Abstract
Selective autophagy underlies many of the important physiological roles that autophagy plays in multicellular organisms, but the mechanisms involved in cargo selection are poorly understood. Here we describe a molecular mechanism that can target conventional endosomes for autophagic degradation. We show that the human transmembrane protein TMEM59 contains a minimal 19-amino-acid peptide in its intracellular domain that promotes LC3 labelling and lysosomal targeting of its own endosomal compartment. Interestingly, this peptide defines a novel protein motif that mediates interaction with the WD-repeat domain of ATG16L1, thus providing a mechanistic basis for the activity. The motif is represented with the same ATG16L1-binding ability in other molecules, suggesting a more general relevance. We propose that this motif may play an important role in targeting specific membranous compartments for autophagic degradation, and therefore it may facilitate the search for adaptor proteins that promote selective autophagy by engaging ATG16L1. Endogenous TMEM59 interacts with ATG16L1 and mediates autophagy in response to Staphylococcus aureus infection.
Keywords: ATG16L1, autophagy, TMEM59, vesicle trafficking, WD-repeat domain
Introduction
Macroautophagy (hereafter referred to as autophagy) is a catabolic process that promotes degradation of bulk cytoplasmic constituents and recycles the resulting basic components as metabolic precursors (Mizushima, 2007; Yang and Klionsky, 2009). This phenomenon involves sequestration of the doomed material into double-membrane vesicles (autophagosomes) that eventually fuse with lysosomes for degradation of their contents (Rubinsztein et al, 2012). Although the autophagic process digests random cytoplasm to maintain nutrient supply during stressful situations (Rabinowitz and White, 2010; Singh and Cuervo, 2011), it can also target specific, superfluous or potentially harmful components, a less characterized phenomenon called selective autophagy (Komatsu and Ichimura, 2010; Kroemer et al, 2010; Mizushima and Komatsu, 2011).
The autophagic pathway is highly conserved in all eukaryotic organisms (Nakatogawa et al, 2009; Mizushima et al, 2011). In mammalian cells, two protein complexes (ULK1/ATG1–mTOR–ATG13–FIP200 and ClassIII Pi3K–BECLIN/ATG6–ATG14) act coordinately to initiate autophagosome nucleation (He and Klionsky, 2009). Membrane elongation and autophagosome closure are driven by two ubiquitin-like modification systems that converge in the lipidation of processed LC3/ATG8 (LC3I) to produce a membrane-bound form called LC3II (Noda et al, 2009; Yang and Klionsky, 2009). ATG12 and LC3 are ubiquitin-like modifiers, and ATG7 is the E1 enzyme for both molecules. In an E2-like step, ATG10 promotes the bonding of ATG12 to ATG5, and ATG3 binds LC3. ATG5–ATG12 then interacts with ATG16L1 to assemble a final E3 system for the conjugation of phosphatidylethanolamine (PE) to LC3I (Tanida, 2011). In this complex, ATG5–ATG12 holds the E3-ligase activity (Hanada et al, 2007), whereas ATG16L1 determines the site of LC3 lipidation (Fujita et al, 2008). Thus, ATG16L1 recruits ATG12–ATG5 to defined membrane localizations, and brings LC3 close to a membranous PE source through interaction between ATG12 and ATG3–LC3I (Fujita et al, 2008). LC3 lipidation and its association with autophagosomes are widely used as autophagic reporter systems (Mizushima et al, 2010).
Autophagy has important implications in processes like tumour suppression, neurodegeneration or native immunity (Cecconi and Levine, 2008; Levine and Kroemer, 2008; Levine et al, 2011), and selective autophagy plays a role in these activities (Mizushima et al, 2008; Mizushima and Komatsu, 2011). For example, tumour suppression might result from a specific elimination of damaged organelles that produce pro-inflammatory and DNA-damaging reactive oxygen species (Mathew et al, 2009; Dikic et al, 2010; Mathew and White, 2011). Atg deletion in the central nervous system (Hara et al, 2006; Komatsu et al, 2006) causes neurodegeneration through accumulation of insoluble protein aggregates (Komatsu et al, 2007), revealing a critical housekeeping role of autophagy in clearing toxic garbage (Garcia-Arencibia et al, 2010; Mizushima and Komatsu, 2011). Elimination of foreign invaders involves their recognition by the autophagic machinery, whether they are loose in the cytoplasm (Deretic and Levine, 2009; Deretic, 2011; Shahnazari and Brumell, 2011) or enclosed in phagosomes (Gutierrez et al, 2004; Sanjuan et al, 2007).
Although the mechanisms that provide cargo specificity in selective autophagy remain poorly understood, some examples are available. Damaged mitochondria (Youle and Narendra, 2011), insoluble protein precipitates (Knaevelsrud and Simonsen, 2010) or cytosolic bacteria (Fujita and Yoshimori, 2011; Randow, 2011) first become ubiquitinated, and adaptor proteins that simultaneously bind ubiquitin and LC3 target them for autophagic degradation (Kirkin et al, 2009b; Johansen and Lamark, 2011). p62 (Pankiv et al, 2007), NBR1 (Kirkin et al, 2009a), NDP52 (Thurston et al, 2009) and Optineurin (Wild et al, 2011) are some of these adaptors. Depolarized mitochondria are also subjected to autophagy by recruiting NIX/bNIP3L, another LC3-binding protein (Novak et al, 2010). Interestingly, all these linker proteins share a common LC3-interacting motif (Noda et al, 2010). In an additional example, phagosomes containing activated TLR2 recruit BECLIN/ATG6 to promote LC3 labelling of this otherwise non-autophagic compartment (Sanjuan et al, 2007). NOD proteins recognize bacteria at the entry site and bind ATG16L1 to cause LC3 activation (Travassos et al, 2010). Thus, a variety of proteins function as adaptor modules that couple the selected substrates directly with the autophagic machinery to promote LC3 decoration of the targeted item. Given the breadth of processes where selective autophagy is involved, one could anticipate the existence of additional linker families that, similar to ubiquitin/LC3 adaptors, might engage specific autophagic effectors through common protein signatures.
Here we show that the human transmembrane molecule TMEM59 defines a novel ATG16L1-binding motif through which the protein promotes labelling of its own endocytic compartment with LC3, in a process that links conventional endocytosis to autophagic degradation. Interestingly, the motif is present with a similar ATG16L1-binding activity in other molecules.
Results
TMEM59 induces LC3 activation
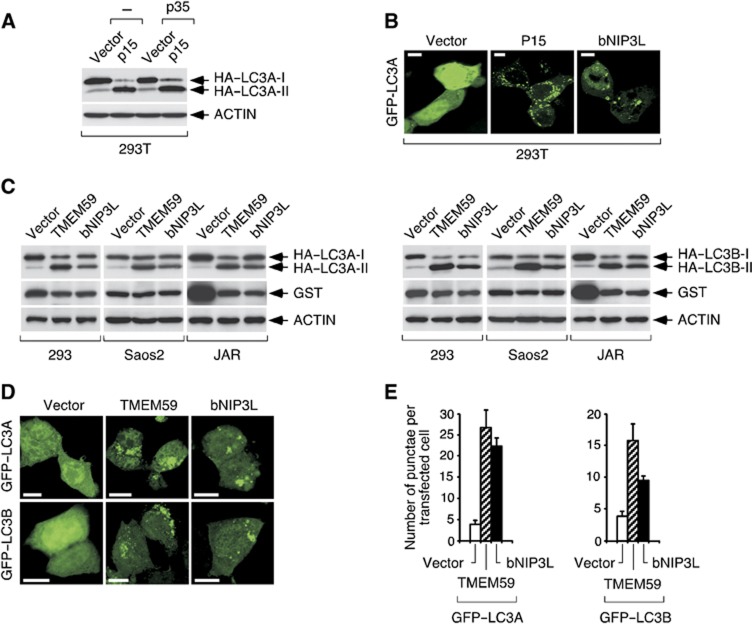
During an extended screening to identify cDNAs whose expression causes cell death (Alcalá et al, 2008), we found clone P15 as able to induce a morphologically atypical death modality (not shown). P15 encoded TMEM59 (Supplementary Figure S1A), a predicted type-I transmembrane protein (C terminus intracellular), known to regulate glycosylation of the amyloid precursor protein (APP) (Ullrich et al, 2010). Unconventional death morphologies have been linked to autophagic (type II) cell death (Shimizu et al, 2004), so to explore a possible pro-autophagic property of TMEM59 we confronted this molecule with reporter systems based on ATG8/LC3 (Mizushima et al, 2010). TMEM59 expression induced HA–LC3 lipidation (Figure 1A and C) and GFP–LC3 redistribution to vesicular structures (Figure 1B, D and E), thus confirming its pro-autophagic capacity. Although functional divergence between different LC3-family members may exist (Chen and Klionsky, 2011), TMEM59 activated the LC3A and B isoforms comparably (see Figure 1C–E).
Figure 1.
TMEM59 induces LC3 activation. (A) Clone P15 induces HA–LC3 lipidation. 293T cells were transfected with P15 plus plasmids expressing HA–LC3A and/or the apoptotic inhibitor p35 (as shown), and lysed for western blotting against the indicated molecules (anti-HA for HA–LC3A). The figure shows that expression of P15 induces HA–LC3A conversion to a lower molecular weight form indicative of protein lipidation. This activity remains unchanged by p35. (B) P15 induces GFP–LC3 translocation to a vesiculated pattern. 293T cells were transfected with the indicated plasmids mixed with vectors expressing GFP–LC3A and p35. The known autophagic inducer bNIP3L constituted a positive control. Representative confocal pictures are shown. (C) TMEM59 induces HA–LC3 lipidation in different cell lines lacking the SV40 large T-antigen plasmid amplification system. Cells were transfected with the shown plasmids, vectors expressing HA–LC3A (left) or HA–LC3B (right) and GST (as transfection control), and lysed for western blotting against the indicated molecules. (D) TMEM59 induces GFP–LC3 activation. 293 cells were transfected with the indicated plasmids and GFP–LC3A (top) or GFP–LC3B (bottom). Representative confocal pictures are shown. (E) Quantification of the phenotype in D. The number of GFP–LC3-positive vesicles per transfected cell was scored for at least fifty cells. Data are expressed as means ±s.d. of one representative experiment of three repetitions.
Source data for this figure is available on the online supplementary information page.
Characterization of endogenous TMEM59
Northern-blot assays revealed two TMEM59-specific mRNAs present in most cell types (Supplementary Figure S1B). Antibodies against the putative extracellular domain (N terminus) recognized a 34–36-kDa band (Supplementary Figure S1C and D) whose diffuse nature is likely due to glycosylation (Supplementary Figure S1E). TMEM59 localized to small cytoplasmic vesicles that were difficult to detect in immunolocalization studies (Supplementary Figure S2A), although the ectopic protein was easily observed (Supplementary Figure S2B). These results suggested low basal levels, perhaps as a consequence of active degradation. Consistently, inhibition of protein synthesis rapidly reduced TMEM59 expression (Supplementary Figure S2C), and lysosome inhibitors induced the protein without altering its mRNA levels (Supplementary Figure S2D–F). Therefore, the low basal expression of TMEM59 is probably due to intense lysosomal degradation. This constitutive degradation is not autophagic, because neither defective autophagy (Supplementary Figure S2G) nor increased autophagic turnover (Supplementary Figure S2H) altered TMEM59 expression levels.
