Abstract
In contrast to the extraordinary body of knowledge gained over the past three decades on the virology, pathogenesis, and immunology of HIV-1 infection, innate sensors that detect HIV-1 had remained elusive until recently. By virtue of integration, retroviridae makes up a substantial portion of our genome. Thus, immune strategies that deal with endogenous retroviruses are, by necessity, those of self-preservation and not of virus elimination. Some of the principles of such strategies may also apply for defense against exogenous retroviruses including HIV-1. Here, I highlight several sensors that have recently been revealed to be capable of recognizing distinct features of HIV-1 infection, while taking into account the host-retrovirus relationship that converges on avoiding pathogenic inflammatory consequences.
Introduction
The immune system protects the host from infections by either eliminating the invading pathogens or by reducing the negative impact of infections on host fitness (Medzhitov et al., 2012). Conversely, pathogens vary in their impact on host fitness, with highly virulent pathogens on one end of the spectrum and beneficial microbes essential for many of the host developmental and physiological processes on the other end of the spectrum. Though this notion has been best appreciated for bacteria, it also applies to other microorganisms colonizing the host, including viruses. Humans are continuously infected by a variety of viruses that do not appear to be associated with any obvious pathological outcomes (Virgin et al., 2009) and perhaps even providing some unrecognized benefits. The extreme example of an intimate association between microorganisms and human hosts are endogenous retroviruses, which form a significant fraction of the human genome. Whatever were the reasons for their “colonization” of our genome, elimination of endogenous retroviruses is no longer an option. Indeed, the only outcome of an immune response against endogenous retroviruses would be a swift destruction of the host. Consequently, our immune system is left with no other choice but to learn to live in peace with the genomic intruders. This, in turn, implies that the human hosts have to tolerate the presence of endogenous retroviruses and refrain from an overt immune response against them.
Although endogenous retroviruses are an extreme case, some of the same strategies may apply to exogenous retroviruses because they all share an integration step, thereby at least transiently becoming part of the host. Thus, one common theme in immunity to retroviruses is the development of a multitude of mechanisms that prevent overt immune reactions directed at them. This may be particularly relevant to our understanding of immunity to HIV-1. This strategy is well appreciated in the case of host immunity to commensal bacteria in the gut, where “beneficial” microbes are the ones that suppress inflammatory responses, whereas “detrimental” bacteria are particularly prone to eliciting a vigorous immunity that can result in colitis and other inflammatory diseases. This notion becomes a central theme of how we might make sense of innate responses to HIV-1, because HIV-1 is a member of the retroviridae, and although it has unique pathogen associated molecular patterns (PAMPs) that can alert the human cells of invasion, it also falls under the multitude of host mechanisms that prevent overt immune reaction to retroviruses in general. Here, I will discuss innate sensors for HIV-1 and other retroviruses that operate in different cell types, in the context of host strategies that have evolved to avoid immune stimulation against retroviruses, and by taking into consideration the cell-to-cell transmission mechanism of HIV-1. The consequences of host antiviral strategies and virus manipulation of host innate immune responses on the outcome of HIV-1 infection and pathogenesis will be discussed.
Innate HIV-1 Sensors
Viruses are detected by the innate immune system primarily by recognition of viral nucleic acids. There are two general modes of sensing viral nucleic acids: the first is the detection of viral genomes present within the virions, the second is detection of nucleic acids produced during viral replication. An example of the first mode of recognition is the Toll-like receptors (TLRs) expressed in the endosomes, which detect nucleic acids of the genomes associated with the virions. Plasmacytoid dendritic cells (pDCs) utilize TLR7 and TLR9 to recognize single-stranded RNA (ssRNA) and double-stranded DNA (dsDNA) viruses, respectively (Pichlmair and Reis e Sousa, 2007). This type of recognition usually does not require productive infection of pDC by viruses. On the other hand, viral sensors present in the cytosol recognize viral nucleic acids generated upon replication. This mode of recognition is engaged only following viral infection of the cell. Thus, infected cells recognize viral RNA through RIG-I-like receptors (RLR) (Takeuchi and Akira, 2008) and viral DNA through a set of receptors including DAI, IFI16, and AIM2 (Thompson et al., 2011). Engagement of these sensors leads to transcriptional activation of proinflammatory cytokines and type I interferons (IFNs). Type I IFNs bind to IFN-abR on infected and neighboring cells and transduce signals that activate expression of hundreds of IFN-stimulated genes (ISGs) that act in concert to block virus replication and spread (Stetson and Medzhitov, 2006).
HIV-1 predominantly infects and replicates in CD4 T cells. In addition, HIV-1 infects a small percentage of CD4+ dendritic cells (DCs) and macrophages. Recent studies highlight distinct sensors of HIV-1 in directly infected cells. In addition, pDCs recognize HIV-1 in the endosomes without becoming productively infected. Important in this regard is the fact that HIV-1 transmission between cells occurs through the virological synapse (Jolly and Sattentau, 2004). Infected cells use virological synapse (Hübner et al., 2009) and filopodia to transmit HIV-1 virions to target cells (Sherer et al., 2007). Based on this new level of understanding of viral transmission mechanism, in some cases, we must rethink the way we interpret innate recognition of HIV-1 and other retroviruses (Jolly, 2011). Recent studies reveal innate sensors that recognize PAMPs associated with HIV-1 or those that are generated during HIV-1 infection (Table 1). Some of these sensors recognize nucleic acids that are associated with retroviral infection in general, whereas others recognize HIV-1 viral proteins in species-specific manner. The engagement of each sensor results in distinct signaling pathways, and they appear to operate in distinct cell types.
Table 1.
Innate HIV-1 Sensors and Effectors
| Type | Sensor | Ligand | Signaling Pathway | Location | Cell Type |
|---|---|---|---|---|---|
| General | TLR7 | ssRNA | IRF7, NF-κB | Endosome | pDC, B |
| TLR8 | ssRNA | IRF7, NF-κB | Endosome | DC | |
| Unknown DNA sensor | ISD DNA (RT product) |
IRF3 | Cytosol | Many cell types (except for transformed cells) |
|
| Species-specific | CypA | Capsid | IRF3 | Cytosol | DC |
| TRIM5, TRIM5-CypA | Capsid | AP-1, NF-κB | Cytosol | DC, myeloid cell | |
|
| |||||
| Type | Effectors/Function | Target or Substrate | End Results | Location | Cell Type |
|
| |||||
| General | SAMHD1/ triphosphohydrolase |
dNTPs | dNs + ppp | Cytosol | DC, macrophage |
| TREX1/exonuclease | ssDNA | dNMPs | Cytosol | Many cell types including DC, macrophage, CD4 T, fibroblast. |
|
| APOBEC3a/Cytidine deaminase |
Negative sense DNA |
Hypermutation | Virion Cytosol | CD4 T, macrophage IFN-inducible in CD4 T, macrophage, hepatocytes |
|
| Tetherin | Budding membrane |
Block virion release |
Cell surface | Type I IFN inducible in many cell types. |
|
| Species-specific | TRIM5 | Capsid | Capsid disruption | Cytosol | CD4 T, macrophage |
Abbreviations; APOBEC, apolipoprotein B mRNA-editing, enzyme-catalytic, polypeptide-like; dNTP, deoxynucleoside triphosphate; dNMP, deoxynucleoside monophosphate; ISD, interferon-stimulatory DNA; ppp, triphosphate; ssDNA, single-stranded DNA.
Detailed functions of effectors APOBEC3 and tetherin are described elsewhere (Blanco-Melo et al., 2012).
