Abstract
The development of a new class of surfactants for membrane protein manipulation, “GNG amphiphiles”, is reported. These amphiphiles display promising behavior for membrane proteins, as demonstrated recently by the high resolution structure of a sodium-pumping pyrophosphatase reported by Kellosalo et al.
Integral membrane proteins (IMPs) are essential biological components, with critical roles in a range of cellular processes including transport, signal transduction and cell-cell recognition. More than 50% of current drugs target membrane proteins, which illustrates the importance of these molecules in human health.1 High resolution IMP structures are difficult to determine because such proteins are difficult to dissolve in aqueous media, a prerequisite for crystallization or NMR analysis.2 Detergents are generally required to extract IMPs from their native lipid bilayers.3 However, conventional detergents cause denaturation and/or aggregation of many membrane proteins.4
N-Dodecyl-β-D-maltoside (DDM), probably the most widely-used detergent in membrane protein research,5 is often superior to other conventional detergents in terms of stabilization of specific IMPs. Crystals generated from DDM-solubilized proteins, however, frequently provide only low-resolution x-ray diffraction data.6 This trend may arise because PDCs formed with DDM tend to be large and therefore expose only small amounts of hydrophilic protein surface area.7 Detergents that tend to form small PDCs, such as n-octyl-β-D-glucopyranoside (OG) and lauryldimethylamine-N-oxide (LDAO), have often enabled the acquisition of high-resolution crystal structures. However, proteins solubilized with these detergents frequently display diminished stability relative to DDM-solubilized forms6a, which tends to limit the use of OG or LDAO to crystallization of membrane proteins that are intrinsically stable. This comparison suggests that new amphiphiles that tend to form small PDCs but are not destabilizing to solubilized proteins could be very useful new tools for membrane protein structural studies.
Diverse strategies have been pursued to develop new tools for solubilization and stabilization of membrane proteins in aqueous solution.8 Examples including amphiphilic polymers (amphipols),8c nanodiscs,8i hemifluorinated surfactants8f and tandem facial amphiphiles8l have displayed promising properties. These amphiphiles, however, were not effective at extracting membrane proteins from biological membranes. Other novel agents, including tripod amphiphiles (TPAs),8d cholesterol-based amphiphiles,8h,j maltose-neopentyl glycol amphiphiles (MNGs),8m and rigid hydrophobic group-bearing detergents,8n,o can both solubilize and stabilize membrane proteins. However, most of these efforts have focused on the amphiphile's ability to stabilize the native protein structure; there is little information regarding PDC size for these agents. As a result, only a very small number of new amphiphiles have been useful for the high-resolution IMP crystal structures.8n,9
MNG amphiphiles have recently been shown to be particularly useful for membrane protein solubilization, stabilization and crystallization.8m,9 We now describe glucose-neopentyl glycol (GNG) amphiphiles. Although these two classes are structurally related, their interactions with membrane proteins could be quite different; such differences are well-known between conventional detergents OG and DDM. In this case, replacement of maltose with glucose leads to smaller PDCs. Thus, we hypothesized that GNG amphiphiles might form smaller PDCs than MNG amphiphiles, a desirable feature for PDC-based membrane protein crystallization. Our findings demonstrate that GNG amphiphiles are useful alternatives to conventional detergents for membrane protein study.
The GNG architecture is illustrated by the four examples in Fig. 1. Each GNG amphiphile features two glucose units comprising the hydrophilic region and two alkyl chains comprising the lipophilic region. The lipophilic unit attachment varies between ether linkages in GNG-1 and GNG-2 and direct connection to the quaternary center in GNG-3 and GNG-4. GNG-4 displayed poor water-solubility (<1%) and was not studied further. The other GNG amphiphiles were highly water-soluble (up to ~20 wt %). Critical micelle concentration (CMC) values and the hydrodynamic radii (Rh) of the micelles were estimated by solubilization experiments employing a hydrophobic fluorescent dye, dicyclohexatriene,10 and dynamic light scattering (DLS), respectively. Data for three GNG amphiphiles and two conventional detergents (DDM and OG) are presented in Table S1. The CMC values of the GNG amphiphiles are smaller than those of glucose-containing conventional detergents such as OG (~25 mM; ~0.73 wt %)11 and OTG (~9 mM; ~0.28 wt %)12. Micelles formed by GNG-1 or GNG-3 are smaller than those formed by DDM but larger than those formed by OG.11 The micelles formed by GNG-2 are significantly larger than those formed by DDM.
