Abstract
Formation of cilia, microtubule-based structures that function in propulsion and sensation, requires Kif3a, a subunit of Kinesin II essential for intraflagellar transport (IFT). We have found that, Kif3a is also required to organize centrioles. In the absence of Kif3a, the subdistal appendages of centrioles are disorganized and lack p150Glued and Ninein. Consequently, microtubule anchoring, centriole cohesion and basal foot formation are abrogated by loss of Kif3a. Kif3a localizes to the mother centriole and interacts with the Dynactin subunit p150Glued. Depletion of p150Glued phenocopies the effects of loss of Kif3a, indicating that Kif3a recruitment of p150Glued is critical for subdistal appendage formation. The transport functions of Kif3a are dispensable for subdistal appendage organization as mutant forms of Kif3a lacking motor activity or the motor domain can restore p150Glued localization. Comparison to cells lacking Ift88 reveals that the centriolar functions of Kif3a are independent of IFT. Thus, in addition to its ciliogenic roles, Kif3a recruits p150Glued to the subdistal appendages of mother centrioles, critical for centrosomes to function as microtubule-organizing centres.
Keywords: centriole cohesion, centrosome, Kif3a, p150Glued, subdistal appendage
Introduction
The centrosome is the major microtubule-organizing centre of most eukaryotic cells, and contains mother and daughter centrioles (Bobinnec et al, 1998). The mother centriole is distinguished from the daughter by accessory structures called the subdistal and distal appendages (Paintrand et al, 1992).
An electron dense cloud of material called the pericentriolar matrix (PCM) surrounds the centriole pair (Bornens, 2002; Takahashi et al, 2002). The PCM provides binding sites for the γ-tubulin ring complex that templates microtubule nucleation at the centrosome (Moritz et al, 2000). Subsequently, these microtubules become preferentially anchored to the subdistal appendages of mother centrioles, augmenting the ability of the centrosome to act as a microtubule-organizing centre (Mogensen et al, 2000; Piel et al, 2000; Chang et al, 2003; Delgehyr et al, 2005).
In addition to its role in anchoring microtubules, the subdistal appendage is required for ciliogenesis. The mother centriole is the foundation upon which the primary cilium is built, at which time it becomes known as the basal body (Ishikawa et al, 2005; Graser et al, 2007). Upon docking of the basal body at the cellular surface, intraflagellar transport (IFT) facilitates the transport of ciliary proteins from the cytoplasm to the cilium (Ishikawa and Marshall, 2011). The anterograde motor for IFT is Kinesin II, which delivers ciliary components to the tip where the axoneme is assembled (Rosenbaum and Witman, 2002; Pedersen et al, 2006). Consequently, disruption of the Kinesin II motor heterodimeric subunits Kif3a or Kif3b blocks mammalian ciliogenesis (Nonaka et al, 1998; Marszalek et al, 1999; Takeda et al, 1999). Kif3a participates in cellular processes apart from IFT, such as immune synapse formation, but its precise role in non-ciliary processes has been unclear (Sfakianos et al, 2007; Corbit et al, 2008; Finetti et al, 2009).
During ciliogenesis, the subdistal appendage of the basal body forms a morphologically distinct structure, the basal foot, upon which cytoplasmic microtubules are anchored (Anderson, 1972). Basal feet project orthogonally from the basal body and, when associated with non-motile cilia, are randomly orientated (Boisvieux-Ulrich and Sandoz, 1991). In contrast, basal feet of motile cilia are oriented in the direction of fluid flow and align the ciliary beat in the direction of the pulling force (Holley and Afzelius, 1986; Mitchell et al, 2007; Kunimoto et al, 2012). Consequently, ciliary basal feet are essential for planarly polarizing the ciliary stroke direction, defects in which can cause primary ciliary dyskinesia (Rayner et al, 1996; Mitchell et al, 2009; Mirzadeh et al, 2010; Kunimoto et al, 2012).
We have found that Kif3a specifically associates with the mother centriole and organizes subdistal appendages. Kif3a interacts with the Dynactin component p150Glued, which facilitates bidirectional transport along microtubules and, in the absence of Kif3a, p150Glued fails to localize to the subdistal appendage, disrupting microtubule anchoring and basal foot formation. These findings indicate that Kif3a is not only required for IFT within cilia, but also for centriole organization and function.
Results
Kif3a associates specifically with the mother centriole
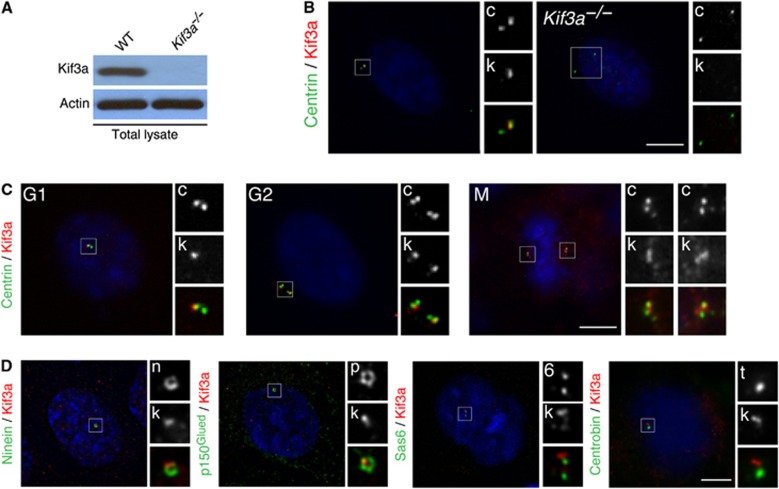
We examined the subcellular localization of Kif3a in mouse embryonic fibroblasts (MEFs) with an antibody against Kif3a. Immunoblotting and immunofluorescence showed a loss of Kif3a in Kif3a−/− lysate and Kif3a−/− cells, confirming antibody specificity (Figure 1A and B). The antibody recognized a prominent focus at centrosomes of wild-type (WT) cells, as previously reported (Haraguchi et al, 2006). In G1 cells, Kif3a was present at one of the paired centrioles containing the centriolar distal lumen protein, Centrin (Figure 1B). Localization of Kif3a to a single centriole was maintained during G2 and mitosis (Figure 1C).
Figure 1.
Kif3a localizes to the subdistal appendage of mother centrioles. (A) Total cell lysates of WT or Kif3a−/− MEFs were analysed by immunoblotting for Kif3a. Actin served as a loading control. 40 μg of protein lysate was loaded per lane. (B) Immunofluorescence microscopy analysis of WT or Kif3a−/− cells co-stained for Centrin (‘c’, green) and Kif3a (‘k’, red). The inset shows magnified images of the boxed region. (C) The cell-cycle-dependent localization of Kif3a was determined by examining the localization of Centrin (‘c’, green) and Kif3a (‘k’, red) in WT MEFs. Centrin and DNA organization were used to determine the cell-cycle stage. (D) Deconvolved immunofluorescence images of WT cells co-stained for Kif3a (‘k’, red), Ninein (‘n’, green) and p150Glued (‘p’, green) to mark the subdistal appendage, Sas6 (‘s’, green) to mark the proximal ends of centrioles and Centrobin (‘t’, green) to mark the daughter centriole. Scale bars indicate 5 μm for images in (B, C), 3 μm for images in (D).
Deconvolution microscopy revealed that at the mother centriole, Kif3a localizes to a single focus (Figure 1D). This focus of Kif3a is at the same point along the proximal–distal axis of the centriole as the ring of subdistal appendages marked by Ninein and p150Glued, two subdistal appendage components (Figure 1D). The localization of Kif3a is similar to ODF2, another mother centriolar protein that localizes to a single centriolar focus close to the subdistal appendages (Nakagawa et al, 2001; Kunimoto et al, 2012). Kif3a did not colocalize with the proximal centriolar component Sas6 or the daughter centriolar protein Centrobin, confirming that it is predominantly near the distal end of the mother centriole (Figure 1D).
