Abstract
Over the last two decades, several transcription factor gene families have been identified with some of them characterized in detail for their roles on transcriptional regulation of plant defense responses against pest or pathogen attack. We have recently added another transcription factor gene family to this list through the characterization of the LATERAL ORGAN BOUNDARIES (LOB) DOMAIN (LBD)-CONTAINING PROTEIN20 (LBD20). We showed LBD20 acts as a repressor of a subset of jasmonate mediated defenses and in susceptibility to the root-infecting fungal pathogen Fusarium oxysporum. However, possible roles for other members of this gene family in plant defense are currently unknown. Here we searched publically available microarray expression data and provide an overview of the expression patterns of selected members of the LBD gene family for their response to other fungal pathogens and soil nematodes. Distinct expression patterns of the LBD genes suggest that certain members of this gene family have previously undescribed roles in plant defense.
Keywords: LBD, jasmonate, transcription, defense, biotic stress, Fusarium oxysporum
The transcriptional control of defense responses following pathogen attack plays a critical role in determining final disease outcome. The amplitude, temporal and spatial expression of these responses is largely controlled through the activity of specific transcription factors, of which some have functions overlapping with other plant processes.1,2
Several well studied plant transcription factor families are associated with defense responses.1 These include the Ethylene Response Factor (ERF), basic-domain leucine-Zipper (bZIP) TGA, MYB, Whirly, WRKY, NAC, and members of the basic helix-loop-helix (bHLH) transcription factors. Members of these families regulate gene expression in response to a range of biotic stimuli including microbes (fungi, oomycetes, bacteria) and insects, and downstream defense signaling hormones such as salicylic acid (SA), jasmonic acid (JA), and ethylene (ET).1,3 Our work focuses on defense and other host responses to the root-infecting fungal pathogen Fusarium oxysporum. This pathogen gains entry into roots through wounds or at points of lateral root formation, moving through the root cortex intercellularly and gaining entry into the xylem. From here the pathogen travels upwards, clogging the vascular system and secreting toxins causing chlorosis, wilting and eventually plant death.4-6
In Arabidopsis, increased resistance to F. oxysporum can be achieved through altering the expression of specific transcriptional activators or repressors.4,7,8 For example, positive regulators of defense against F. oxysporum include the ERF transcription factors ERF1, AtERF2 and AtERF14,9-11 while negative regulators include AtERF4,10 the NAC-domain transcription factor AtAF2,12 and the bHLH AtMYC2.13 Recently, we have identified a member of another transcription factor family, termed LATERAL ORGAN BOUNDARIES (LOB) DOMAIN (LBD),14 which functions as a susceptibility gene to Fusarium disease symptom development.15 Knockouts of this gene, LOB DOMAIN-CONTAINING PROTEIN20 (LBD20), exhibited increased resistance to F. oxysporum and reduced chlorosis following application of F. oxysporum culture filtrate. The lbd20 mutant also showed increased expression of the JA-regulated defense genes THIONIN2.1 (THI2.1) and VEGETATIVE STORAGE PROTEIN2 (VSP2). We further showed that plants overexpressing LBD20 had repressed THI2.1 and VSP2 expression, and exhibited increased susceptibility to F. oxysporum. Together, these results suggested that LBD20 acts as a negative regulator of JA-regulated pathogen defense in Arabidopsis. This is the first time, to our knowledge, that a role for a LBD transcription factor in plant-pathogen interactions has been demonstrated.
To date, 43 LBD genes have been identified in Arabidopsis and classified into two main classes.16 The majority of LBD proteins, including LBD20, belong to class I with LOB domains most similar to the defining LOB member containing a cysteine-repeat motif, a conserved glycine residue and leucine zipper sequence. The smaller and less similar class II group have incomplete leucine zippers and are not predicted to form coiled-coil structures required for protein-protein interactions or homodimerization.14,16 Currently, the function of most LBD family members is not known, although diverse functions have been proposed for those characterized to some extent. These roles include regulating organ development, anthocyanin or nitrogen metabolism, or responses to phytohormones such as cytokinin, auxin and gibberellins.14,16-18 We now demonstrate a role for LBD20 in JA-signaling where it functions downstream of COI1 and MYC2,15 respectively part of the JA receptor complex and a key downstream transcriptional regulator of JA signaling (for a review see ref. 19).