Toll-like Receptor 7 Detects Cell-free and Cell Associated HIV-1
TLR7 is expressed by two specialized cell types in humans. It is constitutively expressed by pDCs, which are a rare population of leukocytes that are specialized in detection of a wide variety of viruses (Gilliet et al., 2008). Although expressed constitutively, TLR7 expression is greatly enhanced by type I IFNs in B cells (Marshak-Rothstein and Rifkin, 2007). TLR7 recognizes ssRNA within the endosome (Diebold et al., 2004; Heil et al., 2004;Lund et al., 2004). Recognition of cell-free HIV-1 by TLR7 in human pDCs requires attachment and endocytosis but not the downstream infection stages, including fusion, reverse transcription, integration, or nucleocapsid production (Beignon et al., 2005). HIV-1 traffics to the early endosome and induces persistent IFN-α stimulation in pDCs (O’Brien et al., 2011). Although purified HIV-1 virions are capable of stimulating human pDC at very high concentrations (Fitzgerald-Bocarsly and Jacobs, 2010), HIV-1 infected CD4 T cells are much more potent at stimulating human pDCs following cell contact (Lepelley et al., 2011). Interestingly, nonfusogenic env mutant HIV-1 elicits much less IFN-α compared to WT HIV-1, indicating that innate recognition of HIV-1-infected cells by pDCs depends on fusion of the virus. Thus, cell-free HIV-1 and cell-associated HIV-1 may be recognized by separate pathways converging on TLR7 (Figure 1). Other retroviruses including murine leukemia virus (MLV) and mouse mammary tumor virus (MMTV) also require TLR7 and MyD88, an adaptor required for TLR signaling, for in vivo recognition and stimulation of humoral immune responses (Browne and Littman, 2009; Kane et al., 2011). TLR7-dependent antibody responses are elicited by live or UV-irradiated MLV, but not heat inactivated MLV, indicating that entry of the virus, but not replication, is required to stimulate TLR7 (Kane et al., 2011). These data are consistent with the requirement for fusion of HIV-1 in recognition of infected cells by pDCs (Lepelley et al., 2011). The exact nature of HIV-1 ligand for TLR7 is unknown. If fusion of the HIV-1 must precede recognition (Lepelley et al., 2011), a mechanism must exist to deliver cytosolic viral RNA into the endosome where TLR7 resides. Other ssRNA viruses are recognized through TLR7 upon delivery of cytosolic RNA replication intermediates through autophagy (Lee et al., 2007). In line with this, knockdown of Atg7 by small interfering RNA (siRNA) in human pDCs results in impaired TLR7-dependent IFN-α secretion in response to HIV-1 (Zhou et al., 2012). Therefore, one can speculate that autophagy may be used to transport cytosolic HIV-1 capsid containing ssRNA into the endosome where TLR7 resides (Figure 1). However, HIV-1 inhibits autophagy (Zhou and Spector, 2008) through env binding to its receptor, inducing mTOR activation and leading to blockade of autophagy in target cells (Blanchet et al., 2010). To what extent TLR7 recognition of HIV-1 is inhibited by env-mediated mTOR activation remains to be determined.
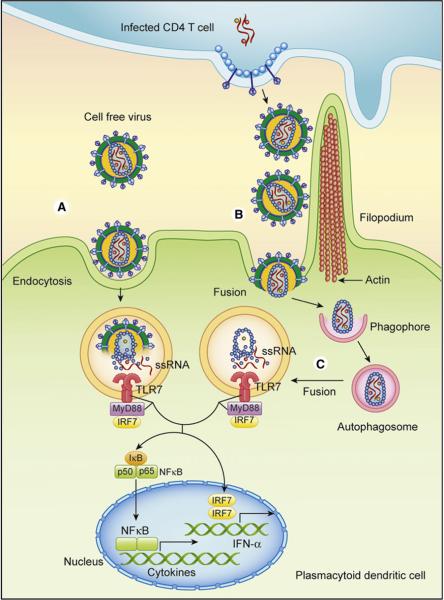
Figure 1. Innate Recognition of HIV-1 by Plasmacytoid Dendritic Cells.
(A) Cell-free virus is taken up by pDCs via endocytosis and is recognized by TLR7. HIV-1 env binds to CD4 molecule expressed on human pDCs and trigger signaling and endocytosis. The ssRNA genome inside the virion stimulates TLR7 upon degradation of envelope and capsid by endosomal hydrolases.
(B) HIV-1-infected CD4 T cells are recognized by pDCs in a process that involves endocytosis and fusion. Based on the HIV-1 cell-to-cell transmission mechanism involving filopodia formation and transport of virions via the virological synapse, the pDCs likely have access to many more virions for recognition through direct cell contact. Once HIV-1 fuses, the ssRNA genome must gain access to endosomes to activate TLR7.
(C) The speculative model is that viral PAMP is introduced to endosomal TLR7 via autophagy. Once TLR7 binds to ssRNA, it signals through MyD88 and IRF7 to trigger activation of type I IFN and cytokine genes, which play critical roles in innate immunity and pathogenesis of HIV-1.
TLR8 Signals Usurped by HIV-1 for Viral Transcription
TLR8 is expressed by DCs but not pDCs in humans and recognizes HIV-1-derived ssRNA (Heil et al., 2004). DCs also express the C-type lectin DC-SIGN, which binds to env and facilitates transmission of HIV-1 to CD4 T cells (Geijtenbeek et al., 2000). Although the majority of human DCs are not productively infected with HIV-1 strains in vitro, a small fraction (1%–3%) of DCs and pDCs do become infected (Smed-Sörensen et al., 2005). On examination of productively infected DCs, paradoxically, TLR8 stimulation by HIV-1 promotes HIV-1 replication in DCs (Gringhuis et al., 2010). Once HIV-1 provirus is integrated into host genome, signals from TLR8 initiate HIV transcription by RNA polymerase II through the recruitment of cyclin-dependent kinase-7, while signals from DC-SIGN lead to transcriptional elongation through the recruitment of P-TEFb (positive transcription elongation factor b). Together, TLR8 and DC-SIGN signaling promotes full transcriptional program for HIV-1 genome and production of HIV-1 proteins. The immunological consequence of productive infection of DCs by HIV-1 is discussed below.
Cytosolic DNA Sensor Hidden by TREX1
Within HIV-1 infected cells, an unknown DNA sensor detects the presence of complementary DNA (cDNA) and DNA intermediates generated by the RT (Figure 2). The sensor triggers activation of IFN-β gene through STING and IRF3 (Table 1). Despite great interest and effort in the field, the putative sensor for DNA in the cytosol responsible for recognizing RT products has not yet been identified and may consist of more than one sensor. The presence and activity of DNA sensor for retroviruses were unknown until recently, because they are concealed by the host exonuclease, TREX1 (Stetson et al., 2008; Yan et al., 2010). TREX1 is a 3′-5′ DNA exonuclease that cleaves ssDNA fragments derived from endogenous retroelements (Stetson et al., 2008). TREX1 represents a good example of how humans have evolved to deal with endogenous retroviruses—by devising ways to avoid generating inflammatory responses against them. Mutations in TREX1 are associated with Aicardi-Goutières syndrome and chilblain lupus, which results from accumulation of endogenous retroelements and overstimulation of a cytosolic DNA sensor (Stetson et al., 2008). This sensor is expressed by many cell types including macrophages, DCs, fibroblasts, and CD4 T cells (Stetson et al., 2008; Yan et al., 2010). In the absence of TREX1, enough ligand for DNA sensor accumulates in HIV-1 infected cells to turn on type I IFN genes. In infected human CD4 T cells, HIV-1 is efficiently controlled when TREX1 is knocked down by siRNA, presumably through the IFNs secreted by the infected cells (Yan et al., 2010) (Figure 3).
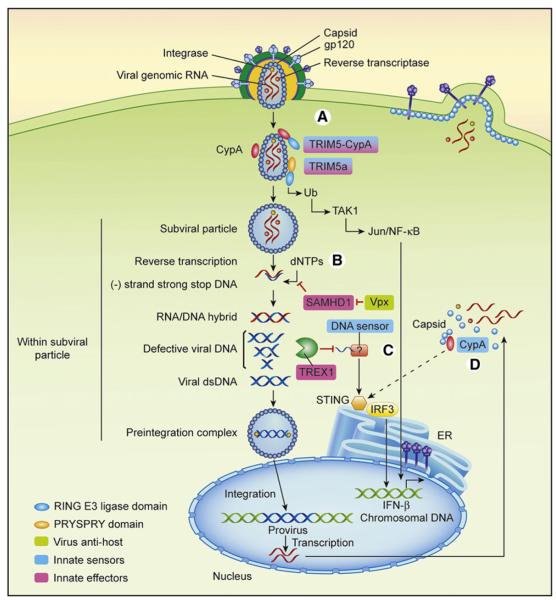
Figure 2. Innate Recognition of HIV-1 by Dendritic Cells and Macrophages.
HIV-1 env binds to CD4 and CCR5 expressed on DCs and macrophages and enables viral fusion.
