Abstract
The ubiquitylation signal promotes trafficking of endogenous and retroviral transmembrane proteins. The signal is decoded by a large set of ubiquitin (Ub) receptors that tether Ub-binding domains (UBDs) to the trafficking machinery. We developed a structure-based procedure to scan the protein data bank for hidden UBDs. The screen retrieved many of the known UBDs. Intriguingly, new potential UBDs were identified, including the ALIX-V domain. Pull-down, cross-linking and E3-independent ubiquitylation assays biochemically corroborated the in silico findings. Guided by the output model, we designed mutations at the postulated ALIX-V:Ub interface. Biophysical affinity measurements using microscale-thermophoresis of wild-type and mutant proteins revealed some of the interacting residues of the complex. ALIX-V binds mono-Ub with a Kd of 119 μM. We show that ALIX-V oligomerizes with a Hill coefficient of 5.4 and IC50 of 27.6 μM and that mono-Ub induces ALIX-V oligomerization. Moreover, we show that ALIX-V preferentially binds K63 di-Ub compared with mono-Ub and K48 di-Ub. Finally, an in vivo functionality assay demonstrates the significance of ALIX-V:Ub interaction in equine infectious anaemia virus budding. These results not only validate the new procedure, but also demonstrate that ALIX-V directly interacts with Ub in vivo and that this interaction can influence retroviral budding.
Keywords: computational analyses, in silico scan, protein trafficking, ubiquitin, virus budding
Introduction
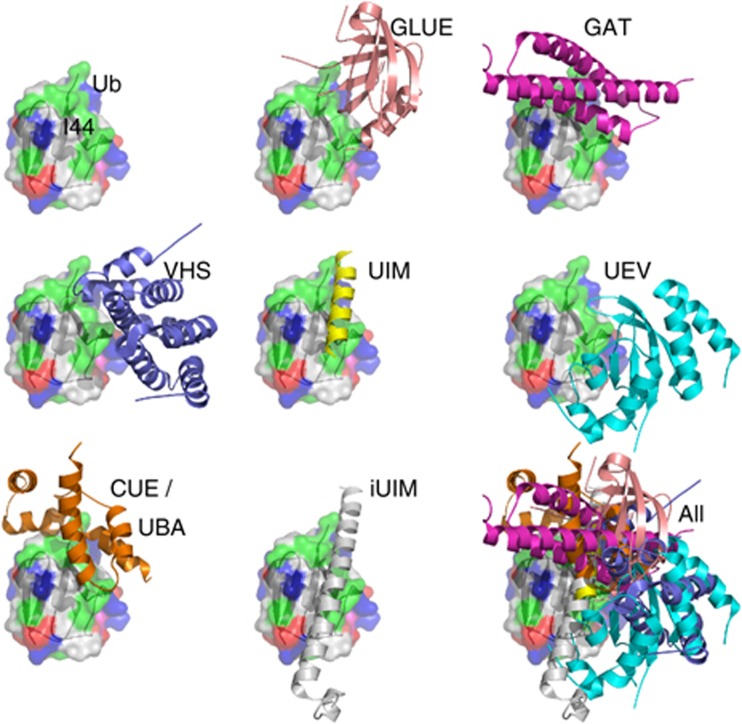
Ubiquitin (Ub) acts as a versatile post-translational signal that regulates a wide range of biological processes. Indeed, more than a third of the eukaryotic proteome is ubiquitylated via a conserved enzymatic cascade (Kim et al, 2011; Ziv et al, 2011). By tethering Ub-binding domains (UBDs) to other functional domains, Ub receptors decode the Ub signal into a cellular response. At least 21 different UBD families have been identified (Hicke et al, 2005; Hurley et al, 2006; Husnjak and Dikic, 2012). The structures of representatives from most of these families have been determined both as apo forms and as complexes with Ub. Interestingly, several biochemical studies revealed a hidden UBD within proteins for which their structures, or those of their homologues, have been determined, as demonstrated in the cases of HECT (French et al, 2009), VHS (Ren and Hurley, 2010) and WD40 (Pashkova et al, 2010). We therefore hypothesized that additional hidden UBDs are likely to exist within the protein data bank (PDB) (Berman et al, 2000). In a recent survey of the PDB, we found that 68 UBDs out of 72 UBD:Ub complexes recognize the same hydrophobic patch on the Ub surface. Interestingly, these UBDs belong to 16 different protein families and do not possess conserved sequences or structures (Figure 1). Thus, we assumed that the binding patches of these UBDs share a similar, distinctive configuration of physico-chemical properties, which can be extracted and used as a signature probe to scan the PDB. Stimulated by these preliminary findings, we scripted a structure-based procedure with the aim of identifying some of the hidden UBDs in silico. Applying this procedure to the PDB revealed the ALIX-V domain as a potential UBD.
Figure 1.
A structural model illustrating the Ub-binding interface of selected representative UBDs. In the bottom, right corner, all the UBD–Ub complexes are superimposed by aligning the Ub on itself. The superimposition clearly illustrates that the considered UBDs bind the same patch on the Ub. The Ub is rendered as molecular surface and the UBDs as a cartoon.
ALIX (apoptosis-linked gene 2 (ALG-2)-interacting protein X) plays a pivotal role in ESCRT (endosomal sorting complex required for transport)-mediated membrane abscission in cytokinesis, and mediates the LYPXnL-motif-driven budding of enveloped viruses. It also has roles in endocytosis and apoptosis. ALIX contains three major functional elements: an N-terminal Bro1 domain, a central V domain and a C-terminal Pro-rich region (PRR). The Bro1 domain recruits and regulates the ESCRT-III machinery that mediates membrane scission. The V domain binds to the LYPXnL motif of retroviral Gag proteins including those of the lentiviruses HIV-1 and EIAV (Fisher et al, 2007; Lee et al, 2007). The PRR binds and regulates several cellular proteins including Tsg101, CIN85 CD2AP, Src, Hck kinases, endophilin, CEP55 and the E3-ligase POSH and Nedd4 (reviewed by Ren and Hurley, 2011). Previous findings had suggested the possibility that the ALIX-V domain binds directly or indirectly to Ub (Joshi et al, 2008). The V-domain could be pulled-down from cell extracts with Ub-agarose beads, and fusion of Ub to the C-terminus of an EIAV Gag protein lacking the LYPXnL motif caused virus release to be sensitive to inhibition by V-domain overexpression (Joshi et al, 2008). Herein, we provide direct evidence that ALIX is a bona fide UBD. The developed structure-based in silico procedure allowed us to pinpoint the interacting residues on both sides of the ALIX:Ub interface and to isolate mutants affecting retrovirus budding.
Results
Structure-based in silico procedure to identify UBDs
We developed a structure-based procedure for in silico search of hidden UBDs within the PDB (Berman et al, 2000). Most UBDs bind the same patch on the Ub surface that is usually composed of L8, R42, I44, G47, H68, V70, R72 and L73 residues. We therefore postulated that these hidden UBDs might contain molecular surface patches with a similar spatial configuration of key physico-chemical properties that are responsible for the UBD interaction with the above-mentioned residues on the Ub surface. A similar logic is at the heart of pharmacophore-based methods in computer-aided drug design (Dror et al, 2004), as well as in the structure-based search for binding sites of a specific drug (Shulman-Peleg et al, 2004).
We therefore scanned the PDB to detect protein structures that contain on their molecular surface a spatial configuration of physico-chemical properties that mimics the configurations that are detected on known Ub-binding proteins. For this purpose, we applied our SiteEngine algorithm (Shulman-Peleg et al, 2004, 2005), which was originally developed for detecting protein functional sites. The algorithm identifies proteins that have similar spatial configurations of physico-chemical properties on their molecular surfaces, even if these proteins differ both in sequence and fold.
The designed UBD search procedure includes four major steps; the first three are computational hypotheses generation steps and the final is an experimental validation step. The flow of the procedure is as follows:
Choice of a ‘template’ UBD to perform the structural PDB scan.
Scan of the molecular surfaces of all the PDB chains with the ‘template’ to detect candidate UBDs.
Superimposition of the Ub on the candidate-binding patch, assessment of the binding energy and search for energetically more favourable binding patches in the immediate vicinity of the detected candidate-binding patch.
For selected high-scoring patches—in vitro and in vivo validation of the binding site by binding assays and mutational analysis.
Below we shall discuss in detail the three computational steps.
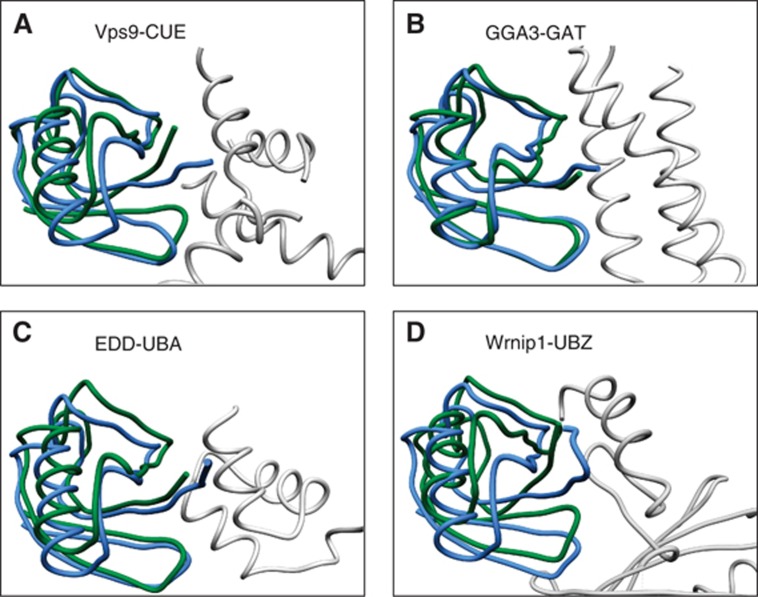
Choice of a ‘template’ UBD. First, a representative set of 54 known UBD:Ub complexes (see Supplementary Table SI) has been selected. Next, each of the UBDs in this set was defined (in turn) as ‘template’, and a physico-chemical ‘signature’ of its residues, which are in bonding distance from the I44 patch on the Ub, was used to structurally scan all the other UBDs in the set using the SiteEngine algorithm (Shulman-Peleg et al, 2004, 2005). For each of the 10 highest-scoring spatial alignments of the ‘template’ binding site to a candidate molecular surface patch on a UBD, the Ub of the ‘template’ was superimposed on the protein and the RMSD was calculated between this ‘template Ub’ and the native protein’s Ub. RMSDs below 6 Å were considered a ‘hit’. Based on the results of this preliminary scan, the PDB protein 3K9P (E2-25K:Ub complex), which scored nine hits, was chosen to be the preferred ‘template’ for the scan of the entire PDB. E2-25K is an E2 that is tethered to a UBD from the UBA family. Figure 2 visually demonstrates the validation process of hits by means of RMSD assessment. Specifically, we provide here an example of four different known UBD:Ub complexes that SiteEngine found to possess a similar Ub recognition surface to the one of 3K9P. Figure 2 demonstrates how well the native and the predicted Ub structures are superimposed (one should also take into account that the structures of the Ub molecules are different from each other since they came from different UBD:Ub complexes). The fact that these UBDs have different folds emphasizes the sequence- and fold-independent nature of the search procedure.
