Abstract
Chronic drug exposure alters gene expression in the brain and produces long-term changes in neural networks that underlie compulsive drug taking and seeking. Exactly how drug-induced changes in synaptic plasticity and subsequent gene expression are translated into persistent neuroadaptations remains unclear. Emerging evidence suggests that complex drug-induced neuroadaptations in the brain are mediated by highly synchronized and dynamic patterns of gene regulation. Recently, it has become clear that epigenetic mechanisms contribute to drug-induced structural, synaptic, and behavioral plasticity by regulating expression of gene networks. Here we review how alterations in histone modifications, DNA methylation, and microRNAs regulate gene expression and contribute to psychostimulant addiction with a focus on the epigenetic mechanisms that regulate brain-derived neurotrophic factor (BDNF) expression following chronic cocaine exposure. Identifying epigenetic signatures that define psychostimulant addiction may lead to novel, efficacious treatments for drug craving and relapse.
Repeated cocaine exposure produces stable gene expression changes (e.g., in brain-derived neurotrophic factor [BDNF]) in discrete brain regions. Many of these changes coincide with drug-seeking behavior.
Drug addiction is a chronic, relapsing disorder that is characterized by compulsive drug seeking and taking despite adverse consequences (Mendelson and Mello 1996). The transition from recreational to chronic drug taking and the persistence of drug addiction are mediated, in part, by drug-induced alterations in gene expression profiles within the reward circuitry of the brain (Nestler 2001; Koob and Volkow 2010; Maze and Nestler 2011). Therefore, elucidating the molecular mechanisms by which chronic drug exposure promotes stable changes in gene expression and ultimately drug-seeking behavior may aid in the development of novel pharmacotherapies for drug addiction. Recent studies indicate that epigenetic mechanisms contribute to drug-induced structural, synaptic, and behavioral plasticity by orchestrating expression of gene networks in discrete brain nuclei (Renthal and Nestler 2008; Russo et al. 2010). In this article, we review how chromatin remodeling, DNA methylation, and microRNAs regulate gene networks and contribute to cocaine addiction. A particular emphasis is placed on the epigenetic mechanisms regulating expression of brain-derived neurotrophic factor (BDNF) in the mesocorticolimbic dopamine system following chronic cocaine exposure as a specific example of the general principles by which chromatin-dependent transcriptional regulation may contribute to drug addiction.
EPIGENETIC MECHANISMS OF CHROMATIN REGULATION
The definition of epigenetics has evolved to include not only heritable changes in gene expression but also stable changes in gene expression within mature, postmitotic neurons that do not include changes in DNA sequence (Bird 2007; Siegmund et al. 2007; Tsankova et al. 2007). Epigenetic mechanisms transduce environmental stimuli to promote stable alterations in chromatin structure that function to activate or repress gene transcription (Jaenisch and Bird 2003). Posttranslational modifications to histones and chromatin remodeling are dynamic epigenetic mechanisms that alter access of transcriptional machinery to promoter regions thereby regulating patterns of gene expression (Cheung et al. 2000; Strahl and Allis 2000; Berger 2007). A growing body of evidence indicates that chromatin remodeling, including stable enzymatic modifications to DNA and histone proteins, is associated with persistent changes in gene expression that may underlie drug addiction (Renthal and Nestler 2008; Maze and Nestler 2011).
Chromatin Structure, Histone Modifications, and Gene Transcription
Chromatin is a highly compact structure that consists of DNA wrapped around octamers of histone proteins. Access of transcription factors and basal transcriptional machinery to DNA sequences including promoter regions is regulated by chromatin structure (Berger 2007; Li et al. 2007a). Chromatin exists in two basic states that are characterized by different levels of condensation. In general, heterochromatin (condensed chromatin) is associated with inactive gene transcription owing to tight packaging of DNA around histone cores, whereas euchromatin (open chromatin) is associated with active gene transcription owing to a more relaxed chromatin structure and accessible DNA sequences (Berger 2007). Complex combinations of posttranslational modifications of histones alter the affinity of DNA sequences for histone proteins, thereby positively or negatively regulating gene transcription (Strahl and Allis 2000). Therefore, chromatin remodeling through covalent modifications of histone proteins is a requisite mechanism of gene transcription.
The amino-terminal tails of histones contain specific amino acid residues that are sites for several posttranslational modifications such as acetylation and methylation. In general, acetylation of lysine residues corresponds with transcriptionally active chromatin, whereas methylation of lysine and arginine residues is associated with transcriptional repression (Strahl and Allis 2000). Other histone modifications that increase gene transcription include phosphorylation and ubiquitination (Renthal and Nestler 2008). In addition, SUMOylation of histone residues has been shown to be associated with decreased gene transcription (Gareau and Lima 2010). Specific enzymes function to add or remove associated histone marks, indicating that these modifications are potentially reversible (Kouzarides 2007). The summation of dynamic histone signatures at single genes and across the genome forms a “Histone Code” that regulates gene expression (Strahl and Allis 2000). Thus, one epigenetic mechanism is the regulation of gene transcription by posttranslational modifications of histones that alter the affinity of DNA sequences for histone residues.
Histone Acetylation and Psychostimulant-Induced Changes in Gene Transcription
Acetylation of basic lysine residues in histone tails decreases the electrostatic interactions between histone proteins and negatively charged DNA (Kouzarides 2007). Hyperacetylation of promoter regions is associated with increased gene expression, whereas hypoacetylation is correlated with decreased gene expression (Kurdistani et al. 2004). Histone acetyltransferases (HATs) are enzymes that catalyze the addition of acetyl moieties to histone proteins creating a more open chromatin configuration that is conducive to gene activation. In contrast, histone deacetylases (HDACs) function to remove acetyl moieties from histone proteins, thereby promoting condensation of chromatin and inactivation of gene transcription (Marks et al. 2003). It should also be noted that specific transcription factors have also been shown to have HAT activity (Doi et al. 2006). Together, HATs and HDACs function in concert to modify chromatin structure and regulate gene transcription.
Increased expression of the immediate early genes Fos and Fosb in the nucleus accumbens following acute cocaine administration is associated with increased histone H4 acetylation at their promoter regions (Kumar et al. 2005; Levine et al. 2005). In addition, global histone H4 acetylation and H3 phosphoacetylation are transiently increased in the striatum following acute cocaine exposure (Brami-Cherrier et al. 2005; Kumar et al. 2005). Furthermore, the time course of histone acetylation following acute cocaine is consistent with the induction kinetics of Fos and Fosb genes (Renthal and Nestler 2008). Chronic cocaine exposure also is associated with increased histone acetylation at distinct promoter regions. For example, repeated cocaine administration produces stable changes in Cdk5 and Bdnf messenger RNA (mRNA) expression as well as increased histone H3 acetylation at their promoters (Kumar et al. 2005). Chronic cocaine exposure also decreases HDAC5 function in the accumbens promoting histone acetylation and increased expression of HDAC5 targeted genes (Renthal et al. 2007). Interestingly, chronic cocaine exposure induces differential epigenetic regulation of Bdnf transcription and these effects are region specific. Recent studies indicate that cocaine-induced increases in BDNF expression are associated with increased acetylation of histone H3 at the promoter encoding Bdnf exon I-containing transcripts in the accumbens (Cleck et al. 2008) and VTA (Schmidt et al. 2011). However, histone H3 acetylation at Bdnf promoter IV, but not promoter I, is preferentially increased in the medial prefrontal cortex (mPFC) following chronic cocaine (Sadri-Vakili et al. 2010). Cocaine-induced alterations in histone H3 acetylation and corresponding changes in gene expression are stable during periods of drug abstinence (Freeman et al. 2008), which suggests that cocaine-induced chromatin remodeling produces persistent changes in gene expression that may underlie drug craving and relapse.
Global histone H3 acetylation levels are significantly enhanced in mice that develop conditioned place preference following repeated methamphetamine administration (Shibasaki et al. 2011). Increased acetylation of histone H3 proteins is associated with genes that regulate synaptic plasticity in the forebrain (Shibasaki et al. 2011). Withdrawal from chronic amphetamine exposure also decreases transcription of the immediate early gene Fos, in part, through mechanisms that recruit HDAC1 to the Fos promoter (Renthal et al. 2008). Future studies are needed to determine the functional significance of amphetamine-induced changes in chromatin structure at gene promoters in the striatum and limbic forebrain.
Histone Acetylation and Psychostimulant-Induced Behavioral Responses
Histone acetylation and chromatin remodeling are functionally relevant as both pharmacological inhibition and genetic manipulation of HDACs alter behavioral responses to cocaine. Systemic and intraaccumbens administration of HDAC inhibitors significantly enhances cocaine-induced locomotor activity and conditioned place preference (Kumar et al. 2005; Renthal et al. 2007). Consistent with these results, viral-mediated overexpression of HDACs in the nucleus accumbens decreases histone acetylation and attenuates cocaine-induced conditioned place preference (Renthal et al. 2007). Mice deficient in the HAT cAMP response element binding (CREB) protein (CBP) have decreased histone H4 acetylation and display reduced sensitivity to cocaine (Levine et al. 2005). Taken together, these results indicate that cocaine-induced behavioral plasticity is mediated, in part, by increased acetylation of gene networks.
Recent studies indicate that the role of histone acetylation in cocaine-taking behavior is complex. Inhibition of HDACs promotes differential behavioral responses in animals self-administering cocaine and these effects are critically dependent on the timing of HDAC inhibitor administration. Systemic administration of an HDAC inhibitor before the initiation of daily cocaine self-administration sessions decreased the number of cocaine infusions self-administered, suggesting that histone acetylation decreases the reinforcing efficacy of cocaine (Romieu et al. 2008). In contrast, cocaine taking increases when animals that are stably self-administering cocaine are pretreated with a HDAC inhibitor, which suggests that histone acetylation in these animals, increases the reinforcing efficacy of cocaine (Sun et al. 2008). Administration of a HDAC inhibitor directly into the accumbens increases an animal’s motivation to self-administer cocaine as measured by a progressive-ratio (PR) schedule of reinforcement and is associated with increased histone H3 acetylation in the accumbens (Wang et al. 2010). Furthermore, overexpressing HDAC4 in the nucleus accumbens shell decreases cocaine self-administration on a PR schedule (Wang et al. 2010). Although these results indicate that increased histone acetylation is one epigenetic mechanism that underlies cocaine-taking behavior, the exact temporal sequence of histone acetylation and gene transcription in relation to cocaine exposure and subsequent behavioral outcomes remains to be determined.
