Abstract
Diverse cellular processes depend on endocytosis, intracellular vesicle trafficking, sorting and exocytosis, processes regulated post-transcriptionally by modifications such as phosphorylation and ubiquitylation. In addition to sorting to the lysosome, cargo is recycled to the plasma membrane via recycling endosomes. Here, we describe a role of the goliath gene family of protease-associated (PA) domain E3 ligases in regulating recycling endosome trafficking. The two Drosophila members of this family—Goliath and GodzillaCG10277—are located on endosomes, and both ectopic expression and loss-of-function lead to the accumulation of Rab5-positive giant endosomes. Furthermore, the human homologue RNF167 exhibits similar behaviour. We show that the soluble N-ethylmaleimide-sensitive fusion attachment protein receptor (SNARE) protein VAMP3 is a target of these ubiquitin ligases, and that recycling endosome trafficking is abrogated in response to their activity. Furthermore, mutation of the Godzilla ubiquitylation target lysines on VAMP3 abrogates the formation of enlarged endosomes induced by either Godzilla or RNF167. Thus, Goliath ubiquitin ligases play a novel role in regulating recycling endosome trafficking via ubiquitylation of the VAMP3 SNARE protein.
Keywords: E3 ubiquitin ligase, goliath, RING domain, SNARE, VAMP3
Introduction
Many important cellular processes such as cell migration (Marin et al, 2010) and neurotransmitter release depend on intracellular vesicle trafficking, sorting and exocytosis (Sudhof and Rothman, 2009). While endocytosis provides a switching-off mechanism for cellular signalling by removing activated receptors from the cell surface, more recent studies suggest signalling from endocytosed receptors is still active and may even be amplified in the endocytic pathway (von Zastrow and Sorkin, 2007; Mazot et al, 2011). Endocytic vesicles are can be roughly divided into early endosomes, late endosomes and recycling endosomes (Platta and Stenmark, 2011) and trafficking is regulated post-transcriptionally by modifications such as phosphorylation and ubiquitylation (Piper and Lehner, 2011; Haglund and Dikic, 2012). In addition to sorting to the lysosome, an estimated two thirds of internalized receptors are recycled back to the plasma membrane via recycling endosomes from early endosome (Steinman et al, 1983; Schmid et al, 1988; Mayor et al, 1993). Generally, vesicles are transported along actin filaments or microtubules prior to docking and fusion with an acceptor membrane. The latter step is largely regulated by soluble N-ethylmaleimide-sensitive fusion attachment protein receptors (SNAREs) (Proux-Gillardeaux et al, 2005; Tayeb et al, 2005; Veale et al, 2010; Zylbersztejn and Galli, 2011).
SNARE proteins are characterized by a conserved stretch of 60–70 amino acids known as the SNARE motif, and most have a carboxyl-terminal transmembrane domain connected by a short linker sequence (Jahn and Scheller, 2006). SNAREs form a complex with two other specific partner SNAREs, leading to specific regulation of membrane fusion. When appropriate sets of SNAREs combined, they form a stable complex consisting of coiled-coil four-helix bundle. While this complex formation is known to be sufficient to drive membrane fusion in vitro, post-transcriptional modifications such as phosphorylation and palmitoylation have also shown to regulate the function and localization of SNAREs (Snyder et al, 2006). A large number of SNARE mammalian proteins have been characterized, including the endosomal SNARE VAMP3/Cellubrevin, which is implicated in recycling of membrane receptors from endosome to the plasma membrane (McMahon et al, 1993; Galli et al, 1994). In Drosophila, two VAMP3/Cellubrevin homologous SNAREs are encoded in the genome; n-Syb is neuronally specific and associated primarily with synaptic vesicles (DiAntonio et al, 1993), while the closely related Syb/dVAMP is more generally expressed (Chin et al, 1993; DiAntonio et al, 1993). These two proteins appear to be functionally interchangeable not only with one another, but also with mammalian VAMP3/cellubrevin (Bhattacharya et al, 2002).
Many studies have established the importance of ubiquitylation in controlling receptor endocytosis and endosomal sorting, and a number of molecular mechanisms and components regulating these processes have been described (Piper and Lehner, 2011; Haglund and Dikic, 2012). Ubiquitylation is considered to be an important signal in regulation of intracellular traficking, offering advantages such as the ability to regulate the modification by removal deubiquitinating enzymes, and the creation of binding sites for ubiquitin binding domain containing proteins. An estimated 300 human genes encode RING domain proteins in keeping with the already comprehensive repertoire of tasks that are undertaken by this class of proteins in the cell (Li et al, 2008). The Drosophila melanogaster Goliath PA domain E3 ligase has been described as expressed in the mesoderm (Bouchard and Cote, 1993), although further analysis of Goliath function in the fruitfly has not been reported. In fact, in Drosophila the goliath gene family comprises two members—goliath and godzillaCG10277. Xenopus GREUL1 (Goliath-related E3 ubiquitin ligase1) has been reported to function in the development of the anterior ectoderm (Borchers et al, 2002). The mouse orthologue G1RP (G1-related protein) was isolated from 32Dcl3 myeloblastic cells and has shown to be induced in apoptosis in response to IL-3 depletion (Baker and Reddy, 2000). Although a human homologue of Goliath, h-Goliath, has been described as expressed in normal and pathological haematopoietic cells, its function remains unknown (Guais et al, 2006). Most functional information comes from studies of mammalian GRAIL/RNF128, which plays a role in immune tolerance (Mueller, 2004).
In this work, we describe the goliath gene family of PA domain E3 ligases as key regulators of the recycling endosome pathway. We show that the two Drosophila members of this family—Goliath and GodzillaCG10277—exhibit an endosomal location, and that both gain- and loss-of-function lead to the accumulation of Rab5-positive giant endosomes, implying a regulatory role in endosomal processes. Furthermore, expression of RNF167, a human member of the Goliath E3-ligase family, exhibits similar behaviour. Here, we identify the SNARE protein, VAMP3, as a prominent ubiquitylation target of these ubiquitin ligases. In agreement with VAMP3 being a target for Goliath family ubiquitin ligases, we show that recycling endosome trafficking is abrogated in response to their activity. Furthermore, loss of VAMP3 reverses both the Godzilla and RNF167 induced Rab5-positive giant endosome phenotype. In conclusion, our data suggest a novel function for the Goliath ubiquitin ligases in regulating recycling endosome trafficking via ubiquitylation of the VAMP3 SNARE protein.
Results
Goliath and Godzilla encode endosomally localized PA-TM-RING domain ubiquitin ligases.
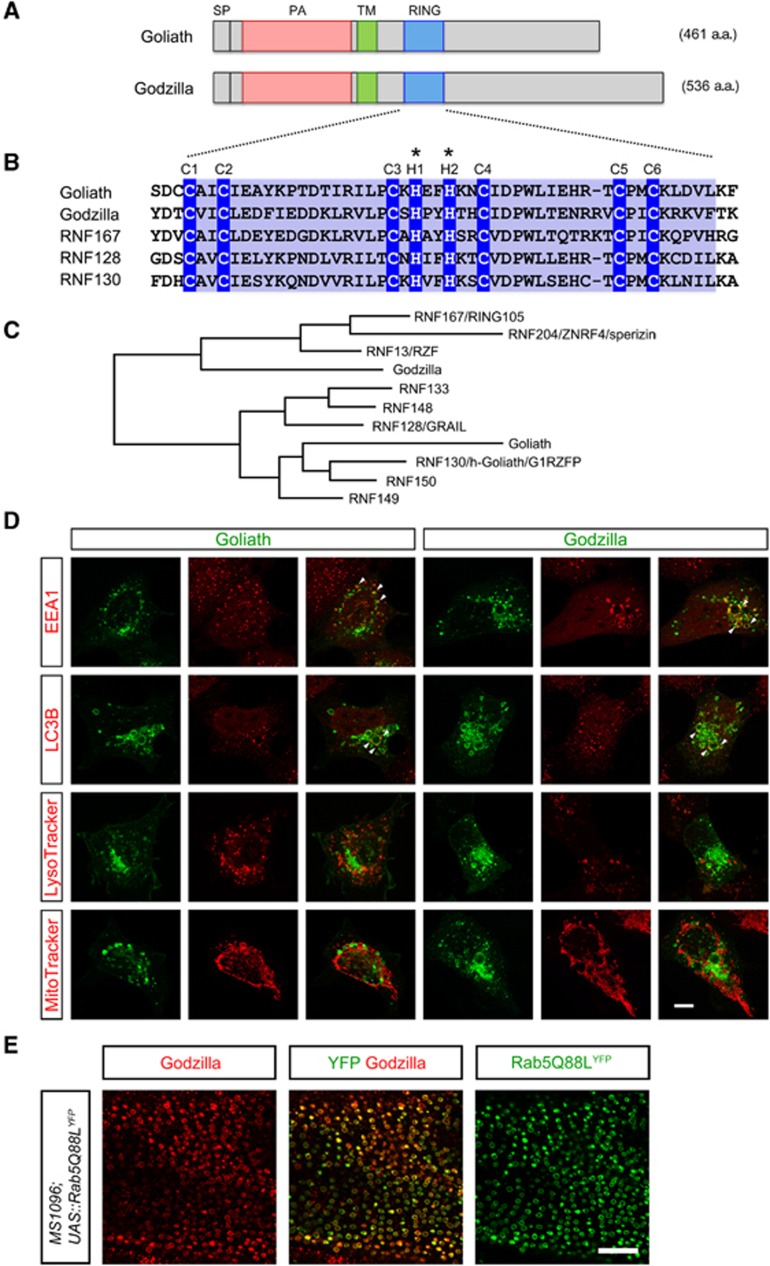
In Drosophila melanogaster, the goliath gene family comprises two members—goliath and godzillaCG10277. Goliath is expressed in muscles (Artero et al, 2003), while the related godzilla exhibits a more general expression pattern. Together, they encode a family of ubiquitin ligases in the fly that exhibit a distinct domain architecture: a signal peptide (a.a. 1–25), a PA domain (a.a. 98–207 Goliath; a.a. 53–155 Godzilla) (Mahon and Bateman, 2000), a transmembrane domain (a.a. 235–257 Goliath; a.a. 173–195 Godzilla) and a RING domain (a.a. 303–343 Goliath; a.a. 235–276 Godzilla) (Deshaies and Joazeiro, 2009; Figure 1A). The unusual presence of a transmembrane domain (a.a. 235–257 Goliath; a.a. 173–195 Godzilla) suggests that Goliath and Godzilla are membrane bound proteins. The PA domain is a highly conserved motif found in members of the protease superfamily and displays 32.3% identity between Goliath and Godzilla (Mahon and Bateman, 2000). Goliath also contains a C3H2C3-type RING domain, which is well-conserved among homologues (Figure 1B). The RING family of E3 ligases is the largest E3-ligase family, containing C3H2C3 or C3HC4 RING domains (Deshaies and Joazeiro, 2009). This domain contains four pairs of metal binding residues with a characteristic linear sequence of Cys-X2-Cys-X9-39-Cys-X1-3His-X2-3Cys/HisX2-Cys-X4-48-Cys-X2-Cys, where X can be any amino acid although there are distinct preferences for particular types of amino acid at a particular position (Lovering et al, 1993). The RING domain of Godzilla is 51% identical to that of Goliath suggesting an important and conserved functional role. This consensus is conserved across species in the larger Goliath family (Figure 1B). In contrast to two members in Drosophila, there are nine predicted homologues in humans (Figure 1C), including GRAIL/RNF128, which plays a role in immune tolerance (Mueller, 2004).