Figure 2.
In silico evaluation of the search procedure. Shown are representative outputs of the search procedure that were superimposed on their known counterpart complexes in the PDB. The complexes were superimposed based on the site-engine-detected structural alignment of the respective UBD molecular surfaces. The resulting Ub orientation in green is shown with respect to the original Ub, which is shown in blue. The Cα RMSD values between the two Ub molecules indicate the accuracy of the complex prediction: (A) Vps9-CUE 3.3 Å, (B) GGA3-GAT 1.7 Å, (C) EDD-UBA 5.0 Å and (D) Wrnip1-UBZ 4.5 Å.
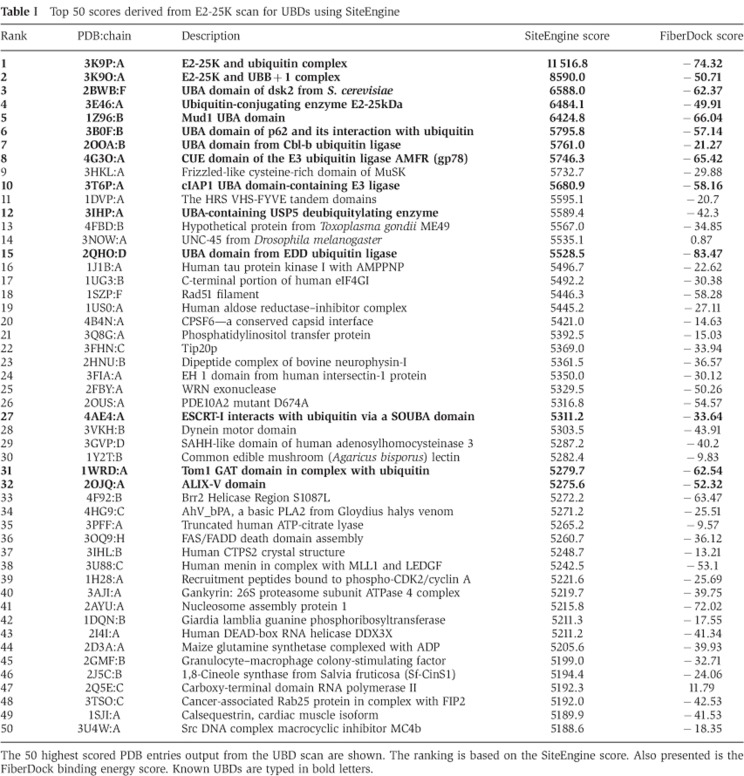
Scan of the PDB with the ‘template’. The procedure described in the paragraph above is now executed with a spatial physico-chemical signature of the 3K9P Ub-binding site to structurally scan the molecular surfaces of the PDB chains. Only ∼30 000 chains of the eukaryotic organisms with <90% sequence identity have been considered. The 50 top scoring results (by the SiteEngine score) of this scan are presented in Table I (the top 1000 results of the scan are given in Supplementary Table SII of the Supplementary data).
Table 1. Top 50 scores derived from E2-25K scan for UBDs using SiteEngine.
The 50 highest scored PDB entries output from the UBD scan are shown. The ranking is based on the SiteEngine score. Also presented is the FiberDock binding energy score. Known UBDs are typed in bold letters.
Assessment and refinement of the Ub binding. For each of the PDB chains, 1000 top SiteEngine solutions for this chain are further evaluated and improved based on their Ub-binding energy. Specifically, the binding site of the template is structurally aligned with its detected counterpart on the protein under examination resulting in a putative Ub–protein complex. This complex is further refined using the FiberDock (Mashiach et al, 2010b) algorithm, which was originally developed for rigid protein–protein docking solution refinement. Since proteins undergo conformational changes upon binding, FiberDock optimizes the interface energy of a candidate complex by allowing limited backbone flexibility in the receptor molecule as well as flexibility in the interface side chain conformations of both molecules and Monte–Carlo-like perturbations of the relative orientations of the molecules. The interface energy score of FiberDock includes a variety of energy terms, such as desolvation energy (ACE), softened van der Waals interactions, partial electrostatics, hydrogen and disulphide bonds, p-stacking, aliphatic interactions and more (as detailed in Andrusier et al, 2007). Supplementary Table SIII presents the top 1000 results sorted by the FiberDock score (both Table I and Supplementary Table SII include the FiberDock-binding energy scores of the presented hypotheses).
Examination of Table I, Supplementary Tables SII and SIII shows that known UBDs usually receive high scores. Significantly, these domains score well both in the SiteEngine score and the FiberDock energy score. The binding energy score that the template achieves when compared to other known UBDs can be used as a gauge to assess the quality of candidate UBDs’ binding scores. Moreover, the output also sheds light on some known apo UBD structures. For example, the procedure ranked the solenoid of overlapping UBAs (SOUBAs) subunit of ESCRT-I at place 27 out of ∼30 000. Superimposing the SOUBA:Ub model derived from our algorithm on known UBA:Ub complexes is well fitted and corroborates the NMR chemical shifts changes analyses data for SOUBA:Ub complex (Agromayor et al, 2012).
Interestingly, the ALIX-V domain achieved a relatively high score of position 31 in Table I. In addition, its binding energy score is quite similar to that of other known UBDs. A literature search did not yield any publication demonstrating a direct interaction between ALIX-V and Ub. Nevertheless, a previous study (Joshi et al, 2008) showed that full-length ALIX and the ALIX-V domain could be pulled-down from cell extracts with Ub–agarose beads. In addition, it was shown that budding of a mutant form of EIAV Gag harbouring a YPDL deletion and a C-terminal Ub fusion (EIAV/ΔYPDL-Ub) is sensitive to depletion of ALIX and to overexpression of ALIX-V (Joshi et al, 2008). These findings suggest that the interaction between the Ub conjugated to Gag and endogenous TSG101, which in turn recruits ALIX, compensates for the absence of a retroviral late domain motif (YPDL) that normally interacts with ALIX-V. Consistent with this hypothesis, mutation of L8 and I44 in the Ub portion of EIAV/ΔYPDL-Ub abrogated the rescue in release conferred by Ub fusion (Joshi et al, 2008). HIV, like a number of other enveloped viruses, hijacks the cellular ESCRT machinery to bud from the cell. This cellular apparatus was first identified based on its ability to promote the budding of ubiquitylated-transmembrane proteins into the multivesicular body (MVB) (Katzmann et al, 2001). Similarly, the function of ALIX in ESCRT-mediated membrane abscission in cytokinesis was also demonstrated. Enveloped viruses have evolved to hijack the ESCRT machinery to drive their budding and release from the infected cell.
Structural model for the ALIX-V domain:Ub complex
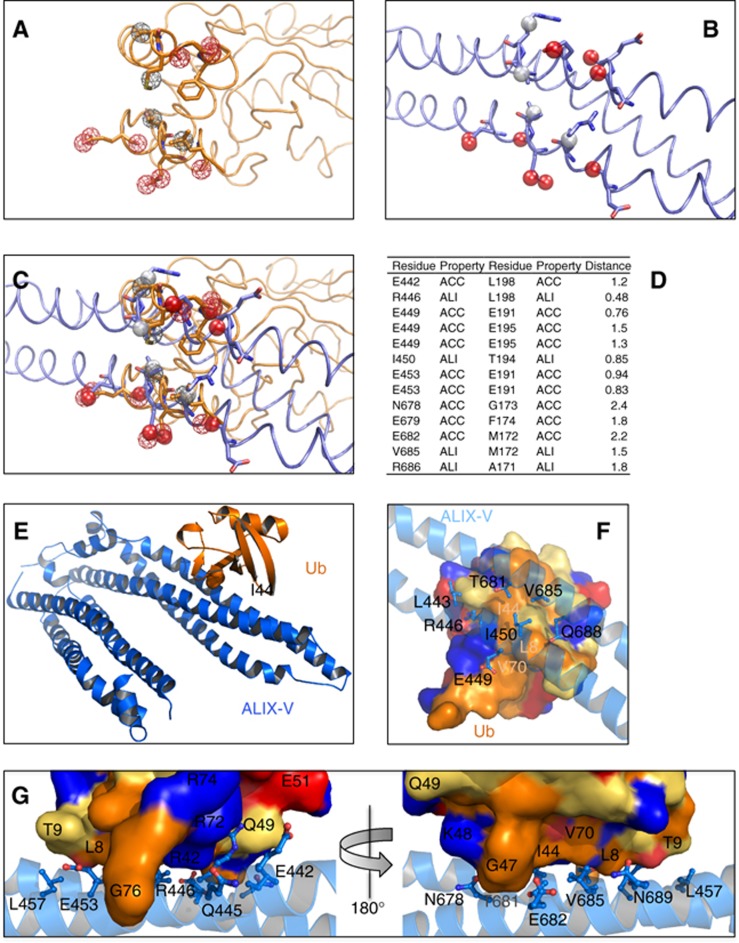
A great advantage of the in silico procedure described above is its immediate structural readout. Specifically, the output provides a high-resolution structural model of the predicted UBD:Ub complex, as explained above in computational step 3. Figure 3 shows the postulated ALIX-V:Ub complex derived from our search followed by energy minimization and structural idealization with Refmac5 (CCP4, 1994). In the complex, Ub residues L8, T9, R42, I44, G47, H68 and V70 form a network of hydrophobic and hydrophilic interactions with ALIX-V residues E442, L443, Q445, R446, E449, I450, E453, R456, N678, T681, E682, V685 and Q688. Overall, the two interfaces seem to be well fitted. Solvent-accessible area calculation indicates a buried interface of 562 and 597 Å2 on the Ub and the ALIX-V surfaces, respectively. A buried surface larger than ∼400 Å2 may indicate specific protein–protein interactions (Nagano et al, 2002). Moreover, a clear relationship between the protein mass and the buried surface interface has been demonstrated (Nagano et al, 2002). Since Ub is a relatively small protein (8.5 kDa), we do not expect to see a large interaction surface. Indeed, all known UBDs present a fairly small interaction surface, varying from 389 to 923 Å2 in the two extreme cases of the Npl4-NZF:Ub (1Q5W) and the Vps9p-CUE:Ub (1P3Q) complexes, respectively (Prag et al, 2003; Alam et al, 2004). The Ub-binding patch is located at the outer side of the V-domain against the HIV-p6-binding site (centred at F676). It seems that similar principles govern the binding of ubiquitylated Gag by ESCRT-I (Tsg101) and ALIX. In both cases, there is no competition between the peptide and Ub binding, suggesting they could interact simultaneously (see model in Figure 8).
Figure 3.