(A) Capsid lattice in the cytosol is bound by TRIM5α and TRIM5-CypA fusion proteins (in certain nonhuman primates) via the PRYSPRY or the CypA domains, respectively, in species-specific manner. This activates E3 ubiquitin ligase function of TRIM5α, to catalyze synthesis of free ubiquitin chains that activate TAK1 kinase complex leading to AP-1 and NF-κB activation. HIV-1 capsid is only weakly recognized by human TRIM5α. Within the subviral particle, cDNA synthesis on HIV-1 genomic RNA begins.
(B) SAMDH1 blocks reverse transcription at the minus strand strong stop DNA stage, by limiting the availability of free dNTPs needed for further cDNA synthesis. However, this is countered by Vpx, which induces SAMDH1 degradation. HIV-1 does not encode Vpx, and thus human DCs are not productively infected.
(C) Defective viral RT-generated DNA products are sensed by a cytosolic DNA sensor. However, accumulation of such DNA intermediates is blocked by TREX1, which degrades such product via its 3′-5′ exonuclease activity.
(D) By introducing Vpx, productively infected DCs stimulate type I IFN synthesis and are properly activated for T cell stimulation, through sensing of de novo synthesized viral capsid via CypA. STING and IRF3 are required to induce type I IFNs downstream of the DNA sensor and the CypA pathways. Viral antihost molecules, innate sensors, and innate effectors are color coded as indicated.
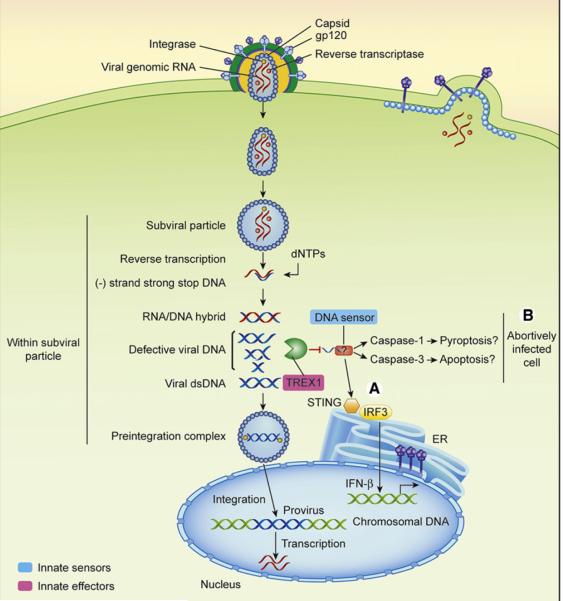
Figure 3. Innate Recognition of HIV-1 in CD4 T Cells.
CD4 T cells are either productively infected or nonpermissive for HIV-1 replication.
(A) In permissive CD4 T cells, HIV-1 recognition by cytosolic DNA sensor is blocked by TREX1 degradation of RT intermediate DNA products.
(B) In CD4 T cells that are abortively infected, HIV-1 triggers death by possibly inducing apoptosis and inflammasome activation (pyroptosis). These pathways of cell death are mediated by a cytosolic DNA sensor that is stimulated by accumulation of defective RT products.
However, it turns out that cytosolic DNA stimulation in T cells may be detrimental, leading to CD4 T cell decline, a hallmark of HIV-1 pathogenesis. Greene and colleagues have shown that accumulation of abortive RT intermediates in resting CD4 T cells following contact with HIV-1 infected cells induces apoptotic and inflammatory programs in nonproductively infected CD4 T cells (Doitsh et al., 2010). Consequently, cytosolic DNA triggers death of “bystander” abortively infected CD4 T cells through the induction of apoptosis and inflammasome activation (Figure 3). The cDNA products in the abortively infected CD4 T cells are presumably not degraded sufficiently by TREX1, possibly because the substrates overwhelm the capacity of TREX1. In addition to possibly stimulating caspase-3 (apoptosis) and caspase-1 (pyroptosis) death pathways, DNA sensor activation in the nonpermissive CD4 T cells also leads to IFN-β induction (Doitsh et al., 2010). Thus, it is possible that the dying bystander CD4 T cells may be a source of type I IFNs in HIV-1 infected individual, especially during the later phase leading to AIDS (von Sydow et al., 1991). As discussed below, chronic secretion of type I IFNs is associated with AIDS progression.
Cyclophilin A in DCs Senses HIV-1 Capsid But Is Masked by SAMHD1
As discussed above, human DCs are not effectively infected by HIV-1. Recent studies revealed that this is because DCs express SAMHD1, which arrests the virus infection at the stage of minus strand strong stop DNA (Figure 2). SAMHD1 hydrolyzes intracellular deoxynucleoside triphosphates (dNTPs), lowering their concentrations and preventing synthesis of the viral DNA by RT in DCs and macrophages (Goldstone et al., 2011) (Table 1). Lentiviruses of the HIV-2 and SIV (simian immunodeficiency virus) sooty mangabey lineage express Vpx, which promotes ubiquitination and degradation of SAMHD1 through recruitment of multisubunit cullin 4A-ring E3 ubiquitin ligase CRL4 (Hrecka et al., 2011; Laguette et al., 2011). Curiously, HIV-1 does not encode Vpx, rendering it incapable of productively infecting DCs. A sensor capable of inducing robust type I IFN responses was discovered by lifting SAMHD1-mediated restriction of HIV-1 in human DCs through introduction of Vpx (Manel et al., 2010). This sensor, cyclophilin A (CypA), is a peptidylpropyl isomerase that is expressed in cytosol. It binds to de novo synthesized HIV-1 capsid and activates IRF3-dependent induction of type I IFN genes (Manel et al., 2010) (Figure 2). Strikingly, direct infection of DCs in the presence of Vpx enabled expression of costimulatory molecules and activation of CD4 T cells in the presence of anti-CD3 antibody. Moreover, HIV-1 infected DCs inhibited trans-enhancement of CD4 T cell infection through secretion of type I IFNs. Whether the capsid itself is the PAMP that activates IRF3 through CypA or whether CypA intersects with nucleic acid recognition pathway to induce type I IFNs remains an intriguing mystery (Manel and Littman, 2011). Of note, even though CD4 T cells are naturally productively infected by HIV-1, CypA is not active in this cell type. In CD4 T cells, the main sensor of HIV-1 appears to be an unknown DNA sensor, because TREX1 knockdown results in enhanced IFN production by HIV-1 infection (Yan et al., 2010) (Figure 3).
Why is Vpx missing in HIV-1? Certain SIV strains possess Vpr that can efficiently degrade SAMHD1, and this preceded the evolutionary emergence of Vpx (Lim et al., 2012). However, phylogenetic analysis revealed that Vpr of HIV-1 or its predecessors (chimpanzee SIV) never possessed the ability to degrade SAMHD1. Thus, lineage of lentiviruses preceding HIV-1 never acquired the ability to degrade SAMHD1, and this may have served HIV-1 well, by disabling DCs from CD4 stimulation and enabling DCs for enhanced trans-infection of CD4 T cells (Manel and Littman, 2011).
TRIM5a Senses Lentiviral Capsid Lattice-Mismatch in Human TRIM5a and HIV-1 Capsid
TRIM5α is well known for its ability to restrict retroviruses in species-specific manner (Joly et al., 1991; Stremlau et al., 2004). TRIM5a binds to incoming retroviral capsid via its PRYSPRY domain and promotes disassembly before completion of reverse transcription (Grütter and Luban, 2012) (Figure 2). Recent study demonstrates that TRIM5-dependent restriction relies on signal transduction. TRIM5α recognition of capsid lattice stimulates its E3 ubiquitin ligase function, to catalyze synthesis of free K63-ubiquitin chains that activate TAK1 kinase complex (Pertel et al., 2011). TAK1 activation by TRIM5a promotes AP-1 and NF-κB signaling in DCs. Ectopic expression of TRIM5α is not sufficient to induce transcription of IFN-β but together with IRF3 overexpression, enhances IFN-β production and mediates antiviral effects (Figure 2). TRIM5α-CypA fusion protein from owl monkey also stimulates the same TAK1 pathway as the full length TRIM5α, even though TRIM5α-CypA lacks the PRYSPRY domain (Pertel et al., 2011), indicating that the RING domain containing the E3 Ub ligase activity is essential for signaling-dependent antiviral activity of TRIM5α. The same study demonstrated that TRIM5α is involved in innate signaling from TLR4 in the absence of capsid recognition. Thus, TRIM5α can act downstream of TLR4 signaling to enhance Jun and NF-κB signaling in macrophages and DCs. However, in lentivirus-infected cells, the presence of the stimulatory capsid structure in the cytosol promotes more efficient lattice formation by TRIM5α (Grütter and Luban, 2012), enhancing its enzymatic activity to promote signaling and to block infection prior to reverse transcription. Because human TRIM5a is only capable of weak binding to HIV-1 capsid and restricting the virus (Joly et al., 1991;Stremlau et al., 2004), innate sensor activity of TRIM5a against HIV-1 is expected to be limited. It would be interesting to determine whether TRIM5α contributes to innate immunity against HIV-1 by acting downstream of TLR7-mediated recognition, akin to the pathway demonstrated for TLR4 (Pertel et al., 2011).
