:809–18. doi: 10.1016/j.molcel.2006.07.018. [DOI] [PubMed] [Google Scholar]
- 33.Qiu H, Hu C, Zhang F, Hwang GJ, Swanson MJ, Boonchird C, et al. Interdependent recruitment of SAGA and Srb mediator by transcriptional activator Gcn4p. Mol Cell Biol. 2005;25:3461–74. doi: 10.1128/MCB.25.9.3461-3474.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu X, Vorontchikhina M, Wang YL, Faiola F, Martinez E. STAGA recruits Mediator to the MYC oncoprotein to stimulate transcription and cell proliferation. Mol Cell Biol. 2008;28:108–21. doi: 10.1128/MCB.01402-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ooi L, Wood IC. Chromatin crosstalk in development and disease: lessons from REST. Nat Rev Genet. 2007;8:544–54. doi: 10.1038/nrg2100. [DOI] [PubMed] [Google Scholar]
- 36.Autran D, Jonak C, Belcram K, Beemster GT, Kronenberger J, Grandjean O, et al. Cell numbers and leaf development in Arabidopsis: a functional analysis of the STRUWWELPETER gene. EMBO J. 2002;21:6036–49. doi: 10.1093/emboj/cdf614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gonzalez D, Bowen AJ, Carroll TS, Conlan RS. The transcription corepressor LEUNIG interacts with the histone deacetylase HDA19 and mediator components MED14 (SWP) and CDK8 (HEN3) to repress transcription. Mol Cell Biol. 2007;27:5306–15. doi: 10.1128/MCB.01912-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Clay NK, Nelson T. The recessive epigenetic swellmap mutation affects the expression of two step II splicing factors required for the transcription of the cell proliferation gene STRUWWELPETER and for the timing of cell cycle arrest in the Arabidopsis leaf. Plant Cell. 2005;17:1994–2008. doi: 10.1105/tpc.105.032771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang W, Chen X. HUA ENHANCER3 reveals a role for a cyclin-dependent protein kinase in the specification of floral organ identity in Arabidopsis. Development. 2004;131:3147–56. doi: 10.1242/dev.01187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gillmor CS, Park MY, Smith MR, Pepitone R, Kerstetter RA, Poethig RS. The MED12-MED13 module of Mediator regulates the timing of embryo patterning in Arabidopsis. Development. 2010;137:113–22. doi: 10.1242/dev.043174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ito J, Sono T, Tasaka M, Furutani M. MACCHI-BOU 2 is required for early embryo patterning and cotyledon organogenesis in Arabidopsis. Plant Cell Physiol. 2011;52:539–52. doi: 10.1093/pcp/pcr013. [DOI] [PubMed] [Google Scholar]
- 42.Kidd BN, Cahill DM, Manners JM, Schenk PM, Kazan K. Diverse roles of the Mediator complex in plants. Semin Cell Dev Biol. 2011;22:741–8. doi: 10.1016/j.semcdb.2011.07.012. [DOI] [PubMed] [Google Scholar]
- 43.Ou B, Yin KQ, Liu SN, Yang Y, Gu T, Wing Hui JM, et al. A high-throughput screening system for Arabidopsis transcription factors and its application to Med25-dependent transcriptional regulation. Mol Plant. 2011;4:546–55. doi: 10.1093/mp/ssr002. [DOI] [PubMed] [Google Scholar]
- 44.Cevik V, Kidd BN, Zhang P, Hill C, Kiddle S, Denby KJ, et al. MEDIATOR25 Acts as an Integrative Hub for the Regulation of Jasmonate-Responsive Gene Expression in Arabidopsis. Plant Physiol. 2012;160:541–55. doi: 10.1104/pp.112.202697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kim YJ, Zheng B, Yu Y, Won SY, Mo B, Chen X. The role of Mediator in small and long noncoding RNA production in Arabidopsis thaliana. EMBO J. 2011;30:814–22. doi: 10.1038/emboj.2011.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Huang L, Jones AM, Searle I, Patel K, Vogler H, Hubner NC, et al. An atypical RNA polymerase involved in RNA silencing shares small subunits with RNA polymerase II. Nat Struct Mol Biol. 2009;16:91–3. doi: 10.1038/nsmb.1539. [DOI] [PubMed] [Google Scholar]
- 47.Barneche F, Steinmetz F, Echeverría M. Fibrillarin genes encode both a conserved nucleolar protein and a novel small nucleolar RNA involved in ribosomal RNA methylation in Arabidopsis thaliana. J Biol Chem. 2000;275:27212–20. doi: 10.1074/jbc.M002996200. [DOI] [PubMed] [Google Scholar]