In silico identification of ALIX-V as a UBD. (A) Representation of physico-chemical properties of the E2-25K UBD. The properties are shown as coloured meshed spheres where hydrogen bond acceptors are in red and hydrophobic/aliphatic property centres in grey. (B) Physico-chemical representation of the ALIX-V:Ub proposed binding site. Properties are shown as opaque spheres with the same colour code. (C) Superimposition of the E2-25K with the ALIX-V-proposed binding site illustrating the physico-chemical property alignment detected by SiteEngine. (D) Alphanumeric presentation of the aligned physico-chemical properties found in ALIX-V with respect to E2-25K domain. ACC, acceptor; and ALI, aliphatic. (E) Cartoon showing overall view of the structural model of ALIX–V:Ub complex derived from the developed search procedure. (F) Top view zooming onto the ALIX-V:Ub interface showing specific interactions. Ub is rendered as a surface and ALIX-V as a cartoon. ALIX-V residues participating in the interaction are rendered as sticks. Colour code for the molecular surface of Ub: blue, positive residues; red, negative residues; orange, hydrophobic residues; and yellow, charged residues. Side chains of ALIX-V are coloured as CPK. (G) Shows two sides of the predicted interacting interfaces rotated by 180 degrees. Rendered and coloured as in (F).
Biochemical assessment of the ALIX:Ub interaction
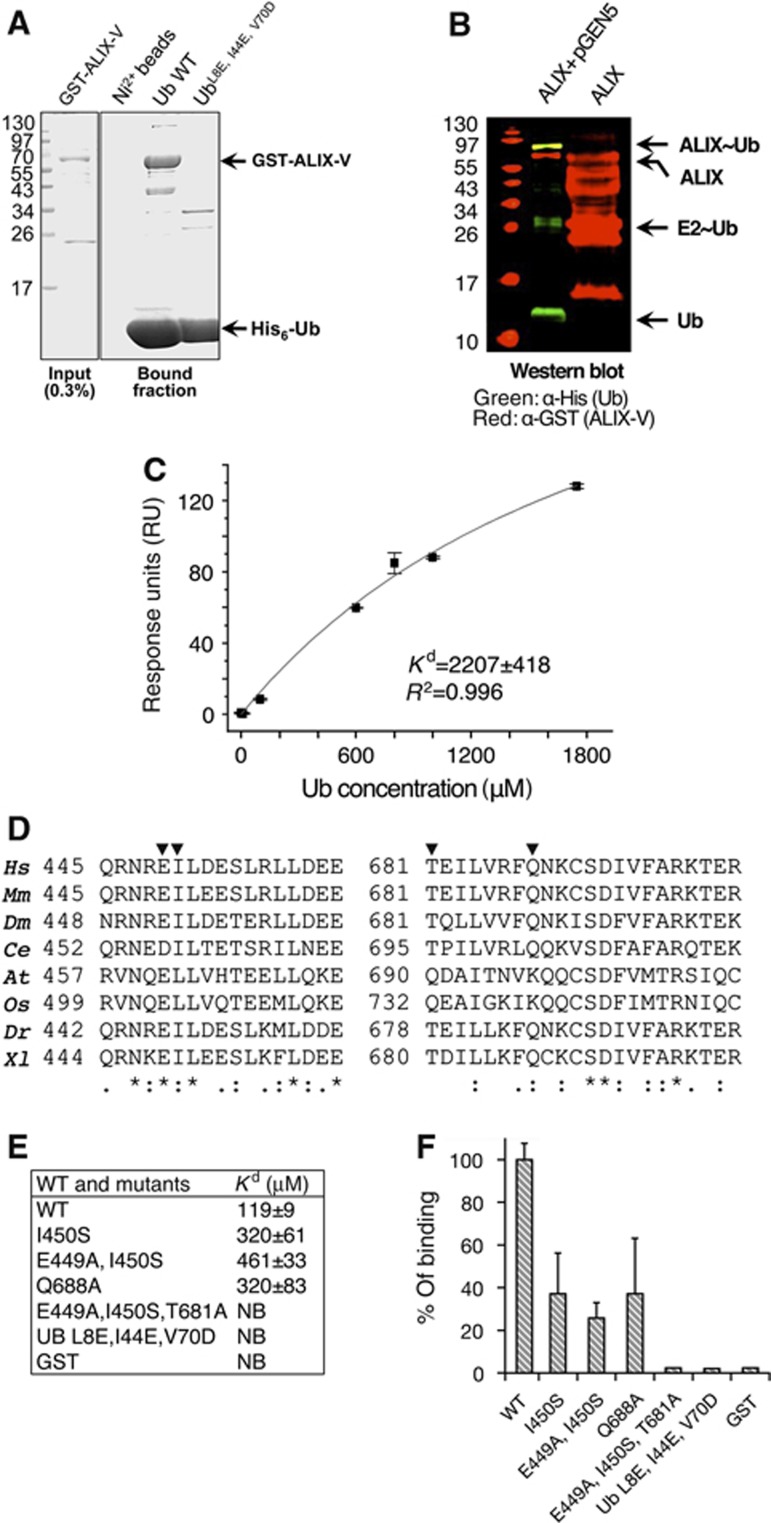
Readouts from the PDB provide an advantage in downstream assessment of their functions as UBDs since expression and purification protocols have been established for these protein constructs. To assess the in silico-based model that ALIX-V binds Ub, we expressed and purified ALIX-V and Ub from bacterial extracts and initiated pull-down assays. As shown in Figure 4A, ALIX-V directly binds Ni2+-immobilized His6-Ub but not His6-UbL8E/I44E/V70D, a defective mutant in the I44 hydrophobic patch. These results support the hypothesis that ALIX-V directly binds Ub at the I44 hydrophobic patch, and corroborates previous observation that the ALIX-V domain interacts, either directly or indirectly, with Ub (Joshi et al, 2008). To the best of our knowledge, the current data demonstrate for the first time that ALIX-V binds Ub directly.
Figure 4.
ALIX-V binds Ub through the Ub I44 hydrophobic patch. (A) Shows Coomassie stained SDS–PAGE of pull-down assay; bacterial recombinant His6-Ub and Ub mutant immobilized to Ni2+-shepharose were used in a pull-down with E. coli lysate expressing pGST-ALIX-V. (B) Shows ALIX-V ubiquitylation in bacteria. Bacterial lysates co-expressing His6-Ub, Uba1, yeast Ubc5 (expressed from pGEN5) and GST-ALIX-V were purified on Ni2+ sepharose beads and subjected to western blot analysis. Only in the presence of the ubiquitylation machinery components, a band corresponding to mono-ubiquitylated ALIX-V is shown (yellow) as detected by both α-GST (red) and α-His6 (green) antibodies (C) SPR binding affinities measured for GST-ALIX-V with Ub. GST-ALIX-V was immobilized using α-GST antibody and binding affinity was measured with increasing concentrations of Ub. Data were analysed and fitted using the Origin software with single-binding site model. (D) Conservation of the Ub-binding patch within species (generated by ClustalW). Multiple sequence alignment of regions within the binding patch is shown. Triangles indicate residues that were mutated. Conservation is shown at the bottom of the alignment: (*)—full conservation, (:)—strongly similar residues and (.)—weakly similar residues. (E–F) Affinity measurements of ALIX-V:Ub WT and mutant proteins. Binding affinity measurements between GST-ALIX-V and Ub proteins were determined by MST. At, Arabidopsis thaliana; Ce, Caenorhabditis elegans; Dm, Drosophila melanogaster; Dr, Danio rerio; Hs, Homo sapiens; Mm, Mus musculus; Os, Oryza sativa; Xl, Xenopus laevis.
ALIX-V undergoes E3-independent ubiquitylation
Ub receptors are regulated by auto-ubiquitylation, presumably to promote intramolecular binding of the UBD to the attached Ub moiety thus leading to a closed, inactive conformation of the receptor (Prag et al, 2003; Shih et al, 2003; Hoeller et al, 2007; Mosesson et al, 2009). Moreover, Dikic and co-workers demonstrated that several Ub receptors, including the signal-transducing adaptor molecule (STAM), undergo auto-mono-ubiquitylation in an E3-independent manner (Hoeller et al, 2007). We therefore postulated that if ALIX possesses a UBD function it might also undergo an E3-independent ubiquitylation. To test this hypothesis, we used a recently developed synthetic system that reconstitutes the entire ubiquitylation cascade in Escherichia coli. We established that mouse STAM2 and yeast Epsin, like other Ub receptors, undergo E3-independent auto-ubiquitylation (Keren-Kaplan et al, 2012). Here, we report that ALIX-V undergoes an E3-independent ubiquitylation (Figure 4B). Specifically, bacterial co-expression of ALIX-V domain with His6-Ub, E1 and E2 yielded a mono-ubiquitylated ALIX-V that could be purified on reduced glutathione (GSH) beads. Western blot analysis of affinity-purified ALIX-V and its modified form could be detected by antibodies against the GST, which was fused to the ALIX-V and against the hexahistidine-tag, which was fused to Ub. This result implies that ALIX-V functions as an E2- or UBD (or both).
Affinity quantification of ALIX:Ub binding
Biophysical studies with purified wild-type (WT) and mutant proteins were performed with surface plasmon resonance (SPR) and microscale-thermophoresis (MST) technology (Wienken et al, 2010). In our SPR experimental setting, a GST fusion of ALIX-V was immobilized on the sensor chip on which a flow of Ub analyte was injected. The SPR measurements showed that ALIX-V domain binds to Ub with a Kd of ∼2200 μM (Figure 4C). Similar weak affinities were previously demonstrated for other UBDs, including the VHS domain of human proteins GGA3 and Hrs and the yeast protein Vps27 (Ren and Hurley, 2010). While the χ2 and s.d. values of the SPR data were very low, comparison of the weak binding derived from the SPR with the pull-down assay (compare Figure 4C with Figure 4A) suggests that the SPR quantification does not accurately represent the apparent affinity. From our experience, ∼2 mM affinity is hardly detected by pull-down assay, and therefore it contradicts our finding. Moreover, while with immobilized Ub, ALIX-V was strongly pulled-down, in the reverse assay in which ALIX-V was immobilized, Ub binding was hardly detected (data not shown). Therefore, we sought to measure the affinity using an assay in which neither binding partner is immobilized, as attainable by MST. This new technology measures the thermophoresis change resulting from a sharp temperature change in a small area beamed from an infrared laser. When the aqueous protein mix in the capillary is heated locally, molecules start moving along the temperature gradient. This molecular flow is opposed by mass diffusion. In steady state, these effects are balanced and a stationary spatial distribution of the molecular concentration is achieved. When the temperature rises, the relative concentration depends solely on the Soret coefficient (ST) of the molecule. Therefore, the ST is a measure of the strength of the thermophoretic molecule flow compared with its ordinary diffusion. The motion of molecules and molecular complexes in temperature gradients is highly sensitive to changes in size, solvation shell and charge. Monitoring the changes in diffusion occurring in the increased temperature gradient enables the estimation of the concentrations of free versus complex ratios and consequently the derivation of the dissociation constant. We used a label-free apo GST-ALIX-V supplemented with increasing concentrations of Ub, and measured the gradient of fluorescent material derived from tryptophan residues of ALIX-V. We found that ALIX-V binds Ub with an affinity value (Kd) of 119+/−9 μM (Figure 4E and F). This Kd is very similar to that of many other characterized UBDs (Alam et al, 2004; Prag et al, 2005) and corroborates the in silico structural model and the biochemical data. Guided by the structural model derived from the in silico procedure, we designed point mutations at the predicted ALIX-V:Ub interface. As shown in Figure 4D, the regions predicted to form the Ub-binding site are conserved in higher eukaryotes. These ALIX-V mutant proteins were purified, using a GSH affinity matrix. The proteins were further subjected to size exclusion chromatography to ensure proper folding and homogeneity. WT Ub and Ub-triple mutant (L8E, IL44E and V70D) defective in the hydrophobic interaction patch were also purified and subjected to the MST measurements. The results show that Ub binds ALIX-V via the I44 hydrophobic patch as the in silico model predicted. Indeed, mutating the patch totally abrogated the binding. The ALIX-V point mutants reduced affinities with Kd values ranging between 320–460 μM. Moreover, we could not detect Ub binding with the ALIX-V-triple mutant (E449A, I450S and T681A). It is worth noting that this triple mutant displayed some minor instability during purification from bacteria, but eventually, a large amount of the protein passed through the size exclusion chromatography column and eluted with identical UV280 absorption peak shape and volume to the WT and the other mutants (see Supplementary Figure S1).
