Abstract
LEC1 acts as a key regulator of embryogenesis in Arabidopsis thaliana, but is involved in a wide range of functions, all the way from embryo morphogenesis to seed maturation. New data show that LEC1, partially in conjunction with abscisic acid, affects auxin synthesis, and both brassinosteroid and light signaling. The phenotype of LEC1 overexpressors confirms LEC1's known participation in the regulation of somatic embryogenesis, but also indicates additional roles in embryonic and extra-embryonic cell elongation. Here we present an integrated model of LEC1 function and suggest potential directions to be taken in future research in this important area of plant science.
Keywords: LEAFY COTYLEDON1, Arabidopsis thaliana, embryogenesis, hypocotyl elongation, abscisic acid, auxin, brassinosteroid, light signaling, somatic embryogenesis
The regulation of transcription by transcription factors (TFs) relies on their specific interaction with sequence motifs in their target gene promoter, a process which triggers the downstream transcriptional machinery. Many TFs have been shown to be involved in multiple developmental events and/or responses to environmental cues, through the variability of their spatio-temporal expression and their interaction with other TFs. The LEAFY COTYLEDON1 (LEC1) TF is a CCAAT-box-binding factor, whose expression in the Arabidopsis thaliana embryo and endosperm peaks at an early stage of seed development and then declines up to the green pre-mature seed stage.1,2 The multifaceted phenotype of LEC1 loss-of-function mutants demonstrates this TF's involvement in the morphogenesis of the embryo, the specification of the suspensor and cotyledons, the synthesis and accumulation of seed storage compounds, and in various desiccation- and dormancy-related processes. Both in vitro and in vivo experiments have established that LEC1 binds with NF-Y subunits,3,4 with TFs of other classes4,5 and with a non-TF protein.6 An additional level of understanding of LEC1 function has been provided by the demonstration that LEC1 has functional relations outside the known seed maturation-related LEC1-AFL/B3 network7,8 and even outside the process of seed development itself.9 Here, we have attempted to integrate its known with its more novel proposed functions to assemble a model for LEC1 function. The picture is one where a crosstalk network involves both transcriptional and hormonal control (Fig. 1).
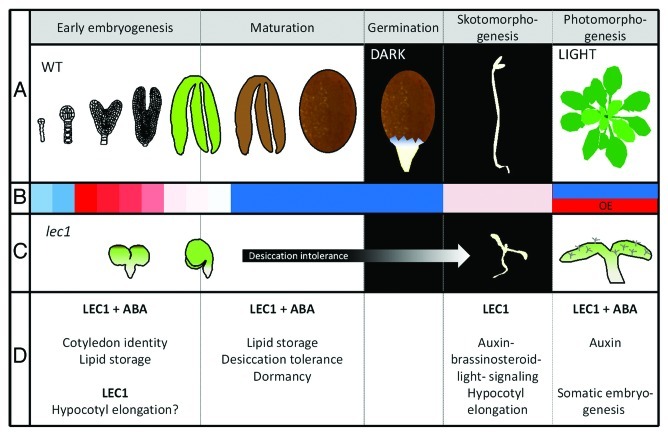
Figure 1. Model of LEC1 functions during the development of Arabidopsis thaliana. (A) Schematic representations of Arabidopsis seed developmental stages starting from the globular stage (early embryogenesis) until seed maturation resulting in a fully desiccated seed. Upon germination in the dark the elongating radicle protrudes the seed coat. In order to reach the light, during etiolation hypocotyl elongation and apical hook formation drive the seedling through the soil and protect the apical meristem from mechanical damage, respectively. (B) LEC1 expression in the respective developmental stages. During embryogenesis LEC1 mRNA can be detected during early stages declining until maturation. Further on weak expression can be found in the etiolated seedling. LEC1 expression is incompatible with vegetative growth and the artificial system of ectopic expression of LEC1 outside of its natural expression domains is indicated by OE (overexpression). (C) Schematic representations of lec1 mutant phenotypes. Lec1 embryos are characterized by round cotyledons that do not curl. The embryonic axis is shorter due to missing hypocotyl elongation. The lec1 embryo is desiccation-intolerant and has to be rescued before maturation in order to continue growth. In the dark lec1 seedlings fail to elongate and light-grown lec1 seedlings develop trichomes on their cotyledons. (D) Functional relations of LEC1.