Figure 1.
Goliath and Godzilla encode endosomally localized PA-TM-RING E3 ligases. (A) Schematic domain structure of Drosophila Goliath and Godzilla. SP, signal peptide; PA, protease-associated domain; TM, transmembrane, and RING domain. (B) Sequence alignment of related RING domains. RNF167, RNF128 (also known as GRAIL) and RNF130 (also known as hGoliath) are human sequences. The RING domain (blue) contains six conserved Cys (C1–C6) and two His (H1 and H2) residues, which coordinate Zn2+ and are important for ligase activity. *For ligase-dead mutants, two His residues were substituted to Arg, corresponding to His323 and His326 in Goliath and His255 and His258 in Godzilla. (C) Phylogenic relationship of PA-TM-RING E3-ligase genes. (D) Subcellular localization of Goliath and Godzilla. Goliath-C-GFP (green, left) or Godzilla-C-GFP (green, right) transfected HEK293 cells were co-stained with cell organelle markers (red). EEA1, early endosome; LC3B, autophagosome; LysoTracker, lysosome; MitoTracker, mitochondria. Goliath-C-GFP and Godzilla-C-GFP-induced enlarged vesicle-like structures colocalize with EEA1 (arrowheads), and partially with LC3B (arrowheads); indicating that Goliath and Godzilla localize on an enlarged endosome membranes, but not on lysosomes or mitochondria. Bar 10 μm. The results shown here are typical images from at least four independent experiments. (E) Endogenous Drosophila protein is localized on endosomes in vivo. Endosomes were enlarged by expression of constitutively active Rab5 (Rab5Q88LYFP) in the Drosophila wing disc with MS1096-Gal4. Endogenous Godzilla is accumulated on the resultant enlarged Rab5-positive endosomes. The results shown here are typical images from at least four independent experiments.
Drosophila goliath null mutants, in which all five coding exons were deleted, are viable and fertile. Given the restricted expression of goliath in embryonic muscles (Artero et al, 2003) we subsequently generated mutants in the locus of the more generally expressed related family member godzillaCG10277 (Supplementary Figure S1). Mutants in this locus—ΔgodzillaCG10277—are larval lethal. In an initial attempt to clarify the mechanism of function of the Goliath family ligases, we examined their subcellular localization. A significant amount of Godzilla clearly colocalizes with endosomal markers, such as EEA1, upon overexpression (Figure 1D; Supplementary Figure S2). While less extensive than that observed with Godzilla, overlap with EEA1 is also observed with the related Goliath (Figures 1D and 2A). In general, we noted that expression of Godzilla results in a stronger enlarged endosome phenotype than Goliath. This was also reflected in the more obvious loss of Mannose-6-Phosphate Receptor staining in cells expressing Godzilla, suggesting that Godzilla expression may lead to perturbations in the endosomal maturation process (Figure 2A). In agreement with these results, endogenous Godzilla protein in Drosophila tissues displays a vesicle localization overlapping with endosomes (Supplementary Figure S1E–G). To confirm the presence of endogenous Godzilla protein in the endosomal compartment, we expressed activated Rab5—Rab5Q88LYFP (Stenmark et al, 1994) to produce enlarged endosomes in a number of tissues. Expression of activated Rab5–Rab5Q88LYFP in the wing disc leads to the accumulation of endogenous Godzilla protein on the resulting enlarged endosomes (Figure 1E). Taken together, these data suggest that Goliath family ubiquitin ligases function in endosomal trafficking processes.
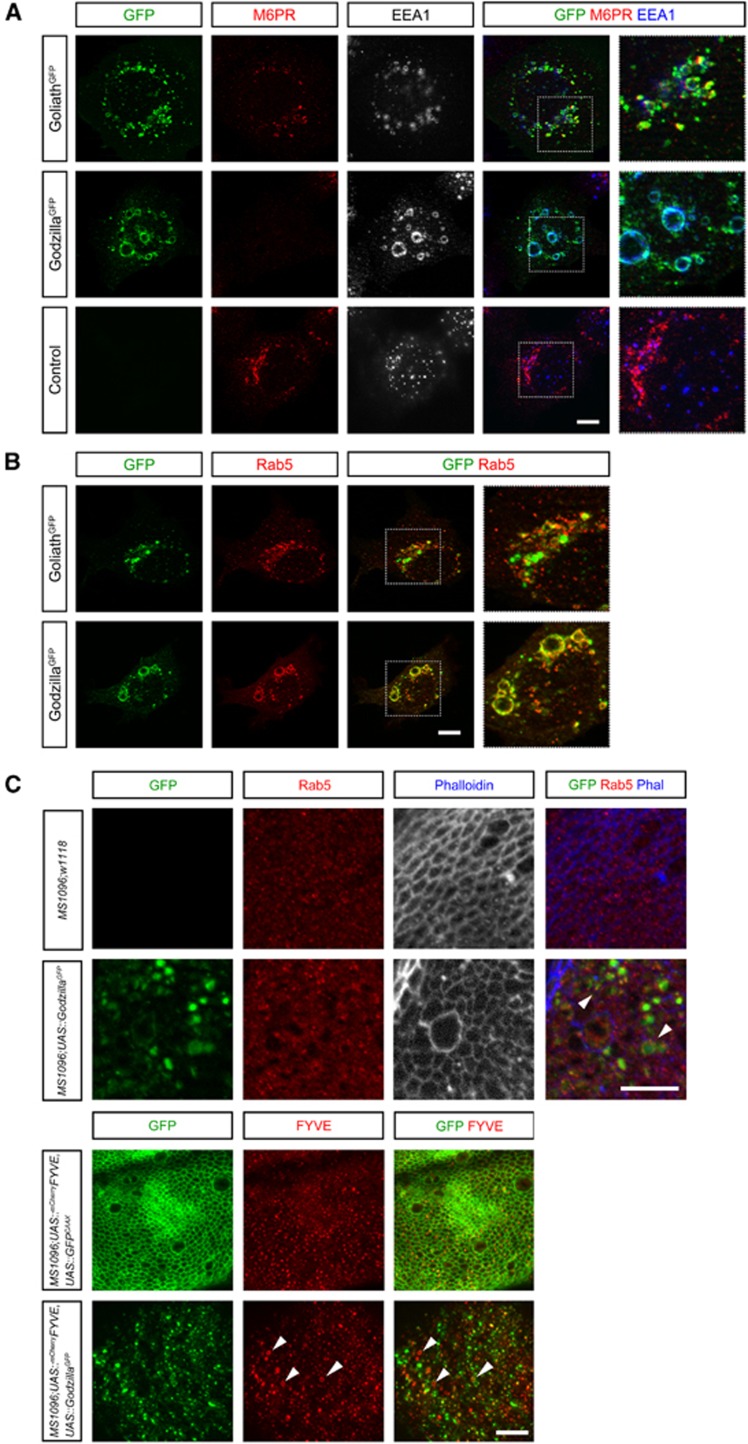
Figure 2.
Goliath and Godzilla ligase activity leads to the accumulation of Rab5-positive giant endosomes. (A) Goliath C-GFP, Godzilla-C-GFP or pCGFP was transfected into HEK293 cells, and stained with antibodies detecting early and late endosome marker proteins (EEA1 and M6PR, respectively). Expression of either Goliath or Godzilla resulted in significantly enlarged endosomes (>3 μm in diameter) as compared with control (∼0.4 μm). Dotted box shows an expanded view of each panel. (B) Rab5 colocalizes on giant vesicles induced by expression of Goliath and Godzilla. HEK293 cells were transfected with Goliath-C-GFP or Godzilla-C-GFP together with pEGFP-DsRed-Rab5. Dotted box shows an expanded view of each panel. (C) Overexpression of Godzilla leads to the formation of giant endosomes in Drosophila. GFP-tagged Godzilla was expressed with the MS1096-Gal4 wing driver. Early endosomes were visualized by anti-Rab5 (upper panel) or by co-expression of mCherry::2xFYVE, an early endosome biomarker (lower panel). Godzilla::GFP (green) is localized on specific microdomains of enlarged mCherry::2xFYVE-positive endosomes. Membrane targeted GFP (GFPCAAX) was used as control. Enlarged endosomes are seen in Godzilla-overexpressing tissue (arrowheads). Bar 10 μm. The results shown here are typical images from at least four independent experiments.