The TMEM59-positive vesicles strongly colocalized with LAMP2 and CD63, and partially with EEA1 (Supplementary Figure S3A and B), suggesting a main localization in late endosomes/lysosomes and a transient presence in early endosomes. Although previous studies with a different antibody (against the C terminus) showed Golgi localization (Ullrich et al, 2010), we found no colocalization with Golgi markers (Supplementary Figure S3A and B). Intriguingly, transfected TMEM59 was expressed at the cell surface (Supplementary Figure S3C) but the endogenous molecule was not found in this location (Supplementary Figure S3D). Some lysosomal membrane proteins transit first through the plasma membrane (Janvier and Bonifacino, 2005; Saftig and Klumperman, 2009), an indirect route revealed by agents that promote their accumulation at the cell surface by inhibiting trafficking to the lysosome (e.g., chloroquine) (Lippincott-Schwartz and Fambrough, 1987). Consistent with this possibility, chloroquine treatment provoked surface exposure of TMEM59 (Supplementary Figure S3D–F). Bafilomycin caused the same effect, although less efficiently (not shown). Importantly, as all these staining procedures were done on unpermeabilized cells and the antibody recognizes the N-terminal part of the molecule, these results confirm a type I topology.
Therefore, TMEM59 is a glycosylated, type I transmembrane protein that mainly localizes to late endosomes/lysosomes. The protein is probably first exported to the cell surface and then actively endocytosed to transiently localize in early endosomes on its way to the late endosomal/lysosomal compartment. In this final destination, TMEM59 becomes quickly degraded, a phenomenon that results in low expression levels.
Since TMEM59 has no sequence features that could provide mechanistic clues about its pro-autophagic activity, we reasoned that the characterization of this function might reveal undescribed mechanisms of autophagic regulation.
TMEM59 induces autophagy through a minimal 19-amino-acid subdomain
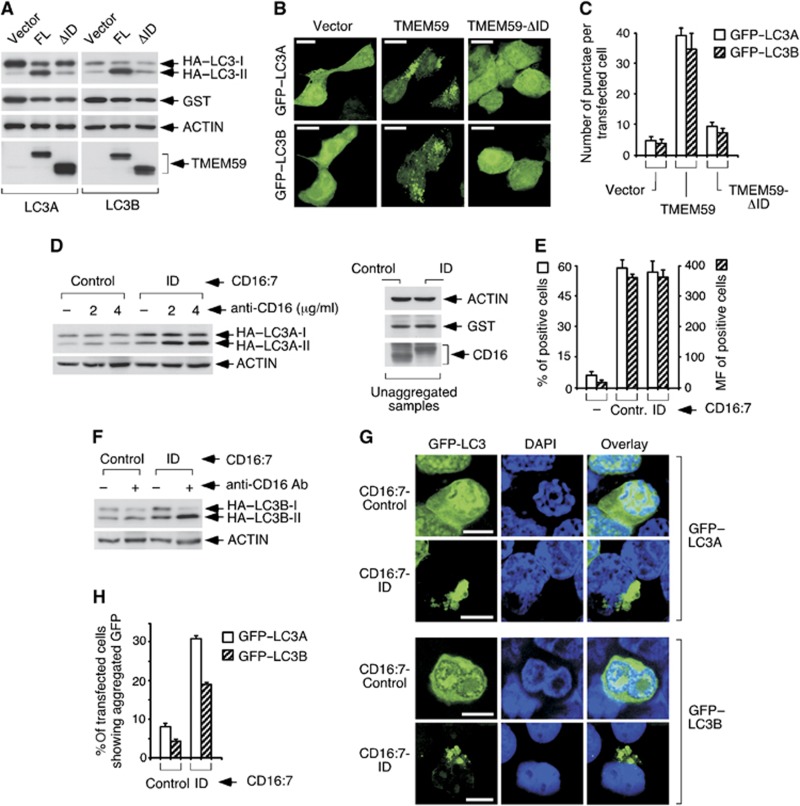
To dissect the autophagic activity of TMEM59, we first determined the signalling region. A deleted version of the molecule lacking the whole intracellular domain (ID) was unable to activate LC3 (Figure 2A–C), thus ascribing a necessary role to this part of the protein. To evaluate whether this domain was also sufficient to induce autophagy, we placed it in a different molecular context that preserves the type I transmembrane configuration. Chimeric molecules containing the extracellular part of CD16 and the transmembrane region of CD7 (CD16:7) fulfil this requirement, and have been used before in functional assays since they can be stimulated by aggregation with anti-CD16 antibodies (Kolanus et al, 1993). Aggregation of the CD16:7 chimera fused to the ID of TMEM59 caused HA–LC3 conversion (Figure 2D–F) and GFP–LC3 redistribution to a perinuclear vesicle cluster (Figure 2G and H). These results indicate that the cytoplasmic region of TMEM59 suffices for autophagy induction, and suggest that an aggregation event can unleash the activity.
Figure 2.
The ID of TMEM59 is necessary and sufficient for LC3 activation. (A) TMEM59 ID is required for HA–LC3 lipidation. 293 cells were transfected with full-length TMEM59 (FL) or a deleted version lacking the ID (ΔID, Δ263–323), HA–LC3A (left) or HA–LC3B (right) and GST. Cells were lysed for western blotting against the shown molecules. (B) The ID is necessary for GFP–LC3 activation. 293 cells were transfected with the indicated TMEM59 constructs and GFP–LC3A or GFP–LC3B (as indicated). Representative confocal pictures are shown. (C) Quantification of the phenotype in B. Scoring and data expression were as in Figure 1E. (D) TMEM59 ID (amino acids 263–323) suffices for HA–LC3A lipidation. 293 cells were transfected with the indicated CD16:7 chimera (Control: empty chimera) and vectors encoding HA–LC3A and GST, subjected to aggregation with the shown amounts of anti-CD16 antibody and lysed for western blotting. The right panel shows control WBs demonstrating equal loading (ACTIN), transfection (GST) and chimera expression (CD16) in unaggregated samples, as all experimental points per chimera derive from a single transfection. (E) Comparable surface levels of CD16:7 constructs. Transfected 293 cells were processed for anti-CD16 flow cytometry. The graph displays percentages of positive cells (left axis) and means of fluorescence of positive cells (MF, right axis) obtained from triplicates. Data are expressed as means ±s.d. of the triplicates. (F) The ID suffices for HA–LC3B lipidation. (G) The ID suffices for GFP–LC3 activation. 293 cells were transfected with the indicated chimeras and GFP–LC3A or GFP–LC3B, aggregated and mounted. Representative confocal pictures are shown. Activated GFP–LC3 appears as a collapsed mass of indiscernible vacuoles. (H) Quantification of the phenotype in G. As individual vesicles could not be counted, quantification was done as the percentage of transfected cells showing redistributed GFP–LC3. At least 10 different fields were counted (about 400 cells). The experiment was repeated three times. Data are expressed as means ±s.d. of the triplicates.
Source data for this figure is available on the online supplementary information page.
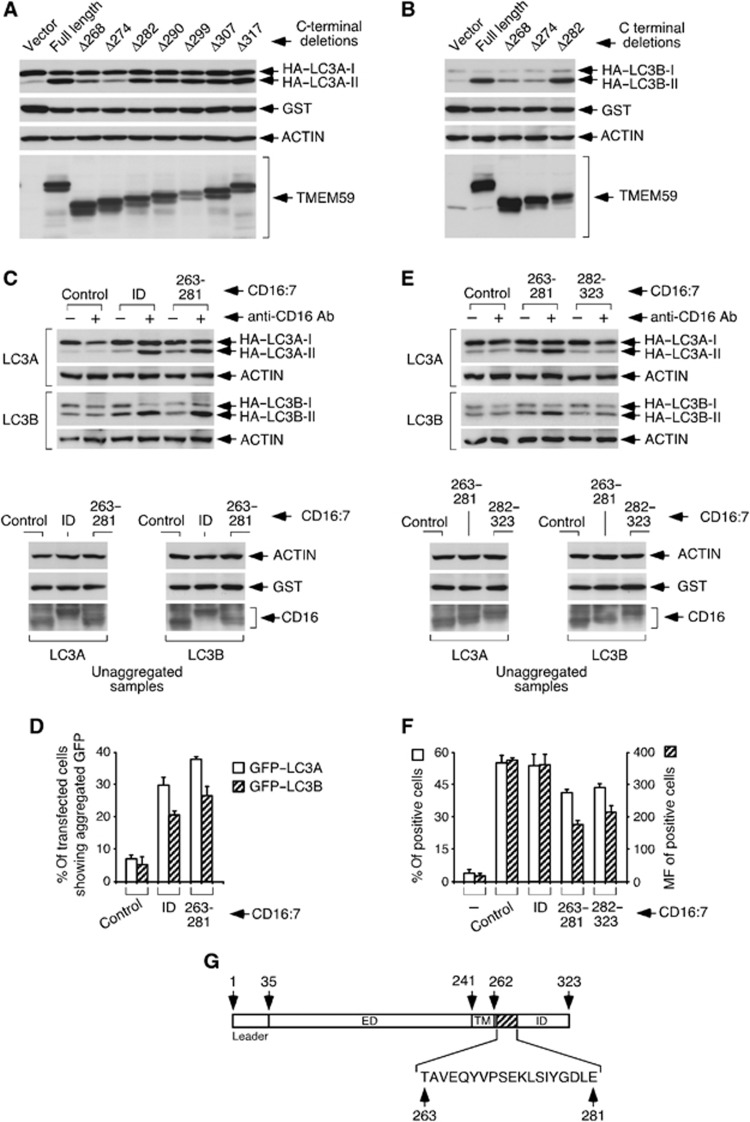
Serial C-terminal deletions of TMEM59 were tested by overexpression to further map the active subdomain. These experiments showed that amino acids 263–281 are necessary for the activity, because deletions beyond Δ282 were no longer functional (Figure 3A and B). When evaluated in the context of the CD16:7 chimera, this subdomain proved sufficient to activate LC3 (Figure 3C, D and F). In fact, this 19-amino-acid stretch retained the full potential of the whole ID to stimulate autophagy when both constructs were evaluated in parallel (Figure 3C, D and F). In contrast, the remaining ID (282–323) was inactive (Figure 3E and F). Therefore, a minimal 19-amino-acid subdomain between amino acids 263 and 281 (Figure 3G) holds the autophagic potential of the molecule.
Figure 3.