Histone acetylation also plays a critical role in the reinstatement of cocaine-seeking behavior. Administration of HDAC inhibitors facilitates extinction of cocaine-conditioned place preference and attenuates reinstatement of cocaine-seeking behavior (Malvaez et al. 2010; Romieu et al. 2011). These behavioral effects coincide with increased acetylation of histone H3 and suggest that chromatin remodeling and altered gene transcription during drug withdrawal may prevent drug craving and relapse (Malvaez et al. 2010).
Recent studies also show a role for histone acetylation in amphetamine-induced behavioral responses. Histone acetylation plays a critical role in behavioral sensitization to the locomotor-activating effects of amphetamine (Kalda et al. 2007; Shen et al. 2008). Chronic amphetamine exposure is associated with increased striatal histone H4 acetylation at the level of the Fosb promoter and increased phosphorylation of CREB (Shen et al. 2008). Furthermore, repeated methamphetamine administration increases histone H3 acetylation at unique gene promoters in the limbic forebrain (Shibasaki et al. 2011). Taken together, these results suggest that amphetamine-induced behavioral plasticity is regulated, in part, by changes in chromatin structure within the striatum that facilitate binding of transcription factors including CREB to promoter sequences to facilitate gene transcription.
Histone Methylation and Psychostimulant-Induced Changes in Gene Transcription
Addition of methyl groups to histone proteins does not change the charge of targeted amino acid residues and these modifications are relatively stable compared to histone acetylation (Rice and Allis 2001). Methylation of lysine and arginine residues on histone tails is complex and can occur in mono-(me), di-(me2), or trimethylated (me3) states with each methylation event having distinct, and often opposite, effects on gene transcription (Rice and Allis 2001). Histone methylation at gene promoters either promotes or represses gene transcription depending on the exact amino acid residues that are methylated and the valence of methylation at these residues (Maze and Nestler 2011); for example, di- and trimethylation of histone H3 lysine residues 9 (H3K9me2/3) and 27 (H3K27me2/3) recruit corepressor proteins that may function to increase chromatin condensation and thereby decrease gene transcription (Rice and Allis 2001). In contrast, trimethylation of histone H3 lysine residues 4 (H3K4me3) and 36 (H3K36me3) correlate with increased levels of gene transcription (Rice and Allis 2001).
Recent evidence indicates that psychostimulant exposure alters gene expression, in part, through changes in histone methylation. Histone H3 methylation is decreased in the mPFC of adult rats that were exposed to cocaine during adolescence and these epigenetic marks coincide with altered gene expression in adulthood (Black et al. 2006). These findings suggest that cocaine exposure during adolescence produces long-lasting changes in gene expression that are mediated by chromatin remodeling. Repeated administration of cocaine in adult mice also reduces histone methylation in the brain and this epigenetic mechanism is associated with decreased expression of the methyltransferase G9a (Maze et al. 2010). Consistent with these findings, expression of G9a target genes is increased in the nucleus accumbens following repeated cocaine administration and viral-mediated knockdown of G9a expression, which mimics the effects of chronic cocaine exposure and facilitates cocaine-induced synaptic and behavioral plasticity (Maze et al. 2010). Moreover, repeated cocaine alters heterochromatic histone H3 methylation in the accumbens and produces long-lasting decreases in heterochromatin, which suggests that cocaine-induced alterations in histone methylation and heterochromatin formation are also an important mechanism in the long-term actions of cocaine (Maze et al. 2011). Amphetamine abstinence is also associated with changes in histone H3 methylation. Histone H3 methylation is increased at the Fos promoter in the striatum following repeated amphetamine exposure and is associated with decreased transcription of this immediate early gene (Renthal et al. 2008). Consistent with these results, expression of the histone H3 methyltransferase KMT1A is increased in the striatum following chronic amphetamine exposure (Renthal et al. 2008).
Histone Methylation and Psychostimulant-Induced Behavioral Responses
Adolescent rats exposed to chronic cocaine develop cognitive impairments in adulthood that are associated with altered histone methylation and gene transcription in the mPFC (Black et al. 2006). Chronic cocaine exposure in adult rats represses G9a expression thereby decreasing global histone methylation in the nucleus accumbens and enhancing cocaine-induced behavioral responses (Maze et al. 2010). The inability of G9a to regulate gene transcription following repeated cocaine results in aberrant synaptic plasticity in the accumbens (Maze et al. 2010) and may correlate with long-term psychostimulant-induced changes in structural plasticity (Robinson and Kolb 1997). G9a regulation of histone methylation in the accumbens also plays a critical role in drug-induced vulnerability to stress (Covington et al. 2011). Taken together, these results suggest that chronic cocaine exposure in adolescence and adulthood regulates expression of gene networks to alter structural plasticity in the brain that, in turn, may contribute to drug-induced behavioral plasticity. The role of histone methylation in regulating distinct gene networks to promote amphetamine-induced behavioral plasticity remains to be determined.
Histone Phosphorylation and Psychostimulant-Induced Changes in Gene Transcription
Histone phosphorylation is another posttranslational modification that is associated with increased gene transcription (Brami-Cherrier et al. 2009). Phosphorylation of serine 10 on histone H3 promotes HAT activity, phosphoacetylation of neighboring amino acid residues, and inhibits repressive methylation marks on H3 (Kouzarides 2007). Acute amphetamine administration transiently increases histone H3 phosphorylation (Rotllant and Armario 2012). Moreover, cocaine administration increases histone H3 phosphorylation and phosphoacetylation at the Fos promoter in the striatum, effects that are mediated by mitogen- and stress-activated protein kinase 1 (MSK1) (Brami-Cherrier et al. 2005; Brami-Cherrier et al. 2009). Constitutive knockdown of MSK1 blocks cocaine-induced increases in histone H3 phosphorylation and Fos expression and alters cocaine-induced behavioral plasticity (Brami-Cherrier et al. 2005). Although histone acetylation and phosphorylation are both associated with increased gene transcription, these epigenetic mechanisms can act in concert or independently to regulate gene expression (Brami-Cherrier et al. 2007). The role of histone phosphorylation in psychostimulant-induced behavioral responses remains to be determined.
DNA Methylation and Psychostimulant-Induced Changes in Gene Transcription
In addition to posttranslational histone modifications, enzymatic modifications to DNA sequences also translate environmental stimuli such as drug exposure into altered patterns of gene expression and enduring behavioral phenotypes. DNA methylation involves the addition of methyl groups to cytosine-guanine dinucleotides (CpG) in the genome by DNA methyltransferases (DNMTs) (Suzuki and Bird 2008). Methylation of CpG islands interferes with transcription factor binding to target DNA sequences through the recruitment of corepressor complexes (Jaenisch and Bird 2003). Methyl-binding domain-containing proteins bind methylated DNA regions and recruit corepressors such as HDACs and methyltransferases to gene promoters. Therefore, it is important to note that DNA methylation and histone modifications are not mutually exclusive. Although originally thought to repress or inhibit gene transcription, DNA methylation is a dynamic process that functions to either promote or repress gene expression (Suzuki and Bird 2008).
Emerging evidence suggests that psychostimulant-induced changes in gene expression are regulated by DNA methylation. The hippocampi of rats exposed to cocaine in utero are characterized by altered global patterns of DNA methylation and corresponding changes in gene transcription (Novikova et al. 2008). Changes in gene expression following cocaine self-administration also correlate with increased expression of the methyl-CpG-binding protein MeCP2 (Host et al. 2011). Further evidence for a role of DNA methylation in cocaine-induced synaptic and behavioral plasticity comes from studies of DNMTs. Acute cocaine administration increases DNA methylation as well as the expression of DNMT3A and DNMT3B in the nucleus accumbens (Anier et al. 2010). Increased DNA methylation following acute cocaine is associated with enhanced binding of MeCP2 to specific gene promoters and corresponding decreases in gene transcription (Anier et al. 2010). Furthermore, pharmacological inhibition of DNMT decreases cocaine-induced DNA hypermethylation and attenuates drug-induced down-regulation of gene expression in the accumbens (Carouge et al. 2010). DNMT3a expression is increased during protracted periods of drug abstinence in cocaine-experienced animals (LaPlant et al. 2010). DNMT3a also plays a critical role in cocaine-induced increases in dendritic spine density, which suggests that DNA methylation is an important epigenetic mechanism in regulating cocaine-induced structural plasticity (LaPlant et al. 2010). Acute and subchronic methamphetamine administration has differential effects on DNMT1 mRNA expression that are brain region specific, suggesting that drug-induced changes in gene transcription are mediated, in part, by DNA methylation (Numachi et al. 2007). However, future studies are required to determine whether chronic methamphetamine increases DNMT1 protein and the functional significance of altered DNA methylation on drug-induced behavioral responses.
DNA Methylation and Psychostimulant-Induced Behavioral Responses
Dynamic changes in DNA methylation may underlie cocaine-induced behavioral responses. Sensitization to the locomotor-activating effects of cocaine is delayed in animals treated with a DNMT inhibitor and coincides with altered DNA methylation at gene promoters (Anier et al. 2010). Decreased DNMT3a function enhances the behavioral response to cocaine supporting the hypothesis that decreased DNA methylation promotes increased gene transcription following repeated cocaine exposure and contributes to drug-induced behavioral plasticity (LaPlant et al. 2010). Together, these results suggest that dynamic changes of DNA methylation may be an important epigenetic mechanism underlying cocaine-induced behavioral effects.