Possible Interplay between Retroviral Innate Sensor Pathways
The host sensing pathways of viral capsid and DNA may be more intimately linked than meets the eye. As suggested by Littman and colleagues, capsid-induced CypA stimulation of IRF3 may involve the DNA sensor (Manel and Littman, 2011). In addition, as mentioned above, in all species of New World owl monkeys, TRIM5α is expressed as a fusion protein between the TRIM5α N-terminal domain and the CypA full-length protein replacing the PRYSPRY domain of TRIM5α. This fusion was mediated by LINE1 retrotransposition of a CypA cDNA into the TRIM5 locus(Sayah et al., 2004). Remarkably, a similar but independent fusion event generated TRIM5α-CypA chimera in some Old World primate species, Asian macaques (Newman et al., 2008). As mentioned above, this fusion protein is capable of supporting TAK1 activation when the CypA portion is engaged by retroviral capsid lattice (Pertel et al., 2011). As long as the TRIM5α domains at the N terminus do not interfere with CypA function, it is expected that such a fusion protein would also be capable of activating IRF3 through the pathway described by Manel et al. (2010). Of note, anti-HIV-1 activity of an engineered human TRIM5α fused to human CypA is more effective than that of the well-characterized rhesus TRIM5α, in blocking HIV-1 infection in primary CD4 T cells and macrophages (Neagu et al., 2009). Perhaps given sufficient time and selective pressure, a similar fusion protein could naturally arise and become selected for in humans in HIV-1 endemic areas. The evolution of TRIM5-CypA fusion proteins in Old World monkeys revealed that the fusion protein acquired several mutations along the way, which enhanced restriction to particular viruses at the cost of broad coverage of lentiviruses (Ylinen et al., 2010). The fascinating convergent evolution of TRIM5-CypA in New World monkeys and Old World monkeys suggests that this fusion protein conferred evolutionary advantage to certain highly pathogenic lentiviruses in the respective species over the preexisting TRIM5α proteins. It is tempting to speculate that the advantage included synergism between the innate sensor pathway of TRIM5α and CypA.
Innate Responses Elicited by HIV-1 in Productively Infected CD4 T Cells
As discussed above, virtually all cell intrinsic innate recognition pathways for HIV-1 are disabled by the host or by the virus. In addition, HIV-1 induces a global disruption of innate signaling pathways in infected cells by degrading interferon-regulatory factor 3 (IRF3) (Doehle et al., 2009). Not surprisingly, infection of CD4 T cells with HIV-1 does not result in typical IFN signatures (Pankiv et al., 2007). Compendium analysis of genes regulated by various stimuli revealed that HIV-1 infection induces very specific changes that are distinct from heat shock stressresponse or IFN response pathways. In contrast, gene profiles induced by type I IFNs and influenza virus infection are mostly overlapping (van ’t Wout et al., 2003). Proteomic analysis of HIV-1-infected CD4 T cells revealed a differential expression of proteins in select biological pathways, including ubiquitin-conjugating enzymes, carrier proteins in nucleocytoplasmic transport, cyclin-dependent kinase in cell-cycle progression, and pyruvate dehydrogenase of the citrate cycle pathways but notably missing in IFN signaling or induction pathways (Chan et al., 2007). However, neither mechanism nor consequences of such distinct gene profiles induced by HIV-1 infection in target CD4 T cells is understood. Whether similar genes are induced by endogenous retroviruses and if so, whether the consequences of turning on such sets of genes assist host tolerance mechanisms for coexistence with endogenous retroviruses is of interest for future studies. Understanding the unique genomic and proteomic signatures associated with retrovirus infection in target cells may provide the key to unlocking the mystery of the “host appropriate” immune response to this group of viruses, and may teach us ways in which we might mimic such responses to avoid pathologic consequences of HIV-1 infection.
Mucosal Barrier as Innate Shield against HIV-1
HIV-1 transmission is extremely inefficient in the female genital mucosa. Successful transmission via heterosexual penilevaginal exposure occurs only 1 in 100 or 1,000 exposures (Powers et al., 2008), whereas male circumcision and existing genital ulcers can dramatically decrease or increase the infection rate, respectively. In contrast, heterosexual anal intercourse between serodiscordant couples has a transmission rate of 1 for every 3–10 episodes. This rate is similar to the transmission risk in penile-anal intercourse between men who have sex with men (Vittinghoff et al., 1999). This highlights the protective barrier provided by the vaginal mucosa compared to rectal mucosa. The vaginal tract is specialized to provide protective mechanisms including its thick epithelial layer, mucus, antimicrobial peptides, low pH, and Lactobacillus-rich endogenous microbiota (Iwasaki, 2010). Instead, HIV-1 enters likely from the endocervix, where epithelial layer is single cell thick and is more prone to disruption (Haase, 2010). The rectal mucosa is covered by a single layer of columnar epithelial cells and contains abundant lymphoid follicles that contain viral target cells (Ribeiro Dos Santos et al., 2011). What clinical approaches might we take in the future to deal with the lack of a protective mucosal barrier in the rectum? Aside from the existing approaches including the use condoms and other safe sex practices, inclusion of antiviral agents or molecules that fortify the epithelial barrier in the lubricant might provide additional protection against rectal HIV-1 exposure.
Role of Type I IFNs in Innate Defense against HIV-1
In humans acutely infected with HIV-1, type I IFNs are detected in the serum starting almost immediately following infection and until 12 days postinfection (von Sydow et al., 1991). This rise in IFN-α is closely followed by a decline in HIV load. Although the exact source of IFN-α is not known, pDCs are a likely candidate as they are equipped with viral sensor TLR7 and are capable of secreting large amount of type I IFNs and cytokines in a rapid response to HIV-1. In addition, as described above, the TLR7 pathway is the only one that is not countered effectively by the virus or the host for IFN synthesis. Type I IFNs induce expression of hundreds of ISGs in responding cells, some of which can contribute to HIV-1 restriction (Florey et al., 2011). However, type I IFNs secreted locally by pDCs in the cervical mucosa paradoxically may have detrimental consequences. Shortly after SIV vaginal challenge, pDCs are recruited just beneath the cervicovaginal epithelial cells, likely through the action of CCL20. Instead of suppressing virus replication, local pDC-secreted type I IFNs and chemokines CCL3 and CCL4 can recruit viral target CCR5+ CD4 T cells (Li et al., 2009).