A minimal 19-amino-acid subdomain between amino acids 263–281 holds the autophagic activity of TMEM59. (A, B) Ability of serial C-terminal deletions of TMEM59 to activate LC3. 293 cells were transfected with the indicated TMEM59 C-terminal deletions, HA–LC3A (A) or HA–LC3B (B) and GST, and lysed for western blotting against the indicated molecules. (C) Amino acids 263–281 retain the full potential of TMEM59 ID to promote HA–LC3 conversion. 293 cells were transfected with the shown CD16:7 chimeras, HA–LC3A or HA–LC3B (as indicated) and GST, and subjected to anti-CD16 aggregation before lysing them for western blotting. The lower panels show control WBs (unaggregated samples). (D) Amino acids 263–281 suffice for GFP–LC3 activation and retain the full potential of TMEM59 ID to promote GFP–LC3 activation. 293 cells were transfected with the indicated CD16:7 chimeras and GFP–LC3A or GFP–LC3B (as indicated), and subjected to anti-CD16 aggregation before fixing them for microscopy. The graph shows percentages of transfected cells exhibiting redistributed GFP–LC3. Quantification and data expression were done as in Figure 2H. (E) Amino acids 282–323 of TMEM59 lack LC3 activation potential. 293 cells were transfected with the shown chimeras, HA–LC3A or HA–LC3B and GST, and processed for western blotting against the indicated molecules. Control WBs of unaggregated samples are shown in the lower panels. (F) Surface expression levels of CD16:7 chimeras. Procedures and data expression were as in Figure 2E. The figure shows that the functional differences observed between CD16:7 chimeras (C–E) are not caused by differential surface expression. (G) Scheme of TMEM59 showing amino-acid positions and the minimal active subdomain (amino acids 263–281). ED, extracellular domain; TM, transmembrane domain.
Source data for this figure is available on the online supplementary information page.
The active subdomain induces LC3 labelling of its own vesicular compartment
To explore the nature of the autophagic response induced by this minimal peptide, we first tested its function in cells depleted of the essential mediators ATG5 or ATG7. Reduced levels of these effectors decreased the autophagic potential of CD16:7 constructs containing the minimal active fragment (CD16:7–263–281; Supplementary Figure S4A–C), indicating that the activity proceeds through an ATG5/ATG7-dependent route.
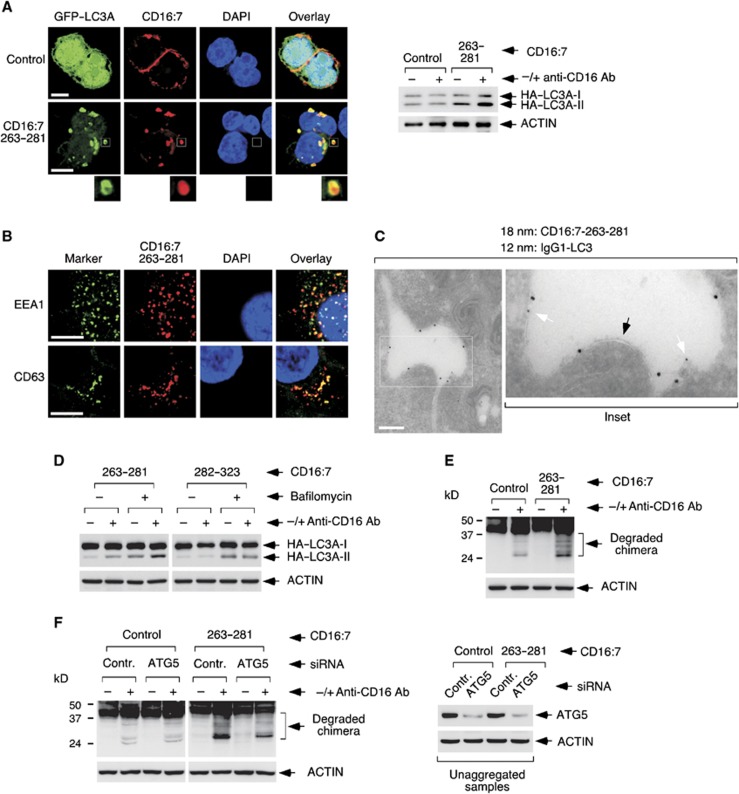
We next determined the subcellular distribution of the autophagic process. Unexpectedly, both overexpressed TMEM59 and aggregated, endocytosed CD16:7–263–281 chimeras tightly colocalized with activated GFP–LC3 (Supplementary Figure S5A and B). Again, 293 cells displayed a clustered signal (see Supplementary Figure S5B), but when the chimera experiment was repeated in JAR cells we detected individual vacuoles stained for both markers (Figure 4A). Most chimera-positive vesicles were labelled with GFP–LC3 in these cells (Supplementary Figure S5C), and GFP–LC3 appeared to stain the vesicle periphery (see inset Figure 4A). These vacuoles colocalized with EEA1 and/or CD63 (Figure 4B), suggesting that, once aggregated, the construct follows the regular endocytic route to the lysosome. In addition, electron microscopy studies showed that the LC3-labelled vesicles containing endocytosed chimera presented single membranes (Figure 4C), and so did the vacuoles induced by straight TMEM59 overexpression (Supplementary Figure S5D). These data point to the notion that single-membrane endosomes become decorated with LC3 in what appears to be an atypical autophagic event not involving canonical double-membrane autophagosomes. Consistent with this idea, TMEM59 does not seem to influence conventional autophagy. For instance, neither aggregation of the active chimera nor TMEM59 overexpression altered the expression levels of three recognized autophagic substrates: p62, NBR1 or GFP–huntingtin-Q74 (Mizushima et al, 2010) (Supplementary Figure S6). Similarly, TMEM59 depletion did not impact on the way the levels of these substrates (or LC3II) were changed by standard modulators of autophagy (bafilomycin or starvation, Supplementary Figures S7 and S8). Therefore, the autophagic activity of TMEM59 seems unrelated to canonical autophagy, and involves LC3 labelling of the same single-membrane endosomes where the molecule becomes activated.
Figure 4.
The minimal active subdomain of TMEM59 promotes LC3 labelling and lysosomal degradation of its own vesicular compartment. (A) Endocytic vesicles containing aggregated CD16:7–263–281 become labelled with GFP–LC3. JAR cells were transfected with vectors expressing the indicated CD16:7 chimeras and GFP–LC3A, and subjected to anti-CD16 aggregation for 8 h before staining for the endocytosed chimera (red). Representative confocal pictures are shown. The inset highlights a vesicle where the peripheral GFP–LC3 staining is particularly distinguishable. The right panel displays control WBs showing that chimera aggregation for 8 h activates HA–LC3A conversion in JAR cells. (B) Colocalization of endocytosed CD16:7–263–281 with EEA1 and CD63. JAR cells were transfected with CD16:7–263–281 and subjected to aggregation before staining them for the endocytosed chimera (red) and EEA1 or CD63 (green; FITC-coupled primary antibodies), as indicated. Representative confocal pictures are shown. (C) Immunoelectron microscopy assay showing that endocytosed CD16:7–263–281 localizes to LC3-labelled, single-membrane vesicles. JAR cells were transfected with the CD16:7–263–281 chimera and a construct expressing human IgG1 fused to LC3A. Cells were subjected to anti-CD16 aggregation for 8 h and processed for immunoelectron microscopy. Thick gold signal (18 nm): aggregated, endocytosed chimera; thin gold signal (12 nm): IgG1–LC3. Arrows indicate single membrane (black) or IgG1–LC3A (white). Scale bar: 400 nm. (D) Lysosomal inhibition increases HA–LC3II levels promoted by CD16:7–263–281. 293 cells were transfected with the indicated chimera and HA–LC3A, and subjected to anti-CD16 aggregation in the absence or presence of bafilomycin (200 nM, added 4 h post aggregation) before lysing them for western blotting against the indicated molecules. (E) Aggregation of CD16:7–263–281 promotes its own degradation. 293 cells were transfected with the indicated constructs, aggregated and lysed for western blotting. Shown are overexposed anti-CD16 WBs. (F) Degradation of CD16:7–263–281 is inhibited by ATG5 depletion. 293 cells were transfected with the indicated siRNAs and subsequently with the shown CD16:7 constructs, aggregated and lysed for anti-CD16 western blotting (left panel). The right panel displays control WBs showing ATG5 depletion.
Source data for this figure is available on the online supplementary information page.
An important question in the field is whether an observed accumulation of active LC3 reflects a real increase in LC3 lipidation and autophagic flux, or it simply represents arrest of autophagosomal degradation (Mizushima and Yoshimori, 2007; Klionsky et al, 2008; Mizushima et al, 2010). To discern between both possibilities, we conducted aggregation studies in the absence or presence of a lysosomal inhibitor (bafilomycin). The presence of this drug increased the levels of LC3II generated by the active CD16:7 construct (CD16:7–263–281; Figure 4D), suggesting that the minimal peptide adds LC3II beyond the degradation blockade imposed by the inhibitor. In addition, the fact that TMEM59 does not promote accumulation of autophagic substrates (see Supplementary Figure S6), argues against the idea that the subdomain blocks autophagic degradation. Taken together, these data suggest that the active subdomain of TMEM59 mainly functions by promoting LC3II synthesis. Since virtually all activated GFP–LC3 colocalized with the endocytosed chimera (see Figure 4A and Supplementary Figure S5B), we interiorized the notion that this synthesis is induced in situ by the aggregation event.
We next examined whether LC3 labelling targets the marked vesicles for degradation. Anti-CD16 western blots (WBs) showed that the stimulated CD16:7–263–281 chimera produced a lower molecular weight smear indicative of protein destruction (Figure 4E). This decay was reduced by E64d and pepstatin (Supplementary Figure S9), implying lysosomal involvement and suggesting again that the active subdomain does not obstruct vesicle maturation. Importantly, the degradation was inhibited by ATG5 depletion (Figure 4F), arguing that it is promoted by the autophagic activity triggered in response to aggregation. Consequently, the active subdomain of TMEM59, if stimulated, suffices to direct its own endocytic compartment for autophagic degradation.