Figure 1.
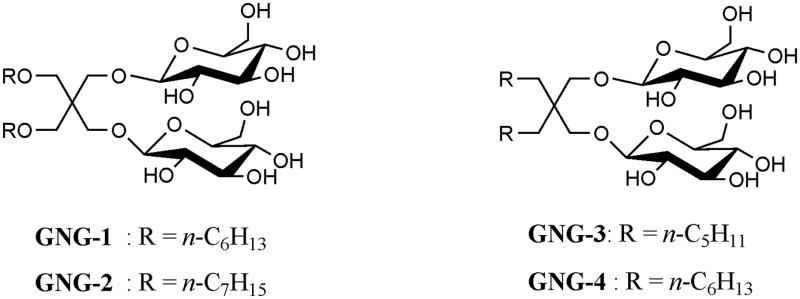
Chemical structures of the new amphiphiles.
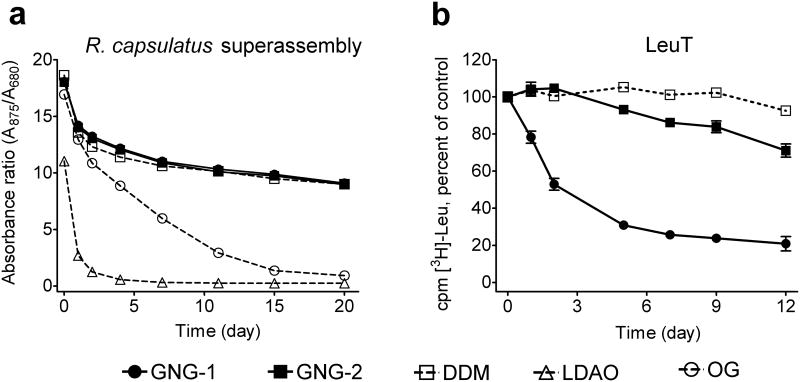
We first evaluated the new amphiphiles using the photosynthetic superassembly of Rhodobacter capsulatus.13 The assay utilized membranes isolated from a R. capsulatus strain expressing both the very labile light harvesting complex I (LHI) and the more stable reaction center complex (RC). Intracytoplasmic R. capsulatus membranes were treated with 1.0 wt % DDM and then purified with DDM at its CMC (0.009 wt %) using a Ni-NTA column. The purified protein solutions were diluted 1:20 with solutions containing individual GNG amphiphiles (GNG-1 and GNG-2) or conventional detergents (DDM, OG and LDAO). The final concentration of detergent/amphiphile in each sample was CMC + 0.04 wt %. Photosynthetic superassembly stability was monitored by measuring the 875/680 absorbance ratio for each preparation over time (Fig. 2a, Fig S1).8k Superassembly solubilized with either GNG-1 or GNG-2 was as stable as a DDM-solubilized protein over a period of 20 days. In contrast, LDAO or OG-solubilized superassembly decomposed rapidly. When we conducted this study at increased detergent/amphiphile concentrations, CMC + 0.2 wt %, similar results were obtained (Fig. S1a).
Figure 2.
Stability time course of (a) R. capsulatus LHI-RC photosynthetic superassembly and (b) LeuT. Detergents were tested at CMC + 0.04 wt % for both systems stored at room temperature. Stability of the superassembly was assessed by measuring the 875/680 absorbance ratio of each sample. Stability of LeuT was assessed based on [3H]-Leu binding using a scintillation proximity assay (SPA). Results are expressed as % activity relative to the day 0 activity (mean ± SEM, n = 2).