LEC1 and Embryogenesis
LEC1 transcript abundance peaks at the globular-to-heart stage transition and then declines steadily toward early seed maturation10 (Fig. 1A, B). During embryo morphogenesis, the most prominent function of LEC1 is the specification of cotyledon identity. The lec1 mutant produces leafy cotyledons,11 whereas the ectopic expression of LEC1 results in the formation of cotyledonary leaves9 (Fig. 1C). During seed maturation, LEC1 expression is required for the synthesis of storage compounds, including components of lipid synthesis and storage (such as oleosins).9,12 As part of the NF-YC2 complex, the LEC1 product activates the promoter of certain storage protein encoding genes.4,13 These seed maturation-related functions are dependent on the presence of abscisic acid (ABA) (Fig. 1D). Since the relevant ABA responsive elements (ABREs) are over-represented in LEC1-regulated target promoters,9 it is conceivable that LEC1 probably confers ABA inducibility to its target genes by the recruitment of additional ABRE binding transcription factors such as bZIP. An example of this type of interaction is represented by the substitutability of bZIP67 for ABA, as a cofactor of LEC1in the regulation of a specific storage protein encoding gene.4 A comprehensive genome-wide analysis of the LEC1 interactome would therefore be useful to allow the identification of further co-operating factors.
LEC1 and Etiolation
The expression of LEC1 has also been detected in etiolated seedlings,6,14 an observation fully consistent with the de-etiolated, short hypocotyl phenotype of dark-grown lec1 seedlings15 (Fig. 1B, C) and the mimicking of etiolation (including hypocotyl elongation and hook formation) shown by LEC1 overexpressing seedlings.9 Given that LEC1 activity regulates genes implicated with auxin, brassinosteroid (BR) and light, the hypothesis has been mooted that both hypocotyl elongation and hook formation may be integrated by LEC1.9 Hypocotyl elongation as well as certain other processes appear to be enhanced via a positive synergy between BR and auxin.16-18 Light signaling factors which form part of the photomorphogenic response have also been implicated with hypocotyl elongation in dark-grown plants, a process which is additionally influenced by the fatty acid reserve of the endosperm.19 These observations are suggestive of a connection between the seed maturation- and seedling elongation-related functions of LEC1. While the global role of ABA in the elongation process is only poorly understood, it has been established that ABA inhibits both BR signaling20 and embryonic stem elongation.21 This elongation-inhibiting role of ABA led us to assume that LEC1 during etiolation may act independent of ABA (Fig. 1D). Discriminating between the ABA-dependent and ABA-independent functions of LEC1 is a priority topic for further experimentation.
LEC1 and Vegetative Plant Development
The formation by the vegetative seedling of cotyledonary leaves and pickle-like root tips able to accumulate storage compounds can be induced by the ectopic expression of LEC1 and the provision of exogenous ABA.9 The supply of auxin induces the initiation of somatic embryogenesis from competent tissues, a phenomenon consistent with the activation of transcription of the auxin synthesis gene YUCCA1022 when LEC1 is expressed in the presence of ABA.9 The seed-specific expression of YUCCA10 may affect the elongation of the embryo downstream of the activity of LEC1, and opposing phenotypes due to varying auxin concentrations have been described. Enhanced auxin signaling is known to inhibit the elongation of the embryonic axis during germination21 and auxin synthesis suppresses hypocotyl growth post germination.23 However, some elongation-promoting effects of auxin have also been described.24
The LEC1 induced and auxin-mediated embryonic differentiation of structures associated with the shoot (cotyledonary leaves) and root (pickle roots) raises the question of their cellular origin. Currently it is unclear whether these structures are either derived from relict undifferentiated stem cells or whether a process of trans-differentiation (possibly via a de-differentiated state) is required. The analysis of stem cell marker expression in a system of controlled activation of LEC1 expression9 will help to answer this question.