Goliath and Godzilla ubiquitin ligase activity induces the formation of giant endosomes
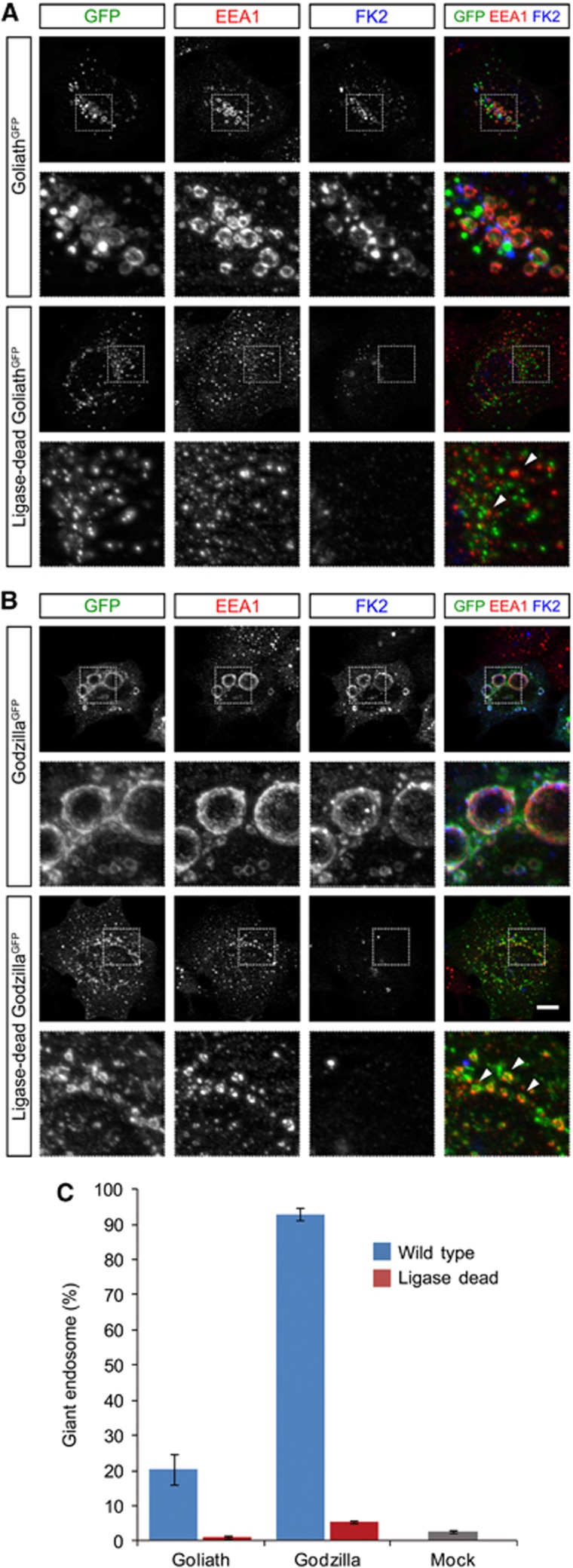
In both Drosophila wing disc expression and vertebrate cell culture experiments, we noticed that the endosomes were abnormally large upon Godzilla expression (>3 μm, up to 15 μm, Figure 2; quantified in Figure 3C). This is not due to the addition of the carboxy-terminal GFP since HA-tagged versions of both Godzilla and Goliath display similar localization and activity as their GFP-tagged counterparts (Supplementary Figure S3). Consistently, we noted that Godzilla was significantly more potent in the production of dramatically enlarged endosomes than Goliath. These enlarged endosomes were Rab5 and mCherry-FYVE positive in their nature (Figure 2B and C; Supplementary Figure S4), leading us to hypothesize that Godzilla/Goliath might play an important role in endosomal maturation perhaps via ubiquitylation of a key component of the endosomal machinery. In keeping with an important role for ubiquitin ligase activity, we were able to show that mutation of key His residues in the RING finger domain, corresponding to His323 and His326 in Goliath and His255 and His258 in Godzilla (Figure 1B; Su et al, 2009), abrogated the ability of either Godzilla or Goliath to generate large endosomes (Figure 3).
Figure 3.
Goliath- or Godzilla-induced endosomal enlargement is dependent on their ubiquitin ligase activity. (A, B) HEK293 cells were transfected with Goliath-C-GFP, ligase-dead Goliath-C-GFP (A), Godzilla-C-GFP, or ligase-dead Godzilla-C-GFP (B), and stained for EEA1 and ubiquitylated proteins (FK2). Strong ubiquitylation was observed on enlarged endosomes upon expression of ligase-active forms of Goliath and Godzilla, but not ligase-dead forms. Lower panels show an expanded view of dotted boxed area of upper image. Bar 10 μm. (C) Quantification of induction of enlarged endosomes by Goliath and Godzilla. The number of transfection-positive (GFP-positive) HEK293 cells containing giant endosomes was quantified. Results are given as means±s.d. of four independent experiments.
Godzilla function is required in vivo for integrity of endosome trafficking
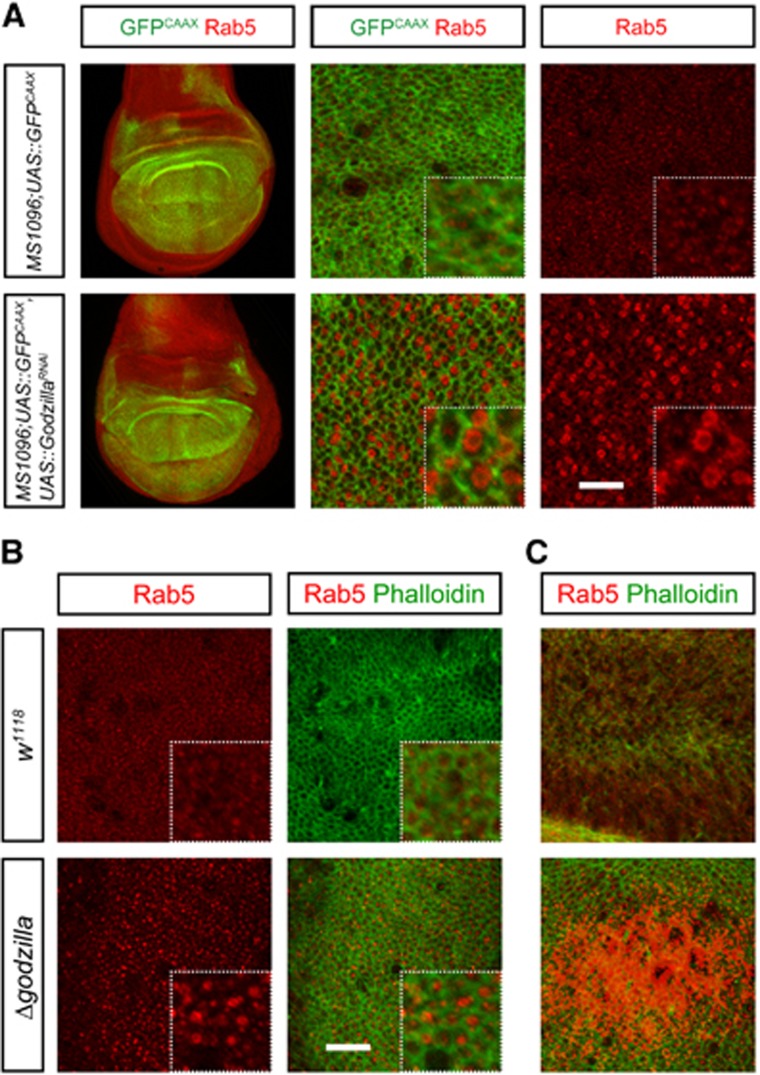
In light of these results, we examined the effect of loss of function of these proteins on endosomes. To test this, we examined endosome accumulation in third instar larvae wing discs lacking Godzilla. RNAi knockdown of Godzilla resulted in a significant decrease in endogenous Godzilla protein (Supplementary Figure S1), and an accompanying robust accumulation of Rab5-positive endosomes (Figure 4A). This robust phenotype was also observed in ΔgodzillaCG10277 mutant animals, where there is a dramatic buildup of Rab5-positive endosomes in third instar larva prior to lethality (Figure 4B and C). Importantly, this endosomal phenotype is rescued by Godzilla genomic rescue (Supplementary Figure S5).
Figure 4.
Loss of Godzilla function leads to a robust accumulation of Rab5 endosomes. (A) RNAi-mediated knockdown of Godzilla driven by MS1096-Gal4 induces the formation of Rab5-positive giant endosomes in the Drosophila wing disc. Control is MS1096-Gal4-driven UAS-GFPCAAX. Rab5 (red) and GFP (green) are shown. Left panels are lower magnification overview of wing discs. Right panels are close-up view of the centre of wing pouch. Enlarged Rab5-positive endosomes are observed with godzilla RNAi but not in control. (B) Godzilla mutant animals exhibit enlarged Rab5-positive endosomes. Wing discs of late third instar larvae were visualized with antibodies towards Rab5 (red) as well as phalloidin to visualize F-actin (green). Dotted box shows an expanded view of each panel. Rab5-positive endosomes are enlarged and accumulated in godzilla mutant wing discs, but not in control (w1118). Bar 10 μm. (C) Rab5 endosomes accumulate to high levels in godzilla mutant wing discs. Both images were taken at identical exposure times. The results shown here are typical images from at least four independent experiments.
Identification of VAMP3 as a target of Godzilla ubiquitin ligase activity
Taken together, our data strongly suggest that Godzilla and Goliath play an important role in endosomal processing, leading to perturbation of signalling processes in vivo. We rationalized that a key substrate for their ubiquitylation activity may be a component of the endosomal trafficking machinery, which may be regulated by this family of ligases. Indeed, expression of constitutively active Rab5 or knockdown of Rab7 has also been documented as generating large endosomes (Stenmark et al, 1994; Poteryaev et al, 2010), suggesting that Godzilla/Goliath may regulate Rab5 activity by some unknown mechanism. With this hypothesis in mind we set out to test a number of candidates, with focus on regulators of Rab5 and Rab7 activity. Those molecules tested included: Rab5, Rab7 (Chavrier et al, 1990), the Rab5-specific guanine nucleotide exchange factor (GEF) Rabex5 (Valsdottir et al, 2001; Penengo et al, 2006) and the Rab GDP dissociation inhibitor RabGDI (Ullrich et al, 1994; Poteryaev et al, 2010). In spite of convincing evidence that expression of either Godzilla or Goliath resulted in a high level of ubiquitylation, as measured by anti-FK2 staining (Figure 3), we were unable to identify a cellular target by this candidate approach.