Ub binding induces ALIX-V oligomerization
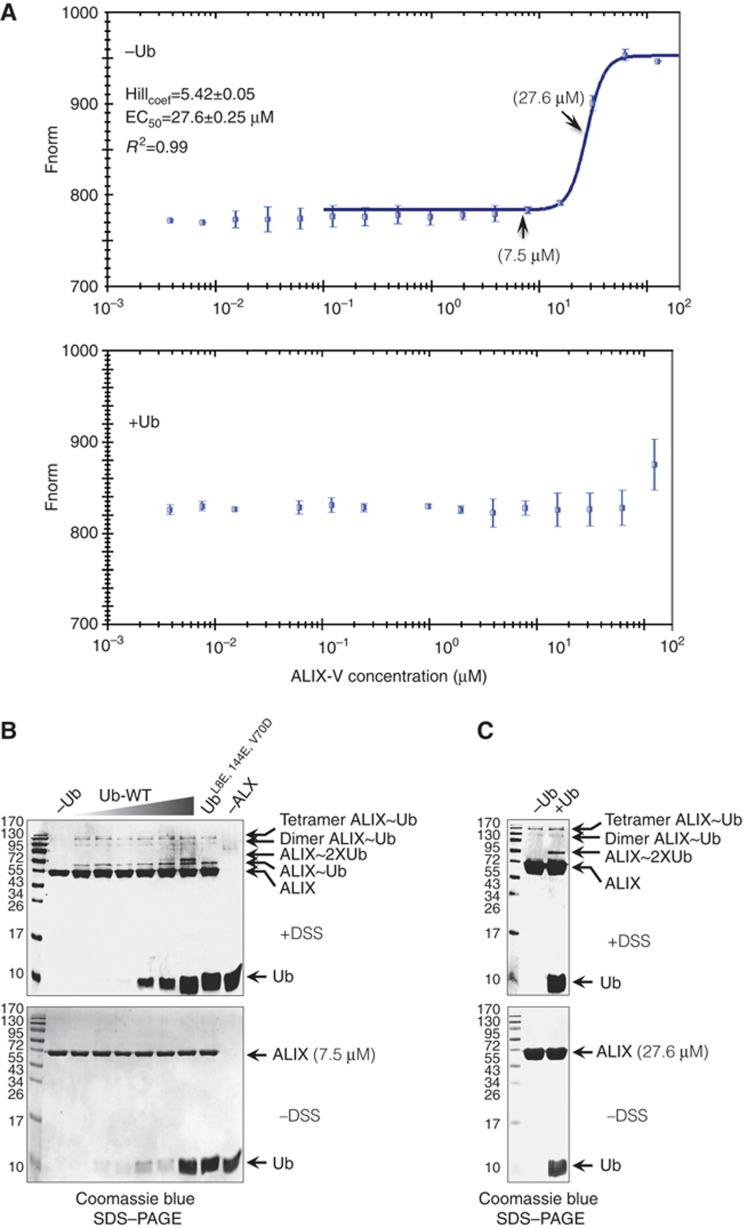
The significant difference in the Kd values that were derived from the SPR compared with those from the MST prompted us to provide a molecular explanation for this discrepancy. One simple (parsimony) postulation is that Ub binding induces ALIX-V oligomerization. The SPR measurements were performed with immobilized ALIX-V and a flow of Ub analyte. In such a setting, we measure the affinity of mono-Ub for ALIX-V that cannot oligomerize. If natural Ub binding drives ALIX-V oligomerization, we expect to obtain a lower affinity in the SPR compared with the MST measurements where both ALIX and Ub are free in the aqueous solution (as they were indeed found to be). To biophysically quantify the oligomerization of ALIX-V and to test the effect of Ub on the oligomerization course, we used MST measurements of ALIX-V in the absence and presence of Ub. To eliminate the possibility that ALIX-V dimerization is driven by the GST fusion, the GST tag was cleaved and removed from GST-ALIX-V to yield a free ALIX-V domain. Low concentration of fluorescently labelled ALIX-V (20 nM) was mixed with increasing concentrations of free ALIX-V in the MST capillaries. The exchange of labelled with unlabelled molecules is quantified in the thermophoresis measurements. Figure 5A shows that in the absence of Ub, ALIX-V underwent oligomerization in a concentration-dependent manner (Figure 5A top). Moreover, Hill coefficient calculation yielded a cooperative value of 5.42 and EC50 value of 27.6±0.25 μM.
Figure 5.
Ub binding induces ALIX-V dimerization in vitro. (A) Shows MST experiment monitoring the oligomerization of ALIX-V in solution. Fluorescently labelled untagged ALIX-V at a constant concentration with increasing concentrations of non-labelled ALIX-V were used and changes in thermophoresis were measured (top). Hill analysis resulted in concentration dependent oligomerization; (bottom) oligomerization in the presence of Ub. (B) Cross-linking assay of ALIX-V. In ALIX-V concentration of 7 μM (below the EC50), oligomerization is not detected (first lane from left). Increasing Ub concentrations 1.5, 4.0, 7.5, 15.0, 40.0 and 75.0 μM promote the oligomerization of ALIX-V (lanes 2–7). Cross-linking assay with 75.0 μM of L8E, I44E and V70D triple Ub mutant is shown (lane 8). Control with maximal Ub concentration but without ALIX-V is shown (lane 9). Control without cross-linker is shown at the bottom gel. (C) Cross-linking assay of ALIX-V in the EC50 concentration (27.6 μM), oligomerization of ALIX-V is shown in the presence and absence of 75.0 μM Ub.
However, in the presence of a saturating concentration of Ub (1 mM) no exchange between the labelled and the unlabelled ALIX-V molecules was observed, suggesting that Ub is stabilizing the oligomeric complex and therefore reducing the exchange between labelled and unlabelled ALIX-V molecules (Figure 5A bottom).
To biochemically test the latter hypothesis, we performed a cross-linking assay. By fixing a dynamic binding, the cross-linking assay provides an excellent biochemical tool to discover weak protein–protein interactions. To eliminate nonspecific cross-linking, a mild cross-linker disuccinimidyl suberate (DSS) was used. Aiming to demonstrate the function of Ub in stabilizing and/or inducing ALIX-V oligomerization, we chose to set up the experiment with 7 μM ALIX-V, a concentration that represents the very beginning of the steep slope in the Hill curve (Figure 5A top) together with increasing Ub concentrations (1.5–70 μM). In accordance with our expectations, we obtained a low yield of Ub binding and oligomerization, but induction of oligomerization by Ub is clearly observed. Figure 5B shows that in the absence of Ub, ALIX-V did not oligomerize in the given concentration (lane 1). Moreover, ALIX-V underwent dimerization and tetramerization even at the lowest Ub concentration, suggesting that mono-Ub induces the oligomerization (compare lane 1 without Ub and lane 2). Nevertheless, increasing the Ub concentration did not yield significant augmentation in the oligomeric ALIX-V:Ub complexes. Due to the low contribution of Ub binding to the mass shift, the SDS–PAGE cannot resolve whether one or two Ub molecules form the complex with dimeric/tetrameric ALIX-V, but the simplest model is that the complex is composed of four ALIX-V and four Ub molecules (or a 2:2 complex, which is also seen to a lesser extent). Tetramerization and higher-order oligomerization of proteins that undergo dimerization is a common phenomenon seen in cross-linking assays and does not necessarily reflect biological significance. Nevertheless, most of the observed complexes are of terameric ALIX:Ub and no higher-order structures were detected, thus implying a specific interaction. Taken together with the Hill value, it is certainly possible that ALIX-V forms tetra or perhaps even hexa-oligomers. To obtain a better view of these possible complexes, we repeated the experiment with an ALIX-V concentration equal to the EC50 (i.e., 27.6 μM; Figure 5C). Indeed, at this concentration, oligomerization is clearly observed without Ub, unlike the cross-linking at 7.5 μM (compare lanes 1 from the marker in Figure 5B and C). Moreover, the majority of the cross-linked complexes seemed to be tetrameric. A very faint but distinct band, likely to be a pentamer, is also observed.
Interestingly, at the high Ub concentrations, two Ub molecules simultaneously bind a single ALIX-V (lanes 6–7 from the marker in Figure 5B), suggesting that ALIX-V possesses a second Ub-binding site with significantly lower affinity. Indeed, in the triple Ub mutant (L8E, I44E and V70D), binding is still observed at the highest Ub concentration, but only a single Ub molecule binding is detected. These results also suggest that additional residues not related to the hydrophobic patch on the Ub surface mediate the interaction with ALIX-V. The two ALIX-V-binding patches on the Ub surface provide a molecular explanation for the mechanism of stabilizing the oligomerization of ALIX-V. It seems that Ub has divalent properties in ALIX-V binding and therefore can induce oligomerization. Finally, the new in silico procedure described above was not able to find this second Ub-binding site not only because it is weaker but also because it probably involves residues other than the I44 hydrophobic patch.