Similar to MEFs, Kif3a localized to a domain close to the subdistal appendages of mother centrioles in HeLa and U2OS cells, non-ciliated human cells, revealed by comparing its distribution with p150Glued and another subdistal appendage component, Cep170 (Guarguaglini et al, 2005; Supplementary Figure 1a). To further confirm that Kif3a is a centrosomal component, we isolated centrosomes from Jurkat cells and found that Kif3a co-fractionated with γ-tubulin and CP110 (Supplementary Figure 1b). Thus, Kif3a is a centrosomal component that localizes to a focus close to the subdistal appendages of mother centrioles throughout the cell cycle.
Kif3a is required for centriole cohesion and organization
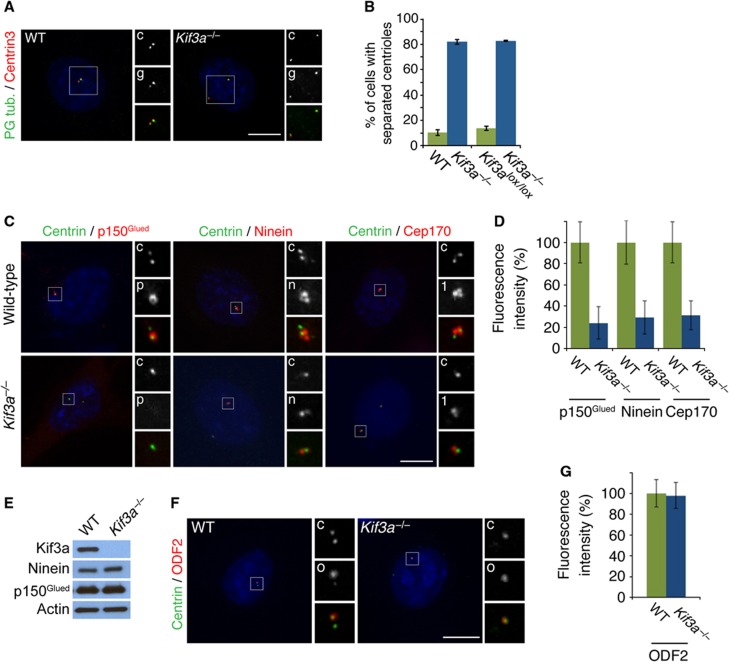
Given its localization at the mother centriole, we explored how Kif3a participates in centrosome organization. In mammalian cells, the mother and daughter centriole remain tightly associated until late G1, when centriole cohesion is broken (Wong and Stearns, 2003). Immunofluorescence analysis of the centriolar components Centrin3 and polyglutamylated tubulin revealed that, without Kif3a, centriole cohesion is broken prematurely during G1 (Figure 2A). Greater than 80% of Kif3a mutant cells displayed separated centrioles in G1, as compared with <15% of WT and conditional Kif3a cells (Figure 2B). We also observed premature centriole separation in MEFs derived from embryonic day 12.5 Kif3aflox/flox embryos and infected with Cre-recombinase to delete Kif3a (Corbit et al, 2008; Figure 2B).
Figure 2.
Kif3a is essential for centriole cohesion and subdistal appendage organization. (A) Immunofluorescence images of WT and Kif3a−/− cells in G1 co-stained for Centrin (red, ‘c’) and polyglutamylated tubulin (‘g’, green) to visualize centrioles. (B) Percentage of split centrioles in G1 for WT, Kif3a−/−, Kif3alox/lox and Kif3a−/− cells generated by Cre-mediated deletion of Kif3a. (C) WT and Kif3a−/− cells in G1 stained for Centrin (‘c’, green), p150Glued (‘p’, red), Ninein (‘n’, red) and Cep170 (‘1’, red), to assess centriole and subdistal appendage organization. (D) Quantification of the mean fluorescence intensities±s.d. of p150Glued, Ninein and Cep170 in WT and Kif3a−/− cells expressed as the mean percentage±s.d. of the fluorescence intensities in WT cells. (E) Total cell lysates of WT or Kif3a−/− MEFs were analysed by immunoblotting for Kif3a, Ninein and p150Glued. Actin served as a loading control. 25 μg of protein lysate was loaded per lane. (F) Immunofluorescence images of WT and Kif3a−/− cells co-stained for Centrin (‘c’, green) and ODF2 (‘o’, red). (G) Quantification of the mean fluorescence intensity percentage±s.d. of ODF2 in WT and Kif3a−/− MEFs. Scale bars indicate 5 μm for all images. For all quantifications at least 100 cells were analysed per experiment (n=3), P<0.001 (paired t-test).
As Kif3a localized to a centriolar region overlapping with the subdistal appendages, we investigated whether loss of Kif3a affects subdistal appendage organization by examining the localization of Centrin and the subdistal appendage proteins p150Glued, Ninein, and Cep170 in WT and Kif3a mutant cells in G1 (Ou et al, 2002; Louie et al, 2004; Guarguaglini et al, 2005). Interestingly, p150Glued, which forms a ring near the subdistal centriole of WT cells, was absent from Kif3a−/− centrioles (24% of WT fluorescence, Figure 2C). Similarly, Ninein was reduced on Kif3a−/− centrioles (31% of WT fluorescence, Figure 2C). On WT mother centrioles, Ninein localized to three foci one of which is proximal (Ishikawa et al, 2005). Kif3a−/− centrioles showed only one strong focus. Cep170, which depends on Ninein to localize to the subdistal appendage was also reduced at the mother centrioles of cells lacking Kif3a (Graser et al, 2007, 29% of WT fluorescence). Loss of Kif3a has no effect on the protein levels of p150Glued or Ninein (Figure 2E), indicating that Kif3a participates in the organization of the subdistal appendages.
Previous studies have shown that another mother centriole component, ODF2, is required for the recruitment of Ninein to the centrosome and the organization of subdistal appendages (Nakagawa et al, 2001; Kunimoto et al, 2012). To assess whether the disruption of the subdistal appendages in Kif3a mutants resulted from mislocalization of ODF2, we examined ODF2 in Kif3a−/− MEFs (Figure 2F). Loss of Kif3a had no effect on the localization of ODF2 to the centrosome (Figure 2G).
To test whether the role of Kif3a in centriole cohesion and subdistal appendage formation are secondary to the established role of Kif3a in IFT, we examined cells that lack Ift88, an IFT protein essential for ciliogenesis (Haycraft et al, 2007; Robert et al, 2007). In contrast to Kif3a−/− MEFs, subdistal appendage organization and centriole cohesion were unaffected in Ift88 mutants (Supplementary Figure 2). To examine whether Kif3a or Ift88 depend on each other for localization to the mother centriole, we examined the localization of Centrin and either Kif3a or Ift88 in WT, Kif3a mutant cells and Ift88 mutant cells. The loss of either Kif3a or Ift88 had no effect on the localization of the other protein to centrioles (Supplementary Figure 3), indicating that Kif3a maintains centriole cohesion and organizes subdistal appendages independent of IFT.