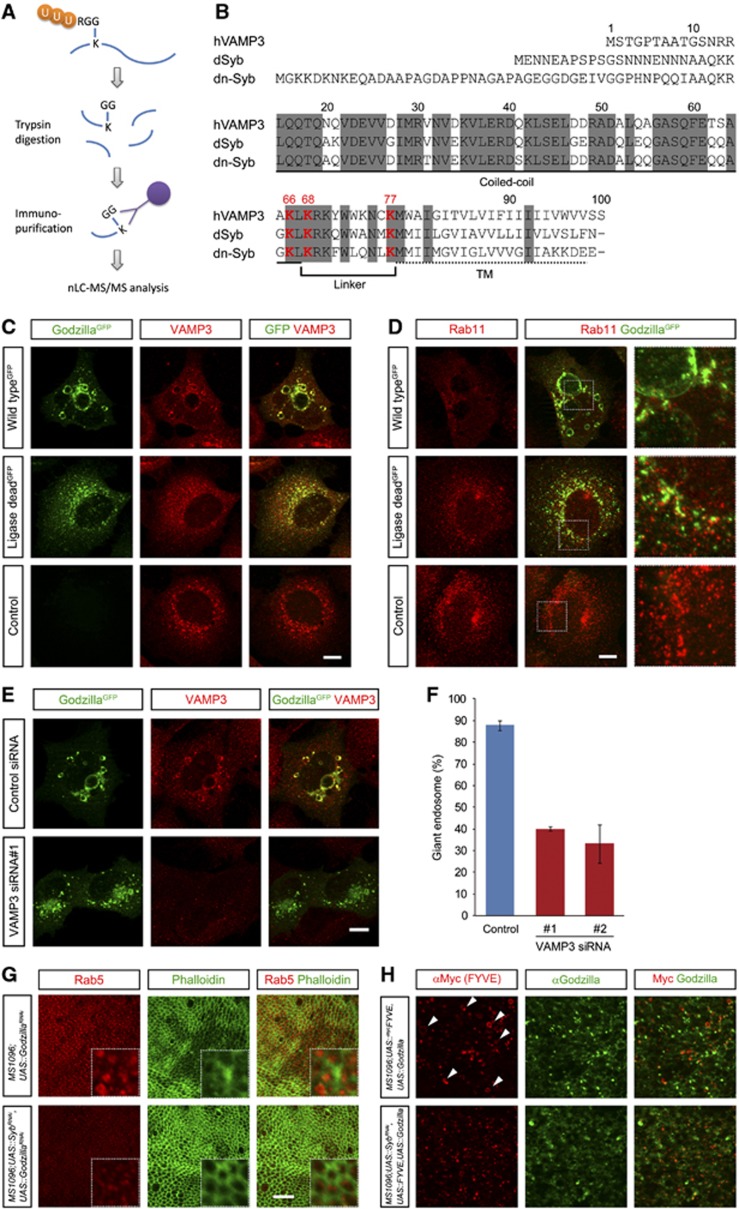
In order to proceed, we carried out a proteomics-based analysis (UbiSCAN), employing anti-GG antibodies to identify conserved cellular targets of Drosophila Godzilla (Xu et al, 2010). This analysis enables the identification, not only of substrate molecules, but also identification of those lysines targeted for ubiquitylation within the substrate. This approach led to the identification of the SNARE protein VAMP3, which is known to be involved in vesicle fusion and trafficking (McMahon et al, 1993) as a substrate for this family of ubiquitin ligases (Figure 5; Supplementary Figure S6; Supplementary Table S1). VAMP3 was heavily ubiquitylated at Lys 66, Lys 68 and Lys 77 in samples containing active Godzilla, but not in Godzilla ligase-dead control lysates. Human VAMP3/Cellubrevin and the Drosophila orthologues Synaptobrevin (Syb, also called dVAMP) and neuronal-Synaptobrevin (n-Syb) show significant identity of 71 and 64%, respectively, and importantly these ubiquitylated Lys residues (Lys 66, 68 and 77) in VAMP3 are completely conserved in the Drosophila homologues (Figure 5B).
Figure 5.
VAMP3, an endosomal SNARE protein, is ubiquitinated by Godzilla. (A) Schematic of identification of ubiquitiylated proteins by anti-GlyGly antibody purification (Xu et al, 2010). Cell extracts containing ubiquitylated proteins were digested by trypsin, resulting in generation of di-glycine conjugated peptides on the ε-amine group of lysine. Peptides were immuno-purified by anti-GlyGly antibody then subjected to nLC-MS/MS analysis. (B) Sequence alignment of VAMP3 and its Drosophila homologue, Synaptobrevin (Syb, dVAMP) and neuronal-Synaptobrevin (n-Syb). Identically conserved residues are highlighted in grey and the transmembrane domain and linker region are marked. Three ubiquitinated lysine residues (Lys66, Lys68 and Lys77) identified in VAMP3 are shown in red. These ubiquitinated lysine residues are completely conserved between human and Drosophila VAMP3 homologues. (C) Localization of VAMP3 on Godzilla-induced giant endosomes. Wild-type or ligase-dead Godzilla expressing HEK293 cells were stained with anti-VAMP3 antibody (red). (D) Rab11-mediated recycling is disrupted in wild-type Godzilla expressing cells, but not in ligase dead. (E) VAMP3 knockdown rescues the formation of giant endosomes induced by Godzilla, quantified in (F). Quantified results are given as mean±s.d. for four independent experiments. Bar 10 μm. (G) Formation of Rab5-positive giant endosomes in godzilla loss-of-function wing discs is suppressed upon knockdown of the Drosophila VAMP3 homologue Synaptobrevin (Syb). Upper panel: late third instar larvae wing disc expressing UAS-godzilla RNAi driven by MS1096-Gal4; bottom panel: MS1096-Gal4 driving both UAS-godzilla RNAi and UAS-syb RNAi wing disc. Wing discs were visualized with Rab5 (red) and phalloidin to visualize F-actin (green). Dotted box shows an expanded view. (H) Formation of FYVE-positive giant endosomes in Godzilla-overexpressing wing discs is suppressed upon knockdown of Syb. Giant endosomes observed upon Godzilla overexpression (upper panel, arrowheads) are suppressed in a Syb knockdown background (bottom panel).
In order to confirm whether VAMP3 is a potential target of Godzilla, we investigated VAMP3 localization in Godzilla-transfected HEK293 cells. VAMP3 is known to localize on both early and recycling endosomes and is involved in endocytic recycling (McMahon et al, 1993; Galli et al, 1994). In both control and ligase-dead Godzilla-transfected cells, VAMP3 displays a similar intracellular distribution, partially overlapping with ligase-dead Godzilla; suggesting an early and recycling endosome distribution (Figure 5C). In contrast, in cells expressing wild-type Godzilla VAMP3 is found on enlarged endosomes, where it colocalizes with Godzilla (Figure 5C).
The Godzilla induced endosomal phenotype peturbs trafficking of both Rab5- and Rab11-positive endosomes and requires VAMP3
As VAMP3 has been shown to play a role in recycling endosome trafficking (Galli et al, 1994; Hong, 2005), we considered the hypothesis that Godzilla may regulate recycling endosome trafficking. In order to test this, we analysed transfected cells for the presence of Rab11-positive recycling endosomes (Ullrich et al, 1994). While cells transfected with ligase-dead Godzilla stain positive for Rab11 recycling endosomes compared with mock-transfected cells, we were unable to detect the presence of Rab11 recycling endosomes in the presence of wild-type ligase active Godzilla (Figure 5D). Interestingly, while the normal pattern of pericentriolar and dispersed Rab11-positive recycling endosomes was not seen, some residual Rab11 staining could still be observed in the vicinity of the Godzilla containing enlarged endosomes, suggesting an inability of Rab11 containing vesicles to be generated (Figure 5D, upper panel). In keeping with this, overexpression of Godzilla in the Drosophila with MS1096-Gal4 also results in a loss of Rab11-positive vesicles in the wing disc (Supplementary Figure S7). To further investigate the significance of VAMP3 as a Godzilla target in the Godzilla-induced giant endosome phenotype, we transfected HEK293 cells either with scrambled control siRNA or with two independently employed siRNAs targeting VAMP3 (Supplementary Figure S8). Indeed, reduction of endogenous VAMP3 expression levels resulted in decreased levels of Rab11 recycling endosomes, while no impact on the presence or vesicle size of EEA1 early endosomes was seen (Supplementary Figure S8). Next, we investigated whether knockdown of VAMP3 was able to block the large endosome phenotype mediated by Godzilla overexpression (Figure 5E and F). Prior to transfection with wild-type Godzilla, endogenous VAMP3 expression levels were reduced by siRNAs targeting VAMP3. A strong reduction in the enlarged endosome phenotype was observed; leading us to conclude that loss of VAMP3 significantly blocks the Godzilla mediated enlarged endosome phenotype. These results suggest that ubiquitylation does not result in a simple inhibition of VAMP3 function, such as inhibition of SNARE complex formation or complex assembly, but that the ubiquitylation of VAMP3 may work as a molecular switch for the recycling endosome pathway.
We then examined the effect of loss of Godzilla function in vivo returning to the Drosophila wing disc. Drosophila has two VAMP3 homologues, the neuronal n-Syb and a more generally expressed Syb (Chin et al, 1993; DiAntonio et al, 1993). Knockdown of Drosophila Syb reverses the godzilla mutant phenotype, resulting in a loss of giant Rab5 endosomes in godzilla mutant wing discs (Figure 5G). Furthermore, Drosophila Syb knockdown blocks the formation of FYVE-positive giant endosomes observed upon Godzilla overexpression in the Drosophila wing disc (Figure 5H). Together, these data provide convincing evidence that VAMP3/Syb is a relevant functional target of the Godzilla ligase in vivo in the fruitfly as well as in mammalian cells.