As the infection progresses, blood pDC numbers decline in HIV-1 infected patients (Chehimi et al., 2002; Soumelis et al., 2001). At later time points of HIV-1 infection, pDC from viremic patients produce little IFN in response to HIV-1 (Machmach et al., 2012) presumably due to downregulation of CD4 molecule in pDCs imposed by viremia (Barblu et al., 2012). This results in overall decline in the plasma IFN levels in most chronically HIV-infected patients. Moreover, high plasma titers of IFN-α during late-stage disease have been shown to correlate with disease progression in humans (von Sydow et al., 1991). This is also supported by the gene profiling studies of SIV-infected nonhuman primates that progress to AIDS (macaques) versus those that harbor high viral titers without developing disease (African green monkeys and sooty mangabeys). Although all groups express similar levels of type I IFNs during the acute infection, only the macaques retained high IFN levels in the chronic phase of infection (Bosinger et al., 2009; Jacquelin et al., 2009). The sustained IFN and cytokine signatures in macaques during chronic SIV infection closely mirror responses in humans infected with HIV-1 (Mandl et al., 2008). Therefore, while the transient type I IFNs in serum of acutely infected individuals may contribute to suppression of systemic level of HIV-1, the initial cervicovaginal IFN response recruits more target cells. On the other hand, chronic expression of IFN fuels HIV-1 pathogenesis. How might the chronic levels of type I IFNs in the susceptible host lead to AIDS? It is thought that natural hosts (sooty mangabeys) are capable of removing viral target cells through maintaining low expression of surface viral receptors (i.e., CD4, CCR5) on T cells, both at steady state and following lentivirus infection, whereas the central memory T cells of the susceptible hosts upregulate CCR5, with the help of chronic levels of IFNs, and fuel the infection of memory T cell pool (Brenchley et al., 2010). Thus, a possible future intervention against AIDS might be to minimize infection of target cells (through blockade of viral receptors) or attenuate chronic activation of the IFN-αβR signaling, preferably in central memory T cells. The latter approach must be carefully weighed against the risk of infection by viruses in general, by interfering with IFN-αβR signaling.
Innate Link to Adaptive Immunity
What innate signals are needed to trigger adaptive immune responses to retroviruses including HIV-1? Recent studies begin to provide clues to this question, but the answer may depend on the immunological outcome. As discussed above, in vivo studies in mice reveal that antibody responses to murine retroviruses, Friend murine leukemia virus (F-MLV), and MMTV require TLR7 and MyD88, as well as DCs (Browne and Littman, 2009; Kane et al., 2011). In contrast, DC-depleted and MyD88-deficient mice develop intact CD8 T cell responses to MLV (Browne and Littman, 2009). In F-MLV-infected TLR7 knockout mice, B cells upregulate CD69 and CD86 early in infection but fail to develop into germinal center B cells (Browne, 2011). TLR7-deficient mice also have impaired CD4 IFN-γ secretion but intact CD8 T cell responses to F-MLV. Deletion of MyD88 in B cells, but not DCs, impaired antibody response to F-MLV, but not CD4 T cell responses. Thus, different innate sensors appear to be needed for different effector functions, namely, that TLR7 in B cells appears to be required for antibody responses, whereas Th1 CD4 T cell responses requires TLR7 expression in non-B cells (likely in DCs) in response to retroviruses in mice. In contrast, CD8 T cell responses develop in the absence of TLR7 or MyD88. However, the importance of these sensors in human HIV-1 infection is not yet determined.
Can HIV-1-infected DCs prime T cell responses? In the case of many if not all viruses, directly infected APCs are rendered incapable of stimulating naive T cells. Viruses can inhibit every aspect of DC biology, from their migration, activation, secretion of cytokines, and presentation of antigens on MHC class I and class II. HIV-1 is no exception. First, HIV-1 does not productively infect DCs. Second, even the rare productively infected DCs would be rendered incapable of stimulating T cells in the presence of nef, which downregulates CD80 and CD86 (Chaudhry et al., 2005), as well as MHC class I (Kenter et al., 2009; Thurston et al., 2009) and class II (Stumptner-Cuvelette et al., 2001). Thus, the likely mode of T cell priming is through cross-priming of viral antigens to naive CD8 T cells by uninfected DCs. However, innate sensors responsible for activating DCs to cross prime HIV-1-specific CD8 T cells remain undefined.
A major hurdle in DC activation of HIV-1-specific CD4 T cells is that interaction between DCs harboring HIV-1 and cognate CD4 T cells results in robust trans-infection of CD4 T cells (Geijtenbeek et al., 2000). DC-SIGN expressed on DCs can bind HIV-1 envelope gp120 and internalize virions through endocytosis. This endosomal location of DC-SIGN protects the virions from degradation, and poises them for infection of CD4 T cells. In addition, DCs and CD4 T cells form syncytia upon HIV-1 infection (Pope et al., 1994) in the tonsil and other lymphoid tissues (Frankel et al., 1996) that provide a breeding ground for HIV-1. Once incorporated into syncytia, HIV-primed CD4 T cells will not be able to escape and perform antiviral activities. Thus, in order for DCs to successfully prime virus-specific CD4 T cells, they would have to take up dead virus or viral antigens, while specifically avoiding live virus, to process and present viral antigens to naive CD4 T cells. This situation may be very rare but must occur, because anti-HIV-1 CD4 T cell responses are robustly generated in HIV-1 infected individuals. HIV-1-specific CD4 T cells may be in fact critical in controlling HIV-1 infection early, which impacts on the disease course. In particular, lytic CD4 T cell phenotype at baseline (high granzyme A expression) and CD107a degranulation phenotype during the early stages of HIV infection (2–6 months) correlate with low viral set point and successful control of HIV-1 infection (Soghoian et al., 2012). Therefore, a mechanism must exist whereby DCs or possibly other APCs sample viral antigens while avoiding infection. Identification of the antigen-presenting cell type and the innate sensors that link HIV-1 recognition to successful priming of virus-specific CD4 and CD8 T cells will provide key knowledge needed to generate robust protective anti-HIV-1 immunity, which may naturally occur in rare controllers. In addition, it is of special interest to determine the mechanism of viral antigen sampling and processing by the key antigen presenting cells that accomplish these tasks without becoming infected or fused to infected cells.
Future Perspectives
Due to the large public and private investments toward HIV-1 research, the field has experienced an explosion of discoveries related to its virology, viral pathogenesis, and immunology of HIV-1 infection. The basic scientific research on HIV-1 has revealed a great deal about host cell biology, innate immunity, and the generation of adaptive immunity, while clinical translation of the basic understanding has lead to over 20 different antiviral agents that are highly effective and are widely used (Deeks et al., 2012). However, it is sobering to realize that all possible innate sensor pathways, perhaps with the exception of TLR7, have been concealed by the virus or the host, leaving the immune system little to work with in terms of generating an early wave of type I IFNs that can limit viral set point and improve the course of HIV-1 disease. Some experts in the field argue that it is the delayed activation of innate immune responses with respect to peak viremia that sets off chronic immune activation and progression to AIDS, and the question of whether an adaptive immune response can be mounted against HIV is irrelevant and should no longer be the focus of vaccine research strategies (Benecke et al., 2012). How might one achieve rapid and robust innate activation following HIV-1 infection that successfully sets viral set point to lowest possible levels? Devising ways of triggering the innate sensors to enable them to generate IFNs and limiting virus replication early during HIV-1 infection may provide an alternative to HIV-1 vaccine based on generating adaptive immunity. Examples of such approach may include knockdown of factors responsible for inhibiting innate sensor activations, such as TREX1, SAMDH1, or introducing “cognate” TRIM5α for HIV-1 capsid. Identification of DNA sensor(s) that recognize RT products in the cytosol will also provide molecular targets for eliciting robust IFN response and adaptive immunity.
Another topic for future investigation is the possible role of innate effectors in the generation of adaptive immunity to HIV-1. As discussed here, innate restriction factors, SAMDH1 and TREX1, control the availability of ligands for DNA sensor during HIV-1 infection, thereby blocking induction of innate and adaptive immune responses. Other restriction factors such as APOBEC3 and tetherin restrict HIV-1 by acting on very different stages of the viral life cycle (see Blanco-Melo et al. [2012] in this issue for in-depth description) (Table 1). Tetherin is an IFN-inducible protein expressed on the cell surface that prevents the release of HIV-1 virions (Neil et al., 2008). APOBEC3G is a member of APOBEC3 family of cytidine deaminase (Sheehy et al., 2002) that is capable of introducing hypermutations in cDNA during the RT process (Yu et al., 2004). Although these effectors have not yet been reported to be involved in modulating the activity of innate sensors, the actions of APOBEC3 and tetherin are predicted to alter the type and magnitude of innate immune responses. For instance, tetherin-expressing HIV-infected cells that accumulate viral products (including PAMPs) may become a factory for release of innate cytokines, as well as better antigen presenting cells or antigen donor cells for bystander DCs. In addition, all HIV-1 restriction factors are expected to modulate adaptive immune response to HIV-1, by reducing viral antigen load, as well as enabling the resistant cells to carry out immune functions. Therefore, immune intervention through exploiting the functions of appropriate innate HIV-1 effectors may not only result in reduce viral load, but possibly also in more efficient priming of T and B cell immune responses.