LBD20 expression was predominantly limited to root tissues, the initial site of F. oxysporum colonization, where its expression increased following infection.15 To determine if other LBD transcription factors may function in root or fungal pathogen interactions, we searched public array data from Arabidopsis transcripts (GENEVESTIGATOR20) for response of LBD genes (26 members with probesets) to leaf necrotrophs or to root pathogens. The expression of 13 LBD genes was either repressed or induced greater than 1.5-fold following inoculation with the leaf fungal necrotrophs Alternaria brassicicola or Botrytis cinerea, or the oomycete root pathogen Phytophthora parasitica, with the greatest response exhibited by LBD41 (Fig. 1). Nearly half of the LBD genes with probesets were responsive to P. parasitica, with six induced and six repressed greater than 1.5-fold. The expression of several LBD genes was also responsive to the root-colonizing parasite, root-knot nematode (Meloidogyne incognita) (Fig. 1). The diverse expression profiles of these LBD genes suggest, in addition to LBD20, other LBD proteins may function during plant defense responses to fungal or root pathogens.
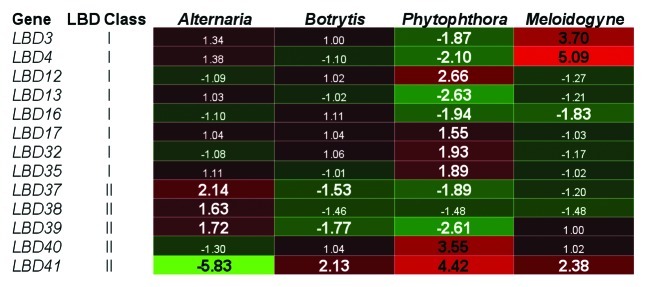
Figure 1.Expression profiles of LBD genes responsive to selected leaf or root pathogens. Public array data examined through GENEVESTIGATOR20 is presented for eight class I and five class II LBD genes in leaf tissue in response to the leaf infecting necrotrophic fungal pathogens Alternaria brassicicola or Botrytis cinerea, or in root tissue in response to the root pathogen Phytophthora parasitica (oomycete) or to the root-knot nematode Meloidogyne incognita. Shown are fold changes relative to controls with those > 1.5-fold highlighted in bold font. For data sets with multiple sampled time-points, time-points with the greatest fold changes were selected. This figure was generated using the HeatMapper tool from The Bio-Array Resource for Plant Biology (http://bar.utoronto.ca/) with green and red representing repressed or induced gene expression, respectively.
Interestingly, the LBD genes whose expression was responsive to multiple pathogens belong to the smaller class II of LBD proteins. These are LBD37 to LBD42 inclusive, with smaller phylogenetically similar subgroups formed between LBD37, 38 and 39, and LBD40, 41 and 42.14,16 In addition, we found the two subgroups showed distinctly opposite expression patterns in response to different pathogens with LBD37, 38 and 39 expression induced by A. brassicicola and repressed by B. cinerea and P. parasitica, and the reverse observed for LBD41 and to a lesser extent LBD40 (Fig. 1). It is plausible that these members play opposing roles in response to these stimuli as a means of maintaining the appropriate expression level of downstream defense responses. In addition to LBD20 expression being induced by F. oxysporum infection, we also found its expression was responsive to the defense signaling hormone JA.15 In GENEVESTIGATOR, we only found LBD genes differentially expressed greater than 1.5-fold from controls to be repressed by JA treatment (data not shown). Again, the class II LBD genes (LBD37, 38, 39 and 41) were most responsive.