GENOME-WIDE STUDIES OF COCAINE-INDUCED CHANGES IN CHROMATIN REGULATION
Drug-induced histone modifications can be identified and characterized across the genome using microarrays and next-generation sequencing methods. Precise genomic loci that are associated with histones modified by drug exposure are identified using genome-wide promoter arrays (ChIP-chip) or massively parallel DNA sequencing platforms (ChIP-Seq) (Renthal et al. 2009; Maze et al. 2011; Zhou et al. 2011). These high-throughput methods characterize complex drug-induced signatures of epigenetic regulation including multifaceted histone modifications that regulate transcription of gene networks and may underlie drug-induced behavioral plasticity. ChIP-chip analyses of nucleus accumbens lysates reveals that chronic cocaine exposure regulates gene transcription by either increasing histone H3 or H4 acetylation (to elevate mRNA levels), or by increasing histone H3 dimethyl-K9/27 (to reduce mRNA expression) (Renthal et al. 2009). Chronic cocaine exposure also decreases repressive histone methylation (H3K9me3) in the accumbens and ChIP-Seq reveals that these histone marks are associated with intergenic genomic regions (Maze et al. 2011). These results suggest that cocaine-induced histone methylation produces heterochromatic derepression and increases expression of retrotransposable elements that in turn regulate gene transcription (Maze et al. 2011). A recent study used whole genome sequencing of mRNA transcripts (RNA-Seq) and ChIP-Seq to characterize histone methylation and gene expression in postmortem hippocampal tissue from cocaine-dependent subjects (Zhou et al. 2011). Interestingly, cocaine-induced changes in histone methylation did not correlate with corresponding changes in gene expression in the hippocampus, which suggests that complex epigenetic pathways act in concert to regulate gene transcription (Zhou et al. 2011). Taken together, these studies show that cocaine acts to alter patterns of gene expression in the nucleus accumbens and hippocampus through epigenetic mechanisms (i.e., histone acetylation and methylation) that promote stable, persistent changes in gene expression. Thus, genome-wide studies identify dynamic chromatin signatures following chronic cocaine exposure and reveal novel gene targets and molecular regulatory pathways that may play critical roles in drug taking and seeking. It is not clear whether other psychostimulants and drugs of abuse exert their behavioral effects through similar or divergent epigenetic regulation of gene networks.
MicroRNAs
Posttranscriptional regulation of gene expression following chronic drug exposure has also been shown to influence drug taking and seeking in laboratory animals (Pietrzykowski 2010; Li and van der Vaart 2011). Specifically, microRNAs (miRNAs) have emerged as a new class of epigenetic regulators that are capable of altering synaptic plasticity and behavior (Guarnieri and DiLeone 2008). miRNAs are a class of nonprotein coding RNA transcripts (∼19–24 nucleotides) that regulate gene expression at the posttranscriptional level (Ambros 2004). It is predicted that there are >800 unique miRNA species in humans (Bentwich et al. 2005; Berezikov et al. 2006), many of which are highly expressed in the brain (Sempere et al. 2004; Lugli et al. 2008). More than 33% of the mammalian genome is subject to miRNA regulation and each miRNA targets on average 200 mRNA transcripts (Lewis et al. 2005; Friedman et al. 2009b). A growing literature indicates that miRNAs have diverse effects on gene expression including mRNA degradation, increased mRNA translation, chromatin remodeling, and DNA methylation.
Initially, miRNAs were thought to be located within intergenic clusters within the genome and regulated by their own promoter regions (Lagos-Quintana et al. 2001; Lau et al. 2001). However, it was determined recently that at least 50% of mammalian miRNAs are located within introns of protein-coding genes, which suggests that concurrent expression of miRNAs and their host genes is regulated by common promoter regions and transcriptional machinery (Rodriguez et al. 2004; Ason et al. 2006; Berezikov et al. 2007; Li et al. 2007b; Okamura et al. 2007; Saini et al. 2007). miRNA genes are transcribed by RNA polymerases to produce immature transcripts (Lee et al. 2002; Lee et al. 2004; Borchert et al. 2006). These immature transcripts are spliced, similar to mRNA, to produce double-stranded, hairpin-loop structures that are several hundred base pairs in length called primary miRNAs (pri-miRNAs) (Lee et al. 2002). Pri-miRNAs are further cleaved in the nucleus by the enzyme Drosha, as part of a protein complex called the microProcessor (Lee et al. 2003; Denli et al. 2004; Gregory et al. 2004; Yeom et al. 2006). The cleaved product is a double-stranded RNA fragment, called precursor miRNA (pre-miRNA), which is ∼70 nucleotides in length and contains a two-nucleotide overhang at the 3′ end. The nuclear membrane protein Exportin 5 binds to the 3′ overhang of pre-miRNAs and transports them from the nucleus into the cytoplasm (Yi et al. 2003; Bohnsack et al. 2004; Lund et al. 2004). Following its translocation into the cytoplasm, pre-miRNA is cleaved by the enzyme Dicer to form an ∼20–25 nucleotide duplex consisting of a mature miRNA strand and its opposite, complementary (“passenger”) strand, miRNA* (Bernstein et al. 2001; Hutvagner et al. 2001). Dicer-mediated cleavage of pre-miRNAs is thought to coincide with unwinding of the duplex to produce single-stranded, active miRNAs (MacRae et al. 2008). Single-stranded miRNAs are preferentially loaded into the microRNA-induced silencing complex (miRISC) (Hutvagner and Zamore 2002). The main protein constituents associated with miRISC complexes are Argonaute (AGO) proteins that bind miRNAs and facilitate cleavage of targeted mRNA transcripts (Baumberger and Baulcombe 2005; Peters and Meister 2007). It is thought that one miRNA is sufficient to direct miRISC to target mRNAs to cleave or silence these transcripts.
miRNAs regulate gene expression by degrading mRNA transcripts, repressing mRNA translation, or both (Jackson and Standart 2007; Pillai et al. 2007). Target specificity is imparted through miRNA recognition and binding to complementary sequences (∼2–7 nucleotides), or “seed regions,” in the 3′-UTR (untranslated region) of mRNA (Lewis et al. 2003; Grimson et al. 2007; Bartel 2009). Originally thought to be nonfunctional by-products of pre-miRNA cleavage (Matranga et al. 2005), miRNA* strands also act at distinct binding sites to regulate gene expression (Tyler et al. 2008; Okamura et al. 2009; Ghildiyal et al. 2010). Emerging evidence indicates that molecular regulation of gene expression by miRNAs is more complex than originally thought. In addition to repressing protein synthesis and directing sequence-specific degradation of complementary mRNA, the miRNA/miRISC complex has also been shown to induce gene expression by activating mRNA translation (Vasudevan et al. 2007; Place et al. 2008; Steitz and Vasudevan 2009). miRNAs also remodel chromatin structure and increase DNA methylation thereby altering expression of target genes (Tan et al. 2009) and, in some cases, inducing gene activation (Place et al. 2008).
MicroRNAs and Psychostimulant Addiction
miRNAs coordinate the expression of networks of related genes involved in synaptic plasticity (Kosik 2006; Schratt et al. 2006). Furthermore, miRNAs have been identified in dendrites, which suggests that miRNAs function, in part, to rapidly translate cellular signals into regulation of local mRNA transcripts (Ashraf and Kunes 2006; Hobert 2008). Chronic drug exposure induces maladaptive changes in neural networks including aberrant synaptic plasticity in the mesocorticolimbic dopamine system (Kalivas et al. 2005; Kauer and Malenka 2007; Thomas et al. 2008; Russo et al. 2010). Given the potential role of drug-evoked synaptic plasticity in the development and persistence of compulsive drug-taking behavior (Hyman et al. 2006; Luscher and Malenka 2011; Mameli and Luscher 2011), it is not surprising that miRNAs play a critical role in drug addiction (Dreyer 2010; Pietrzykowski 2010; Li and van der Vaart 2011).
Recent studies indicate that compulsive cocaine consumption is mediated, in part, by miRNAs. Cocaine self-administration increases expression of miR-212 in the dorsal striatum (Hollander et al. 2010). Furthermore, increased miR-212 expression in the striatum is associated with decreased cocaine self-administration and suggests that up-regulation of striatal miR-212 is a compensatory mechanism that decrease’s the motivational properties of cocaine (Hollander et al. 2010). In contrast, the transcriptional repressor MeCP2 plays a critical role in regulating increased cocaine consumption (Im et al. 2010). MeCP2 attenuates cocaine-induced up-regulation of miR-212 expression in the striatum, whereas miR-212 inhibits MeCP2 expression (Im et al. 2010). Thus, a pivotal balance between MeCP2 and miR-212 levels in the striatum regulates compulsive drug-taking behavior.
Chronic cocaine exposure also increases expression of miR-181a and decreases expression of miR-124 and let-7d in the mesocorticolimbic dopamine system (Chandrasekar and Dreyer 2009). Although increased expression of miR-124 and let-7d in the nucleus accumbens attenuates cocaine-induced conditioned place preference (CPP), increased expression of miR-181a in the accumbens enhances cocaine CPP (Chandrasekar and Dreyer 2011). Differential behavioral effects of miR-124, let-7d, and miR-181a are associated with distinct changes in gene expression in the nucleus accumbens (Chandrasekar and Dreyer 2011). Taken together, these results suggest that complex miRNA regulatory pathways modulate cocaine-induced behavioral plasticity by directing expression of gene networks. It remains to be determined whether other psychostimulants exert similar effects on miRNA expression.
Thus, miRNAs are epigenetic regulators that play a critical role in translating drug-induced changes in synaptic plasticity into persistent neuroadaptations associated with drug addiction. By targeting hundreds of mRNA transcripts, a single miRNA coordinates expression of gene networks that regulate neuronal plasticity and behavior. Although miRNAs may represent promising new targets in the development of novel therapies to treat drug craving and relapse, future studies are needed to determine the precise role of miRNAs and their targets in the molecular mechanisms underlying drug addiction.