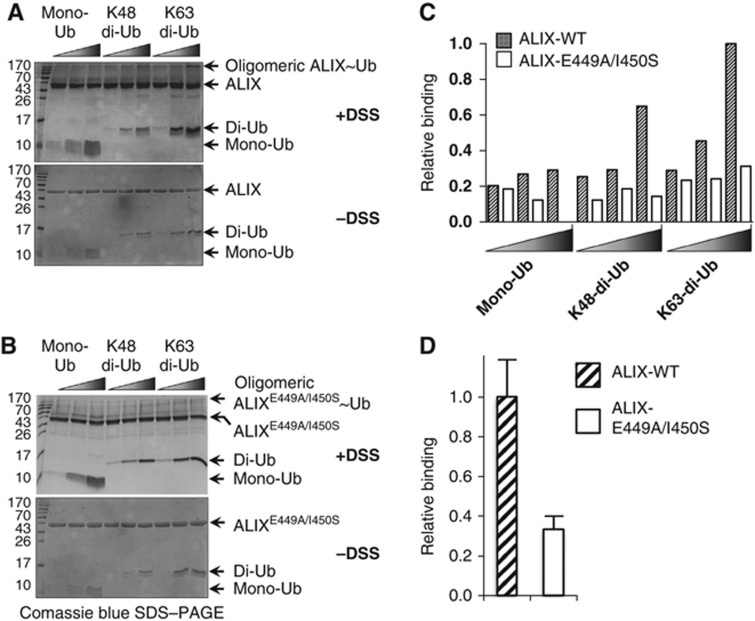
ALIX-V specifically binds K63 di-Ub
ALIX-V is a member of the ESCRT machinery that sorts short K63 poly-ubiquitylated substrates into luminal vesicles of the MVB. We therefore hypothesized that ALIX-V preferentially binds and responds to the K63 poly-Ub signal. This suggests that ALIX-V possesses one or more additional Ub-binding site(s). Why does the newly presented in silico procedure not detect additional binding site(s) on ALIX-V? One possibility is that the additional binding site is significantly weaker and involves fewer interactions below the detection threshold. Alternatively, as mentioned above, it is possible that the interaction of additional Ub is mediated through an unconventional (not I44) binding patch on the Ub surface and therefore cannot be detected by the new procedure. To assess if ALIX-V prefers K63 di-Ub chains over mono-Ub or K48 di-Ub, we performed cross-linking assays with WT and E449A/I450S double-mutant ALIX-V proteins against mono-Ub K48 and K63 di-Ub (Figure 6). Specifically, in these experiments the GST tag has been removed from the ALIX-V derivatives to avoid homodimerization derived from the GST. As shown in Figure 6A and B, the cross-linking assays were performed with increasing concentrations of the different Ub forms, while the ALIX-V concentration remained constant. Notably, these experiments were performed at a concentration of 7.5 μM at which ALIX-V does not undergo self-oligomerization (as shown in Figure 5). As expected, our data clearly show that ALIX-V displayed higher affinity towards K63 di-Ub compared with K48 di-Ub and mono-Ub. While large differences in binding of the Ub forms were noticeable with the WT ALIX-V, the binding of these Ub forms to the ALIX-V double-mutant did not show significant differences and they all seemed to be weak (Figure 6C). This suggests that ALIX-V possesses at least two Ub-binding sites, and that one of these sites is abolished in the double-mutant protein. Hence, the mutant does not constitute preferential affinity towards mono-Ub or di-Ub signals. To quantitatively compare the affinity of WT and the mutant ALIX-V proteins to K63 di-Ub, repetitive cross-linking experiments at the distinctive concentrations (7.5 μM ALIX-V and 40 μM di-Ub) have been performed. Figure 6D shows a reduction of nearly 70% in the affinity of the ALIX-V mutant to K63 di-Ub compared to the WT. Moreover, this result is consistent with our MST measurements, and with the recent findings by Kopito and co-workers that ALIX-V triple mutant at the same Ub-binding site presents about 80% reduction in its affinity to K63 poly-Ub (Dowlatshahi et al, 2012).
Figure 6.
ALIX-V preferentially binds K63 di-Ub. (A) 7.5 μM ALIX-V WT and (B) ALIX-V E449A/I450S double-mutant were incubated with increasing concentrations (5, 20 and 40 μM) of mono-Ub, K48 and K63 di-Ub chains, in the presence (upper panel) and in the absence (lower panel) of 0.5 mM DSS for 60 min. (C) Histogram quantification of the bands was performed by gel analysis with ImageJ and Origin softwares. (D) Normalized ratios of the oligomeric complex to the monomeric ALIX-V bands are presented.
ALIX-V:Ub binding disrupts Ub-dependent retrovirus budding in a dominant-negative fashion
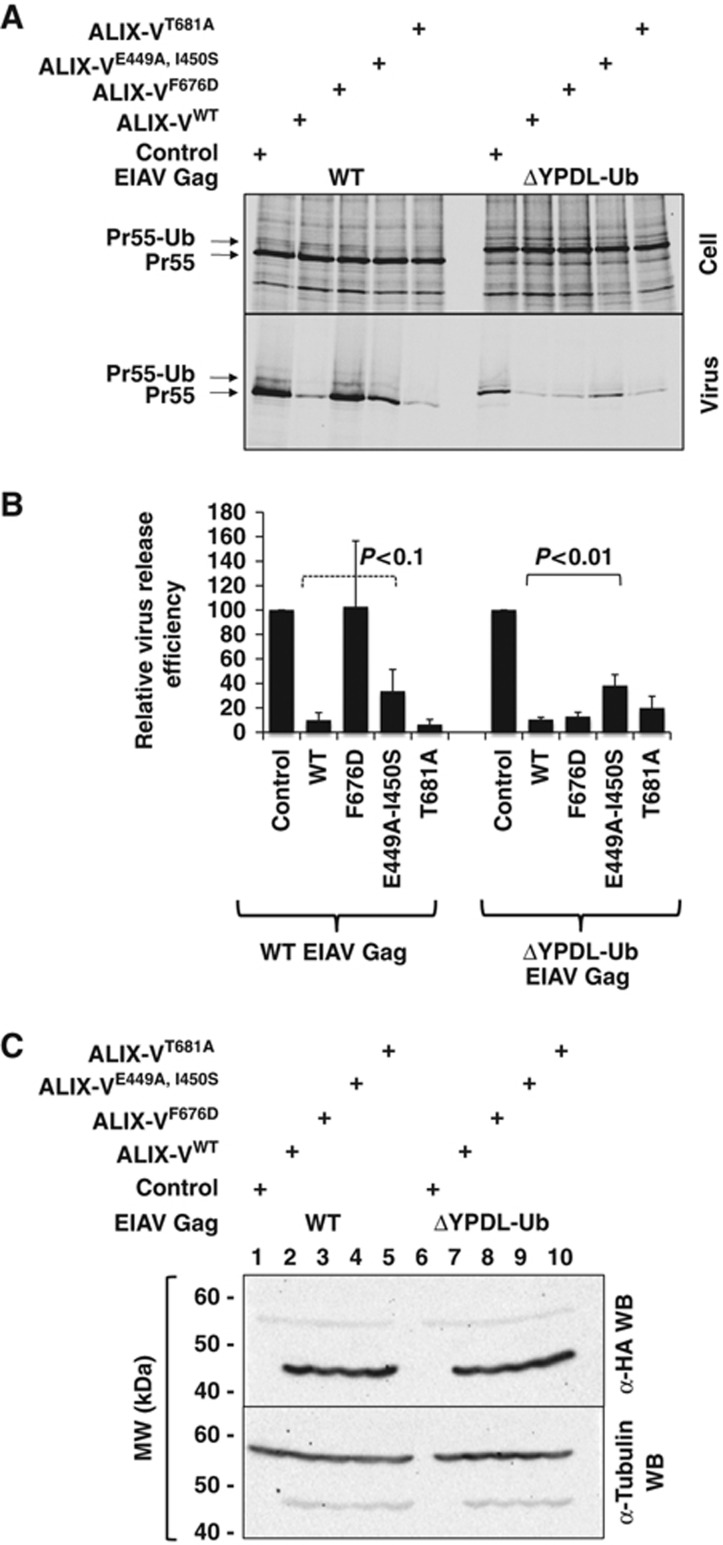
It has been shown that ALIX-V specifically binds retroviral late domains such as those present in HIV-p6 and EIAV-p9. In the case of EIAV, the interaction between YPDL and ALIX is largely sufficient for virus budding (Fujii et al, 2007; Gottlinger, 2007). However, it has been demonstrated that the YPDL late domain of EIAV Gag could be functionally replaced by a C-terminal Ub fusion in a modified Gag construct named EIAV/ΔYPDL-Ub (Joshi et al, 2008). Introduction of L8A/I44A mutations into the Ub fused to EIAV/ΔYPDL Gag abrogated the Ub-mediated rescue of virus release, suggesting that the fused Ub interacts with a UBD in the ESCRT pathway, most likely in Tsg101 and/or perhaps in ALIX-V, to recruit the ESCRT machinery to the site of budding. The release of EIAV/ΔYPDL-Ub could be inhibited by overexpression of ALIX-V. These results led to the proposal that ALIX-V interacted with the fused Ub in EIAV/ΔYPDL-Ub, thus preventing the fused Ub from binding its ESCRT partner. Furthermore, as mentioned above, Ub–agarose pull-downs suggested that ALIX-V binds Ub (Joshi et al, 2008). A prediction from these results is that interfering with the ALIX-V:Ub interaction should mitigate the ability of ALIX-V to block the release of EIAV/ΔYPDL-Ub. This model system thus provides an ideal in vivo system for evaluating the proposed interaction between ALIX-V and Ub. To test this hypothesis, we examined the effect of mutations in the putative ALIX-V UBD on the ability of Alix-V to block the budding of EIAV/ΔYPDL-Ub. 293T cells were transfected with WT EIAV Gag plus vector control or WT or mutant ALIX-V. As we reported earlier (Munshi et al, 2007), WT ALIX-V substantially inhibited release of EIAV virus-like particles (VLPs) (Figure 7A and B). This inhibition was relieved by the F676D mutation, which blocks binding between the YPDL late domain motif in EIAV and ALIX-V (Munshi et al, 2007; Fisher et al, 2007; Lee et al, 2007; Zhai et al, 2008). Mutation of the Ub-binding site of ALIX-V (E449A-I450S or T681A) did not significantly affect ALIX-V-mediated inhibition, consistent with the YPDL motif being the major site for interaction between WT EIAV and ALIX-V. In contrast, the ability of ALIX-V to inhibit release of EIAV/ΔYPDL-Ub VLPs was significantly diminished by the E449A-I450S Ub-binding site mutations. The single mutations had no significant effect on ALIX-V-mediated inhibition, likely due to their smaller effect on Ub binding. All ALIX-V mutants were expressed at comparable levels (Figure 7C). However, the triple mutant (E449A, I450S and T681A) described above was unstable in cell-based assays and was therefore not included in this analysis (data not shown). Taken together, these results demonstrate that the ALIX-V:Ub interaction occurs in vivo and has functional implications for retrovirus budding.
Figure 7.
Mutation of the ALIX-V:Ub interface mitigates the ability of ALIX-V to block the release of EIAV/ΔYPDL-Ub. (A) EIAV Gag VLP release assay. 293T cells were co-transfected with WT EIAV Gag (lanes 1–5) or ΔYPDL-Ub EIAV Gag (lanes 6–10) and WT or mutant HA-ALIX-V. Cell (top) and VLP (bottom) radioimmunoprecipitates were quantitated by phosphorimager analysis. (B) Relative VLP release efficiency was calculated as Pr55Gag in the virus supernatant divided by total Gag (cell+virus). Error bars denote s.d., N=3. GraphPad t-test calculation of statistical significance, dashed lined indicates lack of statistical significance (P<0.1), while solid line indicates statistical significance (P<0.01). (C) Western blot analysis of transfected cells lysates with anti-HA antibody to demonstrate equivalent HA-ALIX-V protein expression and equal sample loading.