The related mammalian RNF167 PA-TM-RING domain ubiquitin ligase induces formation of giant endosomes
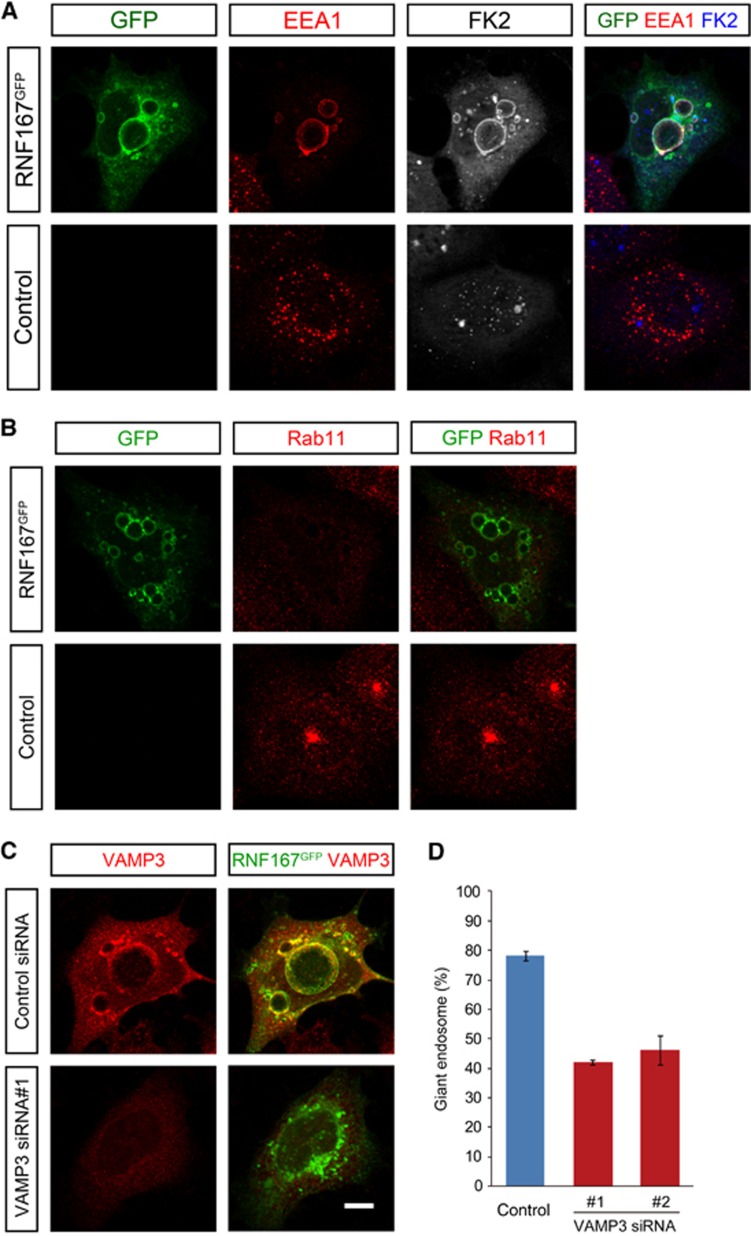
We then asked whether a role in endosomal trafficking may be conserved among the human members of the Goliath family. Consistent with this we found that expression of human RNF167 also led to the accumulation of giant endosomes that were positive for EEA1, FK2 and VAMP3, with a concomitant loss of Rab11-positive recycling endosomes (Figure 6A and B). Moreover, as with the Godzilla induced giant endosome phenotype, we were able to revert the RNF167 induced phenotype upon knockdown of VAMP3 (Figure 6C and D).
Figure 6.
RNF167, a human PA-TM-RING E3 ligase, also affects endosomal trafficking. (A, B) The endosomal phenotype induced by human RNF167 resembles that of Godzilla. HEK293 cells were transfected with RNF167-C-GFP. RNF167 overexpression induces the formation of EEA1 and FK2-positive giant early endosomes (A), as well as loss of Rab11 recycling endosomes (B). (C, D) HEK293 cells expressing human RNF167-C-GFP exhibit robust accumulation of giant endosomes, which are VAMP3 positive. VAMP3 knockdown results in the rescue of the giant endosome phenotype induced by RNF167, quantified in (D). Quantified results are given as mean±s.d. of four independent experiments. Bar 10 μm.
Mutation of Godzilla target lysines in VAMP3 abrogates the ability of Godzilla/RNF167 to induce giant endosomes
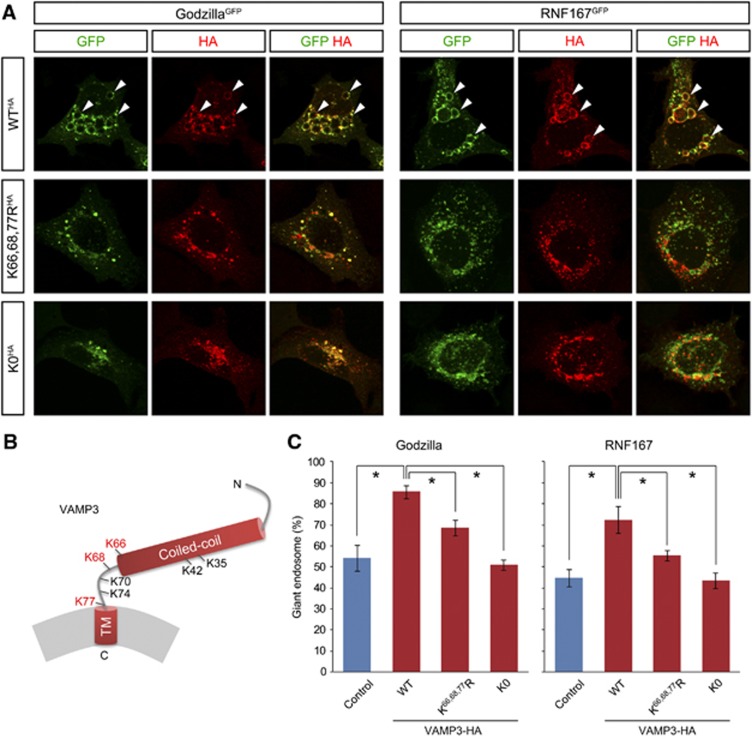
Having established VAMP3 as a target of Godzilla, we wished to examine the importance of ubiquitylation for VAMP3 by mutating the target Lysine residues in the VAMP3 molecule. It has been shown that the linker region, which contains Lysine 66, 68 and 77 targeted by Godzilla (Figure 7B), has a crucial role in SNARE-dependent fusion (McNew et al, 2000a, 2000b; Knecht and Grubmuller, 2003). We generated three carboxyl-terminal HA-tagged VAMP3 mutants (VAMP3-WTHA, VAMP3-K66,68,77RHA and VAMP3-K0HA; Figure 7B) in which target ubiquitylation lysine residues were mutated to arginine. These VAMP3 mutants, which are not targeted by the VAMP3 siRNA reagent employed, were introduced into HEK293 cells together with either Godzilla or RNF167 after knockdown of endogenous VAMP3 protein. First, mutation of the lysine residues in VAMP3 did not appear to affect localization of VAMP3 on endosomes upon cotransfection with Godzilla or RNF167. Second, we observed that the presence of all VAMP3 lysines are required for the formation of giant endosomes by either Godzilla or RNF167 (Figure 7A and C). These data suggest that ubiquiylation of VAMP3 is required to regulate its activity in the fusion process, in agreement with earlier observations of a critical role for the VAMP3 linker region in fusion (McNew et al, 2000a, 2000b; Knecht and Grubmuller, 2003).
Figure 7.
Mutation of Godzilla target ubiquitylation in VAMP3 disrupts VAMP3 protein function. (A) The giant endosome phenotype induced by Godzilla or RNF167 is supported by wild-type VAMP3, but not by Lys-substituted VAMP3 mutants. HA-tagged VAMP3 mutants were co-transfected with Gozilla-C-GFP (left) or RNF167-C-GFP (right) after knockdown of endogenous VAMP3. The giant endosome phenotype is supported by wild-type VAMP3 (WTHA), but only partially by K66, 68, 77RHA and not by K0HA. (B) Schematic structure of VAMP3. hVAMP3 contains seven Lys residues in total; two are in the coiled-coil region, while other five are located in the linker region. The Lys residues identified as ubiquitylated are indicated in red. (C) Statistical analysis of (A). Quantified results are given as mean±s.d. for three independent experiments. Asterisk P<0.05.
Induction of giant endosomes by Godzilla/RNF167 blocks Transferrin receptor recycling
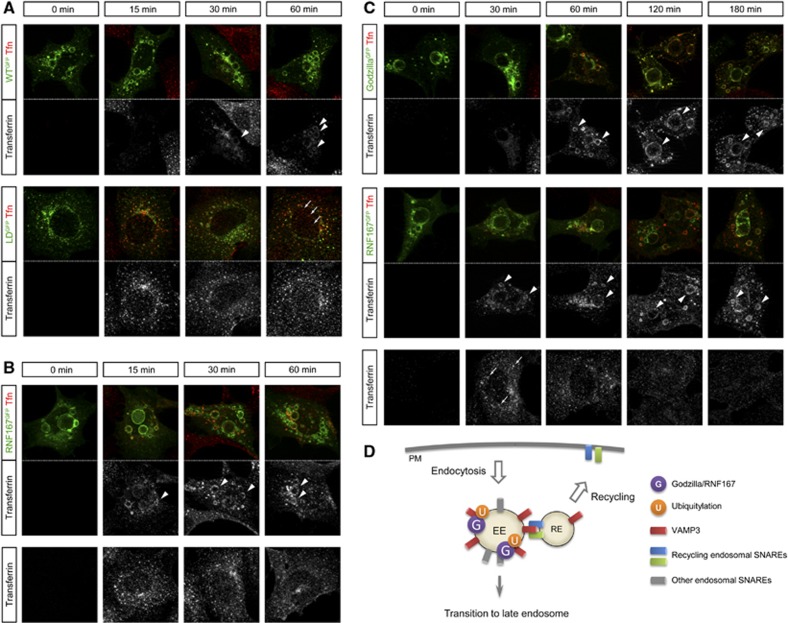
Taken together, these results suggest a model in which Goliath family ubiquitin ligases such as Godzilla regulate trafficking through the recycling endosome pathway. To further support of this hypothesis, we decided to investigate the effect of Godzilla E3-ligase activity on Transferrin receptor trafficking (Figure 8). Transferrin internalization appears to be significantly delayed in Godzilla-transfected cells. While control cells expressing ligase-dead Godzilla displayed efficient trafficking of transferrin, active Godzilla expressing cells accumulated transferrin in enlarged endosomes (Figure 8A, arrowheads). A similar effect on transferrin receptor trafficking was observed with human RNF167 (Figure 8B). In order to investigate if incorporated transferrin is recycled in Godzilla of RNF167-transfected cells, we performed transferrin chasing for longer period up to 3 h (Figure 8C). While control cells displayed efficient trafficking of transferrin with transit of transferrin to endosomes clearly observed at 30 min and almost disappearing to 120 and 180 min, active Godzilla or RNF167-expressing cells accumulated transferrin which was present in enlarged endosomes even at 180 min after addition of transferrin (Figure 8C, arrowheads).
Figure 8.