Although innate intervention in HIV-1-infected individual is clearly needed to prevent AIDS progression, a robust prophylactic vaccine based on adaptive immune memory response is irreplaceable with any other type of approach. Prophylactic vaccine is the only way to provide life-long protection for possibly millions of people worldwide and is the only way infectious agents have been successfully eradicated by human hands in the past. Understanding innate sensor pathways capable of inducing protective immunity to HIV-1 will be a long awaited and highly impactful contribution an immunologist can make in the future years to come.
ACKNOWLEDGMENTS
I wish to thank R. Medzhitov for critical discussion and reading of the manuscript and National Institutes of Health (NIH) for its support of research in my laboratory (AI081884, AI054359, AI062428, and AI064705). Research in my laboratory is supported in part by the NIH/NIAID under Award Number U54AI057160 to the Midwest Regional Center of Excellence for Biodefense and Emerging Infectious Diseases Research (MRCE).
REFERENCES
- Barblu L, Machmach K, Gras C, Delfraissy JF, Boufassa F, Leal M, Ruiz-Mateos E, Lambotte O, Herbeuval JP, for the ANRS EP36 HIV Controllers Study Group Plasmacytoid dendritic cells from HIV controllers produce IFN-alpha and differentiate into functional killer pDC under HIV activation. J. Infect. Dis. 2012;206:790–801. doi: 10.1093/infdis/jis384. [DOI] [PubMed] [Google Scholar]
- Beignon AS, McKenna K, Skoberne M, Manches O, DaSilva I, Kavanagh DG, Larsson M, Gorelick RJ, Lifson JD, Bhardwaj N. Endocytosis of HIV-1 activates plasmacytoid dendritic cells via Toll-like receptor-viral RNA interactions. J. Clin. Invest. 2005;115:3265–3275. doi: 10.1172/JCI26032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benecke A, Gale M, Jr., Katze MG. Dynamics of innate immunity are key to chronic immune activation in AIDS. Curr Opin HIV AIDS. 2012;7:79–85. doi: 10.1097/COH.0b013e32834dde31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchet FP, Moris A, Nikolic DS, Lehmann M, Cardinaud S, Stalder R, Garcia E, Dinkins C, Leuba F, Wu L, et al. Human immunodeficiency virus-1 inhibition of immunoamphisomes in dendritic cells impairs early innate and adaptive immune responses. Immunity. 2010;32:654–669. doi: 10.1016/j.immuni.2010.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco-Melo D, Venkatesh S, Bieniasz PD. Intrinsic Cellular Defenses Against Human Immunodeficiency Viruses. Immunity. 2012;37(this issue):399–411. doi: 10.1016/j.immuni.2012.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosinger SE, Li Q, Gordon SN, Klatt NR, Duan L, Xu L, Francella N, Sidahmed A, Smith AJ, Cramer EM, et al. Global genomic analysis reveals rapid control of a robust innate response in SIV-infected sooty mangabeys. J. Clin. Invest. 2009;119:3556–3572. doi: 10.1172/JCI40115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenchley JM, Silvestri G, Douek DC. Nonprogressive and progressive primate immunodeficiency lentivirus infections. Immunity. 2010;32:737–742. doi: 10.1016/j.immuni.2010.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browne EP. Toll-like receptor 7 controls the anti-retroviral germinal center response. PLoS Pathog. 2011;7:e1002293. doi: 10.1371/journal.ppat.1002293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browne EP, Littman DR. Myd88 is required for an antibody response to retroviral infection. PLoS Pathog. 2009;5:e1000298. doi: 10.1371/journal.ppat.1000298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan EY, Qian WJ, Diamond DL, Liu T, Gritsenko MA, Monroe ME, Camp DG, 2nd, Smith RD, Katze MG. Quantitative analysis of human immunodeficiency virus type 1-infected CD4+ cell proteome: dysregulated cell cycle progression and nuclear transport coincide with robust virus production. J. Virol. 2007;81:7571–7583. doi: 10.1128/JVI.00288-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhry A, Das SR, Hussain A, Mayor S, George A, Bal V, Jameel S, Rath S. The Nef protein of HIV-1 induces loss of cell surface costimulatory molecules CD80 and CD86 in APCs. J. Immunol. 2005;175:4566–4574. doi: 10.4049/jimmunol.175.7.4566. [DOI] [PubMed] [Google Scholar]
- Chehimi J, Campbell DE, Azzoni L, Bacheller D, Papasavvas E, Jerandi G, Mounzer K, Kostman J, Trinchieri G, Montaner LJ. Persistent decreases in blood plasmacytoid dendritic cell number and function despite effective highly active antiretroviral therapy and increased blood myeloid dendritic cells in HIV-infected individuals. J. Immunol. 2002;168:4796–4801. doi: 10.4049/jimmunol.168.9.4796. [DOI] [PubMed] [Google Scholar]
- Deeks SG, Autran B, Berkhout B, Benkirane M, Cairns S, Chomont N, Chun TW, Churchill M, Mascio MD, Katlama C, et al. The International AIDS Society Scientific Working Group on HIV Cure Towards an HIV cure: a global scientific strategy. Nat. Rev. Immunol. 2012;12:607–614. doi: 10.1038/nri3262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diebold SS, Kaisho T, Hemmi H, Akira S, Reis e Sousa C. Innate antiviral responses by means of TLR7-mediated recognition of single-stranded RNA. Science. 2004;303:1529–1531. doi: 10.1126/science.1093616. [DOI] [PubMed] [Google Scholar]
- Doehle BP, Hladik F, McNevin JP, McElrath MJ, Gale M., Jr. Human immunodeficiency virus type 1 mediates global disruption of innate antiviral signaling and immune defenses within infected cells. J. Virol. 2009;83:10395–10405. doi: 10.1128/JVI.00849-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doitsh G, Cavrois M, Lassen KG, Zepeda O, Yang Z, Santiago ML, Hebbeler AM, Greene WC. Abortive HIV infection mediates CD4 T cell depletion and inflammation in human lymphoid tissue. Cell. 2010;143:789–801. doi: 10.1016/j.cell.2010.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzgerald-Bocarsly P, Jacobs ES. Plasmacytoid dendritic cells in HIV infection: striking a delicate balance. J. Leukoc. Biol. 2010;87:609–620. doi: 10.1189/jlb.0909635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Florey O, Kim SE, Sandoval CP, Haynes CM, Overholtzer M. Autophagy machinery mediates macroendocytic processing and entotic cell death by targeting single membranes. Nat. Cell Biol. 2011;13:1335–1343. doi: 10.1038/ncb2363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankel SS, Wenig BM, Burke AP, Mannan P, Thompson LD, Abbondanzo SL, Nelson AM, Pope M, Steinman RM. Replication of HIV-1 in dendritic cell-derived syncytia at the mucosal surface of the adenoid. Science. 1996;272:115–117. doi: 10.1126/science.272.5258.115. [DOI] [PubMed] [Google Scholar]
- Geijtenbeek TB, Kwon DS, Torensma R, van Vliet SJ, van Duijnhoven GC, Middel J, Cornelissen IL, Nottet HS, KewalRamani VN, Littman DR, et al. DC-SIGN, a dendritic cell-specific HIV-1-binding protein that enhances trans-infection of T cells. Cell. 2000;100:587–597. doi: 10.1016/s0092-8674(00)80694-7. [DOI] [PubMed] [Google Scholar]
- Gilliet M, Cao W, Liu YJ. Plasmacytoid dendritic cells: sensing nucleic acids in viral infection and autoimmune diseases. Nat. Rev. Immunol. 2008;8:594–606. doi: 10.1038/nri2358. [DOI] [PubMed] [Google Scholar]
- Goldstone DC, Ennis-Adeniran V, Hedden JJ, Groom HC, Rice GI, Christodoulou E, Walker PA, Kelly G, Haire LF, Yap MW, et al. HIV-1 restriction factor SAMHD1 is a deoxynucleoside triphosphate triphosphohydrolase. Nature. 2011;480:379–382. doi: 10.1038/nature10623. [DOI] [PubMed] [Google Scholar]