Discussion
The linkage between homology and functionality postulates the basis for fundamental biological studies. Indeed, in the genomic and structural-genomic era, in silico sequence alignment algorithms like BLAST (Altschul et al, 1990) or structure alignment algorithms like Geometric Hashing (Wolfson and Rigoutsos, 1977; Nussinov and Wolfson, 1991) and DALI (Holm and Sander, 1993) provide tools to facilitate the downstream biochemical and biophysical functional identification of proteins. However, in some cases, proteins with <5% sequence identity may form identical folds and perform a similar function (Prag et al, 2007; Ren et al, 2009). Alternatively, proteins with identical folds, such as the TIM-barrel, may have very different functions (Nagano et al, 2002). In fact, usually it is the protein surface that provides the framework for the protein’s functional properties. Interestingly, a unique surface patch can be achieved by different sequences and folds as demonstrated in the case of different UBDs that recognize the same surface landscape of the Ub (Figure 1). Generally, proteins are assumed to perform similar functions if they share similar binding patterns and recognize similar binding partners, even if they have different sequences and different (overall) fold homology. Therefore, surface recognition algorithms provide a complementary and highly important tool to facilitate identification of protein function. In this work, we applied a surface recognition approach to investigate the physico-chemical patterns and shape of proteins that bind Ub.
Signalling for protein membrane trafficking is one of the pleiotropic roles of Ub. While in yeast many of the membrane cargo proteins are mono-ubiquitylated, in higher eukaryotes multi-mono-ubiquitylation and short K63 polyubiquitylation are the major signals for membrane protein trafficking (Hicke and Riezman, 1996; Galan and Haguenauer-Tsapis, 1997; Haglund et al, 2003; Staub and Rotin, 2006). Yeast genetics studies demonstrated the linkage between K63-polyubiquitylation and deubiquitylation of transmembrane proteins such as the general amino-acid permease transporter, namely, Gap1 and its Bro1 (the yeast ortholog of ALIX)-dependent MVB trafficking (Springael et al, 1999a, 1999b, 2002). This linkage genetically suggests that BRO1 acts as a Ub receptor at the MVB. Ub receptors have to distinguish between different Ub signals, for example by their ability to recognize the structural differences between K63 and K48 poly-Ub chains. Since ALIX functions on membrane proteins, we expect that it would present a higher affinity towards K63 poly-Ub conjugates compared with K48 poly-Ub conjugates. Indeed, in parallel to this study, Kopito and co-workers showed that ALIX is a bona fide Ub receptor that selectively binds K63 poly-Ub (Dowlatshahi et al, 2012). In this study, we took a different approach to identify novel UBD-containing proteins. Assuming that there are unidentified UBDs within the PDB, we have applied an algorithm that aligns known Ub-binding sites (represented as configurations of physico-chemical properties) to the molecular surfaces of all the protein chains in the PDB, ranking the detected hits by the size of the aligned configurations and the shape compatibility of the aligned patches. The high speed of the algorithm allows screening of the entire PDB and provides an excellent tool for such studies, which can be applied to other cases as well.
The finding of ALIX as a potential Ub receptor was not surprising given the genetic linkage mentioned above and the knowledge of its participation in the trafficking of ubiquitylated-transmembrane proteins and its interaction with the ESCRT machinery. It would be interesting to explore if other domains within ALIX proteins in evolutionary remote organisms bind Ub. In fact, the minimal requirements for binding Ub as presented in the well-defined patches on the UBD surfaces provide a structural explanation for potential convergent evolution mechanisms that may have pushed these domains to acquire the capability to bind Ub (Figure 1). One example is the yeast Vps9 and its human ortholog RABEX5 Ub receptors (Shih et al, 2003; Prag et al, 2005; Lee et al, 2006). Both of these proteins bind Ub and undergo coupled auto-mono-ubiquitylation. However, while the yeast protein binds and is auto-ubiquitylated via a UBD, namely CUE at its C-terminus, the mammalian protein has two distinct UBDs, namely iUIM and A20-like ZnF, that participate in ubiquitylated-cargo recognition and auto-ubiquitylation, respectively. These two domains are totally different from each other and from the CUE domain. Furthermore, they are located at the N-terminus of RABEX5.
It was previously shown that ALIX-V undergoes dimerization (Pires et al, 2009). Moreover, mutating the dimerization interface was shown to decrease the activity of ALIX-V in HIV budding (Carlton et al, 2008). It was also shown that ALIX multimerization is important for binding TGS101 (Carlton et al, 2008). These data suggest that dimerization is important for ALIX activity.
To promote cytokinesis, MVB formation and retrovirus budding, ALIX has to recruit CHMP4/Snf7 subunits of the ESCRT-III complex, which in turn participate in membrane remodelling and fission. A single CHMP4-like C-terminal peptide forms an amphipathic helix that binds the concave surface of the ALIX Bro1 domain in 1:1 stoichiometry (McCullough et al, 2008). However, the ESCRT-III complex is believed to oligomerize into a large spiral shape at the MVB membrane. Our data showing self-oligomerization of ALIX and possible regulation of this process by Ub may shed light on its function in binding and stabilizing/destabilizing the oligomerization of the ESCRT-I and III complexes. Future studies will explore how oligomerization takes place in the presence of a membrane, and specifically if and how Ub regulates this process.
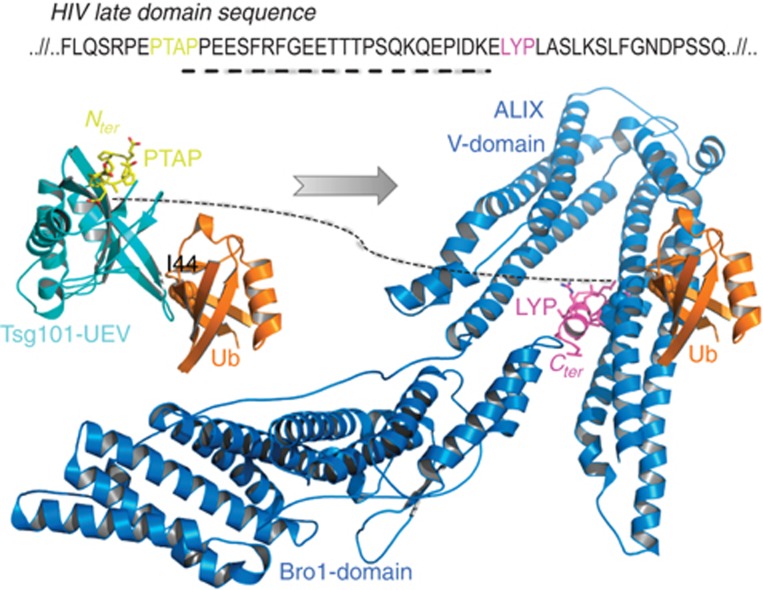
Our results provide a revised view of the mechanisms of retrovirus budding. An alternative route besides the virus LYPXnL:ALIX interaction is achieved by ubiquitylation of the viral late domain followed by direct interaction of this ubiquitylated protein with ALIX-V domain. ALIX recruits the ESCRT-III complex to complete viral membrane abscission. It seems that these two alternatives are redundant and provide robustness to this protein network (Citri and Yarden, 2006; Amit et al, 2007). Figure 8 provides a mechanistic scheme for ubiquitylated versus PTAP/LYPXnL-dependent budding of retroviruses. Our study shows that these two events are complementary since they do not compete on the ALIX-V interface but use different binding patches. Moreover, it is certainly possible that viruses or normal cell division processes utilize these two routes in a synergistic manner. Specifically, the model shows how the ubiquitylated late domain interacts with the UEV domain of TSG101 both by the recognition of the PTAP sequence and the I44 patch on the Ub surface. Similarly, a downstream LYPXnL viral late domain interacts with ALIX-V at the F676-binding site and the newly reported binding site of Ub. In both cases, the Ub-binding patches are at the opposite sides of the peptide-binding sites. In addition to functioning in retroviral budding, the binding of Ub by ALIX-V and Ub-induced multimerization could also function in other activities of ALIX, for example, ESCRT-III recruitment to sites of membrane scission. Given the number of interaction partners that have been reported for the C-terminal ALIX PRR, a wealth of possibilities exist for cellular functions modulated by ALIX-V:Ub binding.
Figure 8.
A model showing how ubiquitylated retroviral late domain is transferred from TSG101-UEV domain to ALIX. The sequence is of HIV p6. Structures of UEV:Ub, UEV:PTAP were superimposed to make the Ub:UEV:PTAP complex. ALIX:Ub complex from this study was superimposed with the ALIX:LYP late domain peptide to create Ub:ALIX:LYP complex. The dashed line represents the sequence between the two interacting peptides PTAP and LYP. ALIX-F676 and Ub-I44 are rendered as space-fill. The PTAP is coloured in yellow and the LYP in magenta.
While this manuscript was under review, Dowlatshahi et al (2012) reported data consistent with the findings presented here. They demonstrated that the ALIX-V domain binds to K63-linked poly-Ub chains. Mutation of the residues in ALIX-V responsible for this poly-Ub binding reduced the ability of ALIX overexpression to rescue the virus release defect conferred by HIV-1 PTAP mutation. Likewise, this mutant ALIX displayed reduced ability to rescue the EIAV release defect imposed by ALIX depletion. These data support the hypothesis that ALIX-V domain binding to poly-Ub plays a physiologically significant function in retrovirus release.
In conclusion, the described in silico search procedure enabled us to identify and to characterize one example of a novel UBD, namely ALIX-V, and provided a number of additional potential hidden UBDs within the PDB. Future in vitro and in vivo studies are required to test these hypothetical complexes. Moreover, the paradigm of this in silico algorithm is not limited to UBD:Ub complexes, but could also be applied to the search of other protein–protein interactions.
Materials and methods
In silico procedure
The SiteEngine (Shulman-Peleg et al, 2005) and FiberDock (Mashiach et al, 2010a) algorithms were applied in order to choose a template Ub-binding site and scan with it all the ∼30 000 single chains of proteins from eukaryotic organisms in the PDB (Berman et al, 2000). The results were ranked by the SiteEngine score, which is a weighted sum taking into account aligned physico-chemical properties as well as molecular surface shape compatibility. In addition the FiberDock score, which represents the binding energy between the Ub and a putative UBD, was used to assess the quality of the results and to model minor conformational changes in the resulting complex.
Cloning
Ub WT and mutant were expressed from pHIS-parallel2 plasmid as hexahistidine fusions (Sheffield et al, 1999). Ub WT or Ub mutant L8E/I44E/V70D ORFs was subcloned between BamHI and EcoRI endonucleases restriction sites downstream to a TEV protease cleavage site.