Analysis of the leucine transporter (LeuT) from Aquifex aeolicus14 showed a markedly different trend. In this case, protein activity was monitored by measuring the ability to bind to a radiolabeled ligand ([3H]leucine) using a scintillation proximity assay (SPA).15 The transporter was initially solubilized from membranes with 1.0 wt % DDM, purified in 0.05 wt % DDM and then transferred into amphiphile- or DDM-containing solutions to generate final concentrations of CMC + 0.04 wt % (the original DDM is well below its CMC at this point). GNG-1, GNG-2 and GNG-3 (last not shown) were inferior to DDM at maintaining LeuT stability over 12 days (Fig. 2b). Of the three, GNG-2 showed the best performance. The activity of GNG-1-solubilized transporter declined over several days; GNG-3-solubilized LeuT was fully inactive after one day. When we increased the amphiphile concentration to CMC + 0.2 wt %, LeuT activity in the presence of a GNG amphiphile declined more rapidly than did activity in the presence of DDM (Fig. S1b).
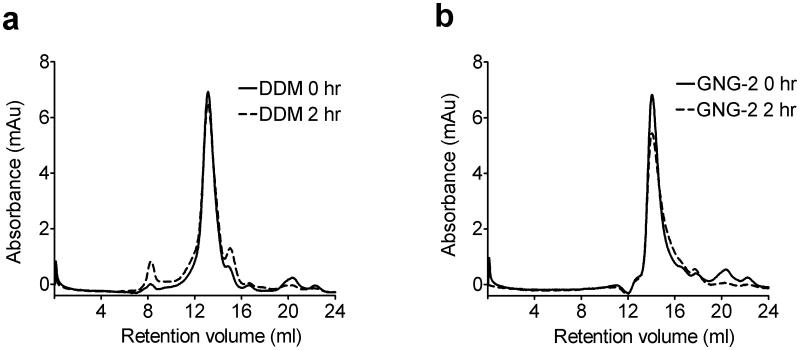
The new amphiphiles were evaluated for their ability to maintain the native states of two prokaryotic proteins, succinate:quinone oxidoreductase (SQR), a respiratory complex,16 and the rhomboid intramembrane serine protease GlpG,17 both of which were expressed in E. coli. The thermostability of each protein was assessed through the N-[4-(7-diethylamino-4-methyl-3-coumarinyl)phenyl]maleimide (CPM) assay.18 For both SQR and GlpG, the assays suggested that all three GNG amphiphiles were slightly more effective than DDM at preserving the native state (Fig. S2a,b). In order to further assess the effects of the GNG amphiphiles, we analysed GlpG using gel filtration before and after 2 hr-incubation at 30 °C (Fig. 3). The DDM-solubilized sample showed a small increase in aggregated protein (peak at ~ 8 ml retention volume) and in degraded protein (peak at ~15 ml retention volume) after this treatment. In contrast, the GNG-2-solubilized sample did not contain any aggregated protein after treatment, and there was little or no degraded protein. The GNG-2-GlpG complex eluted at a higher retention volume than the DDM-GlpG complex (~14.1 ml vs. ~13.1 ml), indicating that GNG-2 forms smaller PDCs than does DDM. The molecular weights of PDCs formed by DDM and GNG-2 were estimated to be 91 kDa and 56 kDa, respectively, suggesting that GlpG is monomeric for both of these solubilizing agents (Fig. S3).
Figure 3.
Gel filtration analysis for GlpG solubilized with (a) DDM or (b) GNG-2. The analysis was performed at a detergent concentration of CMC + 0.04 wt %, before or after incubation of isolated protein at 30 °C for 2 h.
