Abstract
The platelet field is undergoing a radical transformation from reductionist simplification to large scale integration. Following the era of simplification whereby biological processes were dissected at the molecular and atomic level, new technologies have now generated an overwhelming flow of information that can only be comprehended in an integrated approach. High throughput analyses of transcription and translation in megakaryocytes and platelets, individual analyses of membranes and secretory granules, the clustering of pathways for platelet activation and inhibition in signalosomes all add to a complexity that requires platforms for knowledge accumulation. Here we introduce Reactome, a curated knowledgebase of biological pathways with extensive coverage of pathways relevant to megakaryocytes, platelets and haemostasis. This resource is compared with other data resources for platelets, e.g. the Platelet Web.
1. Reactome
Reactome (http://www.reactome.org) is a free open source database and website of human biological pathways built from connected biological ‘reactions’ or pathway steps that encompass all biological events, e.g. binding, phosphorylation, transport, as well as classical biochemical events [1]. Each reaction is derived from literature and includes a citation that experimentally validates the event described. The aim is to represent a consensus view of human biological pathways, as a free reference and core dataset for biologists (see Figures 1a and 1b).
Figure 1a.
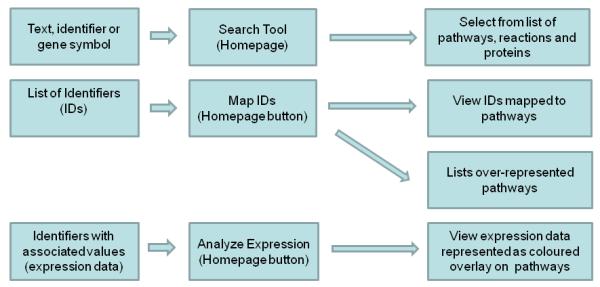
Schematic representing possible paths to details or analysis in Reactome. Users can query using single text names or phrases, database accessions or gene symbols using the Search tool located top left on the homepage. Lists of accession numbers can be submitted to the Map Identifiers (IDs) tool, accessed on the homepage. This has two options, either map IDs to Reactome pathways, or perform over-representation analysis (pathway enrichment). Expression data can be submitted to the Analyze Expression tool, accessed on the homepage. This represents the data as a overlays on Reactome pathways using colour to indicate the expression value.
Figure 1b.
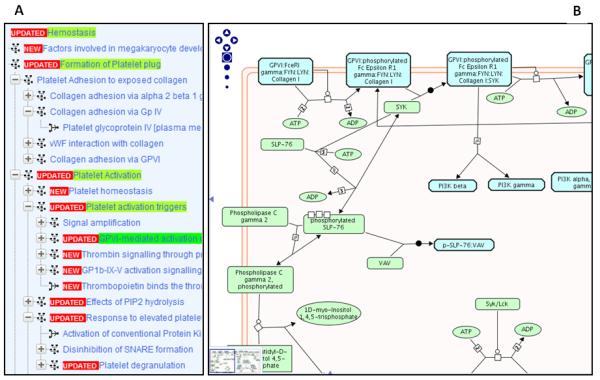
A. Part of the hierarchy of Hemostasis pathways and reactions (pathway steps) in the Reactome Pathway Browser. B. A section of the pathway diagram for GlycoProtein (GP)VI-mediated activation cascade showing proteins (green rectangles) and small molecules (ovals) participating in reactions represented by a central reaction node linked to input and output molecules by lines. Reaction catalysts are connected to the reaction node by a line ending in an open circle. Pathways occur when the output of a reaction is an input or catalyst for another reaction.
