Abstract
Cell (2013) 152: 620–632 doi:; DOI: 10.1016/j.cell.2013.01.006; published online January 31 2013
Although it is known that replication stress causes genetic instability, the underlying mechanisms are not yet fully understood. A new study by Barlow et al (2013) used an elegant genome-wide chromatin immunoprecipitation approach to reveal that DNA lesions induced by replication stress occur predominantly in early replicating and actively transcribed gene clusters. These ‘early replication fragile sites’ (ERFS) can be the source for rearrangements commonly found in cancer, and represent a new type of fragile site, distinct from common fragile sites (CFS).
Genetic instability is a threat to the integrity of DNA and underlies the genomic rearrangements that need to occur early in preneoplastic lesions in order to cause the genetic changes required for development into neoplastic disease. It has been demonstrated that cellular oncogenes, such as mos, cdc6, cyclin E and ras, not only provide proliferation signals but also induce replication stress associated with the formation of DNA double-strand breaks (DSBs). These DSBs lead to the activation of the ATM–p53 tumour barrier that prevents tumour growth, but can also drive genetic instability in cancer (Bartkova et al, 2006; Di Micco et al, 2006). Until now, it has been unclear how DNA damage arises in the context of replication stress. In a new study published in Cell (Barlow et al, 2013), Nussenzweig and co-workers employed chromatin immunoprecipitation with an anti-RPA antibody for genome-wide identification of ssDNA regions after inducing replication stress, which colocalise with newly replicated regions. This approach allowed mapping DNA regions associated with DNA damage. Interestingly, a high proportion of the damaged DNA regions were found within transcriptionally active gene clusters that replicate early. Furthermore, the same DNA regions were damaged both upon hydoxyurea- and oncogene-induced replication stress, and their colocalisation with other DNA-damage response proteins (i.e., BRCA1 and SMC5) further indicated that they were repaired by homologous recombination.
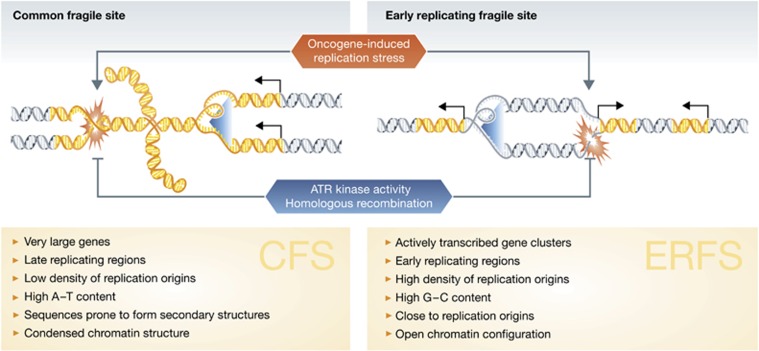
The damaged DNA regions caused by replication stress, termed ‘early replicating fragile sites’ (ERFS), are characterised by their occurrence in early replicating DNA, being close to replication origins, enriched in CpG-rich regions and located in actively transcribed gene clusters (Figure 1). ERFS are therefore clearly distinct from the better studied ‘common fragile sites’ (CFS) CFS occur primarily in late-replicating DNA regions that contain large genes and that exhibit a low density of replication origins in AT-rich sequences, which are prone to form secondary structures (Ozeri-Galai et al, 2012) (Figure 1). Nevertheless, there are also various features shared between CFS and ERFS, such as the stability of both being protected by ATR kinase activity, or the presence of homologous recombination proteins (Barlow et al, 2013). Furthermore, oncogene-induced replication stress triggers instability at both ERFS and CFS (Tsantoulis et al, 2008; Barlow et al, 2013) the underlying reason at CFS could be that the oncogene-induced increase in origin firing results in reduced replication speed (Jones et al, 2012), which should hinder replication completion in those regions with few replication origins or with challenging sequences to replicate. At ERFS, on the other hand, an increase in origin firing near actively transcribed replication clusters would be expected to trigger increased replication–transcription conflicts and thereby contribute to ERFS instability.
Figure 1.
Comparison of CFS with the newly identified ERFS. Oncogene-induced replication stress causes replication fork stalling and collapse at both CFS and ERFS; ATR kinase and homologous recombination prevent collapse and mediate fork restart and repair. Arrows and coloured DNA sequences indicate actively transcribed genes and blue arrowheads denote progressing replication forks.
The concept of impaired transcription as a source of genetic instability derives from extensive studies in S. cerevisae (Aguilera, 2002), showing that it is caused by collisions between DNA replication forks and the RNA polymerase II transcription machinery (Wellinger et al, 2006). Consistent results were also obtained in mammalian cells with reporter constructs to study transcription-associated recombination (Gottipati et al, 2008). The fact that numerous mRNA-processing factors were identified in a screen for suppressors of damage-asscioated γH2AX foci formation (Paulsen et al, 2009) further highlights the importance of removing unprocessed transcripts in order to prevent genetic instability. To what extent active transcription underlies genetic instability had, however, remained somewhat unclear. Our own work recently demonstrated that transcription inhibition can reverse DNA damage caused by oncogene-induced replication stress (Jones et al, 2012). Together with the identification that replication stress causes DNA lesions at actively transcribed genes (Barlow et al, 2013), this allows to safely conclude that collisions between replication and transcription represent a major source of genomic instability.
Both ATR kinase and homologous recombination proteins are required to mediate stability at ERFS (Barlow et al, 2013). Actively transcribed genes were previously found to exhibit an increased rate of homologous recombination (Aguilera, 2002), as a consequence of repair being triggered at replication forks (Wellinger et al, 2006; Gottipati et al, 2008). Increased fragility at ERFS in cells lacking ATR activity or with homologous recombination defects is therefore likely due to the requirement of these pathways for repair and restart of stalled or collapsed replication forks (Petermann and Helleday, 2010).