Transferrin recycling is disrupted in Godzilla- or RNF167-transfected cells. (A) Transferrin is accumulated at enlarged endosomes in Godzilla-transfected, but not in ligase-dead mutant transfected cells. Cells were exposed to Alexa Fluor 555-labelled transferrin in wild-type (top panels) or ligase-dead Godzilla (bottom panels) transfected HEK293 cells for the indicated period. In ligase-dead Godzilla transfected cells, transit of transferrin to endosomes is observed at 15 and 30 min, with appearance in recycling vesicles (segregated signal from ligase-dead Godzilla at 60 min, arrows). In contrast, Alexa Fluor 555-labelled transferrin is accumulated at enlarged endosomes in wild-type Godzilla-transfected cells (arrowheads). (B) Transferrin is accumulated at enlarged endosomes in RNF167-transfected, but not in mock-transfected cells. Cells were exposed to Alexa Fluor 555-labelled transferrin in RNF67 (top panels) or control (bottom panels) transfected HEK293 cells for the indicated time. Alexa Fluor 555-labelled transferrin is accumulated in enlarged endosomes in RNF167-transfected cells. (C) Transferrin recycling is disrupted in Godzilla- or RNF167-transfected cells. Cells were exposed to Alexa Fluor 555-labelled transferrin in Godzilla (top panels) or RNF167 (middle panels) transfected HEK293 cells and chased for the indicated period. In mock-transfected cells (bottom panels), transit of transferrin to endosomes is strongly observed at 30 min (arrows), and has almost disappeared at 120 and 180 min. In contrast, Alexa Fluor 555-labelled transferrin is accumulated at enlarged endosomes in Godzilla- or RNF167-transfected cells (arrowheads). (A–C) The results shown here are typical images from four independent experiments. (D) Schematic model visualizing the role of the Goliath family E3 ligases in regulation of endosomal trafficking. In normal conditions, a significant portion of endocytosed proteins on early endosomes are recycled via recycling endosomes, while the remaining proceed to lysosomal degradation via transition to the late endosome. In the budding process from the early endosome to recycling endosome, VAMP3/Syb on the early endosome forms a SNARE complex with recycling endosome SNAREs, such as Syntaxin1 and SNAP-25. Ubiquitylation of VAMP3/Syb by Godzilla/RNF167 plays a role in the regulation of recycling endosome traficking.
Discussion
Here, we report a number of novel findings concerning the poorly characterized Goliath family of PA-TM-RING domain E3 ligases. First, we show that they are localized at endosomes, where their activity leads to the ubiquitylation of target proteins. Second, manipulation of these ubiquitin ligases leads to a disruption of endosomal trafficking processes, both in cell culture and in vivo in Drosophila tissues. In particular, loss of the ubiquitously expressed Godzilla in the fly leads to a dramatic disruption of endosomal trafficking. Since mammalian endosomal VAMPs have been shown to mediate homotypic fusion of endosomes (Antonin et al, 2000), the robust accumulation of Rab5-positive endosomes we observe in the absence of Godzilla may be a result of enhanced homotypic fusion in the absence of recycling (Figure 8D). Third, our results identify the VAMP3 SNARE protein as a novel ubiquitylated target of these ligases, and show that these target lysines have functional relevance. Previous reports of the involvement of VAMP3 in the regulation of recycling endosomes (McMahon et al, 1993; Galli et al, 1994) are entirely consistent with the results presented here. Furthermore, since loss-of-function of the more generally expressed Syb/dVAMP protein is lethal in Drosophila, a general role in cellular trafficking has been predicted (Bhattacharya et al, 2002). This hypothesis is in keeping with the results presented here, which strongly suggest that Syb/dVAMP has a crucial role in endosome recycling in Drosophila as in mammals. Further, in agreement with our results, we note that several human members of the Goliath family E3 ligases, including RNF167, have been scored as positive hits in a genome-wide RNAi-based study of endocytosis by multiparametric image analysis (Collinet et al, 2010).
Vesicular-SNARE proteins form SNARE complexes with their partner proteins, consisting of a four-helix bundle held together by interacting layers. The current model of this process is that an initial trans-SNARE complex forms between pairing vesicles and further supports a directional zipping from the amino-terminal coiled-coil domain to the carboxyl-terminal transmembrane domain of the complex, resulting in pulling opposing membranes together and completing membrane fusion (Sutton et al, 1998; Stein et al, 2009). It has been shown that the linker region, which contains the Lysines 66, 68 and 77 targeted by Godzilla, has a crucial role in SNARE-dependent fusion (McNew et al, 2000a, 2000b; Knecht and Grubmuller, 2003). Recently, studies of the VAMP2/syntaxin1A/SNAP-25 complex have examined the molecular mechanisms underlying the propagation of ‘zipping’ of the four-helix bundle to the transmembrane domain to complete the fusion (Hernandez et al, 2012). In this work, a VAMP2 mutant at residue Leu84, corresponding to Leu67 of VAMP3, was shown to be unable to fuse large liposomes. It has already been shown that post-transcriptional modifications of SNARE proteins such as phosphorylation and palmitoylation are known to regulate their functions (Snyder et al, 2006; Valdez-Taubas, 2005). However, although ubiquitylation has also been noted in several SNARE proteins including VAMP3 in large-scale proteomics analyses (Supplementary Figure S6; Hornbeck et al, 2012), its role or consequence is not understood. Here, we show that Godzilla/RNF167 regulates endosome recycling by the ubiquitylation of VAMP3 on Lys66, Lys68 and Lys77; namely, two adjacent Lys residues on the both sides of the critical interface of SNARE complex are ubiquitylated. Given the elegant studies on SNARE fusion dynamics (Hernandez et al, 2012) a significant effect of ubiquitylation at these sites on SNARE complex assembly can be envisioned. The endosome phenotypes observed in both loss-of-function and gain-of-function Godzilla suggest that the ubiquitylation of VAMP3 may be employed as a switch in the dynamic regulation endosome recycling, such that both correct spatial and temporal ubiquitylation and deubiquitylation of VAMP3 are important. While we observed ubiquitylation of VAMP3 by Godzilla, we are unable to describe the nature of this ubiquitination, be it mono-ubiquitin or extended ubiquitin chains. Novel tools to address the nature of ubiquitin modifications have recently been developed and may help to elucidate the nature of VAMP3 ubiquitylation in the future (van Wijk et al, 2012). It will also be of interest to examine the functional relevance of VAMP3 ubiquitylation in the fusion process at a structural level.
It follows that if ubiquitylation is employed as a switch in recycling endosome trafficking, that DUB(s) may also play an important function in the regulation of this process. In conclusion, the data presented here demonstrate a novel function for the Goliath PA-TM-RING domain ubiquitin ligase family in regulating recycling endosome trafficking through ubiquitylation of the VAMP3 SNARE protein (Figure 8D), with implications for intracellular recycling and signalling events in both physiological and pathological conditions.
Materials and methods
Recombinant DNA constructs
The following constructs were used for transfection in HEK293 cells: GFP-tagged wild type or ligase-dead Goliath or Godzilla constructs (Goliath WT-C-GFP, Goliath LD-C-GFP, Godzilla WT-C-GFP, Godzilla LD-C-GFP), HA-tagged wild Goliath or Godzilla (pIRES-hrGFP-2a-Goliath WT, pIRES-hrGFP-2a-Godzilla WT) and GFP-tagged human RNF167 (RNF167-C-GFP) (construction details available upon request). For the construction of ligase-dead mutants, two conserved His residues in the RING domain (His323 and His326 in Goliath, His255 and His258 in Godzilla), reported to be involved in the coordination of Zn2+ ions, were substituted to Arg residues (Figure 1B; Su et al, 2009). pEGFP-DsRed-Rab5 is from Addgene (Cambridge, MA). cDNAs for C-terminal HA-tagged VAMP3 mutants (WTHA, K66,68,77KHA, and K0HA) were synthesized (GenScript, NJ) and cloned into pcDNA3 expression vector at BamHI and EcoRI sites.
Cell culture and transfections
HEK293 cells were cultured in Dulbecco’s modified Eagle’s medium, containing 10% foetal bovine serum, penicillin and streptomycin. HEK293 cells were transfected with the above constructs as indicated using Lipofectamine (Invitrogen) according to manufacturer’s protocol prior to fixation and immunostaining.
Transferrin uptake assays
For localization studies on early and recycling endosomes, transfected HEK293 cells were incubated for 2 h in serum-free cell culture medium containing 0.2% BSA at 37°C (Daro et al, 1996). Cells were subsequently incubated with 10 ng/μl Alexa FluoR 555 conjugated transferrin (Molecular Probes) in serum-free cell culture medium+0.2% BSA for 45 min at 4°C, followed by three washes in medium+0.2% BSA. Vesicle recycling was analysed after incubation at 37°C for the indicated times. For the transferrin chase analysis (Figure 8C), transfected HEK293 cells were incubated for 2 h in serum-free medium containing 0.2% BSA at 37°C. Cells were subsequently incubated with 10 ng/μl Alexa Fluor 555 conjugated transferrin for 45 min at 4°C, followed by three washes in 37°C pre-warmed serum containing complete medium. Vesicle recycling was analysed after incubation for up to 180 min at 37°C.
RNA interference
For VAMP3 knockdown, double-stranded RNA duplexes (siMax siRNA, Eurofins MWG Operon, Germany) targeting human VAMP3 were employed. The sequences were the following: siVAMP3 #1: 5′-UCAUUACCUUGUGUGUGAA-3′; siVAMP3 #2: 5′-UCGAAGACUUCAGCAGACA-3′. As a negative control, Stealth RNAi-negative control low CG duplex was used (Invitrogen). At 48 h post transfection, employing Lipofectamine 2000 (Invitrogen), cells were either lysed and knockdown of VAMP3 expression levels were analysed by immunoblotting as described previously (Yang et al, 2007) or fixed and stained for analysis by immunofluorescence. For endogenous VAMP3 depletion, HEK293 cells were transfected with siVAMP3 #1, targeting the 3′-UTR of VAMP3 mRNA on day 1, prior to transfection of Godzilla, RNF167 or VAMP3 on day 2. Cells were subjected to analysis on day 3. Drosophila transgenic RNAi lines to Godzilla (109001KK) and dVAMP/Syb (102922KK) were obtained from Vienna Drosophila Resource Center (Dietzl et al, 2007). Knockdown efficiency of GodzillaRNAi was confirmed with immunofluorescence by anti-Godzilla antibody.