The content of Reactome is based on information provided by expert biologists, converted into reactions and pathways by Reactome curators and peer-reviewed by another expert. Reactions and pathways are extensively cross-referenced to databases such as Ensembl (http://www.ensembl.org/index.html), GO (http://www.ebi.ac.uk/QuickGO), PubMed (http://www.ncbi.nlm.nih.gov/pubmed), ChEBI (http://www.ebi.ac.uk/chebi/index.jsp), UniProt (http://www.uniprot.org) and OMIM (http://www.ncbi.nlm.nih.gov/omim). Pathways are human-centric but may incorporate pathway steps manually inferred to exist in humans, based on data from model organisms. These are clearly differentiated from pathway steps that have been experimentally determined in humans. Pathways for species other than human are computationally inferred by a process based on orthology. Currently over 20 additional species are represented. Tools are available on the Reactome website to allow interactive visualisation of pathways and enable analyses such as pathway over-representation (pathway enrichment), pathway expansion to include protein-protein and protein-small molecule interactions and the overlay of expression data onto pathways enabling pathway differential expression analysis. All of these tools are compatible and designed to operate with user-supplied datasets. Pathways can be exported in a variety of formats including the BioPax and Systems Biology Markup Language (SMBL) standards (for further information see http://sbml.org).
Reactome covers many areas of biology such as DNA replication and repair, membrane trafficking, synaptic transmission and receptor-based signaling pathways. Each of these topics contains relevant biological pathways and associated diagrams. Pathways relevant to megakaryocyte and platelet biology are largely within the major topic of Haemostasis. This currently contains (March 2012) 38 pathways including 293 reactions. Subtopics within Haemostasis include platelet adhesion to exposed collagen, nitric oxide metabolism, platelet sensitisation by low-density lipoprotein, adenosine-di-phosphate signaling through P2Y purinergic receptors, thrombin activation of proteinase activated receptors, glycoprotein (GP)VI (Figure 1b) and αIIbβ3 mediated signaling, platelet calcium regulation, and platelet degranulation.
The Platelet Web (http://plateletweb.bioapps.biozentrum.uni-wuerzburg.de/plateletweb.php) is a dataset with associated website representing a platelet-relevant subset of a generic human protein-protein interaction network derived from Human Protein Reference Database or large-scale yeast two-hybrid studies (Y2H) )[2]. Platelet specificity comes from the representation of proteins with platelet-specific proteomics or transcriptomics data. This set was further annotated by incorporating data concerning the platelet phosphoproteome. This approach is fundamentally different and somewhat complementary to the Reactome one, in that it aims to comprehensively represent all proteins that are known to exist in platelets and presents a network of identified interaction connections between them. It does not however attempt to categorise these into recognisable ‘canonical’ pathways, or explain the context of interactions, or suggest platelet-specific processes that might be of particular interest as opposed to widespread metabolic processes, nor does it distinguish between interactions studied and described in the peer-reviewed literature and unfamiliar interactions that might be novel elements of platelet processes, or artefacts of the technology used to identify the interaction. It is recognized that Y2H technology is a highly artificial measure of protein interactivity that can suggest interactions that have no in vivo relevance. The dataset underlying the Platelet Web site cannot be downloaded, and can only be queried via the website for individual proteins. In contrast, Reactome pathways and the data schema can be downloaded in a variety of re-usable formats including SBML and BioPax standards, or as a list of protein identifiers. There are simple and advanced query interfaces, BioMart representation and an application programming interface offering two alternative methods of bulk querying, and there are several tools that allow the user to analyse their own datasets by comparison or overlay onto Reactome pathways.
2. Linking transcription and translation
The HaemAtlas is a comprehensive compendium of transcripts present in the six main peripheral blood cell elements and in erythroblast and megakaryocytes [3]. It identifies genes that have a significantly higher transcript level in the megakaryocytic lineage than in the seven remaining lineages. The Atlas has recently been expanded with information about changes in the transcriptome for the erythroid and megakaryocytic lineages during differentiation of haematopoietic stem cells [4]. Among the over-expressed category are transcripts for platelet-specific surface receptors in which mutations are known to impair platelet function, such as the receptor for von Willebrand Factor (VWF), GPIbα/Ibβ/IX/V, and the receptor for fibrinogen, vitronectin and VWF, GPIIb/IIIa (integrin αIIbβ3). Information about mutations underlying inherited bleeding disorders of the platelet type like Bernard and Soulier syndrome, Glanzmann’s thrombasthenia and Wiskott Aldrich syndrome are maintained at databases at different institutes and there is a lack of a central portal for all disorders (examples of databases can be found at http://sinaicentral.mssm.edu/intranet/research/glanzmann and http://bioinf.uta.fi/WASbase). The information in these databases is generally not linked to knowledge about signalling pathways as exists in Reactome.