Recently the class II LBD proteins LBD37 and LBD41 were shown to interact with TOPLESS (TPL) and TPL-related (TPR) co-repressors, potentially through an ERF-associated amphiphilic repression-(EAR) motif.21,22 With the exception of LBD39, the remaining class II LBD proteins are also predicted to contain EAR repression domains,23 suggesting that other LBDs may also recruit TPL/TPRs co-repressors to mediate transcriptional repression. We found LBD20 functions as a repressor of a subset of JA-regulated defense genes however, it is not known whether LBD20 directly or indirectly represses their transcription.15 Unlike class I LBD proteins, those characterized so far from class II do not have any reported roles in development.16 Their structural difference, including an incomplete leucine zipper, conserved EAR-repression motifs, and increased response to pathogen attack (Fig. 1), suggest they have distinctly different roles from the class I LBD proteins.
With a role for LBD20 proposed in plant defense and JA-responses,15 and the expression of several other LBD genes responsive to root or fungal pathogens, particularly those of class II (this paper), it is of much interest to determine if other LBD members are transcriptional mediators of resistance or susceptibility to pathogen attack and if so how.
Acknowledgments
L.F.T. was supported by a CSIRO Office of the Chief Executive postdoctoral fellowship.
Footnotes
Previously published online: www.landesbioscience.com/journals/psb/article/22097
References
- 1.Eulgem T. Regulation of the Arabidopsis defense transcriptome. Trends Plant Sci. 2005;10:71–8. doi: 10.1016/j.tplants.2004.12.006. [DOI] [PubMed] [Google Scholar]
- 2.Singh KB, Foley RC, Oñate-Sánchez L. Transcription factors in plant defense and stress responses. Curr Opin Plant Biol. 2002;5:430–6. doi: 10.1016/S1369-5266(02)00289-3. [DOI] [PubMed] [Google Scholar]
- 3.Thatcher LF, Anderson JP, Singh KB. Plant defence responses: what have we learnt from Arabidopsis? Funct Plant Biol. 2005;32:1–19. doi: 10.1071/FP04135. [DOI] [PubMed] [Google Scholar]
- 4.Dombrecht B, Kazan K, Manners JM. Improved resistance to Fusarium wilt through genetic engineering of defense signaling pathways. In: Teixeira da Silva JA, ed. Floriculture, Ornamental and Plant Biotechnology. London: Global Science Books, 2006:388-98. [Google Scholar]
- 5.Czymmek KJ, Fogg M, Powell DH, Sweigard J, Park SY, Kang S. In vivo time-lapse documentation using confocal and multi-photon microscopy reveals the mechanisms of invasion into the Arabidopsis root vascular system by Fusarium oxysporum. Fungal Genet Biol. 2007;44:1011–23. doi: 10.1016/j.fgb.2007.01.012. [DOI] [PubMed] [Google Scholar]
- 6.Michielse CB, Rep M. Pathogen profile update: Fusarium oxysporum. Mol Plant Pathol. 2009;10:311–24. doi: 10.1111/j.1364-3703.2009.00538.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Edgar CI, McGrath KC, Dombrecht B, Manners JM, Schenk PM, Maclean DJ, et al. Salicylic acid mediates resistance to the vascular wilt pathogen Fusarium oxysporum in the model host Arabidopsis thaliana. Australas Plant Pathol. 2006;35:581–91. doi: 10.1071/AP06060. [DOI] [Google Scholar]
- 8.Berrocal-Lobo M, Molina A. Arabidopsis defense response against Fusarium oxysporum. Trends Plant Sci. 2008;13:145–50. doi: 10.1016/j.tplants.2007.12.004. [DOI] [PubMed] [Google Scholar]
- 9.Berrocal-Lobo M, Molina A. Ethylene response factor 1 mediates Arabidopsis resistance to the soilborne fungus Fusarium oxysporum. Mol Plant Microbe Interact. 2004;17:763–70. doi: 10.1094/MPMI.2004.17.7.763. [DOI] [PubMed] [Google Scholar]