The active subdomain defines a novel ATG16L1-binding motif
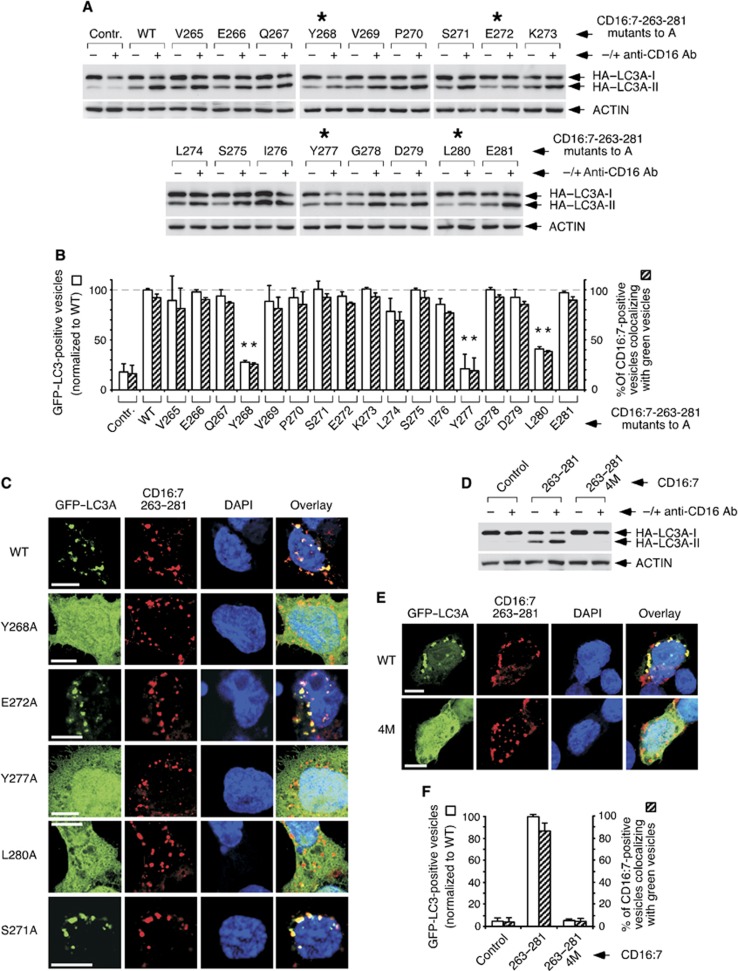
We hypothesized that the active peptide might conceal an underlying protein motif that retains function. To test this idea, we performed an alanine scanning study where all residues were individually mutated to alanine and the resulting derivatives were functionally tested using the CD16:7 chimera system. All constructs exhibited comparable cell surface expression levels (Supplementary Figure S10A), and therefore were equally available for aggregation. Four mutants (Y268A, E272A, Y277A and L280A) showed reduced capacity to lipidate HA–LC3 (Figure 5A), and only three (Y268A, Y277A and L280A) had a diminished potential to promote GFP–LC3 redistribution or colocalization with the endocytosed chimera (Figure 5B and C). Interestingly, E272A activated GFP–LC3 normally (Figure 5B and C). Given that this mutant was inactive in HA–LC3 conversion (see Figure 5A), this result suggests that the colocalizing GFP–LC3 pool is not lipidated. Thus, recruitment of LC3I and its conversion to LC3II might be dissociable events, and E272 would only be involved in the latter activity. Whatever the mechanistic implications of this finding may be, these results show that Y268, E272, Y277 and L280, arranged in a pattern defined by TMEM59 (Y-X3-E-X4-Y-X2-L), are required for in situ LC3II generation in this context. Consistent with their individual importance, mutation of the four critical residues at once (4M versions) prevented LC3 activation by chimera aggregation (Figure 5D–F) or TMEM59 overexpression (Supplementary Figure S10B and C). Although the 4M chimera showed slightly higher surface expression and less endocytosis upon aggregation (Supplementary Figure S10D and E), these partial defects are unlikely to explain the complete signalling blockade observed (see Figure 5D–F). In addition, the signalling impairment was not caused by a major mislocalization of the 4M mutant (Supplementary Figure S10F).
Figure 5.
Alanine scanning approach to identify amino acids in the active subdomain of TMEM59 that are essential for LC3 activation. (A) Identification of critical amino acids for HA–LC3 conversion. 293 cells were transfected with the indicated CD16:7–263–281 mutants and HA–LC3A, subjected to anti-CD16 aggregation and lysed for western blotting against the indicated molecules. Asterisks mark mutations with reduced activity. Shown is one representative experiment of six repetitions. (B) Identification of critical amino acids for GFP–LC3 activation and colocalization with the endocytosed chimera. JAR cells were transfected with the indicated CD16:7–263–281 mutants and GFP–LC3A, aggregated and stained for the endocytosed chimera (red). Preparations were scored by blindly counting the number of red vesicles (chimera) and green vesicles (GFP–LC3A) per cell, as well as the number of red vesicles colocalizing with green ones. The percentage of green vesicles colocalizing with red ones was close to 100% for all mutants, that is, virtually no GFP–LC3A vesicles were unrelated to endocytosed chimera (not shown). At least 50 cells were scored per experimental point. The experiment was repeated three times. The graph shows the number of GFP–LC3A vesicles per cell expressed as the percentage of the value obtained for the wild-type chimera (left axis), and the percentage of chimera vesicles labelled with GFP–LC3A (right axis). Data are expressed as means ±s.d. of the triplicates. Asterisks indicate significant differences with respect to wild-type values (paired Student’s t-test; P<0.01). (C) Representative confocal pictures of the phenotype produced by the indicated mutations. Procedures were as in B. (D) Simultaneous mutation of the four essential amino acids to alanine blocks HA–LC3 conversion induced by CD16:7–263–281. 293 cells were transfected with the indicated chimeras (4M, quadruple mutant) and HA–LC3A, aggregated and lysed for western blotting. (E) Simultaneous mutation of the four essential amino acids blocks GFP–LC3 activation and colocalization with the endocytosed chimera. JAR cells were transfected with the indicated CD16:7–263–281 constructs and GFP–LC3A, aggregated and stained for the endocytosed chimera (red). Representative confocal images are shown. (F) Quantification of the phenotype in E. Data gathering and expression were as in B.
Source data for this figure is available on the online supplementary information page.
Additional studies showed that Y268 can be mutated to W (but not F) without functional loss. Similarly, E272 can be substituted by D, and Y277 by W or F. However, L280 mutation to V depressed the activity (Supplementary Figure S11A and B). These data show that the motif can accommodate some flexibility, and point to the following final formulation: [YW]-X3-[ED]-X4-[YWF]-X2-L.
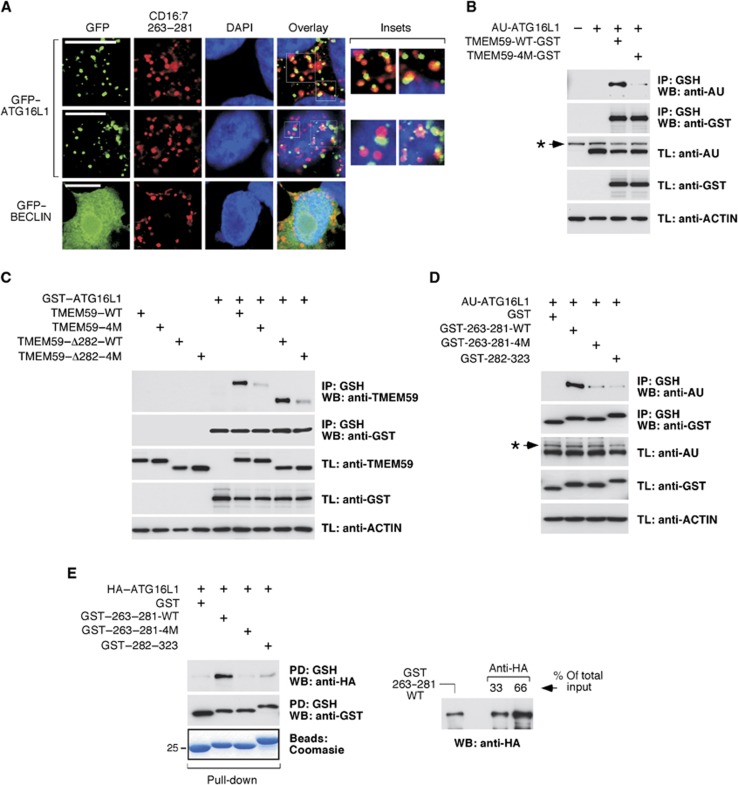
The notion that the active subdomain promotes in situ LC3II synthesis suggests local engagement of the autophagic machinery. Colocalization studies revealed frequent apposition events between the aggregated, endocytosed CD16:7–263–281 chimera and GFP–ATG16L1 (Figure 6A), whereas GFP–BECLIN (Figure 6A) or other GFP–ATG constructs (not shown) were negative. GFP–ATG5 only associated clearly with the internalized chimera when untagged ATG16L1 was also present (Supplementary Figure S12). Therefore, ATG16L1 is recruited at the site where the stimulated chimera is located, and probably brings ATG5 along with it. To explore whether the active subdomain can actually bind ATG16L1, we performed coimmunoprecipitation assays. Consistently, both full-length TMEM59 and its largest active deletion (Δ282, see Figure 3A and B) co-precipitated with ATG16L1 (Figure 6B and C). This interaction was decreased in the case of the 4M derivatives (Figure 6B and C), arguing that it is mediated by the identified motif. Interestingly, the 263–281 peptide fused to GST sufficed to precipitate ATG16L1 and, again, the mutated version was inactive (Figure 6D). This result indicates that the interaction does not require membrane localization of the TMEM59 partner, and opens the possibility to evaluate direct binding in pull-down assays with proteins expressed in bacteria. A purified GST–263–281 fusion protein precipitated ATG16L1 from crude bacterial extracts, whereas the 4M version was inert (Figure 6E). Therefore, the minimal active subdomain of TMEM59 directly interacts with ATG16L1 through the identified protein motif. Consistent with a critical role of this mediator in the autophagic potential of TMEM59, ATG16L1 depletion reduced the ability of the CD16:7 chimera to activate LC3 (Supplementary Figure S13A and B).
Figure 6.
The minimal active subdomain of TMEM59 directly binds ATG16L1 through the identified functional motif. (A) Apposition events between GFP–ATG16L1 and endocytosed CD16:7–263–281. JAR cells were transfected with CD16:7–263–281 and GFP–ATG16L1 or GFP–BECLIN (as indicated), aggregated for 4 h and stained for the endocytosed chimera (red). Representative confocal pictures are shown. Two different examples are provided for GFP–ATG16L1. (B–D) 293T cells were transfected with the indicated constructs, lysed and subjected to GST immunoprecipitation using agarose beads coupled to glutathione (IP, immunoprecipitation; TL, total lysate). Shown are WBs against the indicated molecules. (B) AU–ATG16L1 co-precipitates with wild-type (WT) TMEM59–GST but not with a mutated version where the four essential amino acids were mutated to alanine (TMEM59–4M–GST). (C) Full-length TMEM59 and TMEM59–Δ282 co-precipitate with GST–ATG16L1, whereas the respective 4M versions do not. (D) AU–ATG16L1 co-precipitates with a fusion protein between GST and the minimal active peptide of TMEM59 (GST–263–281), but not with a 4M version (GST–263–281–4M) or a GST fusion protein with the inactive portion of TMEM59–ID (GST–282–323). (E) HA–ATG16L1 expressed in bacteria co-precipitates with a GST–263–281 recombinant protein purified from bacterial cultures, but not with a 4M version of the same construct or GST–282–323. The indicated GST partners were expressed in bacteria and purified using agarose beads coupled to glutathione. The loaded beads were then used for HA–ATG16L1 pull-down from crude bacterial lysates. Shown are WBs against the indicated molecules (PD, pull-down). A Coomasie staining of a protein gel with the purified GST fusion proteins is shown. The right panel compares the amount of HA–ATG16L1 pulled down by GST–263–281 with the signal provided by direct anti-HA immunoprecipitation. This result shows that about 20–25% of the available HA–ATG16L1 protein is precipitated by GST–263–281. Asterisks indicate irrelevant bands in B and D.
Source data for this figure is available on the online supplementary information page.
