Abstract
Angiosperms transport their photoassimilates through sieve tubes, which comprise longitudinally-connected sieve elements. In dicots and also some monocots, the sieve elements contain parietal structural proteins known as phloem proteins or P-proteins. Following injury, P proteins disperse and accumulate as viscous plugs at the sieve plates to prevent the loss of valuable transport sugars. Tobacco (Nicotiana tabacum) P-proteins are multimeric complexes comprising subunits encoded by members of the SEO (sieve element occlusion) gene family. The existence of multiple subunits suggests that P-protein assembly involves interactions between SEO proteins, but this process is largely uncharacterized and it is unclear whether the different subunits perform unique roles or are redundant. We therefore extended our analysis of the tobacco P-proteins NtSEO1 and NtSEO2 to investigate potential interactions between them, and found that both proteins can form homomeric and heteromeric complexes in planta.
Keywords: sieve element occlusion, P-protein, forisome, phloem, wound response, interaction, BiFC
SEO Genes Encode Structural Phloem Proteins in Angiosperms
SEO genes were initially described to encode forisomes, the specialized P-proteins found only in leguminous plants, which are capable of reversible conformational changes.1-3 The discovery of SEO genes in other plants lacking forisomes led to speculation that they might encode the common P-proteins found in all dicots.4-6 We therefore analyzed two tobacco SEO proteins (NtSEO1 and NtSEO2) to determine whether they were P-protein components.7 We found that the NtSEO1 and NtSEO2 promoters restricted reporter gene expression to immature sieve elements, matching the expression profiles determined for these and other SEO genes.3,4,8,9 P-protein subunits first become visible in immature sieve elements and accumulate as large bodies before forming a parietal layer in fully-differentiated sieve elements.10 When expressed as hrGFP fusion proteins in tobacco phloem, both NtSEO1 and NtSEO2 assembled into native P-protein structures revealed as fluorescent protein bodies in immature sieve elements or as parietal agglomerates or large fluorescent plugs at the sieve plates of mature phloem. The heterologous expression of NtSEO1 or NtSEO2 in a non-phloem background (Nicotiana benthamiana epidermal cells) resulted in the formation of large complexes composed of longitudinally arranged fibrillar subunits, clearly resembling native P-protein bodies. Finally, tobacco RNAi plants showing a significant knockdown of both NtSEO1 and NtSEO2 were lacking the typical P-protein structures in sieve elements, finally confirming the identity between SEO proteins and P-proteins in tobacco.7
The generation of NtSEO-knockdown plants lacking P-protein structures also allowed us to test whether P-protein plugs formed at sieve plates as a consequence of wounding can prevent the loss of photosynthate as anticipated. We therefore performed exudation experiments and compared the loss of photoassimilates from NtSEO-RNAi and wild-type tobacco plants. Plants depleted for P-proteins were shown to lose nine times more sucrose than corresponding wild-type plants upon injury, confirming the ability of P-proteins to seal the phloem, although additional functions are certainly conceivable.7
NtSEO1 and NtSEO2 Form Heteromeric Complexes
Many angiosperm genomes contain multiple SEO genes,4,5 raising the question of potential functional redundancy of SEO proteins within one species. This was recently addressed in a study dealing with the Arabidopsis thaliana SEO genes At3g01670 and At3g01680.11 We recommend that the original names (AtSEOR1 and AtSEOR2, for sieve element occlusion related) are replaced with AtSEOa (At3g01670) and AtSEOb (At3g01680) as we proposed earlier,4 because their direct role in sieve tube sealing is now acknowledged.7 The analysis of T-DNA insertion mutants and corresponding complementation lines strongly indicated that interactions between AtSEOa and AtSEOb are necessary to form phloem filaments as no P-protein structures could be observed when either AtSEOa or AtSEOb was knocked out. However, yeast two-hybrid experiments demonstrated strong homomeric but no heteromeric interactions between AtSEOa and AtSEOb, contradicting the in vivo observations.11 To address this inconsistency, we analyzed interactions between NtSEO1 and NtSEO2 in planta using bimolecular fluorescence complementation (BiFC).12,13 Our previous agroinfiltration experiments addressing the assembly of NtSEO complexes demonstrated the suitability of the N. benthamiana background for such experiments.7 To reduce any potential impact of the reporter fragment position, we fused split variants of the monomeric red fluorescent protein mRFP1-Q66T14 to each end of NtSEO1 and NtSEO2 in separate constructs, using GATEWAY-compatible pBatTL vectors (resulting in pBatTL-NmRFP:NtSEO1/2, pBatTL-CmRFP:NtSEO1/2, pBatTL-NtSEO1/2:NmRFP, pBatTL-NtSEO1/2:CmRFP). Agroinfiltration experiments were performed using the 16 possible homomeric and heteromeric combinations, and infiltrated leaf discs were analyzed 4 d post-infiltration by confocal laser scanning microscopy as described before.7 In agreement with our previous work, both NtSEO1 and NtSEO2 formed homomeric complexes in epidermal cells (Fig. 1A, B) but we also observed large heteromeric complexes containing both proteins (Fig. 1C). Indeed, NtSEO interactions were observed in all the combinations we tested.