BDNF AND COCAINE ADDICTION
BDNF is a member of the neurotrophin family that includes nerve growth factor, neurotrophin-3, and neurotrophin 4/5 (Thoenen 1995). BDNF is synthesized as a propeptide (32 KDa) that is proteolytically processed into a smaller (13 KDa), mature form that binds to and activates tropomyosin receptor kinase B (TrkB) receptors (Bibel and Barde 2000). TrkB stimulation results in receptor dimerization and tyrosine phosphorylation that provides docking sites for adapter molecules, internalization, and intracellular signaling leading to changes in gene expression and synaptic plasticity (Sommerfeld et al. 2000; Patapoutian and Reichardt 2001; Lu 2003; Nagappan and Lu 2005). Stimuli that induce neuronal activity in a calcium-dependent manner increase Bdnf mRNA and BDNF protein expression (Shieh et al. 1998). Following transcription, Bdnf mRNA is trafficked to active synapses (Tongiorgi et al. 1997) where long 3′ UTR mRNA transcripts are preferentially localized and translated (An et al. 2008). Synaptic secretion of BDNF and subsequent TrkB receptor activation are associated with increased glutamatergic activity (Jovanovic et al. 2000; Hartmann et al. 2001; Balkowiec and Katz 2002). Furthermore, BDNF promotes both early and late-phase long-term potentiation (LTP), dendritic protein synthesis, and dendritic spine formation (Bramham et al. 1996). BDNF regulates dendritic spine formation and synaptic plasticity by inhibiting miR-134, a miRNA that negatively regulates dendritic spine development and maturation (Schratt et al. 2006). BDNF-mediated inhibition of miR-134 promotes translation of Lim kinase 1, an enzyme that regulates actin filament activity and synaptic plasticity (Schratt et al. 2006).
Many cocaine-induced neuroadaptations that are thought to underlie cocaine seeking are manifested by alterations in the plasticity of mesocorticolimbic circuitry (Schmidt and Pierce 2010). Bdnf mRNA is expressed abundantly in cortical as well as midbrain dopamine neurons and at much lower levels in striatal neurons (Altar et al. 1997; Lipska et al. 2001). In fact, cortical pyramidal neurons are thought to supply ∼80% and dopamine neurons ∼20% of BDNF protein within the striatum (Altar et al. 1997). Endogenous Bdnf mRNA and protein are differentially regulated in mesolimbic and cortical neurons in response to acute and repeated administration of psychostimulants or during extended periods of drug abstinence (Meredith et al. 2002; Le Foll et al. 2005; Filip et al. 2006; Liu et al. 2006; Fumagalli et al. 2007; Saylor and McGinty 2008; Fumagalli et al. 2009). In addition, a persistent BDNF protein response develops in mesolimbic, striatal, and cortical structures and lasts for extended durations during abstinence from cocaine self-administration (Grimm et al. 2003; Im et al. 2010; McGinty et al. 2010). Altered expression of BDNF in this network of reciprocally interconnected structures following cocaine exposure and/or drug abstinence suggests that BDNF may constitute a critical component of cocaine-induced plasticity.
The effects of exogenous BDNF infusion on cocaine seeking are brain region specific and time dependent. Infusion of BDNF into subcortical structures, like the nucleus accumbens and VTA, enhances cocaine-seeking behavior (Lu et al. 2004; Graham et al. 2007). These studies implicate VTA and nucleus accumbens BDNF activity in long-term modulation of cocaine-induced behavior. In contrast, BDNF infusion into the dorsomedial PFC immediately following a final session of cocaine self-administration attenuates the reinstatement of cocaine seeking by normalizing cocaine-induced alterations in phospho-ERK and phospho-CREB expression in the PFC and glutamate transmission in the nucleus accumbens (Berglind et al. 2007; Berglind et al. 2009; Whitfield et al. 2011). In support of the cocaine-suppressing effects of BDNF, knockdown of BDNF in the mPFC augments the intake of cocaine in rats self-administering cocaine (Sadri-Vakili et al. 2010). In contrast, overexpression of BDNF in the dorsal striatum has been implicated in the acceleration of, and loss of control over, compulsive cocaine taking (Im et al. 2010). Moreover, suppression of endogenous BDNF signaling, by infusing a neutralizing antibody to BDNF in the dorsal striatum, decreases cocaine intake (Im et al. 2010). Thus, exogenous infusion or manipulation of endogenous BDNF levels has a selective functional impact in different target areas that are critical to mediating or preventing cocaine-induced dysfunctional neuroadaptations.
EPIGENETIC REGULATION OF BDNF EXPRESSION IN RESPONSE TO COCAINE
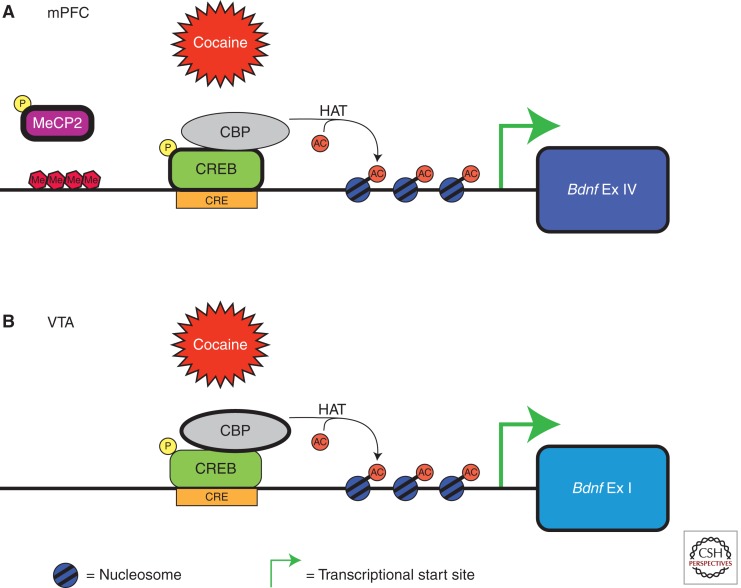
A growing body of evidence suggests that epigenetic mechanisms of regulation are important for the modulation of drug-induced Bdnf transcription (Fig. 1). The Bdnf gene is comprised of nine exons with at least eight alternative promoters that are differentially responsive to cocaine-activated signaling cascades (Liu et al. 2006; Aid et al. 2007). Cocaine-induced increases in Bdnf transcription are associated with increased histone acetylation at several Bdnf promoters (Kumar et al. 2005; Schroeder et al. 2008; Sadri-Vakili et al. 2010; Schmidt et al. 2011). Histone acetylation at specific Bdnf promoters is associated with binding of the histone acetyltransferase CBP (Schmidt et al. 2011), and the histone deacetylases HDAC1 and HDAC2 (Guan et al. 2009) to these promoter regions.
Figure 1.
Differential cocaine-induced effects at specific Bdnf promoters are mediated by distinct epigenetic mechanisms in the mPFC and VTA. (A) Cocaine selectively increases Bdnf exon IV–containing transcript levels in the mPFC. Cocaine-induced increases in mPFC Bdnf transcription are associated with increased CREB phosphorylation and histone H3 acetylation at Bdnf exon IV promoters. Furthermore, MeCP2 binding to Bdnf exon IV promoter regions is decreased following cocaine self-administration. (B) In contrast, cocaine selectively increases Bdnf exon I–containing transcript levels in the VTA. Cocaine-induced increases in VTA Bdnf transcription are associated with recruitment of CBP, an enzyme that catalyzes the addition of acetyl groups to histone proteins, and increased histone H3 acetylation at exon I–containing promoter regions. AC, acetyl group; Bdnf, brain-derived neurotrophic factor; CBP, CREB-binding protein; CRE, cAMP response element; CREB, cAMP response element binding protein; Ex IV, exon IV; Ex I, exon I; HAT, histone acetyltransferase; Me, methyl group; MeCP2: methyl-CpG-binding protein 2; P, phosphate group.
Reduced DNA methylation of various regions across the Bdnf locus has been detected following many stimuli that increase BDNF expression (Lubin et al. 2008; Ma et al. 2009). However, it remains to be determined whether cocaine modulates DNA methylation of the Bdnf gene. Induction of Bdnf transcription in the PFC is correlated with the dissociation of MeCP2 from BDNF promoter IV, consistent with a potential reduction in DNA methylation under these conditions (Sadri-Vakili et al. 2010). Evidence for the functional importance of DNA methylation in the regulation of Bdnf transcription comes largely from studies in which MeCP2 expression has been disrupted. Although MeCP2 is bound widely to DNA across the genome (Skene et al. 2010), decreased MeCP2 expression is associated with surprisingly subtle changes in the expression of a subset of genes (Tudor et al. 2002; Chahrour et al. 2008; Skene et al. 2010). Nonetheless, levels of Bdnf are consistently reduced in the brains of Mecp2 knockout mice (Chang et al. 2006; Fyffe et al. 2008). Interestingly, MeCP2 is a target of regulation by psychostimulant-activated signaling cascades (Deng et al. 2010; Im et al. 2010). Phosphorylation of MeCP2 at Ser421 is induced rapidly and robustly by acute and repeated administration of cocaine or amphetamine, and this phosphorylation is selective for specific populations of neurons in the PFC and nucleus accumbens (Deng et al. 2010). Although the consequences of Ser421 phosphorylation for MeCP2 function are not known, this regulation suggests a mechanism to couple MeCP2-dependent transcription of Bdnf and other genes with psychostimulant exposure. In addition, expression of MeCP2 is up-regulated following chronic cocaine self-administration in the dorsal striatum of rats where knockdown of MeCP2 expression is associated with impaired cocaine-dependent up-regulation of BDNF protein (Im et al. 2010; Host et al. 2011). However, it is unclear whether increased MeCP2 expression is directly acting under these conditions to alter transcriptional regulation of Bdnf.
miRNAs represent a third epigenetic mechanism that may contribute to psychostimulant-induced expression of BDNF. Although a number of miRNAs can bind directly to the 3′-UTR of Bdnf (Mellios et al. 2008; Friedman et al. 2009a; Muinos-Gimeno et al. 2011), the relevance of this epigenetic mechanism for the in vivo regulation of BDNF levels remains to be determined. However, miRNAs may impact cocaine-induced BDNF expression indirectly via regulation of MeCP2 expression and CREB activation. Cocaine self-administration is associated with increased expression of miR-132 and miR-212 in the dorsal striatum, both of which can repress the expression of MeCP2 (Klein et al. 2007; Hollander et al. 2010; Im et al. 2010). MeCP2 appears to exert a complementary repression of miR-132 and miR-212, suggesting that these transcriptional regulators are engaged in a homeostatic feedback loop (Klein et al. 2007; Im et al. 2010). Overexpression of miR-212 in the dorsal striatum attenuates cocaine-induced up-regulation of MeCP2 expression and inhibits cocaine-induced up-regulation of BDNF protein (Im et al. 2010). Interestingly, in addition to its effects on MeCP2 expression, miR-212 has been shown to amplify CREB signaling and to increase cocaine-induced expression of CREB-target genes including Fos (Hollander et al. 2010; Im et al. 2010). Bdnf transcription is also regulated by CREB as well as MeCP2 (Shieh et al. 1998; Tao et al. 1998) and it remains to be determined why the effects of miR-212 overexpression on MeCP2 appear to dominate with respect to BDNF regulation over the effects on CREB. These observations highlight the challenges of interpreting the effects of disrupting single regulatory factors within the context of an interconnected transcriptional network, and suggest that there is a rich world of complexity contributing to the tight spatial and temporal control of BDNF expression that remains to be explored.