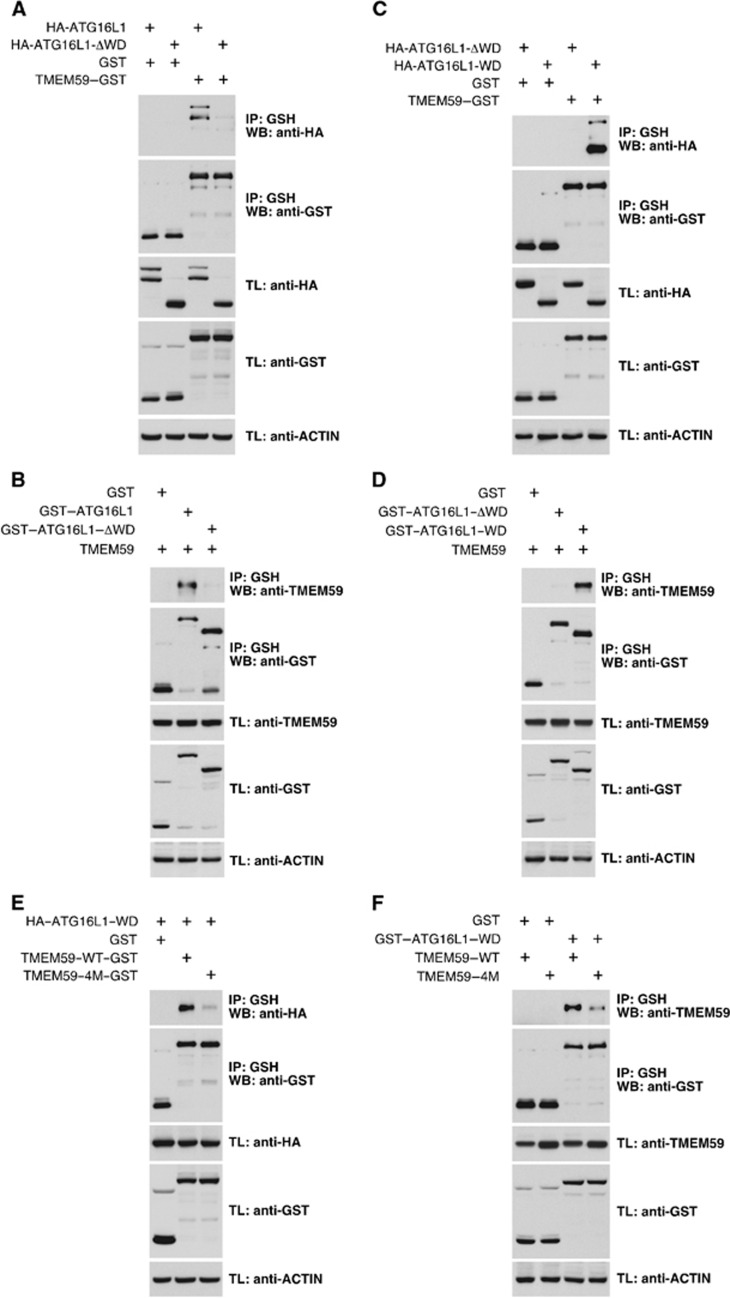
Mammalian ATG16L1 contains a C-terminal WD-repeat domain (WD domain) whose function is unclear. This domain is absent in the yeast ortholog (Mizushima et al, 2003) and is dispensable for conventional autophagy (Fujita et al, 2009), suggesting that it might be involved in specialized forms of autophagy. Coimmunoprecipitation studies showed that the WD domain is both necessary (Figure 7A and B) and sufficient (Figure 7C and D) for interaction with TMEM59 through the ATG16L1-binding motif (Figure 7E and F).
Figure 7.
The ATG16L1-binding motif present in TMEM59 recognizes the WD-repeat domain of ATG16L1. (A–F) 293T cells were transfected with the indicated constructs, lysed and subjected to GST immunoprecipitation with agarose beads coupled to glutathione. Shown are WBs against the indicated molecules. (A) A deleted version of ATG16L1 lacking the WD domain (HA–ATG16L1–ΔWD) does not co-precipitate with TMEM59–GST. (B) TMEM59 does not co-precipitate with ATG16L1–ΔWD fused to GST. (C) The WD domain of ATG16L1 (HA–ATG16L1–WD) suffices to co-precipitate with TMEM59–GST. (D) TMEM59 co-precipitates with ATG16L1–WD fused to GST. (E) HA–ATG16L1–WD does not bind a 4M version of TMEM59–GST. (F) TMEM59–4M does not co-precipitate with the ATG16L1–WD fused to GST.
Source data for this figure is available on the online supplementary information page.
The novel ATG16L1-binding motif includes YXXL (277–280), a signature similar to YXXØ, involved in endocytosis and sorting of transmembrane proteins (Bonifacino and Traub, 2003). In fact, the 4M chimera showed less intracellular signal after aggregation (see Supplementary Figure S10E), suggesting reduced endocytosis. Consistent with a possible role of the motif in trafficking, wild-type TMEM59 co-precipitated with the μ-chains of the clathrin adaptors AP1 (AP1M1) and AP2 (AP2M1), whereas the 4M version did not (Supplementary Figure S14A and B). Since both AP2 and clathrin are known to bind ATG16L1 (Ravikumar et al, 2010), this result raises the possibility that these sorting adaptors (or clathrin) might facilitate binding of the motif to ATG16L1. However, depletion of AP1M1, AP2M1 or the clathrin heavy chain did not prevent interaction between TMEM59 and ATG16L1 (Supplementary Figure S14C), thus arguing against this idea. In addition, a GST–263–281 form where Y277 and L280 were mutated to alanine (2M) did not bind ATG16L1 in a pull-down assay (Supplementary Figure S14D), indicating that, irrespective of its possible involvement in trafficking, the YXXL signature in TMEM59 does participate in direct ATG16L1 recognition. This pattern also resembles WXXL, implicated in LC3 binding (Noda et al, 2010). However, LC3 did not bind the active subdomain in coimmunoprecipitation assays (not shown).
The ATG16L1-binding motif is present in other molecules
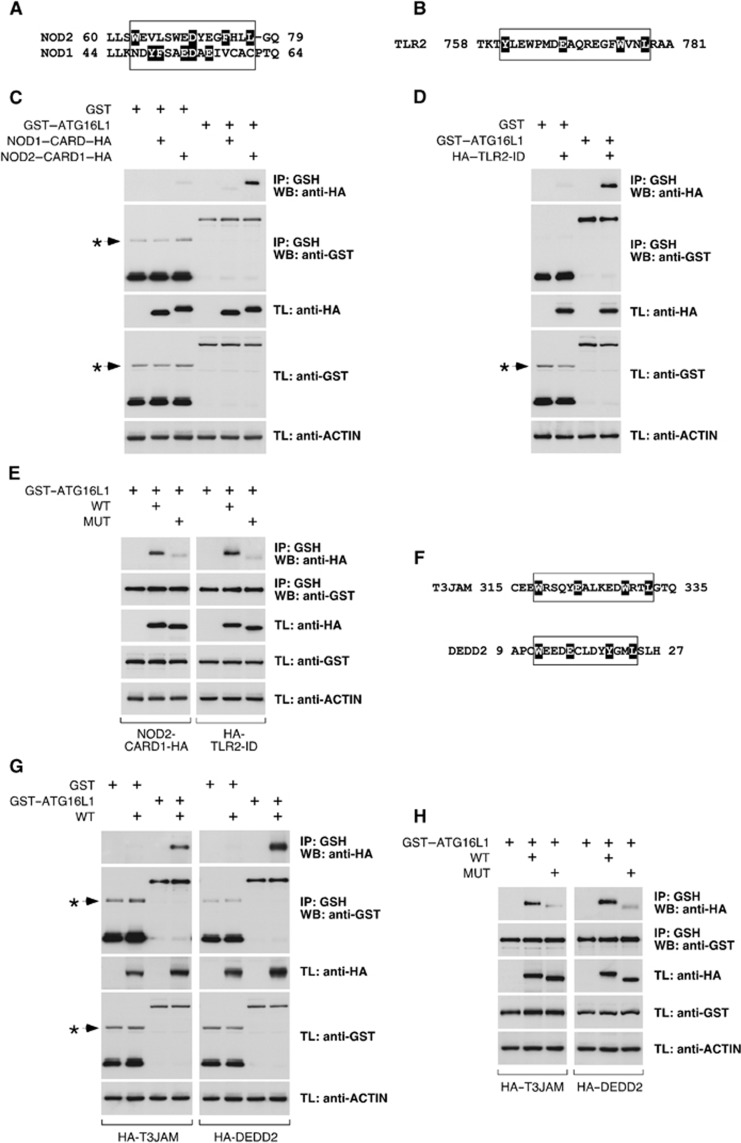
To evaluate whether the identified motif exists in other proteins with a similar ATG16L1-binding functionality, we searched the protein sequence bank using the Prosite algorithm and a version of the motif where the distances between amino acids were flexibilized ([YW]-X(2,6)-[ED]-X(2,6)-[YWF]-X2-L). Interestingly, the pattern was found in molecules previously involved in selective autophagy. Thus, NOD proteins are known to recruit ATG16L1 at the bacterial entry site (Travassos et al, 2010), and the N-terminal CARD of NOD2 (NOD2–CARD1) includes the motif (Figure 8A). Surprisingly, the CARD present in its relative NOD1 (NOD1–CARD) lacks the pattern (Figure 8A). In addition, TLR2 promotes LC3 labelling of conventional phagosomes (Sanjuan et al, 2007), and the motif is present in its intracellular region (TLR2–ID; Figure 8B). Coimmunoprecipitation studies showed that NOD2–CARD1 and TLR2–ID were able to bind ATG16L1 (Figure 8C and D), whereas NOD1–CARD was inactive (Figure 8C). These interactions were again mediated by the WD domain of ATG16L1 (Supplementary Figure S15A and B). Consistent with a role of the identified ATG16L1-binding pattern in these binding events, mutant versions of NOD2–CARD1 and TLR2–ID lacking the motif lost the potential to co-precipitate with ATG16L1 (Figure 8E). Two additional proteins not previously linked to autophagy (T3JAM and DEDD2) also presented the pattern (Figure 8F), and bound ATG16L1 through this region (Figure 8G and H). The minimal peptides harbouring the motif in all these molecules conferred ATG16L1-binding potential to an inactive version of TMEM59 (Supplementary Figure S16), indicating that they suffice to bind ATG16L1. Although establishing the precise functional consequences of these binding events requires additional studies, our results show that the identified ATG16L1-binding motif has a wider representation.
Figure 8.
The ATG16L1-binding motif is present with a similar ATG16L1-binding capacity in other molecules. (A) Alignment of the region that includes the motif in NOD2 with the same area in NOD1. The relevant amino-acid stretch is boxed. Individual amino-acid highlighting (black) in NOD2 indicates the residues identified by the Prosite algorithm as part of the motif. In NOD1, all residues that could be part of an eventual motif are highlighted to indicate that they form an incomplete motif. (B) Amino-acid region in TLR2 that includes the motif. Highlighting indicates residues identified by Prosite as part of the motif. (C, D, E, G, H) 293T cells were transfected with the indicated constructs, lysed and subjected to GST immunoprecipitation with agarose beads coupled to glutathione. Shown are WBs against the indicated molecules. (C) The N-terminal CARD of NOD2 (NOD2–CARD1–HA), but not the CARD of NOD1 (NOD1–CARD–HA), co-precipitates with GST–ATG16L1. (D) The ID of TLR2 (HA–TLR2–ID) co-precipitates with GST–ATG16L1. (E) Mutated versions (MUT) of NOD2–CARD1–HA and HA–TLR2–ID (as shown) do not co-precipitate with GST–ATG16L1. (F) Amino-acid sequences of T3JAM and DEDD2 including the motif. Residues identified by the Prosite algorithm are highlighted. (G) Wild-type (WT) versions of HA–T3JAM and HA–DEDD2 (as indicated) co-precipitate with GST–ATG16L1. (H) Mutated versions (MUT) of HA–T3JAM and HA–DEDD2 do not co-precipitate with GST–ATG16L1. Asterisks indicate irrelevant bands in C, D and G.