Site-directed mutagenesis
Point mutations were introduced using the ExSite (Stratagene) protocol on the pGST-ALIX-V plasmids encoding residues 360–702 of the human ALIX-V (Lee et al, 2007). All constructs were verified by DNA sequencing to ensure introduction of the desired mutations and that no other mutations were introduced.
Protein expression and purification
Competent E. coli cells were transformed with pGST-ALIX-V WT or mutants. Cultures were grown in Terrific Broth medium at 37°C. Each culture was induced by addition of 1 mM isopropyl β-D-1-thiogalactopyranoside (IPTG) and was further grown at 16°C overnight. Cells were harvested by centrifugation and were resuspended in buffer (PBS, pH=7.4) supplemented with lysozyme, AEBSF (4-(2-aminoethyl) benzenesulphonyl fluoride hydrochloride) DNAse and 7 mM β-mercaptoethanol. Complete lysis and DNA shearing were achieved by sonication following centrifugation to isolate the soluble fraction. Proteins were affinity-purified using reduced glutathione (Macherey–Nagel Ltd) according to the manufacturer's instructions. Proteins were further purified by size exclusion chromatography (supedex 200) against buffer (10 mM Hepes pH=7.4, 150 mM NaCl, 0.001% P-20) and concentrated using centricon (Amicon Ultra).
Ub and Ub triple mutant were expressed as His6 fusions. Proteins were affinity-purified using Ni2+ matrix (GE Healthcare Ltd) according to the manufacturer's instructions. Affinity tag was removed with His6-tagged TEV protease. Proteins were further purified by size exclusion (supedex 75) chromatography and concentrated using centricon (Amicon Ultra) to 5–6.25 mM.
K48 and K63 di-Ub were purified from bacterial synthetic system that expresses the ubiquitylation cascade as previously described (Keren-Kaplan et al, 2012). Briefly, Ub was bacterially co-expressed with E1, and E2-25K or Ubc13 and UEV1a to produce K48 or K63 poly-Ub chains, respectively. Poly-Ub chains were purified based on their high stability in perchloric acid followed by ion exchange and size exclusion chromatography steps.
E3-independent ubiquitylation in bacteria
Ubiquitylation assay was carried out as previously described (Keren-Kaplan et al, 2012). Briefly, Rosetta2(λDE3)BL21 E. coli cells were co-transformed with pGEN5 expressing His6 Ub, Wheat E1 and Ubch5 and with the pGST-ALIX-V WT. Cells were grown and lysed as described above. Lysates were affinity purified by Ni2+ and GSH beads and were separated on 12% SDS–PAGE.
Western blotting
Samples were transferred to a nitrocellulose membrane and incubated with rabbit α -His6 epitope tag antibody (1:20 000 dilution, Rockland), mouse α-Ub antibody, or mouse α-GST antibody (1:200, Santa Cruz), and infrared dye coupled goat α-mouse secondary antibody (1:12 000, LI-COR). Scanning was performed with the Odyssey infrared imaging system (LI-COR Biosciences) in accordance with the manufacturer’s instructions at 700 and 800 nm.
Pull-down experiments
Seven hundred and fifty microlitres of lysates expressing different ALIX-V GST- fusions were diluted with 750 μl binding buffer (25 mM Hepes-Na pH 7.4, 125 mM KOAc, 2.5 mM MgOAc, 5 mM EGTA (ethylene glycol tetraacetic acid),1 mM DTT (dithiothreitol)) to a final volume of 1500 μl. The mixture was incubated with 1 μl of Ni2+ beads that were incubated with His6-Ub or His6-Ub mutant L8E/I44E/V70D for 2 h at 4°C. Unbound fraction was removed by centrifugation (3 min, 500 g) and beads were washed six times with 1 ml of binding buffer. Forty microlitres of Laemmli loading buffer was added to each sample. The bound and unbound fractions were analysed by SDS–PAGE.
Cross-linking assays
Cross-linking assays were carried out according to the method described by Azem and co-workers (Azem et al, 1998). Reactions were carried out with 0.5 mM DSS in 25 mM Hepes buffer pH 7.4, 150 mM NaCl. Reactions were stopped after 18 min by boiling the solution in SDS-loading buffer and samples were analysed by SDS–PAGE.
SPR measurements and analysis
SPR measurements were carried out with BiaCoreT200 instrument (GE healthcare) at 25°C. Proteins were purified on size exclusion chromatography prior to the experiment. GST-ALIX ligand was trapped with α-GST antibody that was chemically immobilized on a CM5 chip with GST capture kit according to the manufacturer’s protocol (GE healthcare Ltd). Experiments were carried out in 0.01 M Hepes pH=7.4, 0.15 M NaCl, 3 mM ethylenediaminetetraacetic acid (EDTA) and 0.005% P-20 buffer. Binding level was tracked by measuring change in response units (RUs) following the injection of various Ub concentrations (0.1, 10, 100, 600, 800, 1000 and 1750 μM). Three repeats of the experiment were averaged. Curve fit and Kd were calculated using GraphPad Prism on the basis of a single site-specific binding model.
MST analyses
Binding experiments were carried out with Monolith NT.Label Free (Nano Temper Technologies GMBH). Briefly, 4 μl of sample consisted from 2 μM ALIX-V protein and titrations of Ub or Ub mutants were loaded on standard treated silicon capillary (K002 Monolith NT.115) and thermophoresis in 280 nm was measured. Each measurement was taken twice. Experiments were carried out in 10 mM Hepes pH=7.4, 150 mM NaCl, 0.001% P-20. Analysis was performed with the Monolith software. Dimerization experiments were carried out with Monolith NT.115. ALIX protein was labelled with L001 Monolith NT.115 Protein Labelling Kit RED-NHS (Amine Reactive) dye. Twenty microlitres titrations of non-labelled ALIX-V were mixed with 20-μl labelled ALIX-V in a constant concentration. Four microlitres were mounted on hydrophobic silicon capillary (K003 Monolith NT.115) and used for each measurement. From this stock, 20 μl of each concentration were taken and 4 μl of 2.5 mM Ub was added to a final concentration of 0.5 mM.
EIAV VLP release assay
293T cells were transected with plasmids encoding WT EIAV Gag or DYPDL-Ub EIAV Gag (Li et al, 2002; Shehu-Xhilaga et al, 2004; Joshi et al, 2008) and HA-ALIX-V (Munshi et al, 2007; Joshi et al, 2008) using linear polyethylenimine. At 24 h post transfection, cells were starved in Met/Cys-free medium for 30 min and metabolically labelled with [35S]Met/Cys-Pro-mix (Amersham) for 5 h. Virus pellets were collected upon centrifugation of filtered culture supernatants for 1 h at 100 000 g. VLP and cell lysates were prepared in 0.5% Triton X-100 lysis buffer. Cell and virus lysates were immunoprecipitated with horse anti-EIAV serum (kindly provided by R Montelaro, University of Pittsburgh). Radioimmunoprecipitates were resolved by SDS–PAGE followed by phosphorimager quantitation. Western blotting was performed on cell lysates using anti-HA (clone HA-7, Sigma) and anti-tubulin (clone B-5-1-2, Sigma) antibodies.
Supplementary Material
Acknowledgments
We wish to thank Neta Tanner and Bella Zion for technical help and James Hurley for kindly providing the pGST-ALIX-V vector. This research was supported by grants from the Israeli Science Foundation (grants numbers 1695/08 and 464/11), from the EC FP7 Marie Curie International Reintegration Grant (PIRG03-GA-2008-231079), from the Israeli Ministry of Health (5108), and the Marianna and Jorge Saia Fund for HIV and Parkinson Diseases to GP. The Constantiner Institute for Molecular Genetics for travel support to TKK Research in the EOF laboratory is supported by the Intramural Research Programme of the Center for Cancer Research, National Cancer Institute, NIH, and by the Intramural AIDS Targeted Antiviral Programme. The Research of HJW was partially supported by Israel Science Foundation (ISF grant no. 1403/09) and the Minerva–Minkowski Center for Geometry. EF was supported by the Adams Fellowship Fund.
Author contributions: TKK and IA designed, performed and analysed the biochemical and biophysical affinity measurements, and participated in manuscript writing. ME, EF and AP performed the in silico analysis. LSK performed and analysed the data of the retrovirus budding experiments. MJ-W performed the MST studies. NB purified ALIX proteins. SA performed the E3-independent ubiquitylation. EOF designed and analysed the retrovirus budding experiments and participated in the manuscript writing. HJW designed and analysed the in silico experiments and participated in the manuscript writing. GP conceived the idea, designed the experiments, analysed the in silico data and wrote the manuscript.
Footnotes
The authors declare that they have no conflict of interest.