- 10.McGrath KC, Dombrecht B, Manners JM, Schenk PM, Edgar CI, Maclean DJ, et al. Repressor- and activator-type ethylene response factors functioning in jasmonate signaling and disease resistance identified via a genome-wide screen of Arabidopsis transcription factor gene expression. Plant Physiol. 2005;139:949–59. doi: 10.1104/pp.105.068544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Oñate-Sánchez L, Anderson JP, Young J, Singh KB. AtERF14, a member of the ERF family of transcription factors, plays a nonredundant role in plant defense. Plant Physiol. 2007;143:400–9. doi: 10.1104/pp.106.086637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Delessert C, Kazan K, Wilson IW, Van Der Straeten D, Manners J, Dennis ES, et al. The transcription factor ATAF2 represses the expression of pathogenesis-related genes in Arabidopsis. Plant J. 2005;43:745–57. doi: 10.1111/j.1365-313X.2005.02488.x. [DOI] [PubMed] [Google Scholar]
- 13.Anderson JP, Badruzsaufari E, Schenk PM, Manners JM, Desmond OJ, Ehlert C, et al. Antagonistic interaction between abscisic acid and jasmonate-ethylene signaling pathways modulates defense gene expression and disease resistance in Arabidopsis. Plant Cell. 2004;16:3460–79. doi: 10.1105/tpc.104.025833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shuai B, Reynaga-Peña CG, Springer PS. The lateral organ boundaries gene defines a novel, plant-specific gene family. Plant Physiol. 2002;129:747–61. doi: 10.1104/pp.010926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thatcher LF, Powell JJ, Aitken EA, Kazan K, Manners JM. The Lateral Organ Boundaries Domain transcription factor LBD20 functions in Fusarium wilt susceptibility and jasmonate signaling in Arabidopsis. Plant Physiol. 2012;160:407–18. doi: 10.1104/pp.112.199067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Majer C, Hochholdinger F. Defining the boundaries: structure and function of LOB domain proteins. Trends Plant Sci. 2011;16:47–52. doi: 10.1016/j.tplants.2010.09.009. [DOI] [PubMed] [Google Scholar]
- 17.Borghi L, Bureau M, Simon R. Arabidopsis JAGGED LATERAL ORGANS is expressed in boundaries and coordinates KNOX and PIN activity. Plant Cell. 2007;19:1795–808. doi: 10.1105/tpc.106.047159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bureau M, Rast MI, Illmer J, Simon R. JAGGED LATERAL ORGAN (JLO) controls auxin dependent patterning during development of the Arabidopsis embryo and root. Plant Mol Biol. 2010;74:479–91. doi: 10.1007/s11103-010-9688-2. [DOI] [PubMed] [Google Scholar]
- 19.Pauwels L, Goossens A. The JAZ proteins: a crucial interface in the jasmonate signaling cascade. Plant Cell. 2011;23:3089–100. doi: 10.1105/tpc.111.089300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zimmermann P, Hirsch-Hoffmann M, Hennig L, Gruissem W. GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox. Plant Physiol. 2004;136:2621–32. doi: 10.1104/pp.104.046367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Causier B, Ashworth M, Guo W, Davies B. The TOPLESS interactome: a framework for gene repression in Arabidopsis. Plant Physiol. 2012;158:423–38. doi: 10.1104/pp.111.186999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kazan K. Negative regulation of defence and stress genes by EAR-motif-containing repressors. Trends Plant Sci. 2006;11:109–12. doi: 10.1016/j.tplants.2006.01.004. [DOI] [PubMed] [Google Scholar]
- 23.Kagale S, Links MG, Rozwadowski K. Genome-wide analysis of ethylene-responsive element binding factor-associated amphiphilic repression motif-containing transcriptional regulators in Arabidopsis. Plant Physiol. 2010;152:1109–34. doi: 10.1104/pp.109.151704. [DOI] [PMC free article] [PubMed] [Google Scholar]