3. Sequence variation and platelet phenotypes
Several recent candidate-gene and genome-wide platelet association studies have identified nearly a hundred common coding and non-coding single nucleotide polymorphisms (SNPs) that exert an effect on platelet function [5, 6] and volume and count [4]. About a third of these SNPs are localised in or near genes encoding known regulators of megakaryopoiesis and the formation and survival of platelets. The remaining ones are in or near genes encoding proteins from a diverse array of known functional categories, but their role in megakaryocyte and platelet biology remains to be elucidated [4]. Information about the results of genome-wide association studies (GWAS) is maintained in an on-line catalogue (http://www.genome.gov/gwastudies). Overlaying the GWAS results with pathway knowledge in Reactome can be applied to develop protein-protein interaction networks which will reveal hitherto non-appreciated interactions [4]. It is hoped that the availability of such networks will support researchers in their endeavours to unravel the role and function of this new group of key regulators of megakaryopoiesis and the formation and function of platelets. Knowledge about common sequence variants on platelet phenotypes is of no immediate clinical use because their effect size on the risk of bleeding and thrombotic events is small.
This will change with the increasing use of next generation sequencing technologies (NGST). Global scientific initiatives to decipher the coding fraction (exome) or the entire sequence of hundreds of thousands of human genomes will ultimately lead to a complete catalogue of sequence variants in human populations of different ethnicities [7] and future association studies may identify rare variants with large effects sizes on clinical phenotypes. Several of such variants are likely to become part of the routine diagnostic work-up of patients, particularly those with early onset thrombotic and bleeding disorders. The more immediate application of NGST is in the area of Rare Diseases for which the genetic basis has not yet been resolved. It has now become feasible and affordable to survey the entire coding fraction of the human genome by so-called exome sequencing. This approach has already been successfully applied to identify rare variants and mutations that underlie Rare Diseases. For example the sequencing of the exomes of a relative small number of cases has led to the discovery that NBEAL2 is the causative gene for Grey Platelet syndrome [8, 9] and that the compound inheritance of a low-frequency regulatory SNP and a rare null mutation in the RBM8A gene causes the Thrombocytopenia and Absent Radii syndrome [10], showing the superiority of the exome sequencing approach over linkage studies in large numbers of pedigrees.
4. From databases to clinical care
To allow physicians and patients with rare inherited bleeding, platelet and thrombotic disorders to optimally reap the benefits of the genome revolution carefully curated databases, like Reactome, are a key building block that will allow to visualise clinically relevant information onto cellular pathways, linked to data resources that aim to catalogue the relationships between rare sequence variants and clinical phenotypes. We have commenced in partnership with scientific and clinical experts on rare inherited platelet and bleeding disorders a systematic curation effort that aims to bring information from disparate databases and the literature about causative rare variants and mutations together in a single Locus Reference Genomic (LRG) database (http://www.lrg-sequence.org) and to link this gene-centric information with clinical phenotype descriptions and information about studies of novel treatments at e.g. Orphanet (http://www.orpha.net). This initiative is overseen by the Scientific and Standardization Committee ThromboGenomics (http://www.thrombogenomics.org.uk) of the ISTH. The LRG database is supported by both the European Bioinformatics Institute and the National Center for Biotechnology Information providing a guarantee of a seamless integration with other databases, like dbSNP (http://www.ncbi.nlm.nih.gov/projects/SNP), Ensembl, etc. Similarly the Orphanet database is a long-term and sustainable database initiative and is one of the reference portals for information on Rare Diseases and orphan drugs. Orphanet’s aim is to help improve the diagnosis, care and treatment of these patients by accurately capturing, annotating and cataloguing clinical phenotype information.
In conclusion global collaboration is urgently needed to curate knowledge about the relationship between rare sequence variants with large clinical effect sizes and to integrate the information from disparate disorder-specific databases in a single freely-accessible database environment and related websites.