p150Glued is also required for centriole cohesion and organization
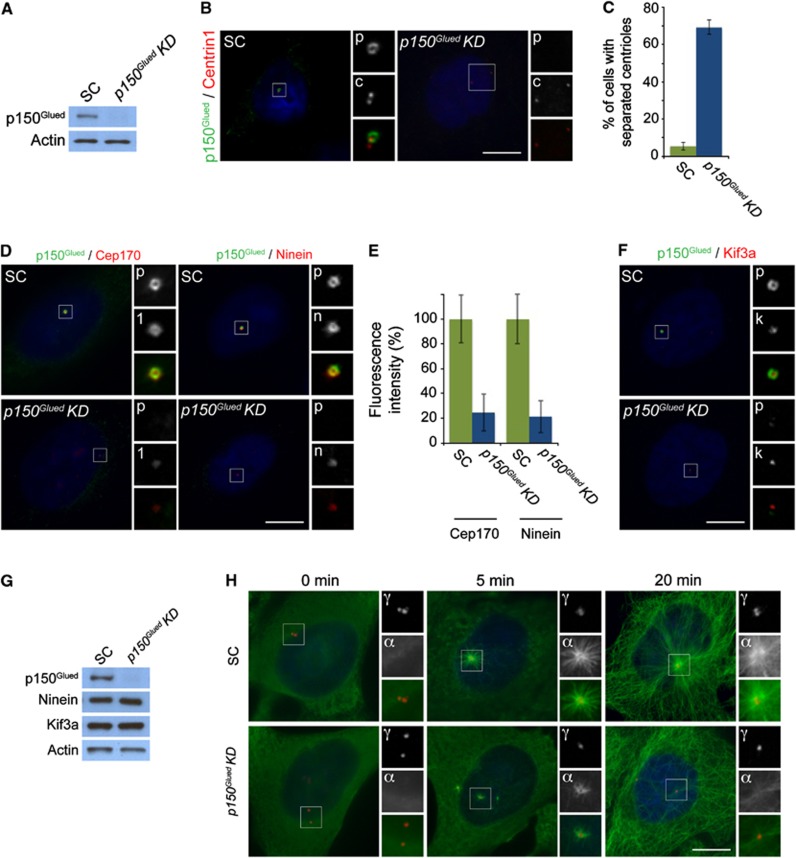
As Kif3a is critical to localize p150Glued to centrosomes, we investigated whether the loss of p150Glued also alters centriole cohesion and subdistal appendage organization. Depletion of p150Glued by siRNA in HeLa cells and examination of Centrin1 revealed that 69% of cells exhibited premature breakage of centriole cohesion during G1 (Figure 3A–C). This premature loss of centriole cohesion is similar to that caused by displacing Dynactin from centrosomes (Quintyne and Schroer, 2002). Together, these findings reveal that Kif3a recruitment of p150Glued to centrioles is essential for the maintenance of cohesion.
Figure 3.
p150Glued is required for centriole cohesion and subdistal appendage organization and function. (A) Total cell lysates of scramble control (SC) or p150Glued siRNA transfected HeLa cells were analysed by immunoblotting for p150Glued. Actin served as a loading control. 25 μg of protein lysate was loaded per lane. (B) HeLa cells in G1 transfected with SC or p150Glued siRNA were co-stained for p150Glued (‘p’, green) and Centrin (‘c’, red) to visualize centrioles. (C) Mean percentage ±s.d. of split centrioles in G1 for SC and p150Glued transfected cells. (D) The depletion of p150Glued (‘p’, green) disrupted the localization of the subdistal appendage proteins Cep170 (‘1’, red) and Ninein (‘n’, red). (E) Quantification of the mean fluorescence intensities±s.d. of Cep170 and Ninein in SC and p150Glued siRNA-treated cells. (F) Immunofluorescence images of SC and p150Glued siRNA-treated HeLa cells co-stained for p150Glued (‘p’, green) and Kif3a (‘k’, red). (G) Total cell lysates of SC or p150GluedsiRNA transfected HeLa cells were analysed by immunoblotting with antibodies to p150Glued, Ninein and Kif3a. Actin served as a loading control. 25 μg of protein lysate was loaded per lane. (H) SC and p150Glued siRNA transfected HeLa cells were subjected to a microtubule regrowth assay, fixed at the indicated time points and stained with antibodies to α-tubulin to visualize microtubules (‘α’, green) and γ-tubulin to mark centrosomes (‘γ’, red). Scale bars indicate 5 μm for all images. For all quantifications at least 100 cells were analysed per experiment (n=3), P<0.001 (paired t-test).
We similarly examined whether loss of p150Glued could account for the subdistal appendage disorganization caused by loss of Kif3a. As in cells lacking Kif3a, p150Glued-depleted centrioles displayed reduced localization of the subdistal appendage proteins Cep170 and Ninein (Figure 3D). The fluorescence intensity of Cep170 and Ninein were decreased by >75% on p150Glued-depleted centrioles (Figure 3E). These findings are similar to disrupting dynein and dynactin function by overexpressing p150Glued, suggesting that the localization of Cep170 and Ninein is dynactin-dependent (Casenghi et al, 2005).
To assess whether the localization of Kif3a to the mother centriole reciprocally depended on p150Glued, we examined the localization of Kif3a after depleting p150Glued. Interestingly, the loss of p150Glued had no effect on the localization of Kif3a to centrosomes (Figure 3F). Thus, Kif3a is required for the localization of p150Glued to the mother centriole, but not vice versa. We confirmed that protein levels of Kif3a and Ninein in p150Glued-depleted cells were unchanged by immunoblot analysis (Figure 3G).
As p150Glued is required for subdistal appendage organization, we assessed whether depletion of p150Glued affected the anchoring of microtubules at the centrosome. We analysed HeLa cells transfected with scrambled or p150Glued siRNA for defects in microtubule nucleation (5-min regrowth) or in microtubule anchoring (20-min regrowth). Five minutes following the removal of nocodozale, control cells nucleated a strong aster of microtubules radiating from the centrosome (Figure 3H). In contrast, the centrosomes of p150Glued-depleted cells formed weak asters at the same time point (Figure 3H). Twenty minutes after nocodozale washout, cytoplasmic microtubules had regrown in control and p150Glued-depleted cells. In contrast to control cells, p150Glued-depleted cells had few detectable microtubules anchored at the centrosome (Figure 3H). Together, these findings demonstrate a requirement for p150Glued in organizing of the subdistal appendage and suggesting that the mislocalization of p150Glued and Ninein accounts for the loss of microtubule anchoring at the mother centriole.
p150Glued interacts with the carboxy-terminal region of Kif3a
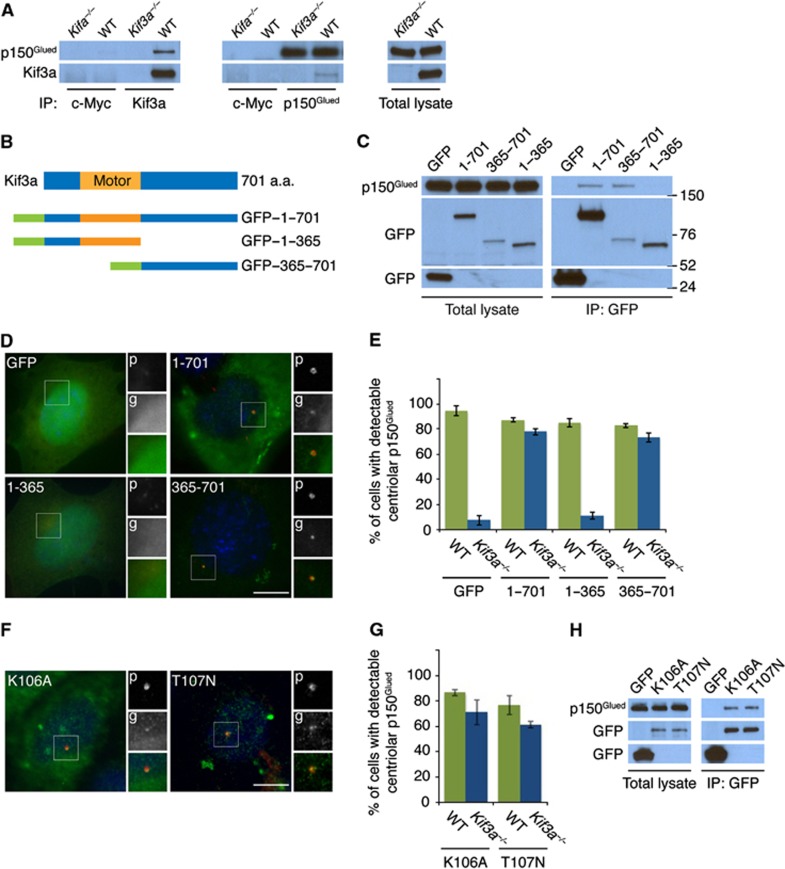
To understand why Kif3a is required to localize p150Glued to the subdistal appendage and prompted by the ability of Xenopus Kinesin II to interact with p150Glued (Deacon et al, 2003), we examined whether murine Kif3a and p150Glued interact. Endogenous p150Glued coimmunoprecipitated endogenous Kif3a, revealing that the two proteins interact biochemically (Figure 4A). Reciprocal immunoprecipitation of p150Glued confirmed the specificity of the interaction (Figure 4A).
Figure 4.