Among the new amphiphiles, GNG-2 showed the most promising behavior in terms of stabilizing various test membrane proteins. We turned next to the question of extracting membrane proteins from bilayers, and we focused on comparing GNG-2 and DDM for this purpose. The R. capsulatus LHI-RC superassembly, LeuT, and CMP-Sia as a fusion with a C-terminal green fluorescent protein (GFP) were used for extraction studies (Fig. S4a,b,c). These studies suggest that GNG-2 is generally comparable to DDM for extraction of membrane proteins from the biological membranes.
We evaluated the ability of GNG-3 to promote PDC-based crystallization of a membrane protein. In preliminary studies, GNG-3 was used for solubilization, purification and crystallization of the E. coli acetate transporter; the resulting crystals diffracted to 4.1-Å resolution (Fig. S5a,b). Although more study is necessary to improve crystal quality, this initial success is consistent with our hypothesis that the ability of GNG-3 to form small PDCs and to stabilize the solubilized protein promotes crystallization. Further support for this hypothesis comes from the very recent report by Kellosalo et al. of the 2.6-Å resolution crystal structure of a sodium-pumping pyrophosphatase, based on crystals grown with GNG-3 (which is now commercially available).19 This is the first success case of novel agents in determination of PDC-based high resolution crystal structure of IMPs with unknown structure.
In conclusion, we have demonstrated that GNG amphiphiles are favorable for solubilization and stabilization of several membrane protein systems, and that these new amphiphiles also have a tendency to form small complexes when bound to a membrane protein. The GNG behavior profile differs from that of classical detergents such as DDM, OG and LDAO, and our findings therefore suggest that GNG amphiphiles may be more conducive to membrane protein crystallization than are classical detergents, at least in some cases. Our previous design, the MNG amphiphile class (maltose headgroups), is generally superior to the GNG class (glucose headgroups) in terms of membrane protein stabilization. This trend mirrors the well-known tendency for membrane proteins to be more stable in the presence of DDM relative to OG. Despite this trend, OG remains very popular for membrane protein crystallization, because protein-detergent complexes formed with OG tend to be smaller than those formed with DDM. By extrapolation, it seems likely that GNG amphiphiles will prove to be as useful as (and complementary to) the MNG amphiphiles for membrane protein crystallization. Specifically, GNG amphiphiles may be particularly useful for PDC-based crystallization, while MNG amphiphiles are more suitable for LCP-based membrane protein crystallization.9
Supplementary Material
Acknowledgments
This work was supported by NIH grant P01 GM75913 (S.H.G.), the National Research Foundation of Korea (NRF) funded by the Ministry of Education, Science and Technology (grant number 2008-0061856 to P.S.C., K.H.C.), NS28471 (B.K.), the European Community's Seventh Framework Programme FP7/2007-2013 under grant agreement n° HEALTH-F4-2007-201924, EDICT Consortium (B.B., K.G., U.G.), the Danish National Research Council (C.J.L., U.G.), the Lundbeck Foundation (S.G.F.R., C.J.L., U.G.). R.R. was funded by the Defence, Science and Technology Laboratories (DSTL), Porton Down, UK. We thank Philip Laible (Argonne National Laboratory) for supplying membrane preparations from R. capsulatus. We also thank Chiara Lee and David Drew (Imperial College London) and, Jonathan Ruprecht (Medical Research Council-Mitochondrial Biology Unit, Cambridge) and Gary Cecchini (University of California, San Francisco) for providing purified GlpG and SQR samples, respectively.
Footnotes
Electronic Supplementary Information (ESI) available: Experimental detail of the amphiphiles' synthesis and their evalution for membrane proteins.
Contributor Information
Pil Seok Chae, Email: pchae@hanyang.ac.kr.
Brian Kobilka, Email: kobilka@stanford.edu.
Claus J. Loland, Email: cllo@sund.ku.dk.
Surajit Banerjee, Email: sbanerjee@anl.gov.
Bernadette Byrne, Email: b.byrne@imperial.ac.uk.
John K. Lee, Email: jklee@umn.edu.
Samuel H. Gellman, Email: gellman@chem.wisc.edu.