Conclusions
Recent insights into the function of LEC1 have revealed the complexity of downstream regulatory interactions involving this TF, acting at various stages of plant development. LEC1 appears to be an integrator of a number of regulatory events, including the action of heterologous TFs as well as both light and hormone signaling. At present it is only possible to investigate a minor part of the full combinatorial potential associated with this TF, which only emphasizes the need for developing more holistic analytical approaches to better understand plant differentiation and development.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/psb/article/22365
References
- 1.Lotan T, Ohto M, Yee KM, West MAL, Lo R, Kwong RW, et al. Arabidopsis LEAFY COTYLEDON1 is sufficient to induce embryo development in vegetative cells. Cell. 1998;93:1195–205. doi: 10.1016/S0092-8674(00)81463-4. [DOI] [PubMed] [Google Scholar]
- 2.Lee HS, Fischer RL, Goldberg RB, Harada JJ. Arabidopsis LEAFY COTYLEDON1 represents a functionally specialized subunit of the CCAAT binding transcription factor. Proc Natl Acad Sci U S A. 2003;100:2152–6. doi: 10.1073/pnas.0437909100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yazawa K, Kamada H. Identification and characterization of carrot HAP factors that form a complex with the embryo-specific transcription factor C-LEC1. J Exp Bot. 2007;58:3819–28. doi: 10.1093/jxb/erm238. [DOI] [PubMed] [Google Scholar]
- 4.Yamamoto A, Kagaya Y, Toyoshima R, Kagaya M, Takeda S, Hattori T. Arabidopsis NF-YB subunits LEC1 and LEC1-LIKE activate transcription by interacting with seed-specific ABRE-binding factors. Plant J. 2009;58:843–56. doi: 10.1111/j.1365-313X.2009.03817.x. [DOI] [PubMed] [Google Scholar]
- 5.Wenkel S, Turck F, Singer K, Gissot L, Le Gourrierec J, Samach A, et al. CONSTANS and the CCAAT box binding complex share a functionally important domain and interact to regulate flowering of Arabidopsis. Plant Cell. 2006;18:2971–84. doi: 10.1105/tpc.106.043299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Warpeha KM, Upadhyay S, Yeh J, Adamiak J, Hawkins SI, Lapik YR, et al. The GCR1, GPA1, PRN1, NF-Y signal chain mediates both blue light and abscisic acid responses in Arabidopsis. Plant Physiol. 2007;143:1590–600. doi: 10.1104/pp.106.089904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Santos-Mendoza M, Dubreucq B, Baud S, Parcy F, Caboche M, Lepiniec L. Deciphering gene regulatory networks that control seed development and maturation in Arabidopsis. Plant J. 2008;54:608–20. doi: 10.1111/j.1365-313X.2008.03461.x. [DOI] [PubMed] [Google Scholar]
- 8.Junker A, Hartmann A, Schreiber F, Bäumlein H. An engineer’s view on regulation of seed development. Trends Plant Sci. 2010;15:303–7. doi: 10.1016/j.tplants.2010.03.005. [DOI] [PubMed] [Google Scholar]
- 9.Junker A, Mönke G, Rutten T, Keilwagen J, Seifert M, Thi TM, et al. Elongation-related functions of LEAFY COTYLEDON1 during the development of Arabidopsis thaliana. Plant J. 2012;71:427–42. doi: 10.1111/j.1365-313X.2012.04999.x. [DOI] [PubMed] [Google Scholar]
- 10.Le BH, Cheng C, Bui AQ, Wagmaister JA, Henry KF, Pelletier J, et al. Global analysis of gene activity during Arabidopsis seed development and identification of seed-specific transcription factors. Proc Natl Acad Sci U S A. 2010;107:8063–70. doi: 10.1073/pnas.1003530107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Meinke DW. A Homeotic Mutant of Arabidopsis-thaliana with Leafy Cotyledons. Science. 1992;258:1647–50. doi: 10.1126/science.258.5088.1647. [DOI] [PubMed] [Google Scholar]