CONCLUDING REMARKS
Increasing evidence suggests that epigenetic mechanisms including histone modifications, DNA methylation, and miRNAs regulate psychostimulant-induced gene expression profiles in discrete brain regions. Many changes in chromatin regulation following chronic psychostimulant exposure correlate in time with the expression of maladaptive behaviors including drug taking and seeking. However, molecular genetic studies have also implicated some of the transcriptional regulatory factors discussed in this review (i.e., miR-212, MeCP2, HDAC5, and BDNF in the mPFC) in the induction of adaptive forms of neuroplasticity that appear to repress or inhibit drug self-administration. Further characterizing the molecular substrates that regulate chromatin remodeling and gene transcription following chronic drug exposure may identify novel drug targets for drug craving and relapse. Given the evidence that BDNF expression in different brain regions has both essential and distinct effects on drug-taking behavior, understanding the transcriptional regulation of this single gene offers an opportunity to discover insights into the role of epigenetic mechanisms of chromatin regulation in drug addiction. However, future studies that more broadly elucidate the epigenetic processes that mediate long-lasting changes in gene expression networks throughout the brain will substantially enhance our understanding of how persistent changes in gene transcription contribute to the development and expression of compulsive drug-taking behaviors.
ACKNOWLEDGMENTS
H.D.S. is supported by an individual K01 award (DA030445). J.F.M. is supported by P50-DA15369 and R01-DA03982. A.E.W is supported by R01-DA022202. G.S-V. is supported by NS063953 and DA022339.
Footnotes
Editors: R. Christopher Pierce and Paul J. Kenny
Additional Perspectives on Addiction available at www.perspectivesinmedicine.org
REFERENCES
- Aid T, Kazantseva A, Piirsoo M, Palm K, Timmusk T 2007. Mouse and rat BDNF gene structure and expression revisited. J Neurosci Res 85: 525–535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altar CA, Cai N, Bliven T, Juhasz M, Conner JM, Acheson AL, Lindsay RM, Wiegand SJ 1997. Anterograde transport of brain-derived neurotrophic factor and its role in the brain. Nature 389: 856–860 [DOI] [PubMed] [Google Scholar]
- Ambros V 2004. The functions of animal microRNAs. Nature 431: 350–355 [DOI] [PubMed] [Google Scholar]
- An JJ, Gharami K, Liao GY, Woo NH, Lau AG, Vanevski F, Torre ER, Jones KR, Feng Y, Lu B, et al. 2008. Distinct role of long 3′ UTR BDNF mRNA in spine morphology and synaptic plasticity in hippocampal neurons. Cell 134: 175–187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anier K, Malinovskaja K, Aonurm-Helm A, Zharkovsky A, Kalda A 2010. DNA methylation regulates cocaine-induced behavioral sensitization in mice. Neuropsychopharmacology 35: 2450–2461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashraf SI, Kunes S 2006. A trace of silence: Memory and microRNA at the synapse. Curr Opin Neurobiol 16: 535–539 [DOI] [PubMed] [Google Scholar]
- Ason B, Darnell DK, Wittbrodt B, Berezikov E, Kloosterman WP, Wittbrodt J, Antin PB, Plasterk RH 2006. Differences in vertebrate microRNA expression. Proc Natl Acad Sci 103: 14385–14389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balkowiec A, Katz DM 2002. Cellular mechanisms regulating activity-dependent release of native brain-derived neurotrophic factor from hippocampal neurons. J Neurosci 22: 10399–10407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP 2009. MicroRNAs: Target recognition and regulatory functions. Cell 136: 215–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumberger N, Baulcombe DC 2005. Arabidopsis ARGONAUTE1 is an RNA Slicer that selectively recruits microRNAs and short interfering RNAs. Proc Natl Acad Sci 102: 11928–11933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentwich I, Avniel A, Karov Y, Aharonov R, Gilad S, Barad O, Barzilai A, Einat P, Einav U, Meiri E, et al. 2005. Identification of hundreds of conserved and nonconserved human microRNAs. Nat Genet 37: 766–770 [DOI] [PubMed] [Google Scholar]
- Berezikov E, Thuemmler F, van Laake LW, Kondova I, Bontrop R, Cuppen E, Plasterk RH 2006. Diversity of microRNAs in human and chimpanzee brain. Nat Genet 3: 1375–1377 [DOI] [PubMed] [Google Scholar]
- Berezikov E, Chung WJ, Willis J, Cuppen E, Lai EC 2007. Mammalian mirtron genes. Mol Cell 28: 328–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger SL 2007. The complex language of chromatin regulation during transcription. Nature 447: 407–412 [DOI] [PubMed] [Google Scholar]
- Berglind WJ, See RE, Fuchs RA, Ghee SM, Whitfield TW Jr, Miller SW, McGinty JF 2007. A BDNF infusion into the medial prefrontal cortex suppresses cocaine seeking in rats. Eur J Neurosci 26: 757–766 [DOI] [PubMed] [Google Scholar]
- Berglind WJ, Whitfield TW Jr, LaLumiere RT, Kalivas PW, McGinty JF 2009. A single intra-PFC infusion of BDNF prevents cocaine-induced alterations in extracellular glutamate within the nucleus accumbens. J Neurosci 29: 3715–3719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein E, Caudy AA, Hammond SM, Hannon GJ 2001. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 409: 363–366 [DOI] [PubMed] [Google Scholar]
- Bibel M, Barde YA 2000. Neurotrophins: Key regulators of cell fate and cell shape in the vertebrate nervous system. Genes Dev 14: 2919–2937 [DOI] [PubMed] [Google Scholar]
- Bird A 2007. Perceptions of epigenetics. Nature 447: 396–398 [DOI] [PubMed] [Google Scholar]
- Black YD, Maclaren FR, Naydenov AV, Carlezon WA Jr, Baxter MG, Konradi C 2006. Altered attention and prefrontal cortex gene expression in rats after binge-like exposure to cocaine during adolescence. J Neurosci 26: 9656–9665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohnsack MT, Czaplinski K, Gorlich D 2004. Exportin 5 is a RanGTP-dependent dsRNA-binding protein that mediates nuclear export of pre-miRNAs. RNA 10: 185–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borchert GM, Lanier W, Davidson BL 2006. RNA polymerase III transcribes human microRNAs. Nat Struct Mol Biol 13: 1097–1101 [DOI] [PubMed] [Google Scholar]
- Bramham CR, Southard T, Sarvey JM, Herkenham M, Brady LS 1996. Unilateral LTP triggers bilateral increases in hippocampal neurotrophin and trk receptor mRNA expression in behaving rats: Evidence for interhemispheric communication. J Comparative Neurol 368: 371–382 [DOI] [PubMed] [Google Scholar]
- Brami-Cherrier K, Valjent E, Herve D, Darragh J, Corvol JC, Pages C, Simon AJ, Girault JA, Caboche J 2005. Parsing molecular and behavioral effects of cocaine in mitogen- and stress-activated protein kinase-1-deficient mice. J Neurosci 25: 11444–11454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brami-Cherrier K, Lavaur J, Pages C, Arthur JS, Caboche J 2007. Glutamate induces histone H3 phosphorylation but not acetylation in striatal neurons: Role of mitogen- and stress-activated kinase-1. J Neurochem 101: 697–708 [DOI] [PubMed] [Google Scholar]
- Brami-Cherrier K, Roze E, Girault JA, Betuing S, Caboche J 2009. Role of the ERK/MSK1 signalling pathway in chromatin remodelling and brain responses to drugs of abuse. J Neurochem 108: 1323–1335 [DOI] [PubMed] [Google Scholar]
- Carouge D, Host L, Aunis D, Zwiller J, Anglard P 2010. CDKL5 is a brain MeCP2 target gene regulated by DNA methylation. Neurobiol Dis 38: 414–424 [DOI] [PubMed] [Google Scholar]
- Chahrour M, Jung SY, Shaw C, Zhou X, Wong ST, Qin J, Zoghbi HY 2008. MeCP2, a key contributor to neurological disease, activates and represses transcription. Science 320: 1224–1229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandrasekar V, Dreyer JL 2009. microRNAs miR-124, let-7d and miR-181a regulate cocaine-induced plasticity. Mol Cell Neurosci 42: 350–362 [DOI] [PubMed] [Google Scholar]
- Chandrasekar V, Dreyer JL 2011. Regulation of MiR-124, Let-7d, and MiR-181a in the accumbens affects the expression, extinction, and reinstatement of cocaine-induced conditioned place preference. Neuropsychopharmacology 36: 1149–1164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang Q, Khare G, Dani V, Nelson S, Jaenisch R 2006. The disease progression of Mecp2 mutant mice is affected by the level of BDNF expression. Neuron 49: 341–348 [DOI] [PubMed] [Google Scholar]
- Chen WG, Chang Q, Lin Y, Meissner A, West AE, Griffith EC, Jaenisch R, Greenberg ME 2003. Derepression of BDNF transcription involves calcium-dependent phosphorylation of MeCP2. Science 302: 885–889 [DOI] [PubMed] [Google Scholar]
- Cheung P, Allis CD, Sassone-Corsi P 2000. Signaling to chromatin through histone modifications. Cell 103: 263–271 [DOI] [PubMed] [Google Scholar]