- Gringhuis SI, van der Vlist M, van den Berg LM, den Dunnen J, Litjens M, Geijtenbeek TB. HIV-1 exploits innate signaling by TLR8 and DC-SIGN for productive infection of dendritic cells. Nat. Immunol. 2010;11:419–426. doi: 10.1038/ni.1858. [DOI] [PubMed] [Google Scholar]
- Grütter MG, Luban J. TRIM5 structure, HIV-1 capsid recognition, and innate immune signaling. Curr. Opin. Virol. 2012;2:142–150. doi: 10.1016/j.coviro.2012.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haase AT. Targeting early infection to prevent HIV-1 mucosal transmission. Nature. 2010;464:217–223. doi: 10.1038/nature08757. [DOI] [PubMed] [Google Scholar]
- Heil F, Hemmi H, Hochrein H, Ampenberger F, Kirschning C, Akira S, Lipford G, Wagner H, Bauer S. Species-specific recognition of single-stranded RNA via toll-like receptor 7 and 8. Science. 2004;303:1526–1529. doi: 10.1126/science.1093620. [DOI] [PubMed] [Google Scholar]
- Hrecka K, Hao C, Gierszewska M, Swanson SK, Kesik-Brodacka M, Srivastava S, Florens L, Washburn MP, Skowronski J. Vpx relieves inhibition of HIV-1 infection of macrophages mediated by the SAMHD1 protein. Nature. 2011;474:658–661. doi: 10.1038/nature10195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hübner W, McNerney GP, Chen P, Dale BM, Gordon RE, Chuang FY, Li XD, Asmuth DM, Huser T, Chen BK. Quantitative 3D video microscopy of HIV transfer across T cell virological synapses. Science. 2009;323:1743–1747. doi: 10.1126/science.1167525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki A. Antiviral immune responses in the genital tract: clues for vaccines. Nat. Rev. Immunol. 2010;10:699–711. doi: 10.1038/nri2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacquelin B, Mayau V, Targat B, Liovat AS, Kunkel D, Petitjean G, Dillies MA, Roques P, Butor C, Silvestri G, et al. Nonpathogenic SIV infection of African green monkeys induces a strong but rapidly controlled type I IFN response. J. Clin. Invest. 2009;119:3544–3555. doi: 10.1172/JCI40093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jolly C. Cell-to-cell transmission of retroviruses: Innate immunity and interferon-induced restriction factors. Virology. 2011;411:251–259. doi: 10.1016/j.virol.2010.12.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jolly C, Sattentau QJ. Retroviral spread by induction of virological synapses. Traffic. 2004;5:643–650. doi: 10.1111/j.1600-0854.2004.00209.x. [DOI] [PubMed] [Google Scholar]
- Joly E, Mucke L, Oldstone MB. Viral persistence in neurons explained by lack of major histocompatibility class I expression. Science. 1991;253:1283–1285. doi: 10.1126/science.1891717. [DOI] [PubMed] [Google Scholar]
- Kane M, Case LK, Wang C, Yurkovetskiy L, Dikiy S, Golovkina TV. Innate immune sensing of retroviral infection via Toll-like receptor 7 occurs upon viral entry. Immunity. 2011;35:135–145. doi: 10.1016/j.immuni.2011.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenter GG, Welters MJ, Valentijn AR, Lowik MJ, Berends-van der Meer DM, Vloon AP, Essahsah F, Fathers LM, Offringa R, Drijfhout JW, et al. Vaccination against HPV-16 oncoproteins for vulvar intraepithelial neoplasia. N. Engl. J. Med. 2009;361:1838–1847. doi: 10.1056/NEJMoa0810097. [DOI] [PubMed] [Google Scholar]
- Laguette N, Sobhian B, Casartelli N, Ringeard M, Chable-Bessia C, Ségéral E, Yatim A, Emiliani S, Schwartz O, Benkirane M. SAMHD1 is the dendritic- and myeloid-cell-specific HIV-1 restriction factor counteracted by Vpx. Nature. 2011;474:654–657. doi: 10.1038/nature10117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HK, Lund JM, Ramanathan B, Mizushima N, Iwasaki A. Autophagy-dependent viral recognition by plasmacytoid dendritic cells. Science. 2007;315:1398–1401. doi: 10.1126/science.1136880. [DOI] [PubMed] [Google Scholar]
- Lepelley A, Louis S, Sourisseau M, Law HK, Pothlichet J, Schilte C, Chaperot L, Plumas J, Randall RE, Si-Tahar M, et al. Innate sensing of HIV-infected cells. PLoS Pathog. 2011;7:e1001284. doi: 10.1371/journal.ppat.1001284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Estes JD, Schlievert PM, Duan L, Brosnahan AJ, Southern PJ, Reilly CS, Peterson ML, Schultz-Darken N, Brunner KG, et al. Glycerol monolaurate prevents mucosal SIV transmission. Nature. 2009;458:1034–1038. doi: 10.1038/nature07831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim ES, Fregoso OI, McCoy CO, Matsen FA, Malik HS, Emerman M. The ability of primate lentiviruses to degrade the monocyte restriction factor SAMHD1 preceded the birth of the viral accessory protein Vpx. Cell Host Microbe. 2012;11:194–204. doi: 10.1016/j.chom.2012.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund JM, Alexopoulou L, Sato A, Karow M, Adams NC, Gale NW, Iwasaki A, Flavell RA. Recognition of single-stranded RNA viruses by Toll-like receptor 7. Proc. Natl. Acad. Sci. USA. 2004;101:5598–5603. doi: 10.1073/pnas.0400937101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machmach K, Leal M, Gras C, Viciana P, Genebat M, Franco E, Boufassa F, Lambotte O, Herbeuval JP, Ruiz-Mateos E. Plasmacytoid dendritic cells reduce HIV production in elite controllers. J. Virol. 2012;86:4245–4252. doi: 10.1128/JVI.07114-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandl JN, Barry AP, Vanderford TH, Kozyr N, Chavan R, Klucking S, Barrat FJ, Coffman RL, Staprans SI, Feinberg MB. Divergent TLR7 and TLR9 signaling and type I interferon production distinguish pathogenic and nonpathogenic AIDS virus infections. Nat. Med. 2008;14:1077–1087. doi: 10.1038/nm.1871. [DOI] [PubMed] [Google Scholar]
- Manel N, Littman DR. Hiding in plain sight: how HIV evades innate immune responses. Cell. 2011;147:271–274. doi: 10.1016/j.cell.2011.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manel N, Hogstad B, Wang Y, Levy DE, Unutmaz D, Littman DR. A cryptic sensor for HIV-1 activates antiviral innate immunity in dendritic cells. Nature. 2010;467:214–217. doi: 10.1038/nature09337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshak-Rothstein A, Rifkin IR. Immunologically active autoantigens: the role of toll-like receptors in the development of chronic inflammatory disease. Annu. Rev. Immunol. 2007;25:419–441. doi: 10.1146/annurev.immunol.22.012703.104514. [DOI] [PubMed] [Google Scholar]
- Medzhitov R, Schneider DS, Soares MP. Disease tolerance as a defense strategy. Science. 2012;335:936–941. doi: 10.1126/science.1214935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neagu MR, Ziegler P, Pertel T, Strambio-De-Castillia C, Grütter C, Martinetti G, Mazzucchelli L, Grütter M, Manz MG, Luban J. Potent inhibition of HIV-1 by TRIM5-cyclophilin fusion proteins engineered from human components. J. Clin. Invest. 2009;119:3035–3047. doi: 10.1172/JCI39354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neil SJ, Zang T, Bieniasz PD. Tetherin inhibits retrovirus release and is antagonized by HIV-1 Vpu. Nature. 2008;451:425–430. doi: 10.1038/nature06553. [DOI] [PubMed] [Google Scholar]