Drosophila strains and genetics
Standard Drosophila husbandry procedures were followed. Drosophila strains were maintained on standard potato-meal medium, and maintained at room temperature unless otherwise stated. Experimental crosses were performed at 25°C. The FRT strains used to generate gl deletion mutants were WHf06772 and RBe01478; FRT strains for the godzillaCG10277 deletion (Δgodzilla1; Supplementary Figure S5) were XPd01485 and WHf07224, which were obtained from the Harvard Exelixis collection (Thibault et al, 2004). Deficiency lines Df(3R)Tpl10 (BL1990), Df(3R)Tpl9 (BL1986) and Df(3R)WIN11 (BL2393) were obtained from the Bloomington stock Center. The P-element line used to generate godzillaCG10277 deletion mutant (Δgodzilla2; Supplementary Figure S5) was EP705, from the Bloomington stock Center. The deletion was confirmed by southern blotting and DNA sequencing. As godzillaCG10277 mutant larva cannot survive on agar-based fly food, mutant larvae were maintained with yeast paste on apple juice plate. UAS::mCherry::FYVEHrs (Velichkova et al, 2010) and UAS-GFPcaax (Finley et al, 1998) were used in subcellular localization studies. For mutant clone analysis, Δgodzilla2 was recombined with P{neoFRT}82B ry[506] (BL2035). The resulting Δgodzilla2, P{neoFRT}82B ry[506] was crossed to P{hsFLP}12; Sco/CyO (BL1929). Progeny was crossed to P{neoFRT}82B P{Ubi-GFP(S65T)nls}3R P{A92}RpS3[Plac92]/TM6B, Tb[1] (BL5627), and larvae were heat-shocked for 30 min at 37°C, prior to analysis.
Genomic rescue and transgenic constructs
The genomic rescue construct harbouring godzillaCG10277 was constructed by ϕ31 integrase-based methodology. Briefly, a Drosophila genomic DNA fosmid clone (FlyFos028285), corresponding to the region containing CG42564, CG42537, CG1024, TfIIFalpha, Godzilla and CG1021 was obtained from FlyFos project (http://transgeneome.mpi-cbg.de/index.php?id=42). To remove CG1021, the fosmid was digested with AatII and religated. The resulting clone was subjected to transgenesis to P[acman] strain 9750 (65B2). Since complementation analyses suggest that Δgodzilla2 harbours an additional lethal mutation on the third chromosome, ΔGodzilla1/ΔGodzilla2 are employed as Godzilla mutant in this study. pUAST-Goliath-GFP and pUAST-Godzilla-GFP constructs were generated from Goliath WT-C-GFP and Godzilla WT-C-GFP by digestion with BglII and BamHI. The obtained fragments were ligated into BglII site in pUAST. The direction of the insert was confirmed by restriction enzyme digestion and the constructed vectors were subjected to embryo injection (BestGene, CA).
Immunostaining and antibodies
The following antibodies were used: goat anti-mouse Cy3 (1:1000, Jackson), goat anti-mouse Cy2 (1:1000, Amersham), goat anti-mouse Cy5 (1:200, Jackson), goat anti-rabbit Cy2 (1:1000, Amersham), and goat anti-rabbit Cy3 (1:1000, Amersham), donkey anti-rabbit Cy5 (1:200, Jackson), donkey anti-guinea pig Cy3 (1:500, Jackson), mouse anti-HA (Covance, 1:1000), rabbit anti-HA (SantaCruz, 1:100), mouse anti-GFP (Clontech, 1:5000), rabbit anti-Myc (Abcam, 1:2000), rabbit anti-EEA1 (1:100, SantaCruz), mouse anti-mannose 6 phosphate receptor (1:500, Abcam), rabbit anti-LC3B (1:400, Cell Signaling), mouse anti-conjugated ubiquitin FK2 (1:200, Enzo Life Science), rabbit anti-Drosophila Rab5 antibody (2 μg/ml, Abcam), guinea-pig anti-Godzilla (1:1,500; this work); rabbit anti-Rab11 (1:8000), anti-pJNK (1:250, Promega). Anti-VAMP3 antibodies were obtained from Synaptic Systems (Synaptic Systems, Germany), rabbit anti-Rab11 (1:100) from Cell Signaling and anti-pan-ERK (1:5000) was obtained from BD Biosciences. MitoTracker and LysoTracker were from Invitrogen. Disc staining was carried out according to Grabbe et al (2004). For immunostaining of HEK cells, cells were grown on collagen-coated coverslips in 24-well plates, fixed with 4% paraformaldehyde/DMEM and blocked with 50 mM NH4Cl/PBS. After permeabilization with 0.3% Triton X-100 and 5% goat serum containing PBS, cells were incubated with primary antibody overnight. For visualization, cells were further incubated with fluorescence-labelled secondary antibody followed by analysis by ApoTome fluorescence microscopy (Zeiss, Germany) and Leica TCS SPE confocal microscope (Leica, Germany).
Antibody generation
Antigen was produced as bacterial recombinant protein in BL21 Star (DE3). Briefly, cDNA corresponding to C-terminal part of the Godzilla protein (residues 291–536) was synthesized (GenScript, NJ) and ligated into NcoI-BamHI site of pETHis1a vector. As the original sequence contains NcoI site in this region, the corresponding nucleotide was replaced to synonymous nucleotide. Purified proteins by using Ni-NTA agarose (Qiagen, Germany) were immunized in guinea pig (Eurogentec, Belgium). Antibody reactivity was verified by western blotting and immunofluorescence (1:1500).
Proteomics
UbiSCAN analysis (Cell Signaling), involving K-GG peptide immunoprecipitation and LCMS/MS, was performed on HEK293 cells expressing either wild-type Godzilla or ligase-dead Godzilla as control. Briefly, Godzilla WT-C-GFP or Godzilla LD-C-GFP was transfected with lipofectamine according to manufacturer’s protocol. After 24 h, cells were harvested with urea lysis buffer. Resulting cell extracts (∼40 mg protein) were used in the subsequent UbiSCAN analysis (Xu et al, 2010).
Bioinformatics sequence analysis
Protein sequence analysis was performed using SMART program from the EMBL, Heidelberg (http://smart.embl-heidelberg.de). For the generation of phylogenetic tree, the multiple nucleotide sequences of ORF were aligned by using ClustalW and then the tree was generated by PhyML.
Supplementary Material
Acknowledgments
We thank the Exelixis Collection at Harvard, the VDRC and the Bloomington Stock Center for fly stocks. The UAS::mCherry::FYVEHrs transgenic fly line and antibodies against Drosophila Rab7 and Rab11 were a kind gift from Dr Amy Kiger (University of California, San Diego) and Dr Akira Nakamura (RIKEN, Japan), respectively. We particularly thank Camilla Sjögren and Tony Hunter for critically reading this manuscript and Karin Ekström for technical assistance. This work has been supported by grants from the Swedish Cancer Society (RHP 11-0336), the Children’s Cancer Foundation (RHP 10/065), the Swedish Research Council (RHP 621-2011-5181), Lions Cancer Society, Umeå, Association for International Cancer Research (RHP 08-0177). RHP is a Swedish Cancer Foundation Research Fellow.
Author contributions: GV initiated the Goliath studies in Drosophila. YY extended these studies and carried out the majority of the experimental analysis. CS assisted with proteomic sample preparation and follow-up analyses. YY and RHP designed the study and wrote the paper with input from CS and BH. All authors discussed the results and commented on the manuscript.
Footnotes
The authors declare that they have no conflict of interest.
References
- Antonin W, Holroyd C, Tikkanen R, Honing S, Jahn R (2000) The R-SNARE endobrevin/VAMP-8 mediates homotypic fusion of early endosomes and late endosomes. Mol Biol Cell 11: 3289–3298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artero R, Furlong EE, Beckett K, Scott MP, Baylies M (2003) Notch and Ras signaling pathway effector genes expressed in fusion competent and founder cells during Drosophila myogenesis. Development 130: 6257–6272 [DOI] [PubMed] [Google Scholar]
- Baker SJ, Reddy EP (2000) Cloning of murine G1RP, a novel gene related to Drosophila melanogaster g1. Gene 248: 33–40 [DOI] [PubMed] [Google Scholar]
- Bhattacharya S, Stewart BA, Niemeyer BA, Burgess RW, McCabe BD, Lin P, Boulianne G, O'Kane CJ, Schwarz TL (2002) Members of the synaptobrevin/vesicle-associated membrane protein (VAMP) family in Drosophila are functionally interchangeable in vivo for neurotransmitter release and cell viability. Proc Natl Acad Sci USA 99: 13867–13872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borchers AG, Hufton AL, Eldridge AG, Jackson PK, Harland RM, Baker JC (2002) The E3 ubiquitin ligase GREUL1 anteriorizes ectoderm during Xenopus development. Dev Biol 251: 395–408 [DOI] [PubMed] [Google Scholar]
- Bouchard ML, Cote S (1993) The Drosophila melanogaster developmental gene g1 encodes a variant zinc-finger-motif protein. Gene 125: 205–209 [DOI] [PubMed] [Google Scholar]
- Chavrier P, Parton RG, Hauri HP, Simons K, Zerial M (1990) Localization of low molecular weight GTP binding proteins to exocytic and endocytic compartments. Cell 62: 317–329 [DOI] [PubMed] [Google Scholar]
- Chin AC, Burgess RW, Wong BR, Schwarz TL, Scheller RH (1993) Differential expression of transcripts from syb, a Drosophila melanogaster gene encoding VAMP (synaptobrevin) that is abundant in non-neuronal cells. Gene 131: 175–181 [DOI] [PubMed] [Google Scholar]
- Collinet C, Stoter M, Bradshaw CR, Samusik N, Rink JC, Kenski D, Habermann B, Buchholz F, Henschel R, Mueller MS, Nagel WE, Fava E, Kalaidzidis Y, Zerial M (2010) Systems survey of endocytosis by multiparametric image analysis. Nature 464: 243–249 [DOI] [PubMed] [Google Scholar]
- Daro E, van der Sluijs P, Galli T, Mellman I (1996) Rab4 and cellubrevin define different early endosome populations on the pathway of transferrin receptor recycling. Proc Natl Acad Sci USA 93: 9559–9564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshaies RJ, Joazeiro CA (2009) RING domain E3 ubiquitin ligases. Annu Rev Biochem 78: 399–434 [DOI] [PubMed] [Google Scholar]
- DiAntonio A, Burgess RW, Chin AC, Deitcher DL, Scheller RH, Schwarz TL (1993) Identification and characterization of Drosophila genes for synaptic vesicle proteins. J Neurosci 13: 4924–4935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietzl G, Chen D, Schnorrer F, Su KC, Barinova Y, Fellner M, Gasser B, Kinsey K, Oppel S, Scheiblauer S, Couto A, Marra V, Keleman K, Dickson BJ (2007) A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature 448: 151–156 [DOI] [PubMed] [Google Scholar]