Acknowledgements
Development of the Reactome database was supported by grants from the National Human Genome Research Institute at the National Institutes of Health (grant number P41 HG003751); the European Union 6th Framework Programme ‘ENFIN’ (grant number LSHG-CT-2005-518254). Funding for open access charge: National Institutes of Health grant number P41 HG003751. WHO is supported by a grant from the National Institute for Health Research England (grant number NIHR:RP-PG-0310-1002).
6. References
- 1.Croft DOKG, Wu G, Haw R, Gillespie M, Matthews L, Caudy M, Garapati P, Gopinath G, Jassal B, Jupe S, Kalatskaya I, Mahajan S, May B, Ndegwa N, Schmidt E, Shamovsky V, Yung C, Birney E, Hermjakob H, D’Eustachio P, Stein L. Reactome: a database of reactions, pathways and biological processes. Nucleic acids research. 2011;39 doi: 10.1093/nar/gkq1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dittrich M, Birschmann I, Mietner S, Sickmann A, Walter U, Dandekar T. Platelet protein interactions: map, signaling components, and phosphorylation groundstate. Arteriosclerosis, thrombosis, and vascular biology. 2008;28:1326–31. doi: 10.1161/ATVBAHA.107.161000. 10.1161/ATVBAHA.107.161000. [DOI] [PubMed] [Google Scholar]
- 3.Watkins NA, Gusnanto A, de Bono B, De S, Miranda-Saavedra D, Hardie DL, Angenent WG, Attwood AP, Ellis PD, Erber W, Foad NS, Garner SF, Isacke CM, Jolley J, Koch K, Macaulay IC, Morley SL, Rendon A, Rice KM, Taylor N, Thijssen-Timmer DC, Tijssen MR, van der Schoot CE, Wernisch L, Winzer T, Dudbridge F, Buckley CD, Langford CF, Teichmann S, Gottgens B, Ouwehand WH. A HaemAtlas: characterizing gene expression in differentiated human blood cells. Blood. 2009;113:e1–9. doi: 10.1182/blood-2008-06-162958. blood-2008-06-162958 [pii] 10.1182/blood-2008-06-162958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gieger C, Radhakrishnan A, Cvejic A, Tang W, Porcu E, Pistis G, Serbanovic-Canic J, Elling U, Goodall AH, Labrune Y, Lopez LM, Magi R, Meacham S, Okada Y, Pirastu N, Sorice R, Teumer A, Voss K, Zhang W, Ramirez-Solis R, Bis JC, Ellinghaus D, Gogele M, Hottenga JJ, Langenberg C, Kovacs P, O’Reilly PF, Shin SY, Esko T, Hartiala J, Kanoni S, Murgia F, Parsa A, Stephens J, van der Harst P, Ellen van der Schoot C, Allayee H, Attwood A, Balkau B, Bastardot F, Basu S, Baumeister SE, Biino G, Bomba L, Bonnefond A, Cambien F, Chambers JC, Cucca F, D’Adamo P, Davies G, de Boer RA, de Geus EJ, Doring A, Elliott P, Erdmann J, Evans DM, Falchi M, Feng W, Folsom AR, Frazer IH, Gibson QD, Glazer NL, Hammond C, Hartikainen AL, Heckbert SR, Hengstenberg C, Hersch M, Illig T, Loos RJ, Jolley J, Tee Khaw K, Kuhnel B, Kyrtsonis MC, Lagou V, Lloyd-Jones H, Lumley T, Mangino M, Maschio A, Mateo Leach I, McKnight B, Memari Y, Mitchell BD, Montgomery GW, Nakamura Y, Nauck M, Navis G, Nothlings U, Nolte IM, Porteous DJ, Pouta A, Pramstaller PP, Pullat J, Ring SM, Rotter JI, Ruggiero D, Ruokonen A, Sala C, Samani NJ, Sambrook J, Schlessinger D, Schreiber S, Schunkert H, Scott J, Smith NL, Snieder H, Starr JM, Stumvoll M, Takahashi A, Tang WH, Taylor K, Tenesa A, Lay Thein S, Tonjes A, Uda M, Ulivi S, van Veldhuisen DJ, Visscher PM, Volker U, Wichmann