- Newman RM, Hall L, Kirmaier A, Pozzi LA, Pery E, Farzan M, O’Neil SP, Johnson W. Evolution of a TRIM5-CypA splice isoform in old world monkeys. PLoS Pathog. 2008;4:e1000003. doi: 10.1371/journal.ppat.1000003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brien M, Manches O, Sabado RL, Baranda SJ, Wang Y, Marie I, Rolnitzky L, Markowitz M, Margolis DM, Levy D, Bhardwaj N. Spatiotemporal trafficking of HIV in human plasmacytoid dendritic cells defines a persistently IFN-α-producing and partially matured phenotype. J. Clin. Invest. 2011;121:1088–1101. doi: 10.1172/JCI44960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pankiv S, Clausen TH, Lamark T, Brech A, Bruun JA, Outzen H, Øvervatn A, BjØrkØy G, Johansen T. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J. Biol. Chem. 2007;282:24131–24145. doi: 10.1074/jbc.M702824200. [DOI] [PubMed] [Google Scholar]
- Pertel T, Hausmann S, Morger D, Züger S, Guerra J, Lascano J, Reinhard C, Santoni FA, Uchil PD, Chatel L, et al. TRIM5 is an innate immune sensor for the retrovirus capsid lattice. Nature. 2011;472:361–365. doi: 10.1038/nature09976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichlmair A, Reis e Sousa C. Innate recognition of viruses. Immunity. 2007;27:370–383. doi: 10.1016/j.immuni.2007.08.012. [DOI] [PubMed] [Google Scholar]
- Pope M, Betjes MG, Romani N, Hirmand H, Cameron PU, Hoffman L, Gezelter S, Schuler G, Steinman RM. Conjugates of dendritic cells and memory T lymphocytes from skin facilitate productive infection with HIV-1. Cell. 1994;78:389–398. doi: 10.1016/0092-8674(94)90418-9. [DOI] [PubMed] [Google Scholar]
- Powers KA, Poole C, Pettifor AE, Cohen MS. Rethinking the heterosexual infectivity of HIV-1: a systematic review and meta-analysis. Lancet Infect. Dis. 2008;8:553–563. doi: 10.1016/S1473-3099(08)70156-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro Dos Santos P, Rancez M, Prétet JL, Michel-Salzat A, Messent V, Bogdanova A, Couëdel-Courteille A, Souil E, Cheynier R, Butor C. Rapid dissemination of SIV follows multisite entry after rectal inoculation. PLoS ONE. 2011;6:e19493. doi: 10.1371/journal.pone.0019493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayah DM, Sokolskaja E, Berthoux L, Luban J. Cyclophilin A retrotransposition into TRIM5 explains owl monkey resistance to HIV-1. Nature. 2004;430:569–573. doi: 10.1038/nature02777. [DOI] [PubMed] [Google Scholar]
- Sheehy AM, Gaddis NC, Choi JD, Malim MH. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature. 2002;418:646–650. doi: 10.1038/nature00939. [DOI] [PubMed] [Google Scholar]
- Sherer NM, Lehmann MJ, Jimenez-Soto LF, Horensavitz C, Pypaert M, Mothes W. Retroviruses can establish filopodial bridges for efficient cell-to-cell transmission. Nat. Cell Biol. 2007;9:310–315. doi: 10.1038/ncb1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smed-Sörensen A, Loré K, Vasudevan J, Louder MK, Andersson J, Mascola JR, Spetz AL, Koup RA. Differential susceptibility to human immunodeficiency virus type 1 infection of myeloid and plasmacytoid dendritic cells. J. Virol. 2005;79:8861–8869. doi: 10.1128/JVI.79.14.8861-8869.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soghoian DZ, Jessen H, Flanders M, Sierra-Davidson K, Cutler S, Pertel T, Ranasinghe S, Lindqvist M, Davis I, Lane K, et al. HIV-specific cytolytic CD4 T cell responses during acute HIV infection predict disease outcome. Sci. Transl. Med. 2012;4:123–125. doi: 10.1126/scitranslmed.3003165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soumelis V, Scott I, Gheyas F, Bouhour D, Cozon G, Cotte L, Huang L, Levy JA, Liu YJ. Depletion of circulating natural type 1 interferon-producing cells in HIV-infected AIDS patients. Blood. 2001;98:906–912. doi: 10.1182/blood.v98.4.906. [DOI] [PubMed] [Google Scholar]
- Stetson DB, Medzhitov R. Type I interferons in host defense. Immunity. 2006;25:373–381. doi: 10.1016/j.immuni.2006.08.007. [DOI] [PubMed] [Google Scholar]
- Stetson DB, Ko JS, Heidmann T, Medzhitov R. Trex1 prevents cell-intrinsic initiation of autoimmunity. Cell. 2008;134:587–598. doi: 10.1016/j.cell.2008.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stremlau M, Owens CM, Perron MJ, Kiessling M, Autissier P, Sodroski J. The cytoplasmic body component TRIM5alpha restricts HIV-1 infection in Old World monkeys. Nature. 2004;427:848–853. doi: 10.1038/nature02343. [DOI] [PubMed] [Google Scholar]
- Stumptner-Cuvelette P, Morchoisne S, Dugast M, Le Gall S, Raposo G, Schwartz O, Benaroch P. HIV-1 Nef impairs MHC class II antigen presentation and surface expression. Proc. Natl. Acad. Sci. USA. 2001;98:12144–12149. doi: 10.1073/pnas.221256498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi O, Akira S. MDA5/RIG-I and virus recognition. Curr. Opin. Immunol. 2008;20:17–22. doi: 10.1016/j.coi.2008.01.002. [DOI] [PubMed] [Google Scholar]
- Thompson MR, Kaminski JJ, Kurt-Jones EA, Fitzgerald KA. Pattern recognition receptors and the innate immune response to viral infection. Viruses. 2011;3:920–940. doi: 10.3390/v3060920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thurston TL, Ryzhakov G, Bloor S, von Muhlinen N, Randow F. The TBK1 adaptor and autophagy receptor NDP52 restricts the proliferation of ubiquitin-coated bacteria. Nat. Immunol. 2009;10:1215–1221. doi: 10.1038/ni.1800. [DOI] [PubMed] [Google Scholar]
- van ’t Wout AB, Lehrman GK, Mikheeva SA, O’Keeffe GC, Katze MG, Bumgarner RE, Geiss GK, Mullins JI. Cellular gene expression upon human immunodeficiency virus type 1 infection of CD4(+)-T-cell lines. J. Virol. 2003;77:1392–1402. doi: 10.1128/JVI.77.2.1392-1402.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virgin HW, Wherry EJ, Ahmed R. Redefining chronic viral infection. Cell. 2009;138:30–50. doi: 10.1016/j.cell.2009.06.036. [DOI] [PubMed] [Google Scholar]
- Vittinghoff E, Douglas J, Judson F, McKirnan D, MacQueen K, Buchbinder SP. Per-contact risk of human immunodeficiency virus transmission between male sexual partners. Am. J. Epidemiol. 1999;150:306–311. doi: 10.1093/oxfordjournals.aje.a010003. [DOI] [PubMed] [Google Scholar]
- von Sydow M, Sönnerborg A, Gaines H, Strannegård O. Interferon-alpha and tumor necrosis factor-alpha in serum of patients in various stages of HIV-1 infection. AIDS Res. Hum. Retroviruses. 1991;7:375–380. doi: 10.1089/aid.1991.7.375. [DOI] [PubMed] [Google Scholar]
- Yan N, Regalado-Magdos AD, Stiggelbout B, Lee-Kirsch MA, Lieberman J. The cytosolic exonuclease TREX1 inhibits the innate immune response to human immunodeficiency virus type 1. Nat. Immunol. 2010;11:1005–1013. doi: 10.1038/ni.1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ylinen LM, Price AJ, Rasaiyaah J, Hué S, Rose NJ, Marzetta F, James LC, Towers GJ. Conformational adaptation of Asian macaque TRIMCyp directs lineage specific antiviral activity. PLoS Pathog. 2010;6:e1001062. doi: 10.1371/journal.ppat.1001062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Q, König R, Pillai S, Chiles K, Kearney M, Palmer S, Richman D, Coffin JM, Landau NR. Single-strand specificity of APOBEC3G accounts for minus-strand deamination of the HIV genome. Nat. Struct. Mol. Biol. 2004;11:435–442. doi: 10.1038/nsmb758. [DOI] [PubMed] [Google Scholar]
- Zhou D, Spector SA. Human immunodeficiency virus type-1 infection inhibits autophagy. AIDS. 2008;22:695–699. doi: 10.1097/QAD.0b013e3282f4a836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou D, Kang KH, Spector SA. Production of interferon a by human immunodeficiency virus type 1 in human plasmacytoid dendritic cells is dependent on induction of autophagy. J. Infect. Dis. 2012;205:1258–1267. doi: 10.1093/infdis/jis187. [DOI] [PMC free article] [PubMed] [Google Scholar]