- Finley KD, Edeen PT, Foss M, Gross E, Ghbeish N, Palmer RH, Taylor BJ, McKeown M (1998) Dissatisfaction encodes a tailless-like nuclear receptor expressed in a subset of CNS neurons controlling Drosophila sexual behavior. Neuron 21: 1363–1374 [DOI] [PubMed] [Google Scholar]
- Galli T, Chilcote T, Mundigl O, Binz T, Niemann H, De Camilli P (1994) Tetanus toxin-mediated cleavage of cellubrevin impairs exocytosis of transferrin receptor-containing vesicles in CHO cells. J Cell Biol 125: 1015–1024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabbe C, Zervas CG, Hunter T, Brown NH, Palmer RH (2004) Focal adhesion kinase is not required for integrin function or viability in Drosophila. Development 131: 5795–5805 [DOI] [PubMed] [Google Scholar]
- Guais A, Siegrist S, Solhonne B, Jouault H, Guellaen G, Bulle F (2006) h-Goliath, paralog of GRAIL, is a new E3 ligase protein, expressed in human leukocytes. Gene 374: 112–120 [DOI] [PubMed] [Google Scholar]
- Haglund K, Dikic I (2012) The role of ubiquitylation in receptor endocytosis and endosomal sorting. J Cell Sci 125: 265–275 [DOI] [PubMed] [Google Scholar]
- Hernandez JM, Stein A, Behrmann E, Riedel D, Cypionka A, Farsi Z, Walla PJ, Raunser S, Jahn R (2012) Membrane fusion intermediates via directional and full assembly of the SNARE complex. Science 336: 1581–1584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong W (2005) SNAREs and traffic. Biochim Biophys Acta 1744: 493–517 [PubMed] [Google Scholar]
- Hornbeck PV, Kornhauser JM, Tkachev S, Zhang B, Skrzypek E, Murray B, Latham V, Sullivan M (2012) PhosphoSitePlus: a comprehensive resource for investigating the structure and function of experimentally determined post-translational modifications in man and mouse. Nucleic Acids Res 40: D261–D270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahn R, Scheller RH (2006) SNAREs--engines for membrane fusion. Nat Rev Mol Cell Biol 7: 631–643 [DOI] [PubMed] [Google Scholar]
- Knecht V, Grubmuller H (2003) Mechanical coupling via the membrane fusion SNARE protein syntaxin 1A: a molecular dynamics study. Biophys J 84: 1527–1547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Bengtson MH, Ulbrich A, Matsuda A, Reddy VA, Orth A, Chanda SK, Batalov S, Joazeiro CA (2008) Genome-wide and functional annotation of human E3 ubiquitin ligases identifies MULAN, a mitochondrial E3 that regulates the organelle's dynamics and signaling. PLoS ONE 3: e1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovering R, Hanson IM, Borden KL, Martin S, O'Reilly NJ, Evan GI, Rahman D, Pappin DJ, Trowsdale J, Freemont PS (1993) Identification and preliminary characterization of a protein motif related to the zinc finger. Proc Natl Acad Sci USA 90: 2112–2116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahon P, Bateman A (2000) The PA domain: a protease-associated domain. Protein Sci 9: 1930–1934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marin O, Valiente M, Ge X, Tsai LH (2010) Guiding neuronal cell migrations. Cold Spring Harb Perspect Biol 2: a001834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayor S, Presley JF, Maxfield FR (1993) Sorting of membrane components from endosomes and subsequent recycling to the cell surface occurs by a bulk flow process. J Cell Biol 121: 1257–1269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazot P, Cazes A, Boutterin MC, Figueiredo A, Raynal V, Combaret V, Hallberg B, Palmer RH, Delattre O, Janoueix-Lerosey I, Vigny M (2011) The constitutive activity of the ALK mutated at positions F1174 or R1275 impairs receptor trafficking. Oncogene 30: 2017–2025 [DOI] [PubMed] [Google Scholar]
- McMahon HT, Ushkaryov YA, Edelmann L, Link E, Binz T, Niemann H, Jahn R, Sudhof TC (1993) Cellubrevin is a ubiquitous tetanus-toxin substrate homologous to a putative synaptic vesicle fusion protein. Nature 364: 346–349 [DOI] [PubMed] [Google Scholar]
- McNew JA, Parlati F, Fukuda R, Johnston RJ, Paz K, Paumet F, Sollner TH, Rothman JE (2000a) Compartmental specificity of cellular membrane fusion encoded in SNARE proteins. Nature 407: 153–159 [DOI] [PubMed] [Google Scholar]
- McNew JA, Weber T, Parlati F, Johnston RJ, Melia TJ, Sollner TH, Rothman JE (2000b) Close is not enough: SNARE-dependent membrane fusion requires an active mechanism that transduces force to membrane anchors. J Cell Biol 150: 105–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller DL (2004) E3 ubiquitin ligases as T cell anergy factors. Nat Immunol 5: 883–890 [DOI] [PubMed] [Google Scholar]
- Penengo L, Mapelli M, Murachelli AG, Confalonieri S, Magri L, Musacchio A, Di Fiore PP, Polo S, Schneider TR (2006) Crystal structure of the ubiquitin binding domains of rabex-5 reveals two modes of interaction with ubiquitin. Cell 124: 1183–1195 [DOI] [PubMed] [Google Scholar]
- Piper RC, Lehner PJ (2011) Endosomal transport via ubiquitination. Trends Cell Biol 21: 647–655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platta HW, Stenmark H (2011) Endocytosis and signaling. Curr Opin Cell Biol 23: 393–403 [DOI] [PubMed] [Google Scholar]
- Poteryaev D, Datta S, Ackema K, Zerial M, Spang A (2010) Identification of the switch in early-to-late endosome transition. Cell 141: 497–508 [DOI] [PubMed] [Google Scholar]
- Proux-Gillardeaux V, Gavard J, Irinopoulou T, Mege RM, Galli T (2005) Tetanus neurotoxin-mediated cleavage of cellubrevin impairs epithelial cell migration and integrin-dependent cell adhesion. Proc Natl Acad Sci USA 102: 6362–6367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid SL, Fuchs R, Male P, Mellman I (1988) Two distinct subpopulations of endosomes involved in membrane recycling and transport to lysosomes. Cell 52: 73–83 [DOI] [PubMed] [Google Scholar]
- Snyder DA, Kelly ML, Woodbury DJ (2006) SNARE complex regulation by phosphorylation. Cell Biochem Biophys 45: 111–123 [DOI] [PubMed] [Google Scholar]
- Stein A, Weber G, Wahl MC, Jahn R (2009) Helical extension of the neuronal SNARE complex into the membrane. Nature 460: 525–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinman RM, Mellman IS, Muller WA, Cohn ZA (1983) Endocytosis and the recycling of plasma membrane. J Cell Biol 96: 1–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenmark H, Parton RG, Steele-Mortimer O, Lutcke A, Gruenberg J, Zerial M (1994) Inhibition of rab5 GTPase activity stimulates membrane fusion in endocytosis. EMBO J 13: 1287–1296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su LL, Iwai H, Lin JT, Fathman CG (2009) The transmembrane E3 ligase GRAIL ubiquitinates and degrades CD83 on CD4 T cells. J Immunol 183: 438–444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudhof TC, Rothman JE (2009) Membrane fusion: grappling with SNARE and SM proteins. Science 323: 474–477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutton RB, Fasshauer D, Jahn R, Brunger AT (1998) Crystal structure of a SNARE complex involved in synaptic exocytosis at 2.4 A resolution. Nature 395: 347–353 [DOI] [PubMed] [Google Scholar]
- Tayeb MA, Skalski M, Cha MC, Kean MJ, Scaife M, Coppolino MG (2005) Inhibition of SNARE-mediated membrane traffic impairs cell migration. Exp Cell Res 305: 63–73 [DOI] [PubMed] [Google Scholar]
- Thibault ST, Singer MA, Miyazaki WY, Milash B, Dompe NA, Singh CM, Buchholz R, Demsky M, Fawcett R, Francis-Lang HL, Ryner L, Cheung LM, Chong A, Erickson C, Fisher WW, Greer K, Hartouni SR, Howie E, Jakkula L, Joo D et al. (2004) A complementary transposon tool kit for Drosophila melanogaster using P and piggyBac. Nat Genet 36: 283–287 [DOI] [PubMed] [Google Scholar]
- Ullrich O, Horiuchi H, Bucci C, Zerial M (1994) Membrane association of Rab5 mediated by GDP-dissociation inhibitor and accompanied by GDP/GTP exchange. Nature 368: 157–160 [DOI] [PubMed] [Google Scholar]
- Valsdottir R, Hashimoto H, Ashman K, Koda T, Storrie B, Nilsson T (2001) Identification of rabaptin-5, rabex-5, and GM130 as putative effectors of rab33b, a regulator of retrograde traffic between the Golgi apparatus and ER. FEBS Lett 508: 201–209 [DOI] [PubMed] [Google Scholar]
- van Wijk SJ, Fiskin E, Putyrski M, Pampaloni F, Hou J, Wild P, Kensche T, Grecco HE, Bastiaens P, Dikic I (2012) Fluorescence-based sensors to monitor localization and functions of linear and K63-linked ubiquitin chains in cells. Mol Cell 47: 797–809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veale KJ, Offenhauser C, Whittaker SP, Estrella RP, Murray RZ (2010) Recycling endosome membrane incorporation into the leading edge regulates lamellipodia formation and macrophage migration. Traffic 11: 1370–1379 [DOI] [PubMed] [Google Scholar]
- Velichkova M, Juan J, Kadandale P, Jean S, Ribeiro I, Raman V, Stefan C, Kiger AA (2010) Drosophila Mtm and class II PI3K coregulate a PI(3)P pool with cortical and endolysosomal functions. J Cell Biol 190: 407–425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Zastrow M, Sorkin A (2007) Signaling on the endocytic pathway. Curr Opin Cell Biol 19: 436–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu G, Paige JS, Jaffrey SR (2010) Global analysis of lysine ubiquitination by ubiquitin remnant immunoaffinity profiling. Nat Biotechnol 28: 868–873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang HL, Eriksson T, Vernersson E, Vigny M, Hallberg B, Palmer RH (2007) The ligand Jelly Belly (Jeb) activates the Drosophila Alk RTK to drive PC12 cell differentiation, but is unable to activate the mouse ALK RTK. J Exp Zool B Mol Dev Evol 308: 269–282 [DOI] [PubMed] [Google Scholar]
- Zylbersztejn K, Galli T (2011) Vesicular traffic in cell navigation. FEBS J 278: 4497–4505 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.