Notes and references
- 1.Overington JP, Al-Lazikani B, Hopkins AL. Nature Reviews Drug Discovery. 2006;5:993. doi: 10.1038/nrd2199. [DOI] [PubMed] [Google Scholar]
- 2.Lacapere JJ, Pebay-Peyroula E, Neumann JM, Etchebest C. Trends Biochem Sci. 2007;32:25. doi: 10.1016/j.tibs.2007.04.001. [DOI] [PubMed] [Google Scholar]
- 3.(a) White SH, Wimley WC. Annu Rev Biophys Biomol Struct. 1999;28:319. doi: 10.1146/annurev.biophys.28.1.319. [DOI] [PubMed] [Google Scholar]; (b) Moller JV, le Maire J. J Biol Chem. 1993;268:18659. [PubMed] [Google Scholar]
- 4.(a) Garavito RM, Ferguson-Miller S. J Biol Chem. 2001;276:32403. doi: 10.1074/jbc.R100031200. [DOI] [PubMed] [Google Scholar]; (b) Bowie JU. Curr Opin Struct Biol. 2001;11:397. doi: 10.1016/s0959-440x(00)00223-2. [DOI] [PubMed] [Google Scholar]; (c) Serrano-Vega MJ, Magnani F, Shibata Y, Tate CG. Proc Natl Acad Sci USA. 2008;105:877. doi: 10.1073/pnas.0711253105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Caffrey M, Li D, Dukkipati A. Biochemistry. 2012;51:6266. doi: 10.1021/bi300010w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.(a) Privé GG. Methods. 2007;41:388. doi: 10.1016/j.ymeth.2007.01.007. [DOI] [PubMed] [Google Scholar]; (b) Zhang Q, Tao H, Hong WX. Methods. 2011;55:318. doi: 10.1016/j.ymeth.2011.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kantardjieff KA, Rupp B. Protein Sci. 2003;12:1865. doi: 10.1110/ps.0350503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.(a) Hjelmeland LM. Proc Natl Acad Sci USA. 1980;77:6368. doi: 10.1073/pnas.77.11.6368. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Schafmeister CE, Miercke LJW, Stroud RM. Science. 1993;262:734. doi: 10.1126/science.8235592. [DOI] [PubMed] [Google Scholar]; (c) Tribet C, Audebert R, Popot JL. Proc Natl Acad Sci USA. 1996;93:15047. doi: 10.1073/pnas.93.26.15047. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) McQuade DT, Quinn MA, Yu SM, Polans AS, Krebs MP, Gellman SH. Angew Chem Int Ed. 2000;39:758. [PubMed] [Google Scholar]; (e) McGregor CL, Chen L, Pomroy NC, Hwang P, Go S, Chakrabartty A, Privé GG. Nat Biotechnol. 2003;21:171. doi: 10.1038/nbt776. [DOI] [PubMed] [Google Scholar]; (f) Breyton C, Chabaud E, Chaudier Y, Pucci B, Popot JL. FEBS Lett. 2004;564:312. doi: 10.1016/S0014-5793(04)00227-3. [DOI] [PubMed] [Google Scholar]; (g) Zhao X, Nagai Y, Reeves PJ, Kiley P, Khorana HG, Zhang S. Proc Natl Acad Sci USA. 2006;103:17707. doi: 10.1073/pnas.0607167103. [DOI] [PMC free article] [PubMed] [Google Scholar]; (h) Zhang Q, Ma X, Ward A, Hong WX, Jaakola VP, Stevens RC, Finn MG, Chang G. Angew Chem Int Ed. 2007;46:7023. doi: 10.1002/anie.200701556. [DOI] [PubMed] [Google Scholar]; (i) Nath A, Atkins WM, Sligar SG. Biochemistry. 2007;46:2059. doi: 10.1021/bi602371n. [DOI] [PubMed] [Google Scholar]; (j) Howell SC, Mittal R, Huang L, Travis B, Breyer RM, Sanders CR. Biochemistry. 2010;49:9572. doi: 10.1021/bi101334j. [DOI] [PMC free article] [PubMed] [Google Scholar]; (k) Chae PS, Wander MJ, Bowling AP, Laible PD, Gellman SH. ChemBioChem. 