Kif3a is required to localize p150Glued to the subdistal appendage. (A) WT and Kif3a−/− total cell lysates were subjected to immunoprecipitation of Kif3a, p150Glued and c-Myc, which served as a negative control. Precipitating proteins were subjected to immunoblotting for Kif3a and p150Glued. (B) Schematic representations of the motor domain and the truncation mutants of GFP-tagged Kif3a. (C) We expressed the indicated GFP-tagged versions of Kif3a in HeLa cells and immunoprecipitated GFP. Endogenous p150Glued and each of the Kif3a fusion proteins were detected by immunoblotting for p150Glued and GFP, respectively. (D) Expression of the full-length GFP-tagged Kif3a and the truncated mutant that interacted with p150Glued (‘g’, green) restored the localization of p150Glued (‘p’, red) to the subdistal appendage in Kif3a−/− cells. (E) Quantification of p150Glued localization to the subdistal appendage. (F) Expression of the motor dead GFP-tagged Kif3a (K106A and T107N) mutants ('g', green) restored the localization of p150Glued (‘p’, red) to the subdistal appendage of Kif3a−/− MEFs. (G) Quantification of p150Glued localization to the subdistal appendage in Kif3a mutants. (H) We immunoprecipitated the GFP tag of the motor dead Kif3a forms and detected endogenous p150Glued and GFP. At least 100 cells were analysed per experiment (n=3), P<0.001 (paired t-test). Scale bars indicate 5 μm for all images.
Source data for this figure is available on the online supplementary information page.
To gain insight into the functional significance of the Kif3a–p150Glued interaction, we investigated whether the Kif3a motor or cargo-binding domain binds p150Glued. We transfected full-length and truncated versions of Kif3a fused to GFP, immunoprecipitated the exogenous Kif3a using an antibody to GFP, and assessed whether endogenous p150Glued co-precipitated (Figure 4B and C). We detected no interaction between p150Glued and either GFP alone or fused to the amino-terminal motor domain (residues 1–365) of Kif3a. However, the carboxy-terminal cargo-binding 347 amino acids of Kif3a (residues 365–701) were necessary and sufficient for its interaction with p150Glued (Figure 4C).
Given that the motor domain and carboxy-terminal domains of Kif3a were differentially required to interact with p150Glued, we assessed whether the Kif3a deletion mutants could localize to the centriole in Kif3a−/− MEFs. As expected, the full-length Kif3a fused to GFP localized to the centriole, similar to endogenous Kif3a (Figure 4D). Like full-length Kif3a, the carboxy-terminal half of Kif3a localized to the centriole, whereas GFP and the amino-terminal half of Kif3a did not. Thus, the p150Glued interaction and subdistal appendage localization of Kif3a is dependent upon its carboxy-terminal domain, but not its motor domain.
We assessed whether the Kif3a deletion mutants could restore the localization of p150Glued to the subdistal appendage of mother centrioles in Kif3a−/− MEFs. The localization of p150Glued to subdistal appendages was not affected by the expression of the truncated forms of Kif3a in WT MEFs (data not shown). Kif3a−/− MEFs transfected with GFP or the amino-terminal motor domain of Kif3a (residues 1–365) did not restore p150Glued localization to the subdistal appendage. However, expression of the carboxy-terminal domain of Kif3a restored p150Glued localization to the subdistal appendage in Kif3a mutant cells as efficiently as full-length Kif3a (Figure 4D and E), suggesting that p150Glued is localized to the subdistal appendage through its interaction with the Kif3a cargo-binding domain and in a manner independent of its motor domain.
To confirm that the motor activity of Kif3a is not required to localize p150Glued to subdistal appendages, we tested the ability of motor dead forms of Kif3a to restore p150Glued localization in Kif3a mutant cells (Wiesner et al, 2010). Kinesins typically contain an amino-terminal P-loop sequence that binds ATP (Sack et al, 1999). Therefore, we expressed motor dead Kif3a containing mutations within its P-loop (lysine 106 and threonine 107 substituted with alanine and asparagine, respectively) in Kif3a−/− MEFs and assessed p150Glued localization (Wiesner et al, 2010). Similar to full-length and the cargo-binding domain, both motor dead forms of Kif3a localized to the centrosome and restored localization of p150Glued to the subdistal appendage (Figure 4F and G). As the motor dead Kif3a could restore p150Glued localization, we assessed whether these mutant constructs could interact with p150Glued. As expected, the motor dead forms of Kif3a interacted with endogenous p150Glued (Figure 4H). These data indicate that Kif3a localizes to the centrosome and brings p150Glued to the subdistal appendages independent of its motor activity.
Kif3a is required for microtubule organization and anchoring
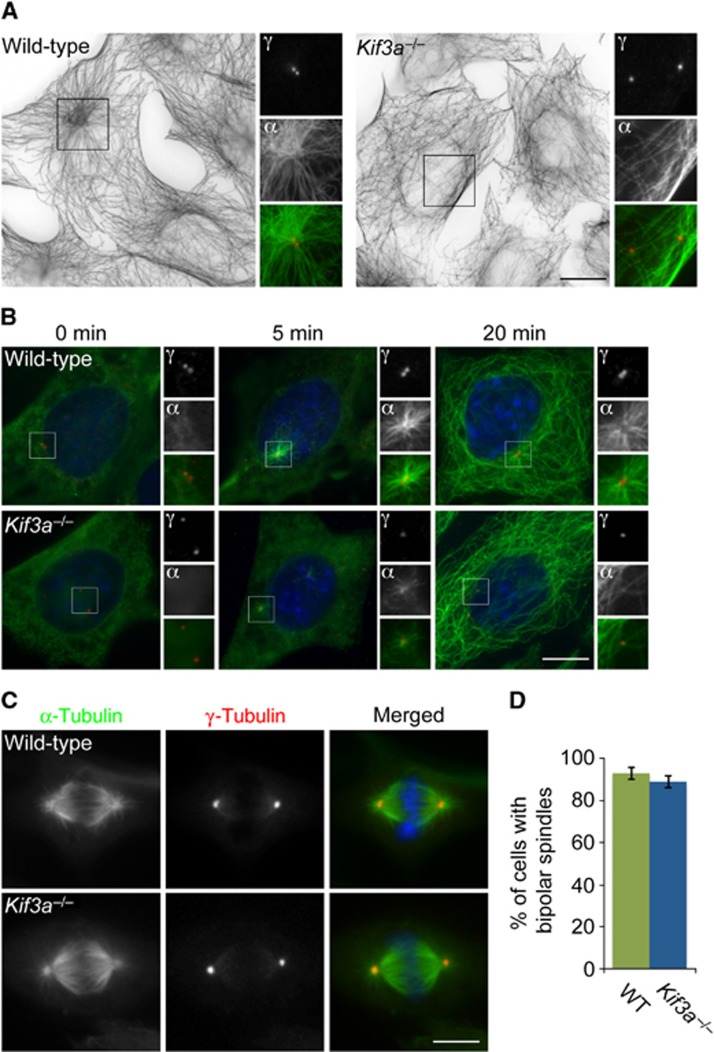
The Dynactin component p150Glued is required for microtubule anchoring at the mother centriole (Quintyne and Schroer, 2002). As Kif3a is required for the localization of p150Glued to the subdistal appendage, we examined whether the anchoring of centrosomal microtubules also depends upon Kif3a. We analysed the microtubule cytoskeleton and PCM of WT and Kif3a−/− MEFs by examining the organization of α-tubulin and γ-tubulin. WT cells displayed radially arranged microtubules, the majority of which originated at the centrosome (Figure 5A, left panel). In contrast, the microtubules of Kif3a mutant cells did not originate from the centrosome and were chaotically oriented (Figure 5A, right panel).
Figure 5.
Kif3a is essential for microtubule anchoring at the mother centriole. (A) Immunofluorescence analysis of WT or Kif3a−/− MEFs co-stained for α-tubulin (‘α’, green) and γ-tubulin (‘γ’, red). Large figures are inverted images of α-tubulin staining. The inset shows magnified images of the boxed region. (B) WT and Kif3a−/− MEFs were subjected to a microtubule regrowth assay, fixed at the indicated time points and stained with antibodies to α-tubulin to visualize microtubules (‘α’, green) and γ-tubulin to mark centrosomes (‘γ’, red). (C) Mitotic WT and Kif3a−/− MEFs were stained for α-tubulin and γ-tubulin to visualize mitotic spindles and poles, respectively. (D) Percentage of WT and Kif3a−/− cells with bipolar mitotic spindles. At least 100 cells were analysed per experiment (n=3). Scale bars indicate 5 μm for images in a, 3 μm for images in (B).