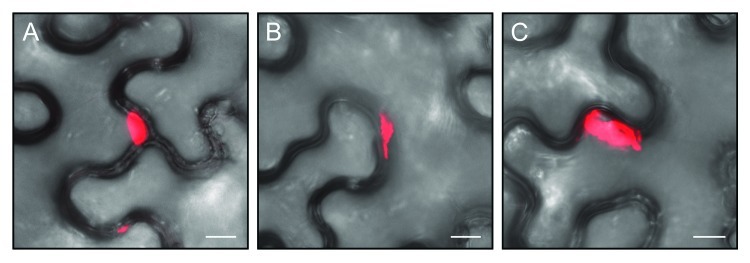
Figure 1.NtSEO-derived interaction complexes in epidermal cells of agroinfiltrated N. benthamiana leaves. BiFC analysis confirmed homomeric interactions for both NtSEO1 (A, the combination NmRFP:NtSEO1 + NtSEO1:CmRFP is shown as an example) and NtSEO2 (B, the combination NmRFP:NtSEO2 + CmRFP:NtSEO2 is shown as an example) as well as heteromeric interactions between NtSEO1 and NtSEO2 (C, the combination NtSEO1:NmRFP + CmRFP:NtSEO2 is shown as an example). The images are overlays of mRFP fluorescence and the corresponding transmitted light pictures. Scale bars: 10 µm.
In summary, NtSEO proteins are highly interactive, forming both homomeric and (more notably) heteromeric complexes in N. benthamiana leaf epidermal cells. However, the formation of heteromers could not be demonstrated for AtSEO proteins.11 A potential explanation for the successful demonstration of SEO heteromer formation in this study might be the plant background. To examine whether this is the case or if the differing observations are species-specific, BiFC experiments should be performed with AtSEOa and AtSEOb (preferably in an Arabidopsis background).15 The mode of interaction is an interesting aspect regarding the further characterization of SEO proteins as this would provide further insight into the functional mechanisms of P-proteins and forisomes.
Acknowledgments
We thank Dr. G. Jach (Phytowelt Green Technologies GmbH) and Dr. H. Uhrig (University of Cologne) for kindly providing the pBatTL vector system and Dr. Marion Rehers for helpful discussion. This work was supported in part by German Federal Ministry of Education and Research Grant 0312014, the Fraunhofer MAVO program, and Volkswagen Foundation Contract I/82 075.