Source data for this figure is available on the online supplementary information page.
Endogenous TMEM59 mediates autophagy during Staphylococcus aureus infection
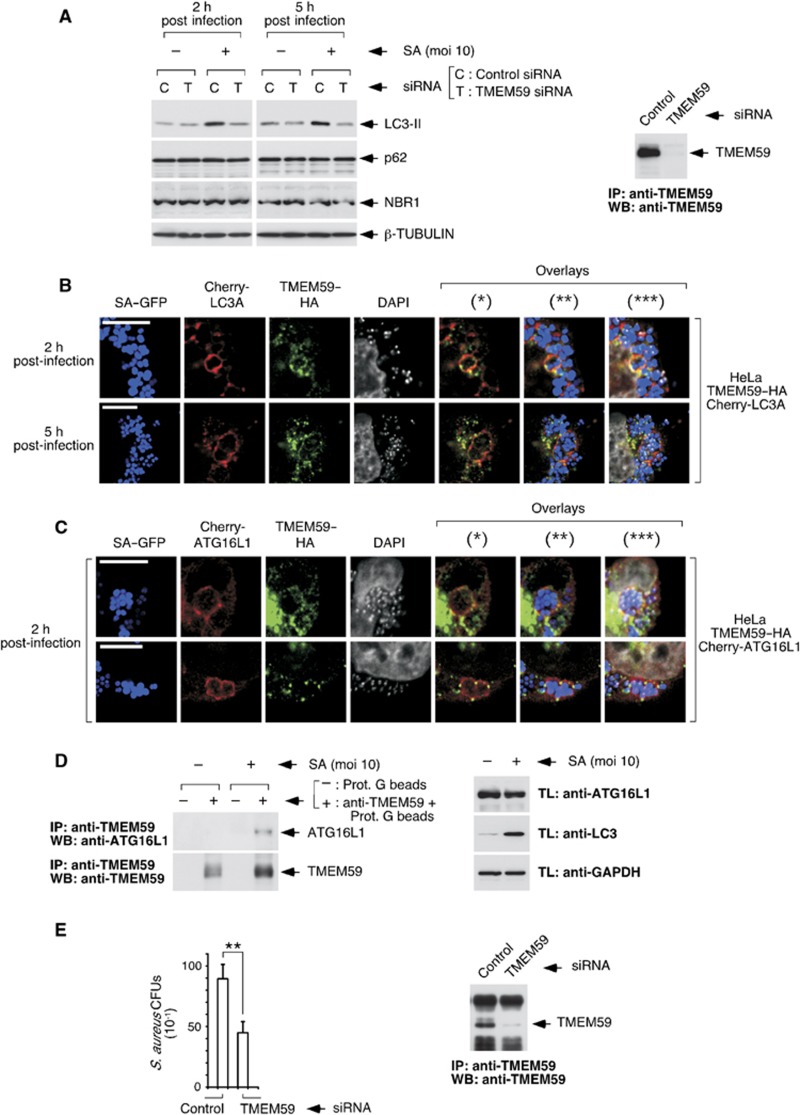
Taken together, our studies with the recombinant molecule suggest that endogenous TMEM59 might participate in promoting autophagic targeting of the membranous compartment where it becomes activated by aggregation. Autophagy directed to membrane-enclosed compartments occurs during some bacterial infections (Campoy and Colombo, 2009; Deretic and Levine, 2009; Shahnazari and Brumell, 2011). For example, phagosomes containing Mycobacterium tuberculosis and Staphylococcus aureus undergo an autophagic process that, in the first case, helps fight infection (Gutierrez et al, 2004), and in the second case is required for bacterial replication (Schnaith et al, 2007). In fact, the initial export of endogenous TMEM59 to the plasma membrane and its subsequent trafficking through endosomes to the lysosome suggest a function along this route, a possibility that fits well with the phagosomal niche subjected to autophagy during bacterial invasion. Consistent with this view, depletion of TMEM59 blocked LC3II induction provoked by one of these bacteria (S. aureus, SA) at early infection times (Figure 9A; Supplementary Figure S17A and B). The pathway that generates LC3II at this stage has unconventional features, since it does not impact on the levels of conventional autophagic substrates (p62 or NBR1; see Figure 9A, Supplementary Figure S17C) and it does not rely on BECLIN-1 or VPS34 (Supplementary Figure S17D). We also detected colocalization events between TMEM59, bacteria and Cherry–LC3 in what appeared to be bacterial sacs (Figure 9B) that were limited by single membranes (Supplementary Figure S17E). Intriguingly, TMEM59 expression was upregulated without mRNA contribution (Supplementary Figure S17F), suggesting that the protein becomes stabilized early during infection. Triple colocalization events between TMEM59, bacteria and Cherry–ATG16L1 were also found (Figure 9C), arguing that TMEM59 might recruit ATG16L1. In agreement with this idea, the number of TMEM59/ATG16L1 complexes increased in the presence of bacteria (Figure 9D). Taken together, these results point to a scenario where, in response to SA infection, TMEM59 becomes activated and promotes LC3 labelling of single-membrane bacterial phagosomes by directly engaging ATG16L1. This activity appears to have physiological consequences, since TMEM59 depletion resulted in reduced bacterial recovery from infected cells (Figure 9E), a result consistent with the known requirement of autophagy for SA replication (Schnaith et al, 2007). These data confirm that endogenous TMEM59 can perform an in situ autophagic function whose mechanistic details resemble those established for the recombinant molecule.
Figure 9.
Endogenous TMEM59 mediates LC3 activation and interacts with ATG16L1 in response to S. aureus (SA) infection. (A) Depletion of TMEM59 inhibits LC3II generation by SA at early infection times. HeLa cells were transfected with the indicated siRNAs and, 48 h later, infected with the bacteria (SA) for the shown times before lysing them for western blotting against the indicated molecules. The right panel shows successful TMEM59 depletion (rabbit anti-TMEM59 immunoprecipitation plus chicken anti-TMEM59 WB). (B) Colocalization events between bacteria (GFP labelled), TMEM59–HA and Cherry–LC3A in infected cells. HeLa cells stably expressing TMEM59–HA and Cherry–LC3A were infected for the indicated times (moi=10), and processed for anti-HA immunofluorescence. Representative confocal images are shown. The GFP, anti-HA (Cy5) and DAPI signals were pseudocolored in blue, green and white, respectively. Asterisks indicate overlay images: (*), Cherry–LC3A/TMEM59; (**), Cherry–LC3A/TMEM59/bacteria; (***), Cherry–LC3A/TMEM59/bacteria/DAPI. (C) Colocalization between bacteria, TMEM59–HA and Cherry–ATG16L1. HeLa cells stably expressing TMEM59–HA and Cherry–ATG16L1 were infected with SA (2 h, moi=10), and processed for anti-HA immunofluorescence. Shown are confocal pictures displaying two representative examples. Asterisks indicate overlays as in B. (D) SA promotes co-precipitation of endogenous ATG16L1 with endogenous TMEM59. HeLa cells were infected with SA (2 h, moi=10), lysed and subjected to immunoprecipitation with anti-TMEM59 antibodies or irrelevant protein G beads (as indicated). Immunoprecipitates were processed for western blotting. The right panel shows control WBs of the lysates used for immunoprecipitation. (E) Depletion of TMEM59 reduces recovery of SA from infected cells. HeLa cells were transfected with the indicated siRNAs, infected with SA 96 h later (5 h, moi=0.1) and lysed for CFU evaluation. The experiment was carried out three independent times. The graph displays average CFU counts obtained for the 10−1 dilution of the relevant extracts, ±s.d. of the three data sets. Asterisks indicate significant differences (P<0.01, paired Student’s t-test). The right panel shows successful TMEM59 depletion (anti-TMEM59 immunoprecipitation plus anti-TMEM59 WB).
Source data for this figure is available on the online supplementary information page.
Discussion
Selective autophagy underlies many of the important roles that autophagy plays in mammalian organisms, and therefore identification of the mechanisms that control this phenomenon and help define cargo selection becomes a relevant theme. Results presented here contribute to this general goal by identifying a novel protein motif that binds ATG16L1 and promotes an autophagic process directed to the endocytic compartment where the motif undergoes activation.
We discovered this minimal signature by studying TMEM59, a protein initially found as an inducer of atypical death. In retrospect, it seems plausible that this death process resulted from excessive autophagy caused by overexpression of the molecule, since exaggerated autophagy can provoke cell death (Gozuacik and Kimchi, 2007; Eisenberg-Lerner et al, 2009). TMEM59 has been involved in the complex glycolsylation of APP (Ullrich et al, 2010), but how this phenomenon relates to the function described here is unknown.
To characterize the active peptide in TMEM59, we used CD16:7 chimeric molecules. This artificial system preserves the native topology of the protein, and provides an activation switch to test activities exclusively triggered by aggregation. In addition, it nicely recapitulates the migration route followed by the endogenous molecule. Thus, TMEM59 is exported to the plasma membrane and subsequently endocytosed to be finally degraded in lysosomes, just like the active chimeric constructs do if activated by aggregation. These two parallel routes suggest that TMEM59 could have an in situ LC3II-generating activity in any of these compartments, provided that a proper aggregating stimulus is present. The nature of such stimulus is currently unknown, although it certainly becomes available during SA infection. Since the process described here involves LC3 labelling of non-autophagic vesicular compartments, it is reminiscent of the TLR2-mediated LC3 decoration of conventional phagosomes that has been previously described (Sanjuan et al, 2007). Stimulus-dependent LC3 labelling of single-membrane compartments might therefore constitute a general mechanism to promote a more efficient vesicle delivery to the lysosome, as it has been shown for LC3-positive phagosomes (Sanjuan et al, 2007). This notion is also suggested by our data showing autophagy-dependent degradation of the active CD16:7 chimera (see Figure 4F). The need of an activating stimulus to promote LC3 labelling of these single-membrane vesicles has perhaps precluded their previous identification as unconventional autophagic structures.
ATG16L1 defines the site of LC3 lipidation by recruiting the LC3II synthesis complex (ATG12–ATG5) to defined membrane localizations (Hanada et al, 2007; Fujita et al, 2008). Therefore, ATG16L1 constitutes a suitable target for selective autophagy against membranous compartments and, in fact, the existence of adaptor proteins that recruit ATG16L1 to specific sites has been anticipated (Fujita et al, 2008; Kageyama et al, 2011; Mizushima et al, 2011). We propose that TMEM59 may be one of these adaptors, and the ATG16L1-binding motif described here could facilitate identification of additional members of this group. Consistently, we found that the motif is conserved with the same ATG16L1-binding function in other molecules, confirming a wider representation. NODs are known to recruit ATG16L1 to the bacterial entry site, and therefore act as ATG16L1 adaptor molecules (Travassos et al, 2010). Intriguingly, we show that the N-terminal CARD of NOD2 binds ATG16L1 through the identified motif. However, NOD1 also binds ATG16L1 (Travassos et al, 2010) and its CARD lacks the motif, suggesting that other ATG16L1-binding sites probably exist in these molecules. Clathrin also interacts with ATG16L1 (Ravikumar et al, 2010), but its role as an adaptor molecule for selective autophagy remains unexplored. Given the important roles that selective autophagy plays in multicellular organisms and the relevance that ATG16L1 seems to have in some of these directed processes, more molecules are expected to join the list of ATG16L1 adaptors in the near future.