In another test of microtubule anchoring, we assessed whether Kif3a mutant cells could anchor microtubules at the mother centriole following microtubule depolymerization. Five minutes following removal of nocodazole, WT cells radiated a strong microtubule aster from the centrosome (Figure 5B). In contrast, Kif3a mutant cells only supported weak asters at the same time point. Twenty minutes after nocodazole removal, microtubules had similarly regrown in WT and Kif3a−/− cells. However, in contrast to the centrosomally anchored microtubules observed in WT cells, the microtubules of Kif3a mutant cells were not anchored at centrosomes (Figure 5B). The defect in microtubule organization is similar to that caused by p150Glued depletion, suggesting that Kif3a localization of p150Glued and Ninein to the subdistal appendages supports microtubule assembly and anchoring at the centrosome.
Because previous studies suggested that Kinesin II function was required for chromosomal stability and bipolar spindle formation (Haraguchi et al, 2006), we examined whether Kif3a−/− MEFs undergo abnormal mitoses. Visualization of spindle microtubules and poles revealed that WT and Kif3a mutants both form bipolar spindles that properly align chromosomes at the metaphase plate (Figure 5C). Quantification indicated that there was no change in the number of abnormal spindles in WT and Kif3a−/− cells (Figure 5D).
Ninein is required for centriole organization and microtubule anchoring
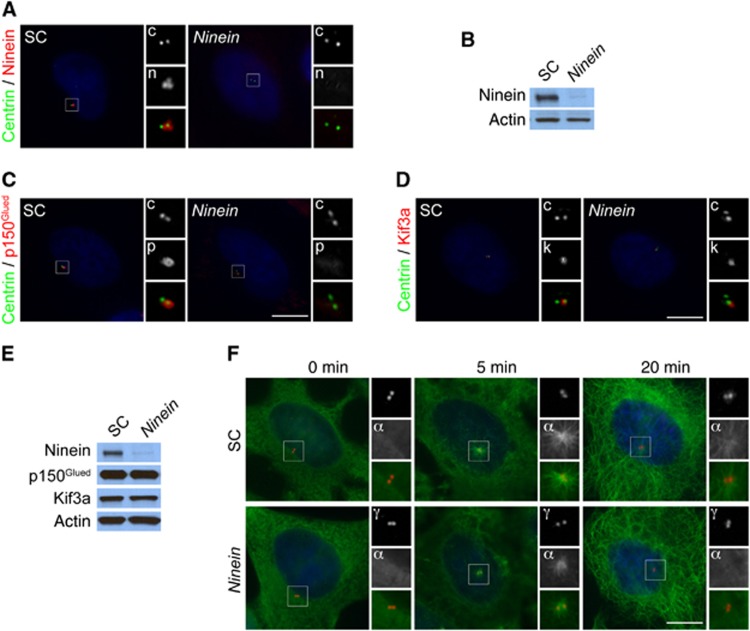
As Kif3a and p150Glued are required for the localization of Ninein to the subdistal appendage and for centriole cohesion, we examined whether centriole attachment depends upon Ninein. Depletion of Ninein by siRNA in HeLa cells and examination of centrioles revealed that centriole cohesion remained intact (Figure 6A and B), indicating that Kif3a and p150Glued maintain centriole cohesion independent of Ninein.
Figure 6.
Ninein is required for subdistal appendage organization and microtubule anchoring. (A) Immunofluorescence microscopy analysis of SC or Ninein siRNA transfected HeLa cells co-stained for Ninein (‘n’, red) and Centrin (‘c’, green) to visualize centrioles. (B) Total cell lysates of scramble control (SC) or Ninein siRNA transfected HeLa cells were analysed by immunoblotting for Ninein. Actin served as a loading control. 25 μg of protein lysate was loaded per lane. (C) The depletion of Ninein disrupted the localization of the subdistal appendage protein p150Glued (‘p’, red). Cells were stained for Centrin (‘c’, green) to visualize centrioles in G1. (D) Immunofluorescence images of SC and Ninein siRNA-treated HeLa cells co-stained for Centrin (‘c’, green) and Kif3a (‘k’, red). (E) Total cell lysates of SC or Ninein-depleted cells were analysed by immunoblotting with antibodies to Ninein, p150Glued and Kif3a. Actin served as a loading control. 25 μg of protein lysate was loaded per lane. (F) SC and Ninein siRNA transfected HeLa cells were subjected to a microtubule regrowth assay, fixed at the indicated time points and stained with antibodies to α-tubulin to visualize microtubules (‘α’, green) and γ-tubulin to mark centrosomes (‘γ’, red). Scale bars indicate 5 μm for all images.
To assess whether the localization of p150Glued and Kif3a to the mother centriole reciprocally depend on Ninein, we examined the localization of these proteins after depleting Ninein. Interestingly, p150Glued was absent from centrosomes in Ninein-depleted cells, suggesting that Ninein and p150Glued require each other to localize to the subdistal appendage (Figure 6C). In contrast, the centrosome localization of Kif3a was unaltered in Ninein-depleted cells (Figure 6D). We confirmed that protein levels of Kif3a and p150Glued in Ninein-depleted cells were unchanged by immunoblot analysis (Figure 6E). Thus, Kif3a is required for the localization of p150Glued and Ninein to the mother centriole, but not vice versa.
Given the mislocalization of Ninein in Kif3a mutants and p150Glued-depleted cells, we examined whether the siRNA depletion of Ninein was sufficient to evoke a loss of microtubule anchoring at the centrosome. We assessed whether microtubule anchoring is abrogated in Ninein-depleted cells by performing a microtubule regrowth assay. Similar to the loss of Kif3a and p150Glued, Ninein-depleted cells nucleated weak microtubule asters 5 min after nocodazole removal and failed to anchor microtubules at the centrosome after 20 min (Figure 6F). Taken together, our data indicate that Kif3a recruits p150Glued to the centrosome. In turn, p150Glued recruits Ninein to the subdistal appendage to facilitate microtubule anchoring at the mother centriole.