Altogether, it appears that faithful timing of origin firing is critical for coordinating DNA replication with transcription thus preventing genetic instability. Two other recent landmark papers provide clues on the molecular mechanisms of how replication and transcription are coordinated to prevent collisions in S. cerevisae. Alzu et al (2012) found the Sen1/Senataxin helicase associated with the replication machinery, allowing efficient progression through transcribed genes by removing RNA–DNA structures. Duch et al (2013) showed that the stress-activated protein kinase Hog1 phosphorylates Mrc1, a subunit of the replication complex, to delay Cdc45 origin loading and replication firing. Further work now lies ahead to uncover how conflicts between replication and transcription are avoided in mammalian cells; here, topoisomerase I activity was previously found to be critical for maintaining effective replication fork speed through transcribed regions and for preventing formation of RNA–DNA hybrids (Tuduri et al, 2009). Further elucidating the intrinsic process that coordinates replication origin firing and transcription to prevent genetic instability is of high interest, and such knowledge is essential to fully understand the impact of oncogene-induced transcription and deregulated origin firing in the context of genetic instability in cancer.
Footnotes
The authors declare that they have no conflict of interest.
References
- Aguilera A (2002) The connection between transcription and genomic instability. Embo J 21: 195–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alzu A, Bermejo R, Begnis M, Lucca C, Piccini D, Carotenuto W, Saponaro M, Brambati A, Cocito A, Foiani M, Liberi G (2012) Senataxin associates with replication forks to protect fork integrity across RNA-polymerase-II-transcribed genes. Cell 151: 835–846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlow J, Faryabi RB, Callen E, Wong N, Malhowski A, Tang Chen H, Gutierez-Cruz G, Sun H-W, McKinnon P, Wright G, Casellas R, Robbiani DF, Staudt L, Fernandez-Capetillo O, Nussenzweig A (2013) Identification of early replicating fragile sites that contribute to genome instability. Cell 152: 620–632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartkova J, Rezaei N, Liontos M, Karakaidos P, Kletsas D, Issaeva N, Vassiliou LV, Kolettas E, Niforou K, Zoumpourlis VC, Takaoka M, Nakagawa H, Tort F, Fugger K, Johansson F, Sehested M, Andersen CL, Dyrskjot L, Orntoft T, Lukas J et al. (2006) Oncogene-induced senescence is part of the tumorigenesis barrier imposed by DNA damage checkpoints. Nature 444: 633–637 [DOI] [PubMed] [Google Scholar]
- Di Micco R, Fumagalli M, Cicalese A, Piccinin S, Gasparini P, Luise C, Schurra C, Garre M, Nuciforo PG, Bensimon A, Maestro R, Pelicci PG, d’Adda di Fagagna F (2006) Oncogene-induced senescence is a DNA damage response triggered by DNA hyper-replication. Nature 444: 638–642 [DOI] [PubMed] [Google Scholar]
- Duch A, Felipe-Abrio I, Barroso S, Yaakov G, Garcia-Rubio M, Aguilera A, de Nadal E, Posas F (2013) Coordinated control of replication and transcription by a SAPK protects genomic integrity. Nature 493: 116–119 [DOI] [PubMed] [Google Scholar]
- Gottipati P, Cassel TN, Savolainen L, Helleday T (2008) Transcription-associated recombination is dependent on replication in mammalian cells. Mol Cell Biol 28: 154–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones RM, Mortusewicz O, Afzal I, Lorvellec M, García P, Helleday T, Petermann E (2012) Increased replication initiation and conflicts with transcription underlie Cyclin E-induced replication stress. Oncogene (advance online publication 3 September 2012; doi: 10.1038/onc.2012.387) [DOI] [PubMed] [Google Scholar]
- Ozeri-Galai E, Bester AC, Kerem B (2012) The complex basis underlying common fragile site instability in cancer. Trends Genet 28: 295–302 [DOI] [PubMed] [Google Scholar]
- Paulsen RD, Soni DV, Wollman R, Hahn AT, Yee MC, Guan A, Hesley JA, Miller SC, Cromwell EF, Solow-Cordero DE, Meyer T, Cimprich KA (2009) A genome-wide siRNA screen reveals diverse cellular processes and pathways that mediate genome stability. Mol Cell 35: 228–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petermann E, Helleday T (2010) Pathways of mammalian replication fork restart. Nat Rev Mol Cell Biol 11: 683–687 [DOI] [PubMed] [Google Scholar]
- Tsantoulis PK, Kotsinas A, Sfikakis PP, Evangelou K, Sideridou M, Levy B, Mo L, Kittas C, Wu XR, Papavassiliou AG, Gorgoulis VG (2008) Oncogene-induced replication stress preferentially targets common fragile sites in preneoplastic lesions. A genome-wide study. Oncogene 27: 3256–3264 [DOI] [PubMed] [Google Scholar]
- Tuduri S, Crabbe L, Conti C, Tourriere H, Holtgreve-Grez H, Jauch A, Pantesco V, De Vos J, Thomas A, Theillet C, Pommier Y, Tazi J, Coquelle A, Pasero P (2009) Topoisomerase I suppresses genomic instability by preventing interference between replication and transcription. Nat Cell Biol 11: 1315–1324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellinger RE, Prado F, Aguilera A (2006) Replication fork progression is impaired by transcription in hyperrecombinant yeast cells lacking a functional THO complex. Mol Cell Biol 26: 3327–3334 [DOI] [PMC free article] [PubMed] [Google Scholar]