Our results show that the motif recognizes the C-terminal WD-repeat domain in ATG16L1, and therefore help answer the standing question of what the function of this region might be (Mizushima et al, 2003; Fujita et al, 2009). We propose that the WD repeats likely provide a docking platform for protein adaptors that are able to engage ATG16L1. This interaction would leave the N-terminal part of the molecule free to interact with ATG5 (Mizushima et al, 2003) and form the functional complex that drives LC3 lipidation. Since the ATG161-binding motif is present in proteins involved in the cellular response to bacterial invasion (NOD2, TLR2 and TMEM59), our data are consistent the idea that the WD domain has evolved to fulfil more sophisticated autophagic functions that are exclusive of higher eukaryotes (Cadwell et al, 2009). Interestingly, the motif might have two overlapping functions in transmembrane proteins like TMEM59: intracellular trafficking and binding to ATG16L1. This coincidence of both signals in one amino-acid signature suggests a complex interaction between the two activities that might help integrate the final destination of the protein.
Materials and methods
DNA constructs
The source of all cDNAs and DNA manipulation methods are described in Supplementary Materials and Methods. Constructs expressing NOD2–CARD1–HA included amino acids 1–122 of the whole molecule (Uniprot, Q9HC29), NOD1–CARD–HA amino acids 1–105 (Uniprot, Q9Y239), HA–TLR2–ID amino-acids 610–784 (Uniprot, O60603), ATG16L1–ΔWD amino acids 1–319 (Uniprot, Q676U5) and ATG16L1–WD amino acids 320–607. Point mutants were generated by site-directed mutagenesis (QuikChange, Stratagene). Mutant NOD2–CARD1–HA contains nine mutations to alanine in all residues potentially participating in an ATG16L1-binding motif (W63, E64, W68, E69, D70, Y71, E72, F74, and L77), since several amino-acid combinations can build different motif versions in the same region. HA–TLR2–ID, HA–T3JAM and HA–DEDD2 mutants were deleted versions lacking the motif (boxed regions in Figure 8B and F).
Immunofluorescence
Cells were seeded onto poly-L-lysine-treated coverslips (Sigma). Coverslips were fixed (4% paraformaldehyde) and stained with the relevant antibodies (1 h, RT) as described (Klee and Pimentel-Muiños, 2005; Klee et al, 2009). To stain for endocytosed CD16:7 chimeras, unpermeabilized cells were quenched in vivo with unlabelled goat anti-mouse κ-chain (Jackson Immunoresearch, 40 min, 4°C, 0.1% azide) to remove cell surface signal. Cells were fixed, permeabilized and stained with a goat anti-mouse κ-antibody conjugated to Cy3. Confocal pictures were taken using the 488-nm (Alexa-488), 561-nm (Cy3) or 633-nm (Cy5) laser bands of a Leica SP5 confocal microscope. Scale bars represent 10 μm in all micrographs. Quantifications were carried out using conventional microscopy. The number of GFP–LC3 dots per cell was evaluated on pictures taken with a Zeiss Axiophot-2 microscope equipped with a Hamamatsu ORCA–ER CCD camera and the Openlab software. To quantify transfected 293 cells showing GFP–LC3 activated by chimera aggregation, cells were centrifuged (cytospin) onto poly-L-lysine coated slides before fixation.
Stimulation of CD16:7 chimeras
Cells were transfected, split next day to as many wells as experimental points were to be used (control plus activation points) and stimulated 36 h post transfection with the anti-CD16 antibody (4 μg/ml, mAb, NA/LE formulation, BD) plus 10 μg/ml of a rabbit anti-mouse polyclonal serum (Jackson Immunoresearch) for further aggregation. As all experimental points per construct derive from a single transfected well, control WBs can be carried out with unaggregated samples (the aggregating antibodies impaired protein detection in certain molecular weights). Aggregation was done for 16 h in 293 cells, for 8 h in JAR cells (vesicle formation and colocalization with GFP–LC3 were best detected at this latter time point) and for 4 h to evaluate colocalization with GFP–ATG16L1 or GFP–ATG5. ‘Control’ CD16:7 chimera refers to empty chimera.
Bacteria infection assays
HeLa cells were infected with GFP-expressing Staphylococcus aureus RN6390 (Kahl et al, 2000), kindly provided by Dr A. Cheung (Dartmouth Medical School, New Hampshire, USA). Cells were incubated with the bacteria for 45 min, washed and treated with 100 μg/ml gentamicin (1 h) to inhibit extracellular proliferation of S. aureus. This was considered the 2 h infection time point. Cells were washed and incubated for additional time-points in the presence of 10 μg/ml gentamicin. Experiments involving siRNAs were done in HeLa cells expressing HA–LC3A. However, changes in LC3 lipidation were best detected in this setting using an anti-LC3 antibody that does not recognize ectopic HA–LC3A, and therefore WBs display endogenous LC3 levels. Stainings for TMEM59–HA were done using an IgG1 anti-HA mAb (Covance). This isotype has low affinity for protein A, thus minimizing direct binding to S. aureus (known to produce protein A; Sinha and Fraunholz, 2010). Preparations did not show detectable direct bacterial staining by immunofluorescence. For determination of CFUs, infected HeLa cells were lysed in 0.1% Triton X-100 and the lysates serially diluted for plating in BHI-agar and subsequent colony quantification.
Supplementary Material
Acknowledgments
We thank Drs J.M. de Pereda, T. Yoshimori, N. Mizushima, B. Levine, G. Núñez, A. Cheung, I. Dikic, D. Rubinsztein and B. Seed for their help and/or kindly sharing reagents. We appreciate Judit Fernández’s experimental help. This work was funded by grants from the Ministerio de Ciencia e Innovación of the Spanish Government (Refs SAF2008-00350 and SAF2011-23714), Fundación Solórzano, Junta de Castilla y León (Consejería de Educación, Ref. CSI001A10-2, and Consejería de Sanidad) and Consejo Superior de Investigaciones Científicas (CSIC; Ref. 200720I026). Additional funding comes from the FEDER programme of the European Union. EB is a graduate student funded by a predoctoral fellowship from the FPU programme (Ministerio de Educación, MEC, Spanish Government). ML is funded by JAE-Doc and Juan de la Cierva postdoctoral contracts (MEC and Social European Fund of the European Union, 2007–2013). AF is funded by a long-term EMBO postdoctoral fellowship and a Juan de la Cierva contract. KP is a graduate student funded by an FPI fellowship (MEC). CB is an undergraduate master student. FXP holds a tenured position at the CSIC.
Author contributions: EB, ML, AF and KP performed the experiments. CR contributed with DNA construct generation. FXP designed the experiments and wrote the manuscript.
Footnotes
The authors declare that they have no conflict of interest.
References
- Alcalá S, Klee M, Fernandez J, Fleischer A, Pimentel-Muiños FX (2008) A high-throughput screening for mammalian cell death effectors identifies the mitochondrial phosphate carrier as a regulator of cytochrome c release. Oncogene 27: 44–54 [DOI] [PubMed] [Google Scholar]
- Bonifacino JS, Traub LM (2003) Signals for sorting of transmembrane proteins to endosomes and lysosomes. Annu Rev Biochem 72: 395–447 [DOI] [PubMed] [Google Scholar]
- Cadwell K, Stappenbeck TS, Virgin HW (2009) Role of autophagy and autophagy genes in inflammatory bowel disease. Curr Top Microbiol Immunol 335: 141–167 [DOI] [PubMed] [Google Scholar]
- Campoy E, Colombo MI (2009) Autophagy in intracellular bacterial infection. Biochim Biophys Acta 1793: 1465–1477 [DOI] [PubMed] [Google Scholar]
- Cecconi F, Levine B (2008) The role of autophagy in mammalian development: cell makeover rather than cell death. Dev Cell 15: 344–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Klionsky DJ (2011) The regulation of autophagy-unanswered questions. J Cell Sci 124: 161–170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deretic V (2011) Autophagy in immunity and cell-autonomous defense against intracellular microbes. Immunol Rev 240: 92–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deretic V, Levine B (2009) Autophagy, immunity, and microbial adaptations. Cell Host Microbe 5: 527–549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dikic I, Johansen T, Kirkin V (2010) Selective autophagy in cancer development and therapy. Cancer Res 70: 3431–3434 [DOI] [PubMed] [Google Scholar]
- Eisenberg-Lerner A, Bialik S, Simon HU, Kimchi A (2009) Life and death partners: apoptosis, autophagy and the cross-talk between them. Cell Death Differ 16: 966–975 [DOI] [PubMed] [Google Scholar]
- Fujita N, Itoh T, Omori H, Fukuda M, Noda T, Yoshimori T (2008) The Atg16L complex specifies the site of LC3 lipidation for membrane biogenesis in autophagy. Mol Biol Cell 19: 2092–2100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita N, Saitoh T, Kageyama S, Akira S, Noda T, Yoshimori T (2009) Differential involvement of Atg16L1 in Crohn disease and canonical autophagy: analysis of the organization of the Atg16L1 complex in fibroblasts. J Biol Chem 284: 32602–32609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita N, Yoshimori T (2011) Ubiquitination-mediated autophagy against invading bacteria. Curr Opin Cell Biol 23: 492–497 [DOI] [PubMed] [Google Scholar]
- Garcia-Arencibia M, Hochfeld WE, Toh PP, Rubinsztein DC (2010) Autophagy, a guardian against neurodegeneration. Semin Cell Dev Biol 21: 691–698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gozuacik D, Kimchi A (2007) Autophagy and cell death. Curr Top Dev Biol 78: 217–245 [DOI] [PubMed] [Google Scholar]
- Gutierrez MG, Master SS, Singh SB, Taylor GA, Colombo MI, Deretic V (2004) Autophagy is a defense mechanism inhibiting BCG and Mycobacterium tuberculosis survival in infected macrophages. Cell 119: 753–766 [DOI] [PubMed] [Google Scholar]
- Hanada T, Noda NN, Satomi Y, Ichimura Y, Fujioka Y, Takao T, Inagaki F, Ohsumi Y (2007) The Atg12-Atg5 conjugate has a novel E3-like activity for protein lipidation in autophagy. J Biol Chem 282: 37298–37302 [DOI] [PubMed] [Google Scholar]
- Hara T, Nakamura K, Matsui M, Yamamoto A, Nakahara Y, Suzuki-Migishima R, Yokoyama M, Mishima K, Saito I, Okano H, Mizushima N (2006) Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature 441: 885–889 [DOI] [PubMed] [Google Scholar]