- Cleck JN, Ecke LE, Blendy JA 2008. Endocrine and gene expression changes following forced swim stress exposure during cocaine abstinence in mice. Psychopharmacology (Berl) 201: 15–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covington HE III, Maze I, Sun H, Bomze HM, DeMaio KD, Wu EY, Dietz DM, Lobo MK, Ghose S, Mouzon E, et al. 2011. A role for repressive histone methylation in cocaine-induced vulnerability to stress. Neuron 71: 656–670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng JV, Rodriguiz RM, Hutchinson AN, Kim IH, Wetsel WC, West AE 2010. MeCP2 in the nucleus accumbens contributes to neural and behavioral responses to psychostimulants. Nat Neurosci 13: 1128–1136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denli AM, Tops BB, Plasterk RH, Ketting RF, Hannon GJ 2004. Processing of primary microRNAs by the Microprocessor complex. Nature 432: 231–235 [DOI] [PubMed] [Google Scholar]
- Doi M, Hirayama J, Sassone-Corsi P 2006. Circadian regulator CLOCK is a histone acetyltransferase. Cell 125: 497–508 [DOI] [PubMed] [Google Scholar]
- Dreyer JL 2010. New insights into the roles of microRNAs in drug addiction and neuroplasticity. Genome Med 2: 92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filip M, Faron-Gorecka A, Kusmider M, Golda A, Frankowska M, Dziedzicka-Wasylewska M 2006. Alterations in BDNF and trkB mRNAs following acute or sensitizing cocaine treatments and withdrawal. Brain Res 1071: 218–225 [DOI] [PubMed] [Google Scholar]
- Freeman WM, Patel KM, Brucklacher RM, Lull ME, Erwin M, Morgan D, Roberts DC, Vrana KE 2008. Persistent alterations in mesolimbic gene expression with abstinence from cocaine self-administration. Neuropsychopharmacology 33: 1807–1817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman LM, Dror AA, Mor E, Tenne T, Toren G, Satoh T, Biesemeier DJ, Shomron N, Fekete DM, Hornstein E, et al. 2009a. MicroRNAs are essential for development and function of inner ear hair cells in vertebrates. Proc Natl Acad Sci 106: 7915–7920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman RC, Farh KK, Burge CB, Bartel DP 2009b. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res 19: 92–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fumagalli F, Di Pasquale L, Caffino L, Racagni G, Riva MA 2007. Repeated exposure to cocaine differently modulates BDNF mRNA and protein levels in rat striatum and prefrontal cortex. Eur J Neurosci 26: 2756–2763 [DOI] [PubMed] [Google Scholar]
- Fumagalli F, Caffino L, Racagni G, Riva MA 2009. Repeated stress prevents cocaine-induced activation of BDNF signaling in rat prefrontal cortex. Eur Neuropsychopharmacol 19: 402–408 [DOI] [PubMed] [Google Scholar]
- Fyffe SL, Neul JL, Samaco RC, Chao HT, Ben-Shachar S, Moretti P, McGill BE, Goulding EH, Sullivan E, Tecott LH, et al. 2008. Deletion of Mecp2 in Sim1-expressing neurons reveals a critical role for MeCP2 in feeding behavior, aggression, and the response to stress. Neuron 59: 947–958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gareau JR, Lima CD 2010. The SUMO pathway: Emerging mechanisms that shape specificity, conjugation and recognition. Nat Rev Mol Cell Biol 11: 861–871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghildiyal M, Xu J, Seitz H, Weng Z, Zamore PD 2010. Sorting of Drosophila small silencing RNAs partitions microRNA* strands into the RNA interference pathway. RNA 16: 43–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham DL, Edwards S, Bachtell RK, DiLeone RJ, Rios M, Self DW 2007. Dynamic BDNF activity in nucleus accumbens with cocaine use increases self-administration and relapse. Nat Neurosci 10: 1029–1037 [DOI] [PubMed] [Google Scholar]
- Gregory RI, Yan KP, Amuthan G, Chendrimada T, Doratotaj B, Cooch N, Shiekhattar R 2004. The Microprocessor complex mediates the genesis of microRNAs. Nature 432: 235–240 [DOI] [PubMed] [Google Scholar]
- Grimm JW, Lu L, Hayashi T, Hope BT, Su TP, Shaham Y 2003. Time-dependent increases in brain-derived neurotrophic factor protein levels within the mesolimbic dopamine system after withdrawal from cocaine: Implications for incubation of cocaine craving. J Neurosci 23: 742–747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP 2007. MicroRNA targeting specificity in mammals: Determinants beyond seed pairing. Mol Cell 27: 91–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan JS, Haggarty SJ, Giacometti E, Dannenberg JH, Joseph N, Gao J, Nieland TJ, Zhou Y, Wang X, Mazitschek R, et al. 2009. HDAC2 negatively regulates memory formation and synaptic plasticity. Nature 459: 55–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarnieri DJ, DiLeone RJ 2008. MicroRNAs: A new class of gene regulators. Ann Med 40: 197–208 [DOI] [PubMed] [Google Scholar]
- Hartmann M, Heumann R, Lessmann V 2001. Synaptic secretion of BDNF after high-frequency stimulation of glutamatergic synapses. EMBO J 20: 5887–5897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobert O 2008. Gene regulation by transcription factors and microRNAs. Science 319: 1785–1786 [DOI] [PubMed] [Google Scholar]
- Hollander JA, Im HI, Amelio AL, Kocerha J, Bali P, Lu Q, Willoughby D, Wahlestedt C, Conkright MD, Kenny PJ 2010. Striatal microRNA controls cocaine intake through CREB signalling. Nature 466: 197–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Host L, Dietrich JB, Carouge D, Aunis D, Zwiller J 2011. Cocaine self-administration alters the expression of chromatin-remodelling proteins; modulation by histone deacetylase inhibition. J Psychopharmacol 25: 222–229 [DOI] [PubMed] [Google Scholar]
- Hutvagner G, Zamore PD 2002. A microRNA in a multiple-turnover RNAi enzyme complex. Science 297: 2056–2060 [DOI] [PubMed] [Google Scholar]
- Hutvagner G, McLachlan J, Pasquinelli AE, Balint E, Tuschl T, Zamore PD 2001. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science 293: 834–838 [DOI] [PubMed] [Google Scholar]
- Hyman SE, Malenka RC, Nestler EJ 2006. Neural mechanisms of addiction: The role of reward-related learning and memory. Annu Rev Neurosci 29: 565–598 [DOI] [PubMed] [Google Scholar]
- Im HI, Hollander JA, Bali P, Kenny PJ 2010. MeCP2 controls BDNF expression and cocaine intake through homeostatic interactions with microRNA-212. Nat Neurosci 13: 1120–1127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson RJ, Standart N 2007. How do microRNAs regulate gene expression? Sci STKE 2007: re1. [DOI] [PubMed] [Google Scholar]
- Jaenisch R, Bird A 2003. Epigenetic regulation of gene expression: How the genome integrates intrinsic and environmental signals. Nat Genet 33 (Suppl): 245–254 [DOI] [PubMed] [Google Scholar]
- Jovanovic JN, Czernik AJ, Fienberg AA, Greengard P, Sihra TS 2000. Synapsins as mediators of BDNF-enhanced neurotransmitter release. Nat Neurosci 3: 323–329 [DOI] [PubMed] [Google Scholar]
- Kalda A, Heidmets LT, Shen HY, Zharkovsky A, Chen JF 2007. Histone deacetylase inhibitors modulates the induction and expression of amphetamine-induced behavioral sensitization partially through an associated learning of the environment in mice. Behav Brain Res 181: 76–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalivas PW, Volkow N, Seamans J 2005. Unmanageable motivation in addiction: A pathology in prefrontal-accumbens glutamate transmission. Neuron 45: 647–650 [DOI] [PubMed] [Google Scholar]
- Kauer JA, Malenka RC 2007. Synaptic plasticity and addiction. Nat Rev Neurosci 8: 844–858 [DOI] [PubMed] [Google Scholar]
- Klein ME, Lioy DT, Ma L, Impey S, Mandel G, Goodman RH 2007. Homeostatic regulation of MeCP2 expression by a CREB-induced microRNA. Nat Neurosci 10: 1513–1514 [DOI] [PubMed] [Google Scholar]
- Koob GF, Volkow ND 2010. Neurocircuitry of addiction. Neuropsychopharmacology 35: 217–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosik KS 2006. The neuronal microRNA system. Nat Rev Neurosci 7: 911–920 [DOI] [PubMed] [Google Scholar]
- Kouzarides T 2007. Chromatin modifications and their function. Cell 128: 693–705 [DOI] [PubMed] [Google Scholar]
- Kumar A, Choi KH, Renthal W, Tsankova NM, Theobald DE, Truong HT, Russo SJ, Laplant Q, Sasaki TS, Whistler KN, et al. 2005. Chromatin remodeling is a key mechanism underlying cocaine-induced plasticity in striatum. Neuron 48: 303–314 [DOI] [PubMed] [Google Scholar]
- Kurdistani SK, Tavazoie S, Grunstein M 2004. Mapping global histone acetylation patterns to gene expression. Cell 117: 721–733 [DOI] [PubMed] [Google Scholar]
- Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T 2001. Identification of novel genes coding for small expressed RNAs. Science 294: 853–858 [DOI] [PubMed] [Google Scholar]
- LaPlant Q, Vialou V, Covington HE III, Dumitriu D, Feng J, Warren BL, Maze I, Dietz DM, Watts EL, Iniguez SD, et al. 2010. Dnmt3a regulates emotional behavior and spine plasticity in the nucleus accumbens. Nat Neurosci 13: 1137–1143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau NC, Lim LP, Weinstein EG, Bartel DP 2001. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science 294: 858–862 [DOI] [PubMed] [Google Scholar]
- Le Foll B, Diaz J, Sokoloff P 2005. A single cocaine exposure increases BDNF and D3 receptor expression: Implications for drug-conditioning. Neuroreport 16: 175–178 [DOI] [PubMed] [Google Scholar]
- Lee Y, Jeon K, Lee JT, Kim S, Kim VN 2002. MicroRNA maturation: Stepwise processing and subcellular localization. EMBO J 21: 4663–4670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, Lee J, Provost P, Radmark O, Kim S, et al. 2003. The nuclear RNase III Drosha initiates microRNA processing. Nature 425: 415–419 [DOI] [PubMed] [Google Scholar]