References
- Agromayor M, Soler N, Caballe A, Kueck T, Freund SM, Allen MD, Bycroft M, Perisic O, Ye Y, McDonald B, Scheel H, Hofmann K, Neil SJ, Martin-Serrano J, Williams RL (2012) The UBAP1 subunit of ESCRT-I interacts with ubiquitin via a SOUBA domain. Structure 20: 414–428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alam SL, Sun J, Payne M, Welch BD, Blake BK, Davis DR, Meyer HH, Emr SD, Sundquist WI (2004) Ubiquitin interactions of NZF zinc fingers. EMBO J 23: 1411–1421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215: 403–410 [DOI] [PubMed] [Google Scholar]
- Amit I, Wides R, Yarden Y (2007) Evolvable signaling networks of receptor tyrosine kinases: relevance of robustness to malignancy and to cancer therapy. Mol Syst Biol 3: 151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrusier N, Nussinov R, Wolfson HJ (2007) FireDock: fast interaction refinement in molecular docking. Proteins 69: 139–159 [DOI] [PubMed] [Google Scholar]
- Azem A, Weiss C, Goloubinoff P (1998) Structural analysis of GroE chaperonin complexes using chemical cross-linking. Methods Enzymol 290: 253–268 [DOI] [PubMed] [Google Scholar]
- Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE (2000) The protein data bank. Nucleic Acids Res 28: 235–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlton JG, Agromayor M, Martin-Serrano J (2008) Differential requirements for Alix and ESCRT-III in cytokinesis and HIV-1 release. Proc Natl Acad Sci USA 105: 10541–10546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- CCP4. (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr 50: 760–763 [DOI] [PubMed] [Google Scholar]
- Citri A, Yarden Y (2006) EGF-ERBB signalling: towards the systems level. Nat Rev Mol Cell Biol 7: 505–516 [DOI] [PubMed] [Google Scholar]
- Dowlatshahi DP, Sandrin V, Vivona S, Shaler TA, Kaiser SE, Melandri F, Sundquist WI, Kopito RR (2012) ALIX is a Lys63-specific polyubiquitin binding protein that functions in retrovirus budding. Dev Cell 23: 1247–1254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dror O, Shulman-Peleg A, Nussinov R, Wolfson HJ (2004) Predicting molecular interactions in silico: I. A guide to pharmacophore identification and its applications to drug design. Curr Med Chem 11: 71–90 [DOI] [PubMed] [Google Scholar]
- Fisher RD, Chung HY, Zhai Q, Robinson H, Sundquist WI, Hill CP (2007) Structural and biochemical studies of ALIX/AIP1 and its role in retrovirus budding. Cell 128: 841–852 [DOI] [PubMed] [Google Scholar]
- French ME, Kretzmann BR, Hicke L (2009) Regulation of the RSP5 ubiquitin ligase by an intrinsic ubiquitin-binding site. J Biol Chem 284: 12071–12079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii K, Hurley JH, Freed EO (2007) Beyond Tsg101: the role of Alix in 'ESCRTing' HIV-1. Nat Rev Microbiol 5: 912–916 [DOI] [PubMed] [Google Scholar]
- Galan JM, Haguenauer-Tsapis R (1997) Ubiquitin lys63 is involved in ubiquitination of a yeast plasma membrane protein. EMBO J 16: 5847–5854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottlinger HG (2007) How HIV-1 hijacks ALIX. Nat Struct Mol Biol 14: 254–256 [DOI] [PubMed] [Google Scholar]
- Haglund K, Sigismund S, Polo S, Szymkiewicz I, Di Fiore PP, Dikic I (2003) Multiple monoubiquitination of RTKs is sufficient for their endocytosis and degradation. Nat Cell Biol 5: 461–466 [DOI] [PubMed] [Google Scholar]
- Hicke L, Riezman H (1996) Ubiquitination of a yeast plasma membrane receptor signals its ligand-stimulated endocytosis. Cell 84: 277–287 [DOI] [PubMed] [Google Scholar]
- Hicke L, Schubert HL, Hill CP (2005) Ubiquitin-binding domains. Nat Rev Mol Cell Biol 6: 610–621 [DOI] [PubMed] [Google Scholar]
- Hoeller D, Hecker CM, Wagner S, Rogov V, Dotsch V, Dikic I (2007) E3-independent monoubiquitination of ubiquitin-binding proteins. Mol Cell 26: 891–898 [DOI] [PubMed] [Google Scholar]
- Holm L, Sander C (1993) Protein structure comparison by alignment of distance matrices. J Mol Biol 233: 123–138 [DOI] [PubMed] [Google Scholar]
- Hurley JH, Lee S, Prag G (2006) Ubiquitin-binding domains. Biochem J 399: 361–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husnjak K, Dikic I (2012) Ubiquitin-binding proteins: decoders of ubiquitin-mediated cellular functions. Annu Rev Biochem 81: 291–322 [DOI] [PubMed] [Google Scholar]
- Joshi A, Munshi U, Ablan SD, Nagashima K, Freed EO (2008) Functional replacement of a retroviral late domain by ubiquitin fusion. Traffic 9: 1972–1983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katzmann DJ, Babst M, Emr SD (2001) Ubiquitin-dependent sorting into the multivesicular body pathway requires the function of a conserved endosomal protein sorting complex, ESCRT-I. Cell 106: 145–155 [DOI] [PubMed] [Google Scholar]
- Keren-Kaplan T, Attali I, Motamedchaboki K, Davis BA, Tanner N, Reshef Y, Laudon E, Kolot M, Levin-Kravets O, Kleifeld O, Glickman M, Horazdovsky BF, Wolf DA, Prag G (2012) Synthetic biology approach to reconstituting the ubiquitylation cascade in bacteria. EMBO J 31: 378–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim W, Bennett EJ, Huttlin EL, Guo A, Li J, Possemato A, Sowa ME, Rad R, Rush J, Comb MJ, Harper JW, Gygi SP (2011) Systematic and quantitative assessment of the ubiquitin-modified proteome. Mol Cell 44: 325–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Joshi A, Nagashima K, Freed EO, Hurley JH (2007) Structural basis for viral late-domain binding to Alix. Nat Struct Mol Biol 14: 194–199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Tsai YC, Mattera R, Smith WJ, Kostelansky MS, Weissman AM, Bonifacino JS, Hurley JH (2006) Structural basis for ubiquitin recognition and autoubiquitination by Rabex-5. Nat Struct Mol Biol 13: 264–271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Chen C, Puffer BA, Montelaro RC (2002) Functional replacement and positional dependence of homologous and heterologous L domains in equine infectious anemia virus replication. J Virol 76: 1569–1577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mashiach E, Nussinov R, Wolfson HJ (2010a) FiberDock: a web server for flexible induced-fit backbone refinement in molecular docking. Nucleic Acids Res 38: W457–W461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mashiach E, Nussinov R, Wolfson HJ (2010b) FiberDock: flexible induced-fit backbone refinement in molecular docking. Proteins 78: 1503–1519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCullough J, Fisher RD, Whitby FG, Sundquist WI, Hill CP (2008) ALIX-CHMP4 interactions in the human ESCRT pathway. Proc Natl Acad Sci USA 105: 7687–7691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosesson Y, Chetrit D, Schley L, Berghoff J, Ziv T, Carvalho S, Milanezi F, Admon A, Schmitt F, Ehrlich M, Yarden Y (2009) Monoubiquitinylation regulates endosomal localization of Lst2, a negative regulator of EGF receptor signaling. Dev Cell 16: 687–698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munshi UM, Kim J, Nagashima K, Hurley JH, Freed EO (2007) An Alix fragment potently inhibits HIV-1 budding: characterization of binding to retroviral YPXL late domains. J Biol Chem 282: 3847–3855 [DOI] [PubMed] [Google Scholar]
- Nagano N, Orengo CA, Thornton JM (2002) One fold with many functions: the evolutionary relationships between TIM barrel families based on their sequences, structures and functions. J Mol Biol 321: 741–765 [DOI] [PubMed] [Google Scholar]
- Nussinov R, Wolfson HJ (1991) Efficient detection of three-dimensional structural motifs in biological macromolecules by computer vision techniques. Proc Natl Acad Sci USA 88: 10495–10499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pashkova N, Gakhar L, Winistorfer SC, Yu L, Ramaswamy S, Piper RC (2010) WD40 repeat propellers define a ubiquitin-binding domain that regulates turnover of F box proteins. Mol Cell 40: 433–443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pires R, Hartlieb B, Signor L, Schoehn G, Lata S, Roessle M, Moriscot C, Popov S, Hinz A, Jamin M, Boyer V, Sadoul R, Forest E, Svergun DI, Gottlinger HG, Weissenhorn W (2009) A crescent-shaped ALIX dimer targets ESCRT-III CHMP4 filaments. Structure 17: 843–856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prag G, Lee S, Mattera R, Arighi CN, Beach BM, Bonifacino JS, Hurley JH (2005) Structural mechanism for ubiquitinated-cargo recognition by the Golgi-localized, gamma-ear-containing, ADP-ribosylation-factor-binding proteins. Proc Natl Acad Sci USA 102: 2334–2339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prag G, Misra S, Jones EA, Ghirlando R, Davies BA, Horazdovsky BF, Hurley JH (2003) Mechanism of ubiquitin recognition by the CUE domain of Vps9p. Cell 113: 609–620 [DOI] [PubMed] [Google Scholar]
- Prag G, Watson H, Kim YC, Beach BM, Ghirlando R, Hummer G, Bonifacino JS, Hurley JH (2007) The Vps27/Hse1 complex is a GAT domain-based scaffold for ubiquitin-dependent sorting. Dev Cell 12: 973–986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren X, Hurley JH (2010) VHS domains of ESCRT-0 cooperate in high-avidity binding to polyubiquitinated cargo. EMBO J 29: 1045–1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren X, Hurley JH (2011) Proline-rich regions and motifs in trafficking: from ESCRT interaction to viral exploitation. Traffic 12: 1282–1290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren X, Kloer DP, Kim YC, Ghirlando R, Saidi LF, Hummer G, Hurley JH (2009) Hybrid structural model of the complete human ESCRT-0 complex. Structure 17: 406–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheffield P, Garrard S, Derewenda Z (1999) Overcoming expression and purification problems of RhoGDI using a family of “parallel” expression vectors. Protein Expr Purif 15: 34–39 [DOI] [PubMed] [Google Scholar]
- Shehu-Xhilaga M, Ablan S, Demirov DG, Chen C, Montelaro RC, Freed EO (2004) Late domain-dependent inhibition of equine infectious anemia virus budding. J Virol 78: 724–732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih SC, Prag G, Francis SA, Sutanto MA, Hurley JH, Hicke L (2003) A ubiquitin-binding motif required for intramolecular monoubiquitylation, the CUE domain. EMBO J 22: 1273–1281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shulman-Peleg A, Nussinov R, Wolfson HJ (2004) Recognition of functional sites in protein structures. J Mol Biol 339: 607–633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shulman-Peleg A, Nussinov R, Wolfson HJ (2005) SiteEngines: recognition and comparison of binding sites and protein-protein interfaces. Nucleic Acids Res 33: W337–W341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springael JY, De Craene JO, Andre B (1999a) The yeast Npi1/Rsp5 ubiquitin ligase lacking its N-terminal C2 domain is competent for ubiquitination but not for subsequent endocytosis of the gap1 permease. Biochem Biophys Res Commun 257: 561–566 [DOI] [PubMed] [Google Scholar]
- Springael JY, Galan JM, Haguenauer-Tsapis R, Andre B (1999b) NH4+-induced down-regulation of the Saccharomyces cerevisiae Gap1p permease involves its ubiquitination with lysine-63-linked chains. J Cell Sci 112: Pt 91375–1383 [DOI] [PubMed] [Google Scholar]
- Springael JY, Nikko E, Andre B, Marini AM (2002) Yeast Npi3/Bro1 is involved in ubiquitin-dependent control of permease trafficking. FEBS Lett 517: 103–109 [DOI] [PubMed] [Google Scholar]
- Staub O, Rotin D (2006) Role of ubiquitylation in cellular membrane transport. Physiol Rev 86: 669–707 [DOI] [PubMed] [Google Scholar]
- Wienken CJ, Baaske P, Rothbauer U, Braun D, Duhr S (2010) Protein-binding assays in biological liquids using microscale thermophoresis. Nat Commun 1: 100. [DOI] [PubMed] [Google Scholar]
- Wolfson HJ, Rigoutsos I (1977) Geometric hashing: an overview. Comput Sci Eng IEEE 4: 10–21 [Google Scholar]
- Zhai Q, Fisher RD, Chung HY, Myszka DG, Sundquist WI, Hill CP (2008) Structural and functional studies of ALIX interactions with YPX(n)L late domains of HIV-1 and EIAV. Nat Struct Mol Biol 15: 43–49 [DOI] [PubMed] [Google Scholar]
- Ziv I, Matiuhin Y, Kirkpatrick DS, Erpapazoglou Z, Leon S, Pantazopoulou M, Kim W, Gygi SP, Haguenauer-Tsapis R, Reis N, Glickman MH, Kleifeld O (2011) A perturbed ubiquitin landscape distinguishes between ubiquitin in trafficking and in proteolysis. Mol Cell Proteomics 10: M111009753 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.