- 48.Lecharny A, Boudet N, Gy I, Aubourg S, Kreis M. Introns in, introns out in plant gene families: a genomic approach of the dynamics of gene structure. J Struct Funct Genomics. 2003;3:111–6. doi: 10.1023/A:1022614001371. [DOI] [PubMed] [Google Scholar]
- 49.Kato Y, Habas R, Katsuyama Y, Näär AM, He X. A component of the ARC/Mediator complex required for TGF beta/Nodal signalling. Nature. 2002;418:641–6. doi: 10.1038/nature00969. [DOI] [PubMed] [Google Scholar]
- 50.Kim YH, Kim MD, Choi YI, Park SC, Yun DJ, Noh EW, et al. Transgenic poplar expressing Arabidopsis NDPK2 enhances growth as well as oxidative stress tolerance. Plant Biotechnol J. 2011;9:334–47. doi: 10.1111/j.1467-7652.2010.00551.x. [DOI] [PubMed] [Google Scholar]
- 51.Yin Y, Wang ZY, Mora-Garcia S, Li J, Yoshida S, Asami T, et al. BES1 accumulates in the nucleus in response to brassinosteroids to regulate gene expression and promote stem elongation. Cell. 2002;109:181–91. doi: 10.1016/S0092-8674(02)00721-3. [DOI] [PubMed] [Google Scholar]
- 52.Goda H, Shimada Y, Asami T, Fujioka S, Yoshida S. Microarray analysis of brassinosteroid-regulated genes in Arabidopsis. Plant Physiol. 2002;130:1319–34. doi: 10.1104/pp.011254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nemhauser JL, Mockler TC, Chory J. Interdependency of brassinosteroid and auxin signaling in Arabidopsis. PLoS Biol. 2004;2:E258. doi: 10.1371/journal.pbio.0020258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chung HS, Howe GA. A critical role for the TIFY motif in repression of jasmonate signaling by a stabilized splice variant of the JASMONATE ZIM-domain protein JAZ10 in Arabidopsis. Plant Cell. 2009;21:131–45. doi: 10.1105/tpc.108.064097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Knight H, Mugford SG, Ulker B, Gao D, Thorlby G, Knight MR. Identification of SFR6, a key component in cold acclimation acting post-translationally on CBF function. Plant J. 2009;58:97–108. doi: 10.1111/j.1365-313X.2008.03763.x. [DOI] [PubMed] [Google Scholar]
- 56.Yang T, Chaudhuri S, Yang L, Du L, Poovaiah BW. A calcium/calmodulin-regulated member of the receptor-like kinase family confers cold tolerance in plants. J Biol Chem. 2010;285:7119–26. doi: 10.1074/jbc.M109.035659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Deyholos MK. Making the most of drought and salinity transcriptomics. Plant Cell Environ. 2010;33:648–54. doi: 10.1111/j.1365-3040.2009.02092.x. [DOI] [PubMed] [Google Scholar]
- 58.Xiong L, Lee H, Ishitani M, Zhu JK. Regulation of osmotic stress-responsive gene expression by the LOS6/ABA1 locus in Arabidopsis. J Biol Chem. 2002;277:8588–96. doi: 10.1074/jbc.M109275200. [DOI] [PubMed] [Google Scholar]
- 59.Goda H, Sasaki E, Akiyama K, Maruyama-Nakashita A, Nakabayashi K, Li W, et al. The AtGenExpress hormone and chemical treatment data set: experimental design, data evaluation, model data analysis and data access. Plant J. 2008;55:526–42. doi: 10.1111/j.1365-313X.2008.03510.x. [DOI] [PubMed] [Google Scholar]
- 60.Mizuno T, Yamashino T. Comparative transcriptome of diurnally oscillating genes and hormone-responsive genes in Arabidopsis thaliana: insight into circadian clock-controlled daily responses to common ambient stresses in plants. Plant Cell Physiol. 2008;49:481–7. doi: 10.1093/pcp/pcn008. [DOI] [PubMed] [Google Scholar]
- 61.Leivar P, Monte E, Oka Y, Liu T, Carle C, Castillon A, et al. Multiple phytochrome-interacting bHLH transcription factors repress premature seedling photomorphogenesis in darkness. Curr Biol. 2008;18:1815–23. doi: 10.1016/j.cub.2008.10.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jiao Y, Lau OS, Deng XW. Light-regulated transcriptional networks in higher plants. Nat Rev Genet. 2007;8:217–30. doi: 10.1038/nrg2049. [DOI] [PubMed] [Google Scholar]
- 63.Peschke F, Kretsch T. Genome-wide analysis of light-dependent transcript accumulation patterns during early stages of Arabidopsis seedling deetiolation. Plant Physiol. 2011;155:1353–66. doi: 10.1104/pp.110.166801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Deato MD, Marr MT, Sottero T, Inouye C, Hu P, Tjian R. MyoD targets TAF3/TRF3 to activate myogenin transcription. Mol Cell. 2008;32:96–105. doi: 10.1016/j.molcel.2008.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.