- Lee Y, Kim M, Han J, Yeom KH, Lee S, Baek SH, Kim VN 2004. MicroRNA genes are transcribed by RNA polymerase II. EMBO J 23: 4051–4060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine AA, Guan Z, Barco A, Xu S, Kandel ER, Schwartz JH 2005. CREB-binding protein controls response to cocaine by acetylating histones at the fosB promoter in the mouse striatum. Proc Natl Acad Sci 102: 19186–19191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB 2003. Prediction of mammalian microRNA targets. Cell 115: 787–798 [DOI] [PubMed] [Google Scholar]
- Lewis BP, Burge CB, Bartel DP 2005. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 120: 15–20 [DOI] [PubMed] [Google Scholar]
- Li MD, van der Vaart AD 2011. MicroRNAs in addiction: Adaptation’s middlemen? Mol Psychiatry 16: 1159–1168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Carey M, Workman JL 2007a. The role of chromatin during transcription. Cell 128: 707–719 [DOI] [PubMed] [Google Scholar]
- Li SC, Tang P, Lin WC 2007b. Intronic microRNA: Discovery and biological implications. DNA Cell Biol 26: 195–207 [DOI] [PubMed] [Google Scholar]
- Lipska BK, Khaing ZZ, Weickert CS, Weinberger DR 2001. BDNF mRNA expression in rat hippocampus and prefrontal cortex: Effects of neonatal ventral hippocampal damage and antipsychotic drugs. Eur J Neurosci 14: 135–144 [DOI] [PubMed] [Google Scholar]
- Liu QR, Lu L, Zhu XG, Gong JP, Shaham Y, Uhl GR 2006. Rodent BDNF genes, novel promoters, novel splice variants, and regulation by cocaine. Brain Res 1067: 1–12 [DOI] [PubMed] [Google Scholar]
- Lu B 2003. BDNF and activity-dependent synaptic modulation. Learn Mem 10: 86–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu L, Dempsey J, Liu SY, Bossert JM, Shaham Y 2004. A single infusion of brain-derived neurotrophic factor into the ventral tegmental area induces long-lasting potentiation of cocaine seeking after withdrawal. J Neurosci 24: 1604–1611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubin FD, Roth TL, Sweatt JD 2008. Epigenetic regulation of BDNF gene transcription in the consolidation of fear memory. J Neurosci 28: 10576–10586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lugli G, Torvik VI, Larson J, Smalheiser NR 2008. Expression of microRNAs and their precursors in synaptic fractions of adult mouse forebrain. J Neurochem 106: 650–661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund E, Guttinger S, Calado A, Dahlberg JE, Kutay U 2004. Nuclear export of microRNA precursors. Science 303: 95–98 [DOI] [PubMed] [Google Scholar]
- Luscher C, Malenka RC 2011. Drug-evoked synaptic plasticity in addiction: From molecular changes to circuit remodeling. Neuron 69: 650–663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma DK, Jang MH, Guo JU, Kitabatake Y, Chang ML, Pow-Anpongkul N, Flavell RA, Lu B, Ming GL, Song H 2009. Neuronal activity-induced Gadd45b promotes epigenetic DNA demethylation and adult neurogenesis. Science 323: 1074–1077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacRae IJ, Ma E, Zhou M, Robinson CV, Doudna JA 2008. In vitro reconstitution of the human RISC-loading complex. Proc Natl Acad Sci 105: 512–517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malvaez M, Sanchis-Segura C, Vo D, Lattal KM, Wood MA 2010. Modulation of chromatin modification facilitates extinction of cocaine-induced conditioned place preference. Biol Psychiatry 67: 36–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mameli M, Luscher C 2011. Synaptic plasticity and addiction: Learning mechanisms gone awry. Neuropharmacology 61: 1052–1059 [DOI] [PubMed] [Google Scholar]
- Marks PA, Miller T, Richon VM 2003. Histone deacetylases. Curr Opin Pharmacol 3: 344–351 [DOI] [PubMed] [Google Scholar]
- Matranga C, Tomari Y, Shin C, Bartel DP, Zamore PD 2005. Passenger-strand cleavage facilitates assembly of siRNA into Ago2-containing RNAi enzyme complexes. Cell 123: 607–620 [DOI] [PubMed] [Google Scholar]
- Maze I, Nestler EJ 2011. The epigenetic landscape of addiction. Ann NY Acad Sci 1216: 99–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maze I, Covington HE III, Dietz DM, LaPlant Q, Renthal W, Russo SJ, Mechanic M, Mouzon E, Neve RL, Haggarty SJ, et al. 2010. Essential role of the histone methyltransferase G9a in cocaine-induced plasticity. Science 327: 213–216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maze I, Feng J, Wilkinson MB, Sun H, Shen L, Nestler EJ 2011. Cocaine dynamically regulates heterochromatin and repetitive element unsilencing in nucleus accumbens. Proc Natl Acad Sci 108: 3035–3040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGinty JF, Whitfield TW Jr, Berglind WJ 2010. Brain-derived neurotrophic factor and cocaine addiction. Brain Res 1314: 183–193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellios N, Huang HS, Grigorenko A, Rogaev E, Akbarian S 2008. A set of differentially expressed miRNAs, including miR-30a-5p, act as post-transcriptional inhibitors of BDNF in prefrontal cortex. Hum Mol Genet 17: 3030–3042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendelson JH, Mello NK 1996. Management of cocaine abuse and dependence. N Engl J Med 334: 965–972 [DOI] [PubMed] [Google Scholar]
- Meredith GE, Callen S, Scheuer DA 2002. Brain-derived neurotrophic factor expression is increased in the rat amygdala, piriform cortex and hypothalamus following repeated amphetamine administration. Brain Res 949: 218–227 [DOI] [PubMed] [Google Scholar]
- Muinos-Gimeno M, Espinosa-Parrilla Y, Guidi M, Kagerbauer B, Sipila T, Maron E, Pettai K, Kananen L, Navines R, Martin-Santos R, et al. 2011. Human microRNAs miR-22, miR-138–2, miR-148a, and miR-488 are associated with panic disorder and regulate several anxiety candidate genes and related pathways. Biol Psychiatry 69: 526–533 [DOI] [PubMed] [Google Scholar]
- Nagappan G, Lu B 2005. Activity-dependent modulation of the BDNF receptor TrkB: Mechanisms and implications. Trends Neurosci 28: 464–471 [DOI] [PubMed] [Google Scholar]
- Nestler EJ 2001. Molecular basis of long-term plasticity underlying addiction. Nat Rev Neurosci 2: 119–128 [DOI] [PubMed] [Google Scholar]
- Novikova SI, He F, Bai J, Cutrufello NJ, Lidow MS, Undieh AS 2008. Maternal cocaine administration in mice alters DNA methylation and gene expression in hippocampal neurons of neonatal and prepubertal offspring. PLoS ONE 3: e1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Numachi Y, Shen H, Yoshida S, Fujiyama K, Toda S, Matsuoka H, Sora I, Sato M 2007. Methamphetamine alters expression of DNA methyltransferase 1 mRNA in rat brain. Neurosci Lett 414: 213–217 [DOI] [PubMed] [Google Scholar]
- Okamura K, Hagen JW, Duan H, Tyler DM, Lai EC 2007. The mirtron pathway generates microRNA-class regulatory RNAs in Drosophila. Cell 130: 89–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura K, Liu N, Lai EC 2009. Distinct mechanisms for microRNA strand selection by Drosophila Argonautes. Mol Cell 36: 431–444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patapoutian A, Reichardt LF 2001. Trk receptors: Mediators of neurotrophin action. Curr Opin Neurobiol 11: 272–280 [DOI] [PubMed] [Google Scholar]
- Peters L, Meister G 2007. Argonaute proteins: Mediators of RNA silencing. Mol Cell 26: 611–623 [DOI] [PubMed] [Google Scholar]
- Pietrzykowski AZ 2010. The role of microRNAs in drug addiction: A big lesson from tiny molecules. Int Rev Neurobiol 91: 1–24 [DOI] [PubMed] [Google Scholar]
- Pillai RS, Bhattacharyya SN, Filipowicz W 2007. Repression of protein synthesis by miRNAs: How many mechanisms? Trends Cell Biol 17: 118–126 [DOI] [PubMed] [Google Scholar]
- Place RF, Li LC, Pookot D, Noonan EJ, Dahiya R 2008. MicroRNA-373 induces expression of genes with complementary promoter sequences. Proc Natl Acad Sci 105: 1608–1613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renthal W, Nestler EJ 2008. Epigenetic mechanisms in drug addiction. Trends Mol Med 14: 341–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renthal W, Maze I, Krishnan V, Covington HE III, Xiao G, Kumar A, Russo SJ, Graham A, Tsankova N, Kippin TE, et al. 2007. Histone deacetylase 5 epigenetically controls behavioral adaptations to chronic emotional stimuli. Neuron 56: 517–529 [DOI] [PubMed] [Google Scholar]
- Renthal W, Carle TL, Maze I, Covington HE III, Truong HT, Alibhai I, Kumar A, Montgomery RL, Olson EN, Nestler EJ 2008. ΔFosB mediates epigenetic desensitization of the c-fos gene after chronic amphetamine exposure. J Neurosci 28: 7344–7349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renthal W, Kumar A, Xiao G, Wilkinson M, Covington HE III, Maze I, Sikder D, Robison AJ, LaPlant Q, Dietz DM, et al. 2009. Genome-wide analysis of chromatin regulation by cocaine reveals a role for sirtuins. Neuron 62: 335–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice JC, Allis CD 2001. Histone methylation versus histone acetylation: New insights into epigenetic regulation. Curr Opin Cell Biol 13: 263–273 [DOI] [PubMed] [Google Scholar]