2008;9:1706. doi: 10.1002/cbic.200800169. [DOI] [PMC free article] [PubMed] [Google Scholar]; (l) Chae PS, et al. J Am Chem Soc. 2010;132:16750. doi: 10.1021/ja1072959. [DOI] [PMC free article] [PubMed] [Google Scholar]; (m) Chae PS, et al. Nat Methods. 2010;7:1003. doi: 10.1038/nmeth.1526. [DOI] [PMC free article] [PubMed] [Google Scholar]; (n) Hovers J, et al. Mol Membr Biol. 2011;28:171. doi: 10.3109/09687688.2011.552440. [DOI] [PMC free article] [PubMed] [Google Scholar]; (o) Chae PS, et al. Chem Eur J. 2012;18:9485. [Google Scholar]
- 9.(a) Rasmussen SGF, et al. Nature. 2011;469:236. doi: 10.1038/nature09665. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Rosenbaum DM, et al. Nature. 2011;469:175. doi: 10.1038/nature09648. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Rasmussen SGF, et al. Nature. 2011;477:540. [Google Scholar]; (d) Kruse AC, et al. Nature. 2012;482:552. doi: 10.1038/nature10867. [DOI] [PMC free article] [PubMed] [Google Scholar]; (e) Haga K, et al. Nature. 2012;482:547. doi: 10.1038/nature10753. [DOI] [PMC free article] [PubMed] [Google Scholar]; (f) Manglik A, et al. Nature. 2012;485:321. doi: 10.1038/nature10954. [DOI] [PMC free article] [PubMed] [Google Scholar]; (g) Granier S, Manglik A, Kruse AC, Kobilka TS, Thian FS, Weis WI, Kobilka BK. Nature. 2012;485:400. doi: 10.1038/nature11111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chattopadhyay A, London E. Anal Biochem. 1984;139:408. doi: 10.1016/0003-2697(84)90026-5. [DOI] [PubMed] [Google Scholar]
- 11.Lorber B, Bishop JB, Delucas L. J Biochim Biophys Acta. 1990;1023:254. doi: 10.1016/0005-2736(90)90421-j. [DOI] [PubMed] [Google Scholar]
- 12.Saito S, Tsuchiya T. Biochem J. 1984;222:829. doi: 10.1042/bj2220829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Laible PD, Kirmaier C, Udawatte CS, Hofman SJ, Holten D, Hanson DK. Biochemistry. 2003;42:1718. doi: 10.1021/bi026959b. [DOI] [PubMed] [Google Scholar]
- 14.Deckert G, et al. Nature. 1998;392:353. doi: 10.1038/32831. [DOI] [PubMed] [Google Scholar]
- 15.Quick M, Javitch JA. Proc Natl Acad Sci USA. 2007;104:3603. doi: 10.1073/pnas.0609573104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Horsefield R, Iwata S, Byrne B. Curr Protein Pept Sci. 2004;5:107. doi: 10.2174/1389203043486847. [DOI] [PubMed] [Google Scholar]
- 17.Urban S. Biochem J. 2010;425:501. doi: 10.1042/BJ20090861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Alexandrov A, Mileni M, Chien EY, Hanson MA, Stevens RC. Structure. 2008;16:351. doi: 10.1016/j.str.2008.02.004. [DOI] [PubMed] [Google Scholar]
- 19.(a) Kellosalo J, Kajander T, Kogan K, Pokharel K, Goldman A. Science. 2012;337:473. doi: 10.1126/science.1222505. [DOI] [PubMed] [Google Scholar]; (b) Kellosalo J, Kajander T, Honkanen R, Goldman A. Mol Membr Biol. 2012 doi: 10.3109/09687688.2012.712162. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.