Kif3a is required for the formation of ciliary basal feet
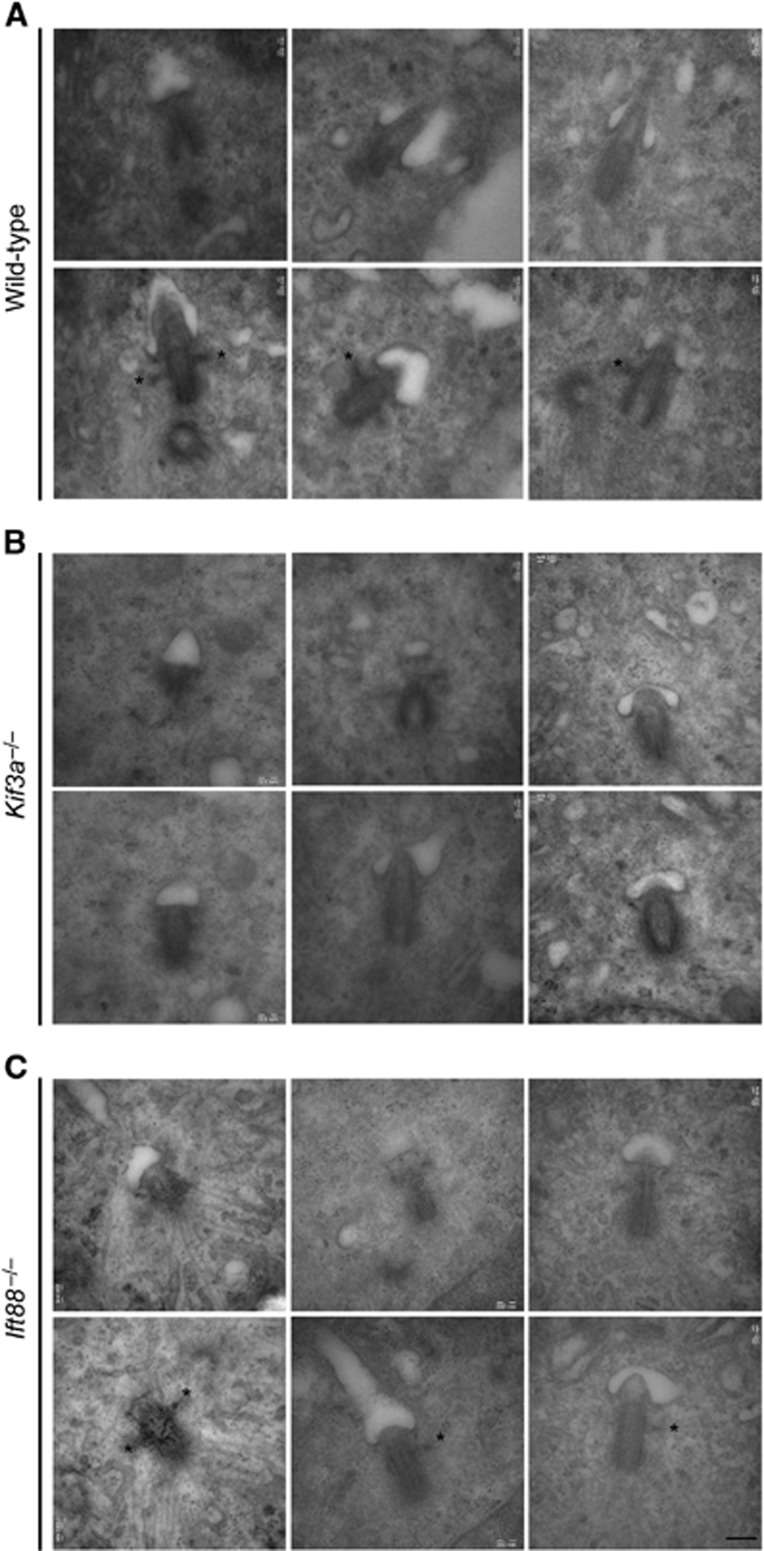
The relationship of subdistal appendages to basal feet is unclear, but they form at similar positions along the proximal–distal length of the centriole and are comprised of some of the same proteins, suggesting that they have similar etiologies (Anderson, 1972). We examined centrosomes in over 50 serum deprived WT, Kif3a−/− and Ift88−/− MEFs by serial, ultrathin section electron microscopy. WT cells formed primary cilia with associated basal bodies bearing one or two basal feet (Figure 7A, top panel). Although basal bodies docked at the plasma membrane in Kif3a−/− cells, these basal bodies lacked both primary cilia and basal feet (Figure 7B, middle panel). Like Kif3a−/− cells, Ift88−/− cells lack primary cilia and basal bodies dock to the plasma membrane, but unlike Kif3a−/− cells, Ift88−/− cells have basal bodies with intact subdistal appendages (Pazour et al, 2000; Supplementary Figure 2A). Interestingly, basal bodies in Ift88−/− cells also form basal feet (Figure 7C, bottom panel). These findings indicate that, as with centriole cohesion and subdistal appendage formation, Kif3a has an indispensable role in basal foot formation that is independent of IFT and ciliogenesis.
Figure 7.
Kif3a is required for ciliary basal foot formation. Serial ultrathin sections electron micrographs of basal bodies in (A) WT, (B) Kif3a−/− and (C) Ift88−/− MEFs. Two consecutive sections reveal the presence of a primary cilium in three WT cells. Primary cilia are absent in Kif3a and Ift88 mutant MEFs. Ciliary basal feet (asterisks) are detected in the subdistal region of basal bodies in WT and Ift88−/− MEFs, but are not found in Kif3a−/− MEFs. Basal feet in at least 10 serially sectioned samples were analysed per experiment (n=3). Scale bars indicate 200 nm for all images.
Discussion
We have found that Kif3a is a component of the mother centriole that regulates the organization and function of the subdistal appendage. Kif3a interacts with p150Glued through its carboxy-terminal cargo-binding domain to bring p150Glued to the subdistal appendages of the mother centriole. As loss of Kif3a or depletion of p150Glued both inhibit the localization of Ninein and Cep170 to subdistal appendages, it is likely that Kif3a recruits p150Glued to the mother centriole, which in turn recruits other subdistal appendage components such as Ninein and Cep170.
However, Kif3a and p150Glued do not extensively colocalize at the mother centriole. Kif3a localizes to a single focus at the same proximal–distal position along the mother centriole as subdistal appendages, whereas p150Glued forms a ring consistent with localization to the subdistal appendages themselves. These observations raise the possibility that the interaction of Kif3a with p150Glued is transient, with Kif3a bringing p150Glued to the centriole, but not anchoring p150Glued at the subdistal appendages. Given that the localization of Kif3a to the centrosome precedes both p150Glued and Ninein, Kif3a may provide a centrosomal docking site on the mother centriole for the subdistal appendage to reform upon exit from mitosis.
Like Kif3a, the mother centriolar protein ODF2 localizes to a single focus on mother centrioles and is required to recruit Ninein to the subdistal appendage (Nakagawa et al, 2001; Kunimoto et al, 2012). Given that the localization of Kif3a and ODF2 is distinct from that of the subdistal appendage proteins that depend on them, it remains unclear how Kif3a and ODF2 coordinate the recruitment of subdistal appendage components. One possibility is that Dynactin must bind to both Dynein and Kinesin II to move to the minus ends of microtubules where unloading of subdistal appendage components onto the centrioles occurs.
Kif3a is also required for centriole cohesion during G1, similar to the Dynactin component, p150Glued (Quintyne et al, 1999; Quintyne and Schroer, 2002). The cargo-binding domain of Kif3a interacts with p150Glued to localize it to the subdistal appendages. Curiously, the depletion of Ninein leads to the mislocalization of p150Glued, but does not lead to the loss of centriole cohesion. One possibility is that a Kif3a–p150Glued complex functions outside of the centrosome to recruit factors that maintain centriole cohesion. It will be interesting to investigate how Kif3a and p150Glued complex interact with other proteins involved in centriole attachment.
We found that Kif3a, p150Glued and Ninein are required for microtubule anchoring. Furthermore, we found that Kif3a localization does not require Ninein, suggesting that the microtubule anchoring defect in Kif3a mutants is due to the mislocalization of Ninein.
Complex cellular events, including basal body organization and the assembly of functional IFT particles, must be orchestrated to support ciliogenesis (Marshall and Rosenbaum, 2001; Song and Dentler, 2001; Graser et al, 2007; Vladar and Stearns, 2007). We found that subdistal appendage and basal foot formation require Kif3a, but not Ift88, indicating that the ciliogenic function of Kinesin II is not limited to IFT, but that Kif3a also participates in centriole organization. The genetic requirement for Kif3a in the formation of both subdistal appendages and basal feet may underscore many other commonalities between them, including composition and origin (Seeley and Nachury, 2010; Kunimoto et al, 2012). Future experiments to determine the components of the basal foot and their relationship to Kif3a and the subdistal appendage may uncover the molecular mechanism underlying basal foot formation. In summary, we demonstrate that Kif3a is an essential component of the mother centriole ensuring the integrity of the subdistal appendage and that the loss of Kif3a abrogates microtubule anchoring and ciliary basal foot formation.
Materials and methods
Antibodies
Antibodies used in this study are the following: anti-Cep170 (Invitrogen and gift of G Guarguaglini, Sapienza, University of Rome, Italy), anti-Ninein (Millipore, Bethyl and gift of S Tsukita, Osaka University, Japan), anti-Centrin1 (Proteintech), anti-Centrin 20H5 (Millipore and gift of J Salisbury, Mayo Clinic), anti-Centrin3 (gift of M Bornens, Centre National de la Recherche Scientifique–Pierre Fabre, France), anti-polyglutamylated tubulin (gift of C Janke, Centre de Recherche de Biochimie Macromoléculaire, France), anti-p150Glued (BD Biosciences, Santa Cruz Biotechnology and gift of E Holzbaur, University of Pennsylvania), anti-Kif3a (Sigma, Proteintech, BD Biosciences and Covance), anti-Centrobin (Abcam), anti-α-tubulin and γ-tubulin (Sigma), anti-Ift88 and CP110 (Proteintech), anti-GFP (Abcam, Santa Cruz Biotechnologies and Invitrogen). Alexa-conjugated secondary antibodies, Hoechst 33342 and DAPI were obtained from Molecular Probes (Invitrogen).