- Robinson TE, Kolb B 1997. Persistent structural modifications in nucleus accumbens and prefrontal cortex neurons produced by previous experience with amphetamine. J Neurosci 17: 8491–8497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez A, Griffiths-Jones S, Ashurst JL, Bradley A 2004. Identification of mammalian microRNA host genes and transcription units. Genome Res 14: 1902–1910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romieu P, Host L, Gobaille S, Sandner G, Aunis D, Zwiller J 2008. Histone deacetylase inhibitors decrease cocaine but not sucrose self-administration in rats. J Neurosci 28: 9342–9348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romieu P, Deschatrettes E, Host L, Gobaille S, Sandner G, Zwiller J 2011. The inhibition of histone deacetylases reduces the reinstatement of cocaine-seeking behavior in rats. Curr Neuropharmacol 9: 21–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotllant D, Armario A 2012. Brain pattern of histone H3 phosphorylation after acute amphetamine administration: Its relationship to brain c-fos induction is strongly dependent on the particular brain area. Neuropharmacology 62: 1073–1081 [DOI] [PubMed] [Google Scholar]
- Russo SJ, Dietz DM, Dumitriu D, Morrison JH, Malenka RC, Nestler EJ 2010. The addicted synapse: Mechanisms of synaptic and structural plasticity in nucleus accumbens. Trends Neurosci 33: 267–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadri-Vakili G, Kumaresan V, Schmidt HD, Famous KR, Chawla P, Vassoler FM, Overland RP, Xia E, Bass CE, Terwilliger EF, et al. 2010. Cocaine-induced chromatin remodeling increases brain-derived neurotrophic factor transcription in the rat medial prefrontal cortex, which alters the reinforcing efficacy of cocaine. J Neurosci 30: 11735–11744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saini HK, Griffiths-Jones S, Enright AJ 2007. Genomic analysis of human microRNA transcripts. Proc Natl Acad Sci 104: 17719–17724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saylor AJ, McGinty JF 2008. Amphetamine-induced locomotion and gene expression are altered in BDNF heterozygous mice. Genes Brain Behav 7: 906–914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt HD, Pierce RC 2010. Cocaine-induced neuroadaptations in glutamate transmission: Potential therapeutic targets for craving and addiction. Ann NY Acad Sci 1187: 35–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt HD, Sangrey GR, Darnell SB, Schassburger RL, Cha JH, Pierce RC, Sadri-Vakili G 2011. Increased BDNF expression in the ventral tegmental area during cocaine abstinence is associated with increased histone acetylation at BDNF exon i-containing promoters. J Neurochem 120: 202–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schratt GM, Tuebing F, Nigh EA, Kane CG, Sabatini ME, Kiebler M, Greenberg ME 2006. A brain-specific microRNA regulates dendritic spine development. Nature 439: 283–289 [DOI] [PubMed] [Google Scholar]
- Schroeder FA, Penta KL, Matevossian A, Jones SR, Konradi C, Tapper AR, Akbarian S 2008. Drug-induced activation of dopamine D(1) receptor signaling and inhibition of class I/II histone deacetylase induce chromatin remodeling in reward circuitry and modulate cocaine-related behaviors. Neuropsychopharmacology 33: 2981–2992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sempere LF, Freemantle S, Pitha-Rowe I, Moss E, Dmitrovsky E, Ambros V 2004. Expression profiling of mammalian microRNAs uncovers a subset of brain-expressed microRNAs with possible roles in murine and human neuronal differentiation. Genome Biol 5: R13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen HY, Kalda A, Yu L, Ferrara J, Zhu J, Chen JF 2008. Additive effects of histone deacetylase inhibitors and amphetamine on histone H4 acetylation, cAMP responsive element binding protein phosphorylation and ΔFosB expression in the striatum and locomotor sensitization in mice. Neuroscience 157: 644–655 [DOI] [PubMed] [Google Scholar]
- Shibasaki M, Mizuno K, Kurokawa K, Ohkuma S 2011. L-type voltage-dependent calcium channels facilitate acetylation of histone H3 through PKCγ phosphorylation in mice with methamphetamine-induced place preference. J Neurochem 118: 1056–1066 [DOI] [PubMed] [Google Scholar]
- Shieh PB, Hu SC, Bobb K, Timmusk T, Ghosh A 1998. Identification of a signaling pathway involved in calcium regulation of BDNF expression. Neuron 20: 727–740 [DOI] [PubMed] [Google Scholar]
- Siegmund KD, Connor CM, Campan M, Long TI, Weisenberger DJ, Biniszkiewicz D, Jaenisch R, Laird PW, Akbarian S 2007. DNA methylation in the human cerebral cortex is dynamically regulated throughout the life span and involves differentiated neurons. PLoS ONE 2: e895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skene PJ, Illingworth RS, Webb S, Kerr AR, James KD, Turner DJ, Andrews R, Bird AP 2010. Neuronal MeCP2 is expressed at near histone-octamer levels and globally alters the chromatin state. Mol Cell 37: 457–468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommerfeld MT, Schweigreiter R, Barde YA, Hoppe E 2000. Down-regulation of the neurotrophin receptor TrkB following ligand binding. Evidence for an involvement of the proteasome and differential regulation of TrkA and TrkB. J Biol Chemistry 275: 8982–8990 [DOI] [PubMed] [Google Scholar]
- Steitz JA, Vasudevan S 2009. miRNPs: Versatile regulators of gene expression in vertebrate cells. Biochem Soc Trans 37: 931–935 [DOI] [PubMed] [Google Scholar]
- Strahl BD, Allis CD 2000. The language of covalent histone modifications. Nature 403: 41–45 [DOI] [PubMed] [Google Scholar]
- Sun J, Wang L, Jiang B, Hui B, Lv Z, Ma L 2008. The effects of sodium butyrate, an inhibitor of histone deacetylase, on the cocaine- and sucrose-maintained self-administration in rats. Neurosci Lett 441: 72–76 [DOI] [PubMed] [Google Scholar]
- Suzuki MM, Bird A 2008. DNA methylation landscapes: Provocative insights from epigenomics. Nat Rev Genet 9: 465–476 [DOI] [PubMed] [Google Scholar]
- Tan Y, Zhang B, Wu T, Skogerbo G, Zhu X, Guo X, He S, Chen R 2009. Transcriptional inhibiton of Hoxd4 expression by miRNA-10a in human breast cancer cells. BMC Mol Biol 10: 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao X, Finkbeiner S, Arnold DB, Shaywitz AJ, Greenberg ME 1998. Ca2+ influx regulates BDNF transcription by a CREB family transcription factor-dependent mechanism. Neuron 20: 709–726 [DOI] [PubMed] [Google Scholar]
- Thoenen H 1995. Neurotrophins and neuronal plasticity. Science 270: 593–598 [DOI] [PubMed] [Google Scholar]
- Thomas MJ, Kalivas PW, Shaham Y 2008. Neuroplasticity in the mesolimbic dopamine system and cocaine addiction. Br J Pharmacol 154: 327–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tongiorgi E, Righi M, Cattaneo A 1997. Activity-dependent dendritic targeting of BDNF and TrkB mRNAs in hippocampal neurons. J Neurosci 17: 9492–9505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsankova N, Renthal W, Kumar A, Nestler EJ 2007. Epigenetic regulation in psychiatric disorders. Nat Rev Neurosci 8: 355–367 [DOI] [PubMed] [Google Scholar]
- Tudor M, Akbarian S, Chen RZ, Jaenisch R 2002. Transcriptional profiling of a mouse model for Rett syndrome reveals subtle transcriptional changes in the brain. Proc Natl Acad Sci 99: 15536–15541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyler DM, Okamura K, Chung WJ, Hagen JW, Berezikov E, Hannon GJ, Lai EC 2008. Functionally distinct regulatory RNAs generated by bidirectional transcription and processing of microRNA loci. Genes Dev 22: 26–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasudevan S, Tong Y, Steitz JA 2007. Switching from repression to activation: MicroRNAs can up-regulate translation. Science 318: 1931–1934 [DOI] [PubMed] [Google Scholar]
- Wang L, Lv Z, Hu Z, Sheng J, Hui B, Sun J, Ma L 2010. Chronic cocaine-induced H3 acetylation and transcriptional activation of CaMKIIα in the nucleus accumbens is critical for motivation for drug reinforcement. Neuropsychopharmacology 35: 913–928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitfield TW Jr, Shi X, Sun WL, McGinty JF 2011. The suppressive effect of an intra-prefrontal cortical infusion of BDNF on cocaine-seeking is Trk receptor and extracellular signal-regulated protein kinase mitogen-activated protein kinase dependent. J Neurosci 31: 834–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeom KH, Lee Y, Han J, Suh MR, Kim VN 2006. Characterization of DGCR8/Pasha, the essential cofactor for Drosha in primary miRNA processing. Nucleic Acids Res 34: 4622–4629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi R, Qin Y, Macara IG, Cullen BR 2003. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev 17: 3011–3016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z, Yuan Q, Mash DC, Goldman D 2011. Substance-specific and shared transcription and epigenetic changes in the human hippocampus chronically exposed to cocaine and alcohol. Proc Natl Acad Sci 108: 6626–6631 [DOI] [PMC free article] [PubMed] [Google Scholar]