HE, Wiggins KL, Willemsen G, Yang TP, Hua Zhao J, Zitting P, Bradley JR, Dedoussis GV, Gasparini P, Hazen SL, Metspalu A, Pirastu M, Shuldiner AR, Joost van Pelt L, Zwaginga JJ, Boomsma DI, Deary IJ, Franke A, Froguel P, Ganesh SK, Jarvelin MR, Martin NG, Meisinger C, Psaty BM, Spector TD, Wareham NJ, Akkerman JW, Ciullo M, Deloukas P, Greinacher A, Jupe S, Kamatani N, Khadake J, Kooner JS, Penninger J, Prokopenko I, Stemple D, Toniolo D, Wernisch L, Sanna S, Hicks AA, Rendon A, Ferreira MA, Ouwehand WH, Soranzo N. New gene functions in megakaryopoiesis and platelet formation. Nature. 2011;480:201–8. doi: 10.1038/nature10659. 10.1038/nature10659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jones CI, Bray S, Garner SF, Stephens J, de Bono B, Angenent WG, Bentley D, Burns P, Coffey A, Deloukas P, Earthrowl M, Farndale RW, Hoylaerts MF, Koch K, Rankin A, Rice CM, Rogers J, Samani NJ, Steward M, Walker A, Watkins NA, Akkerman JW, Dudbridge F, Goodall AH, Ouwehand WH. A functional genomics approach reveals novel quantitative trait loci associated with platelet signaling pathways. Blood. 2009;114:1405–16. doi: 10.1182/blood-2009-02-202614. blood-2009-02-202614 [pii] 10.1182/blood-2009-02-202614. [DOI] [PubMed] [Google Scholar]
- 6.Johnson AD, Yanek LR, Chen MH, Faraday N, Larson MG, Tofler G, Lin SJ, Kraja AT, Province MA, Yang Q, Becker DM, O’Donnell CJ, Becker LC. Genome-wide meta-analyses identifies seven loci associated with platelet aggregation in response to agonists. Nat Genet. 2010;42:608–13. doi: 10.1038/ng.604. 10.1038/ng.604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Durbin R, Abecasis GR, Altshuler DL, Auton A, Brooks LD, Gibbs RA, Hurles ME, McVean GA. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Albers CA, Cvejic A, Favier R, Bouwmans EE, Alessi MC, Bertone P, Jordan G, Kettleborough RN, Kiddle G, Kostadima M, Read RJ, Sipos B, Sivapalaratnam S, Smethurst PA, Stephens J, Voss K, Nurden A, Rendon A, Nurden P, Ouwehand WH. Exome sequencing identifies NBEAL2 as the causative gene for gray platelet syndrome. Nat Genet. 2011;43:735–7. doi: 10.1038/ng.885. 10.1038/ng.885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kahr WH, Hinckley J, Li L, Schwertz H, Christensen H, Rowley JW, Pluthero FG, Urban D, Fabbro S, Nixon B, Gadzinski R, Storck M, Wang K, Ryu GY, Jobe SM, Schutte BC, Moseley J, Loughran NB, Parkinson J, Weyrich AS, Di Paola J. Mutations in NBEAL2, encoding a BEACH protein, cause gray platelet syndrome. Nat Genet. 2011;43:738–40. doi: 10.1038/ng.884. 1038/ng.884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Albers CA, Paul DS, Schulze H, Freson K, Stephens JC, Smethurst PA, Jolley JD, Cvejic A, Kostadima M, Bertone P, Breuning MH, Debili N, Deloukas P, Favier R, Fiedler J, Hobbs CM, Huang N, Hurles ME, Kiddle G, Krapels I, Nurden P, Ruivenkamp CA, Sambrook JG, Smith K, Stemple DL, Strauss G, Thys C, van Geet C, Newbury-Ecob R, Ouwehand WH, Ghevaert C. Compound inheritance of a low-frequency regulatory SNP and a rare null mutation in exon-junction complex subunit RBM8A causes TAR syndrome. Nat Genet. 2012 doi: 10.1038/ng.1083. 10.1038/ng.1083.7. [DOI] [PMC free article] [PubMed] [Google Scholar]