Cell culture, plasmid and siRNA transfections
Kif3a−/− and Ift88−/− MEFs were derived from embryonic days 8.5 and 10.5 littermate embryos, respectively, along with MEFs from littermate WT controls. MEFs were grown in DMEM (UCSF tissue culture facility) supplemented with 15% FBS (Invitrogen), Glutamax-I (Invitrogen), sodium pyruvate, non-essential amino acids, penicillin and streptomycin. MEFs were subsequently immortalized with SV40 large T antigen. MEFs conditional for Kif3a were cultured as described previously (Corbit et al, 2008). HeLa cells (UCSF tissue culture facility) were cultured in Advanced DMEM (Invitrogen) supplemented with 2% FBS and Glutamax-I. For transfection of HeLa cells and MEFs, Fugene6 (Roche) or Lipofectamine PLUS (Invitrogen) transfection reagents were used according to the manufacturer’s instructions. The following siRNA sequences were used: Scramble control (SC): 5′-AAUUCUCCGAACGUGUCACGUdtdt-3′ (Qiagen), human p150Glued: 5′-GCCUUGAACAGUUCCAUCAdtdt-3′ (Delgehyr et al, 2005) human Stealth Ninein #1: 5′-GCCAGGGUUAGUAAUGUCUUCUUGU-3′ (Invitrogen) and human Stealth Ninein #2: 5′-GGAGGCGGAGCUCUCUGAAGUUAAA-3′ (Invitrogen). HeLa cells were transfected with siRNA using Oligofectamine (Invitrogen) and analysed 48 h later.
Immunofluorescence
To visualize centrosome proteins in MEFs, cells were fixed in −20 °C methanol for 3 min. To detect Kif3a in MEFs, cells were pre-extracted for 30 s in PEM-P buffer (100 mM Pipes, 10 mM EGTA, 1 mM MgCl and 0.1% Triton-X-100) followed by fixation in 4% paraformaldehyde in PBS for 8 min and then postfixed in methanol for 30 s. Following fixation, cells were incubated in blocking solution (0.1% Triton-X-100, 0.01% NaN3 and 2.5% BSA in PBS) overnight at 4 °C. Primary and secondary antibodies were diluted in blocking solution and incubated with cells at room temperature for 1 h. Coverslips were mounted using Gelvatol mounting media and imaged with an inverted Axio Observer D1 (Zeiss) or a Deltavision deconvolution microscope (Applied Precision), image processing and measurements were completed with Fiji image processing software and Adobe Photoshop.
Immunoprecipitation and immunoblotting
Cells were harvested using a cell scraper and lysed on ice for 10 min in lysis buffer (50 mM Tris–HCl pH7.4, 150 mM NaCl, 1% NP-40) supplemented with a protease inhibitor cocktail (Calbiochem). Lysates were clarified (13 000 r.p.m., 4 °C, 10 min) and the protein concentrations were determined using the Bradford assay (Bio-Rad). For each immunoprecipitation reaction, 500 μg of total lysate was incubated with 2 μg of antibody for 2 h and then incubated with protein G-Agarose (Invitrogen) for an additional hour at 4 °C. Immunocomplexes were washed three times in lysis buffer and subsequently boiled in 2 × Laemmli reducing buffer. Samples were separated by SDS–PAGE, transferred onto nitrocellulose (Whattman) and then subjected to immunoblot analysis using ECL Lightening Plus (Perkin-Elmer).
Transmission electron microscopy
Cells were plated on two-well Permanox slides (Nunc), fixed in 3.5% glutaraldehyde in 0.1 M phosphate buffer (PB) for 1 h at room temperature, then washed three times in 0.1 M PB. Cells were postfixed in 2% osmium tetroxide for 2 h, dehydrated and embedded in Durcupan (Fluka). Serial ultrathin sections (70 nm) were cut with a diamond knife to reconstruct whole cilia, stained with lead citrate and examined using a FEI Tecnai Spirit electron microscope.
Supplementary Material
Acknowledgments
This work was funded by grants from NIH (T32HL007731) to AK, the NIH (R01AR054396, R01GM095941), the Burroughs Wellcome Fund, the Packard Foundation, and the Sandler Family Supporting Foundation to JFR.
Author contributions: AK and JFR designed the experiments. AK and AS derived wild-type, Kif3a−/−, and Ift88−/− mouse embryonic fibroblasts. JFR cloned the Kif3a fusion constructs. ATK, MSSP, and JMGV prepared and imaged the electron microscopy samples. AK and JFR interpreted the data and wrote the paper.
Footnotes
The authors declare that they have no conflict of interest.
References
- Anderson RG (1972) The three-dimensional structure of the basal body from the rhesus monkey oviduct. J Cell Biol 54: 246–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bobinnec Y, Khodjakov A, Mir LM, Rieder CL, Edde B, Bornens M (1998) Centriole disassembly in vivo and its effect on centrosome structure and function in vertebrate cells. J Cell Biol 143: 1575–1589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boisvieux-Ulrich E, Sandoz D (1991) Determination of ciliary polarity precedes differentiation in the epithelial cells of quail oviduct. Biol Cell 72: 3–14 [DOI] [PubMed] [Google Scholar]
- Bornens M (2002) Centrosome composition and microtubule anchoring mechanisms. Curr Opin Cell Biol 14: 25–34 [DOI] [PubMed] [Google Scholar]
- Casenghi M, Barr FA, Nigg EA (2005) Phosphorylation of Nlp by Plk1 negatively regulates its dynein-dynactin-dependent targeting to the centrosome. J Cell Sci 118: 5101–5108 [DOI] [PubMed] [Google Scholar]
- Chang P, Giddings TH Jr., Winey M, Stearns T (2003) Epsilon-tubulin is required for centriole duplication and microtubule organization. Nat Cell Biol 5: 71–76 [DOI] [PubMed] [Google Scholar]
- Corbit KC, Shyer AE, Dowdle WE, Gaulden J, Singla V, Chen MH, Chuang PT, Reiter JF (2008) Kif3a constrains beta-catenin-dependent Wnt signalling through dual ciliary and non-ciliary mechanisms. Nat Cell Biol 10: 70–76 [DOI] [PubMed] [Google Scholar]
- Deacon SW, Serpinskaya AS, Vaughan PS, Lopez Fanarraga M, Vernos I, Vaughan KT, Gelfand VI (2003) Dynactin is required for bidirectional organelle transport. J Cell Biol 160: 297–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delgehyr N, Sillibourne J, Bornens M (2005) Microtubule nucleation and anchoring at the centrosome are independent processes linked by ninein function. J Cell Sci 118: 1565–1575 [DOI] [PubMed] [Google Scholar]
- Finetti F, Paccani SR, Riparbelli MG, Giacomello E, Perinetti G, Pazour GJ, Rosenbaum JL, Baldari CT (2009) Intraflagellar transport is required for polarized recycling of the TCR/CD3 complex to the immune synapse. Nat Cell Biol 11: 1332–1339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graser S, Stierhof YD, Lavoie SB, Gassner OS, Lamla S, Le Clech M, Nigg EA (2007) Cep164, a novel centriole appendage protein required for primary cilium formation. J Cell Biol 179: 321–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarguaglini G, Duncan PI, Stierhof YD, Holmstrom T, Duensing S, Nigg EA (2005) The forkhead-associated domain protein Cep170 interacts with Polo-like kinase 1 and serves as a marker for mature centrioles. Mol Biol Cell 16: 1095–1107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haraguchi K, Hayashi T, Jimbo T, Yamamoto T, Akiyama T (2006) Role of the kinesin-2 family protein, KIF3, during mitosis. J Biol Chem 281: 4094–4099 [DOI] [PubMed] [Google Scholar]
- Haycraft CJ, Zhang Q, Song B, Jackson WS, Detloff PJ, Serra R, Yoder BK (2007) Intraflagellar transport is essential for endochondral bone formation. Development 134: 307–316 [DOI] [PubMed] [Google Scholar]