Footnotes
Previously published online: www.landesbioscience.com/journals/psb/article/22452
References
- 1.Knoblauch M, Peters WS, Ehlers K, van Bel AJE. Reversible calcium-regulated stopcocks in legume sieve tubes. Plant Cell. 2001;13:1221–30. doi: 10.1105/tpc.13.5.1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Knoblauch M, Noll GA, Müller T, Prüfer D, Schneider-Hüther I, Scharner D, et al. ATP-independent contractile proteins from plants. Nat Mater. 2003;2:600–3. doi: 10.1038/nmat960. [DOI] [PubMed] [Google Scholar]
- 3.Noll GA, Fontanellaz ME, Rüping B, Ashoub A, van Bel AJE, Fischer R, et al. Spatial and temporal regulation of the forisome gene for1 in the phloem during plant development. Plant Mol Biol. 2007;65:285–94. doi: 10.1007/s11103-007-9217-0. [DOI] [PubMed] [Google Scholar]
- 4.Rüping B, Ernst AM, Jekat SB, Nordzieke S, Reineke AR, Müller B, et al. Molecular and phylogenetic characterization of the sieve element occlusion gene family in Fabaceae and non-Fabaceae plants. BMC Plant Biol. 2010;10:219. doi: 10.1186/1471-2229-10-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ernst AM, Rüping B, Jekat SB, Nordzieke S, Reineke AR, Müller B, et al. The sieve element occlusion gene family in dicotyledonous plants. Plant Signal Behav. 2011;6:151–3. doi: 10.4161/psb.6.1.14308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang B, Tolstikov V, Turnbull C, Hicks LM, Fiehn O. Divergent metabolome and proteome suggest functional independence of dual phloem transport systems in cucurbits. Proc Natl Acad Sci U S A. 2010;107:13532–7. doi: 10.1073/pnas.0910558107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ernst AM, Jekat SB, Zielonka S, Müller B, Neumann U, Rüping B, et al. Sieve element occlusion (SEO) genes encode structural phloem proteins involved in wound sealing of the phloem. Proc Natl Acad Sci U S A. 2012;109:E1980–9. doi: 10.1073/pnas.1202999109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Noll GA, Rüping B, Ernst AM, Bucsenez M, Twyman RM, Fischer R, et al. The promoters of forisome genes MtSEO2 and MtSEO3 direct gene expression to immature sieve elements in Medicago truncatula and Nicotiana tabacum. Plant Mol Biol Rep. 2009;27:526–33. doi: 10.1007/s11105-009-0120-5. [DOI] [Google Scholar]
- 9.Bucsenez M, Rüping B, Behrens S, Twyman RM, Noll GA, Prüfer D. Multiple cis-regulatory elements are involved in the complex regulation of the sieve element-specific MtSEO-F1 promoter from Medicago truncatula. Plant Biol (Stuttg) 2012 doi: 10.1111/j.1438-8677.2011.00556.x. In press. [DOI] [PubMed] [Google Scholar]
- 10.Cronshaw J, Esau K. Tubular and fibrillar components of mature and differentiating sieve elements. J Cell Biol. 1967;34:801–15. doi: 10.1083/jcb.34.3.801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Anstead JA, Froelich DR, Knoblauch M, Thompson GA. Arabidopsis P-protein filament formation requires both AtSEOR1 and AtSEOR2. Plant Cell Physiol. 2012;53:1033–42. doi: 10.1093/pcp/pcs046. [DOI] [PubMed] [Google Scholar]
- 12.Bracha-Drori K, Shichrur K, Katz A, Oliva M, Angelovici R, Yalovsky S, et al. Detection of protein-protein interactions in plants using bimolecular fluorescence complementation. Plant J. 2004;40:419–27. doi: 10.1111/j.1365-313X.2004.02206.x. [DOI] [PubMed] [Google Scholar]
- 13.Walter M, Chaban C, Schütze K, Batistic O, Weckermann K, Näke C, et al. Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J. 2004;40:428–38. doi: 10.1111/j.1365-313X.2004.02219.x. [DOI] [PubMed] [Google Scholar]
- 14.Jach G, Pesch M, Richter K, Frings S, Uhrig JF. An improved mRFP1 adds red to bimolecular fluorescence complementation. Nat Methods. 2006;3:597–600. doi: 10.1038/nmeth901. [DOI] [PubMed] [Google Scholar]
- 15.Tsuda K, Qi Y, Nguyen V, Bethke G, Tsuda Y, Glazebrook J, et al. An efficient Agrobacterium-mediated transient transformation of Arabidopsis. Plant J. 2012;69:713–9. doi: 10.1111/j.1365-313X.2011.04819.x. [DOI] [PubMed] [Google Scholar]