- He C, Klionsky DJ (2009) Regulation mechanisms and signaling pathways of autophagy. Annu Rev Genet 43: 67–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janvier K, Bonifacino JS (2005) Role of the endocytic machinery in the sorting of lysosome-associated membrane proteins. Mol Biol Cell 16: 4231–4242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansen T, Lamark T (2011) Selective autophagy mediated by autophagic adapter proteins. Autophagy 7: 279–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kageyama S, Omori H, Saitoh T, Sone T, Guan JL, Akira S, Imamoto F, Noda T, Yoshimori T (2011) The LC3 recruitment mechanism is separate from Atg9L1-dependent membrane formation in the autophagic response against Salmonella. Mol Biol Cell 22: 2290–2300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahl BC, Goulian M, van Wamel W, Herrmann M, Simon SM, Kaplan G, Peters G, Cheung AL (2000) Staphylococcus aureus RN6390 replicates and induces apoptosis in a pulmonary epithelial cell line. Infect Immun 68: 5385–5392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkin V, Lamark T, Sou YS, Bjorkoy G, Nunn JL, Bruun JA, Shvets E, McEwan DG, Clausen TH, Wild P, Bilusic I, Theurillat JP, Overvatn A, Ishii T, Elazar Z, Komatsu M, Dikic I, Johansen T (2009a) A role for NBR1 in autophagosomal degradation of ubiquitinated substrates. Mol Cell 33: 505–516 [DOI] [PubMed] [Google Scholar]
- Kirkin V, McEwan DG, Novak I, Dikic I (2009b) A role for ubiquitin in selective autophagy. Mol Cell 34: 259–269 [DOI] [PubMed] [Google Scholar]
- Klee M, Pallauf K, Alcalá S, Fleischer A, Pimentel-Muiños FX (2009) Mitochondrial apoptosis induced by BH3-only molecules in the exclusive presence of endoplasmic reticular Bak. EMBO J 28: 1757–1768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klee M, Pimentel-Muiños FX (2005) Bcl-X(L) specifically activates Bak to induce swelling and restructuring of the endoplasmic reticulum. J Cell Biol 168: 723–734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky DJ, Abeliovich H, Agostinis P, Agrawal DK, Aliev G, Askew DS, Baba M, Baehrecke EH, Bahr BA, Ballabio A, Bamber BA, Bassham DC, Bergamini E, Bi X, Biard-Piechaczyk M, Blum JS, Bredesen DE, Brodsky JL, Brumell JH, Brunk UT et al. (2008) Guidelines for the use and interpretation of assays for monitoring autophagy in higher eukaryotes. Autophagy 4: 151–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knaevelsrud H, Simonsen A (2010) Fighting disease by selective autophagy of aggregate-prone proteins. FEBS Lett 584: 2635–2645 [DOI] [PubMed] [Google Scholar]
- Kolanus W, Romeo C, Seed B (1993) T cell activation by clustered tyrosine kinases. Cell 74: 171–183 [DOI] [PubMed] [Google Scholar]
- Komatsu M, Ichimura Y (2010) Selective autophagy regulates various cellular functions. Genes Cells 15: 923–933 [DOI] [PubMed] [Google Scholar]
- Komatsu M, Ueno T, Waguri S, Uchiyama Y, Kominami E, Tanaka K (2007) Constitutive autophagy: vital role in clearance of unfavorable proteins in neurons. Cell Death Differ 14: 887–894 [DOI] [PubMed] [Google Scholar]
- Komatsu M, Waguri S, Chiba T, Murata S, Iwata J, Tanida I, Ueno T, Koike M, Uchiyama Y, Kominami E, Tanaka K (2006) Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature 441: 880–884 [DOI] [PubMed] [Google Scholar]
- Kroemer G, Marino G, Levine B (2010) Autophagy and the integrated stress response. Mol Cell 40: 280–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine B, Kroemer G (2008) Autophagy in the pathogenesis of disease. Cell 132: 27–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine B, Mizushima N, Virgin HW (2011) Autophagy in immunity and inflammation. Nature 469: 323–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippincott-Schwartz J, Fambrough DM (1987) Cycling of the integral membrane glycoprotein, LEP100, between plasma membrane and lysosomes: kinetic and morphological analysis. Cell 49: 669–677 [DOI] [PubMed] [Google Scholar]
- Mathew R, Karp CM, Beaudoin B, Vuong N, Chen G, Chen HY, Bray K, Reddy A, Bhanot G, Gelinas C, Dipaola RS, Karantza-Wadsworth V, White E (2009) Autophagy suppresses tumorigenesis through elimination of p62. Cell 137: 1062–1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathew R, White E (2011) Autophagy in tumorigenesis and energy metabolism: friend by day, foe by night. Curr Opin Genet Dev 21: 113–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N (2007) Autophagy: process and function. Genes Dev 21: 2861–2873 [DOI] [PubMed] [Google Scholar]
- Mizushima N, Komatsu M (2011) Autophagy: renovation of cells and tissues. Cell 147: 728–741 [DOI] [PubMed] [Google Scholar]
- Mizushima N, Kuma A, Kobayashi Y, Yamamoto A, Matsubae M, Takao T, Natsume T, Ohsumi Y, Yoshimori T (2003) Mouse Apg16L, a novel WD-repeat protein, targets to the autophagic isolation membrane with the Apg12-Apg5 conjugate. J Cell Sci 116: 1679–1688 [DOI] [PubMed] [Google Scholar]
- Mizushima N, Levine B, Cuervo AM, Klionsky DJ (2008) Autophagy fights disease through cellular self-digestion. Nature 451: 1069–1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N, Yoshimori T (2007) How to interpret LC3 immunoblotting. Autophagy 3: 542–545 [DOI] [PubMed] [Google Scholar]
- Mizushima N, Yoshimori T, Levine B (2010) Methods in mammalian autophagy research. Cell 140: 313–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N, Yoshimori T, Ohsumi Y (2011) The role of atg proteins in autophagosome formation. Annu Rev Cell Dev Biol 27: 107–132 [DOI] [PubMed] [Google Scholar]
- Nakatogawa H, Suzuki K, Kamada Y, Ohsumi Y (2009) Dynamics and diversity in autophagy mechanisms: lessons from yeast. Nat Rev Mol Cell Biol 10: 458–467 [DOI] [PubMed] [Google Scholar]
- Noda NN, Ohsumi Y, Inagaki F (2010) Atg8-family interacting motif crucial for selective autophagy. FEBS Lett 584: 1379–1385 [DOI] [PubMed] [Google Scholar]
- Noda T, Fujita N, Yoshimori T (2009) The late stages of autophagy: how does the end begin? Cell Death Differ 16: 984–990 [DOI] [PubMed] [Google Scholar]
- Novak I, Kirkin V, McEwan DG, Zhang J, Wild P, Rozenknop A, Rogov V, Lohr F, Popovic D, Occhipinti A, Reichert AS, Terzic J, Dotsch V, Ney PA, Dikic I (2010) Nix is a selective autophagy receptor for mitochondrial clearance. EMBO Rep 11: 45–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pankiv S, Clausen TH, Lamark T, Brech A, Bruun JA, Outzen H, Overvatn A, Bjorkoy G, Johansen T (2007) p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem 282: 24131–24145 [DOI] [PubMed] [Google Scholar]
- Rabinowitz JD, White E (2010) Autophagy and metabolism. Science 330: 1344–1348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randow F (2011) How cells deploy ubiquitin and autophagy to defend their cytosol from bacterial invasion. Autophagy 7: 304–309 [DOI] [PubMed] [Google Scholar]
- Ravikumar B, Moreau K, Jahreiss L, Puri C, Rubinsztein DC (2010) Plasma membrane contributes to the formation of pre-autophagosomal structures. Nat Cell Biol 12: 747–757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinsztein DC, Shpilka T, Elazar Z (2012) Mechanisms of autophagosome biogenesis. Curr Biol 22: R29–R34 [DOI] [PubMed] [Google Scholar]
- Saftig P, Klumperman J (2009) Lysosome biogenesis and lysosomal membrane proteins: trafficking meets function. Nat Rev Mol Cell Biol 10: 623–635 [DOI] [PubMed] [Google Scholar]
- Sanjuan MA, Dillon CP, Tait SW, Moshiach S, Dorsey F, Connell S, Komatsu M, Tanaka K, Cleveland JL, Withoff S, Green DR (2007) Toll-like receptor signalling in macrophages links the autophagy pathway to phagocytosis. Nature 450: 1253–1257 [DOI] [PubMed] [Google Scholar]
- Schnaith A, Kashkar H, Leggio SA, Addicks K, Kronke M, Krut O (2007) Staphylococcus aureus subvert autophagy for induction of caspase-independent host cell death. J Biol Chem 282: 2695–2706 [DOI] [PubMed] [Google Scholar]
- Shahnazari S, Brumell JH (2011) Mechanisms and consequences of bacterial targeting by the autophagy pathway. Curr Opin Microbiol 14: 68–75 [DOI] [PubMed] [Google Scholar]
- Shimizu S, Kanaseki T, Mizushima N, Mizuta T, Arakawa-Kobayashi S, Thompson CB, Tsujimoto Y (2004) Role of Bcl-2 family proteins in a non-apoptotic programmed cell death dependent on autophagy genes. Nat Cell Biol 6: 1221–1228 [DOI] [PubMed] [Google Scholar]
- Singh R, Cuervo AM (2011) Autophagy in the cellular energetic balance. Cell Metab 13: 495–504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha B, Fraunholz M (2010) Staphylococcus aureus host cell invasion and post-invasion events. Int J Med Microbiol 300: 170–175 [DOI] [PubMed] [Google Scholar]
- Tanida I (2011) Autophagosome formation and molecular mechanism of autophagy. Antioxid Redox Signal 14: 2201–2214 [DOI] [PubMed] [Google Scholar]
- Thurston TL, Ryzhakov G, Bloor S, von Muhlinen N, Randow F (2009) The TBK1 adaptor and autophagy receptor NDP52 restricts the proliferation of ubiquitin-coated bacteria. Nat Immunol 10: 1215–1221 [DOI] [PubMed] [Google Scholar]
- Travassos LH, Carneiro LA, Ramjeet M, Hussey S, Kim YG, Magalhaes JG, Yuan L, Soares F, Chea E, Le Bourhis L, Boneca IG, Allaoui A, Jones NL, Nunez G, Girardin SE, Philpott DJ (2010) Nod1 and Nod2 direct autophagy by recruiting ATG16L1 to the plasma membrane at the site of bacterial entry. Nat Immunol 11: 55–62 [DOI] [PubMed] [Google Scholar]
- Ullrich S, Munch A, Neumann S, Kremmer E, Tatzelt J, Lichtenthaler SF (2010) The novel membrane protein TMEM59 modulates complex glycosylation, cell surface expression, and secretion of the amyloid precursor protein. J Biol Chem 285: 20664–20674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wild P, Farhan H, McEwan DG, Wagner S, Rogov VV, Brady NR, Richter B, Korac J, Waidmann O, Choudhary C, Dotsch V, Bumann D, Dikic I (2011) Phosphorylation of the autophagy receptor optineurin restricts Salmonella growth. Science 333: 228–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Klionsky DJ (2009) An overview of the molecular mechanism of autophagy. Curr Top Microbiol Immunol 335: 1–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youle RJ, Narendra DP (2011) Mechanisms of mitophagy. Nat Rev Mol Cell Biol 12: 9–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.