- Holley MC, Afzelius BA (1986) Alignment of cilia in immotile-cilia syndrome. Tissue Cell 18: 521–529 [DOI] [PubMed] [Google Scholar]
- Ishikawa H, Kubo A, Tsukita S (2005) Odf2-deficient mother centrioles lack distal/subdistal appendages and the ability to generate primary cilia. Nat Cell Biol 7: 517–524 [DOI] [PubMed] [Google Scholar]
- Ishikawa H, Marshall WF (2011) Ciliogenesis: building the cell’s antenna. Nat Rev Mol Cell Biol 12: 222–234 [DOI] [PubMed] [Google Scholar]
- Kunimoto K, Yamazaki Y, Nishida T, Shinohara K, Ishikawa H, Hasegawa T, Okanoue T, Hamada H, Noda T, Tamura A, Tsukita S (2012) Coordinated ciliary beating requires Odf2-mediated polarization of basal bodies via basal feet. Cell 148: 189–200 [DOI] [PubMed] [Google Scholar]
- Louie RK, Bahmanyar S, Siemers KA, Votin V, Chang P, Stearns T, Nelson WJ, Barth AI (2004) Adenomatous polyposis coli and EB1 localize in close proximity of the mother centriole and EB1 is a functional component of centrosomes. J Cell Sci 117: 1117–1128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall WF, Rosenbaum JL (2001) Intraflagellar transport balances continuous turnover of outer doublet microtubules: implications for flagellar length control. J Cell Biol 155: 405–414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marszalek JR, Ruiz-Lozano P, Roberts E, Chien KR, Goldstein LS (1999) Situs inversus and embryonic ciliary morphogenesis defects in mouse mutants lacking the KIF3A subunit of kinesin-II. Proc Natl Acad Sci USA 96: 5043–5048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirzadeh Z, Han YG, Soriano-Navarro M, Garcia-Verdugo JM, Alvarez-Buylla A (2010) Cilia organize ependymal planar polarity. J Neurosci 30: 2600–2610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell B, Jacobs R, Li J, Chien S, Kintner C (2007) A positive feedback mechanism governs the polarity and motion of motile cilia. Nature 447: 97–101 [DOI] [PubMed] [Google Scholar]
- Mitchell B, Stubbs JL, Huisman F, Taborek P, Yu C, Kintner C (2009) The PCP pathway instructs the planar orientation of ciliated cells in the Xenopus larval skin. Curr Biol 19: 924–929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mogensen MM, Malik A, Piel M, Bouckson-Castaing V, Bornens M (2000) Microtubule minus-end anchorage at centrosomal and non-centrosomal sites: the role of ninein. J Cell Sci 113: Pt 173013–3023 [DOI] [PubMed] [Google Scholar]
- Moritz M, Braunfeld MB, Guenebaut V, Heuser J, Agard DA (2000) Structure of the gamma-tubulin ring complex: a template for microtubule nucleation. Nat Cell Biol 2: 365–370 [DOI] [PubMed] [Google Scholar]
- Nakagawa Y, Yamane Y, Okanoue T, Tsukita S (2001) Outer dense fiber 2 is a widespread centrosome scaffold component preferentially associated with mother centrioles: its identification from isolated centrosomes. Mol Biol Cell 12: 1687–1697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonaka S, Tanaka Y, Okada Y, Takeda S, Harada A, Kanai Y, Kido M, Hirokawa N (1998) Randomization of left-right asymmetry due to loss of nodal cilia generating leftward flow of extraembryonic fluid in mice lacking KIF3B motor protein. Cell 95: 829–837 [DOI] [PubMed] [Google Scholar]
- Ou YY, Mack GJ, Zhang M, Rattner JB (2002) CEP110 and ninein are located in a specific domain of the centrosome associated with centrosome maturation. J Cell Sci 115: 1825–1835 [DOI] [PubMed] [Google Scholar]
- Paintrand M, Moudjou M, Delacroix H, Bornens M (1992) Centrosome organization and centriole architecture: their sensitivity to divalent cations. J Struct Biol 108: 107–128 [DOI] [PubMed] [Google Scholar]
- Pazour GJ, Dickert BL, Vucica Y, Seeley ES, Rosenbaum JL, Witman GB, Cole DG (2000) Chlamydomonas IFT88 and its mouse homologue, polycystic kidney disease gene tg737, are required for assembly of cilia and flagella. J Cell Biol 151: 709–718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen LB, Geimer S, Rosenbaum JL (2006) Dissecting the molecular mechanisms of intraflagellar transport in chlamydomonas. Curr Biol 16: 450–459 [DOI] [PubMed] [Google Scholar]
- Piel M, Meyer P, Khodjakov A, Rieder CL, Bornens M (2000) The respective contributions of the mother and daughter centrioles to centrosome activity and behavior in vertebrate cells. J Cell Biol 149: 317–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintyne NJ, Gill SR, Eckley DM, Crego CL, Compton DA, Schroer TA (1999) Dynactin is required for microtubule anchoring at centrosomes. J Cell Biol 147: 321–334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintyne NJ, Schroer TA (2002) Distinct cell cycle-dependent roles for dynactin and dynein at centrosomes. J Cell Biol 159: 245–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayner CF, Rutman A, Dewar A, Greenstone MA, Cole PJ, Wilson R (1996) Ciliary disorientation alone as a cause of primary ciliary dyskinesia syndrome. Am J Respir Crit Care Med 153: 1123–1129 [DOI] [PubMed] [Google Scholar]
- Robert A, Margall-Ducos G, Guidotti JE, Bregerie O, Celati C, Brechot C, Desdouets C (2007) The intraflagellar transport component IFT88/polaris is a centrosomal protein regulating G1-S transition in non-ciliated cells. J Cell Sci 120: 628–637 [DOI] [PubMed] [Google Scholar]
- Rosenbaum JL, Witman GB (2002) Intraflagellar transport. Nat Rev Mol Cell Biol 3: 813–825 [DOI] [PubMed] [Google Scholar]
- Sack S, Kull FJ, Mandelkow E (1999) Motor proteins of the kinesin family. Structures, variations, and nucleotide binding sites. Eur J Biochem 262: 1–11 [DOI] [PubMed] [Google Scholar]
- Seeley ES, Nachury MV (2010) The perennial organelle: assembly and disassembly of the primary cilium. J Cell Sci 123: 511–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sfakianos J, Togawa A, Maday S, Hull M, Pypaert M, Cantley L, Toomre D, Mellman I (2007) Par3 functions in the biogenesis of the primary cilium in polarized epithelial cells. J Cell Biol 179: 1133–1140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song L, Dentler WL (2001) Flagellar protein dynamics in Chlamydomonas. J Biol Chem 276: 29754–29763 [DOI] [PubMed] [Google Scholar]
- Takahashi M, Yamagiwa A, Nishimura T, Mukai H, Ono Y (2002) Centrosomal proteins CG-NAP and kendrin provide microtubule nucleation sites by anchoring gamma-tubulin ring complex. Mol Biol Cell 13: 3235–3245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda S, Yonekawa Y, Tanaka Y, Okada Y, Nonaka S, Hirokawa N (1999) Left-right asymmetry and kinesin superfamily protein KIF3A: new insights in determination of laterality and mesoderm induction by kif3A-/- mice analysis. J Cell Biol 145: 825–836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vladar EK, Stearns T (2007) Molecular characterization of centriole assembly in ciliated epithelial cells. J Cell Biol 178: 31–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiesner C, Faix J, Himmel M, Bentzien F, Linder S (2010) KIF5B and KIF3A/KIF3B kinesins drive MT1-MMP surface exposure, CD44 shedding, and extracellular matrix degradation in primary macrophages. Blood 116: 1559–1569 [DOI] [PubMed] [Google Scholar]
- Wong C, Stearns T (2003) Centrosome number is controlled by a centrosome-intrinsic block to reduplication. Nat Cell Biol 5: 539–544 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.