- 12.Mu JY, Tan HL, Zheng Q, Fu FY, Liang Y, Zhang JA, et al. LEAFY COTYLEDON1 is a key regulator of fatty acid biosynthesis in Arabidopsis. Plant Physiol. 2008;148:1042–54. doi: 10.1104/pp.108.126342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kagaya Y, Toyoshima R, Okuda R, Usui H, Yamamoto A, Hattori T. LEAFY COTYLEDON1 controls seed storage protein genes through its regulation of FUSCA3 and ABSCISIC ACID INSENSITIVE3. Plant Cell Physiol. 2005;46:399–406. doi: 10.1093/pcp/pci048. [DOI] [PubMed] [Google Scholar]
- 14.Siefers N, Dang KK, Kumimoto RW, Bynum WE, 4th, Tayrose G, Holt BF., 3rd Tissue-specific expression patterns of Arabidopsis NF-Y transcription factors suggest potential for extensive combinatorial complexity. Plant Physiol. 2009;149:625–41. doi: 10.1104/pp.108.130591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brocard-Gifford IM, Lynch TJ, Finkelstein RR. Regulatory networks in seeds integrating developmental, abscisic acid, sugar, and light signaling. Plant Physiol. 2003;131:78–92. doi: 10.1104/pp.011916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hardtke CS. Transcriptional auxin-brassinosteroid crosstalk: who’s talking? Bioessays. 2007;29:1115–23. doi: 10.1002/bies.20653. [DOI] [PubMed] [Google Scholar]
- 17.Clouse SD. Molecular genetic studies confirm the role of brassinosteroids in plant growth and development. Plant J. 1996;10:1–8. doi: 10.1046/j.1365-313X.1996.10010001.x. [DOI] [PubMed] [Google Scholar]
- 18.Romano CP, Robson PRH, Smith H, Estelle M, Klee H. Transgene-mediated auxin overproduction in Arabidopsis: hypocotyl elongation phenotype and interactions with the hy6-1 hypocotyl elongation and axr1 auxin-resistant mutants. Plant Mol Biol. 1995;27:1071–83. doi: 10.1007/BF00020881. [DOI] [PubMed] [Google Scholar]
- 19.Penfield S, Rylott EL, Gilday AD, Graham S, Larson TR, Graham IA. Reserve mobilization in the Arabidopsis endosperm fuels hypocotyl elongation in the dark, is independent of abscisic acid, and requires PHOSPHOENOLPYRUVATE CARBOXYKINASE1. Plant Cell. 2004;16:2705–18. doi: 10.1105/tpc.104.024711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang S, Cai Z, Wang X. The primary signaling outputs of brassinosteroids are regulated by abscisic acid signaling. Proc Natl Acad Sci U S A. 2009;106:4543–8. doi: 10.1073/pnas.0900349106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Belin C, Megies C, Hauserová E, Lopez-Molina L. Abscisic acid represses growth of the Arabidopsis embryonic axis after germination by enhancing auxin signaling. Plant Cell. 2009;21:2253–68. doi: 10.1105/tpc.109.067702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cheng YF, Dai XH, Zhao YD. Auxin synthesized by the YUCCA flavin monooxygenases is essential for embryogenesis and leaf formation in Arabidopsis. Plant Cell. 2007;19:2430–9. doi: 10.1105/tpc.107.053009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stone SL, Braybrook SA, Paula SL, Kwong LW, Meuser J, Pelletier J, et al. Arabidopsis LEAFY COTYLEDON2 induces maturation traits and auxin activity: Implications for somatic embryogenesis. Proc Natl Acad Sci U S A. 2008;105:3151–6. doi: 10.1073/pnas.0712364105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhao YD, Christensen SK, Fankhauser C, Cashman JR, Cohen JD, Weigel D, et al. A role for flavin monooxygenase-like enzymes in auxin biosynthesis. Science. 2001;291:306–9. doi: 10.1126/science.291.5502.306. [DOI] [PubMed] [Google Scholar]