Abstract
Probiotics are beneficial components of the microbiota that have been used for centuries because of the health benefits they confer to the host. Only recently, however, has the contribution of probiotics to modulation of immunological, respiratory, and gastrointestinal functions started to be fully appreciated and scientifically evaluated. Probiotics such as Escherichia coli Nissle 1917 and lactic acid bacteria are currently used to, or have been evaluated for use to, prevent or treat a range of intestinal maladies including inflammatory bowel disease, constipation, and colon cancer. Engineering these natural probiotics to produce immunomodulatory molecules may help to further increase the benefit to the host. In this article, we will discuss some of the mechanisms of action of probiotics as well as advances in the rational design of probiotics.
Probiotic bacteria benefit immunological, respiratory, and gastrointestinal functions. Engineered probiotics are an excellent tool to deliver molecules (e.g., cytokines) within a host.
Probiotics are defined as living bacteria that, when administered in adequate amounts, confer a health benefit on the host (FAO/WHO 2001). The original observation of the beneficial properties conferred by some bacteria is attributed to the Nobel Prize winner Eli Metchnikoff, who is regarded as the grandfather of modern probiotics. In the early 20th century, Metchnikoff discovered that “healthy bacteria,” especially lactic acid bacteria (LAB), can have a positive influence on digestion and the immune system (Anukam and Reid 2008). Most microorganisms recognized to date as probiotics are Gram-positive, with Lactobacillus and Bifidobacterium being the main species used as treatments of intestinal dysfunctions (Marco et al. 2006). However, some Gram-negatives are also used as probiotics. The best example of this group is Escherichia coli Nissle 1917 (EcN) (Nissle 1959), also known as “Mutaflor,” which has been used in Germany for many years in the treatment of chronic constipation (Mollenbrink and Bruckschen 1994) and colitis (Schutz 1989).
In the last century, many studies have reported probiotic bacteria to play important roles in the modulation of immunological, respiratory, and gastrointestinal functions (Floch et al. 2011). Furthermore, probiotics have been shown to play a protective role by directly competing with intestinal pathogens through the release of antibacterial substances such as bacteriocins (Cotter et al. 2005) or metabolites such as acetic acid and lactic acid (Servin 2004). Although most studies on probiotics have been empirical, new advancements may originate from research on the interactions between commensal microorganisms (termed the microbiota), pathogens, and the host. Understanding the mechanisms of gut colonization in both normal and inflammatory conditions is essential to designing probiotics for a specific use. In this article, we will discuss some of the recent developments on the mechanisms of action of probiotics and their utilization for delivering molecules to specific sites within the host.
THE HUMAN DIGESTIVE TRACT AND THE MICROBIOTA
The human digestive tract comprises a variety of ecological niches populated by several bacterial species that have established a symbiotic relationship with the host. This bacterial population, also called the intestinal microbiota, plays an important role in the development of the gut immune system, digestion of food, production of short-chain fatty acids and essential vitamins, and resistance to colonization from pathogenic microorganisms (Hooper and Gordon 2001). The human gut microbiome consists of many different species of bacteria, some of which are nonculturable and therefore not well known or characterized. Indeed, it has only been through the advent of deep sequencing, genomics, and metagenomics in the last decade that the complexity of the microbiota has been fully appreciated.
The distribution of the intestinal microbiota varies along three main locations in the digestive tract: (1) the stomach, populated by <102 colony-forming units (cfu)/ml, including lactobacilli and streptococci; (2) the ileum and distal ileum, populated by 102–103 cfu/mL of bacteria, including E. coli, Klebsiella, Enterococcus, and Bacteroides; and (3) the large intestine, which constitutes the largest microbial population of the body, with 1010–1012 cfu/mL (DiBaise et al. 2008). Remarkably, each individual organism presents a specific “bacterial fingerprint,” which is influenced by a variety of factors including the maternal environment, host genotype, diet, and antibiotic treatment (Spor et al. 2011). But even though the composition of the microbiota differs from person to person, it clusters in three distinct groups, so-called enterotypes. These human enterotypes are enriched in Bacteroides, Prevotella, or Ruminococcus and use different routes to generate energy from fermentable substrates available in the colon: Bacteroides uses carbohydrates; Prevotella, mucins; and Ruminococcus, mucins and sugars (Arumugam et al. 2011). Enterotypes have also been associated with long-term diets (Wu et al. 2011). Despite this heterogeneity, Firmicutes and Bacteroidetes are the most common intestinal phyla across all vertebrates, representing >90% of the microbiota, followed by Actinobacteria and Proteobacteria (Mahowald et al. 2009). Members of the microbiota from phylum Bacteroidetes are represented by a variety of species, including Bacteroides fragilis, which was recently shown to possess immunomodulatory capabilities via its polysaccharide capsule (Cobb et al. 2004; Coyne et al. 2005; Mazmanian et al. 2005; Liu et al. 2008). In contrast, phylum Firmicutes is mainly represented by species belonging to class Clostridia, which are known for their abilities to metabolize fiber and produce butyrate, a short-chain fatty acid with immunomodulatory activity (Gophna et al. 2006; Vinolo et al. 2011). Bacteria belonging to class Bacilli, including Lactobacillus acidophilus and Enterococcus faecalis, constitute the rest of the Firmicutes phylum.
The advent of germ-free mice gave rise to a better understanding of the impact the intestinal microbiota has on the host. Studies have shown that these mice exhibit a thin intestinal epithelium (Jervis and Biggers 1964), loss of short-chain fatty acid production (Høverstad and Midtvedt 1986), and alterations to the immune system (Maslowski et al. 2009; Tlaskalova-Hogenova et al. 2011). Remarkably, administration of the microbiota restores full functionality of the gut (Aureli et al. 2011). Moreover, some components of the microbiota have been shown to induce the development of T-cell subsets (Ivanov et al. 2009; Feng et al. 2010; Atarashi et al. 2011; Shaw et al. 2012) and the release of defensins (Bevins and Salzman 2011). Another notable aspect is that in normal individuals, the host establishes tolerance to commensal bacteria while maintaining its capacity to mount an immune response against pathogens. This balance between tolerance and immunity, when disrupted, can lead to the development of a number of intestinal pathologies including inflammatory bowel disease (IBD) and intestinal cancer (Karin et al. 2006; Artis 2008). Components of the immune response that normally orchestrate the mucosal barrier to the microbiota can, if altered, also contribute to the development of IBD. For example, an inflammatory response mounted against the microbiota can lead to a change in the host environment that in turn affects the composition and quantity of the microbiota, which further exacerbates intestinal inflammation (Fava and Danese 2011). Intestinal inflammation results in dramatic alterations to the microbiota, with loss of the most abundant species (Bacteroidetes and Clostridiales) and enhanced growth of Enterobacteriaceae (Fava and Danese 2011; Mukhopadhya et al. 2012). Similarly, in many cases of self-limiting gastrointestinal infections, antibiotic treatment is not recommended because of its limited efficacy and notable side effects, including alterations to the microbiota that can result in antibiotic-associated colitis (Hogenauer et al. 1998).
In light of these observations, it becomes apparent that restoring balance to or augmenting the microbiota can potentially provide beneficial resolution of a variety of diseases. Moreover, as we discuss later, probiotics can be engineered to better mitigate the conditions arising from a particular intestinal pathology by cytokine and enzyme delivery or through direct competition with pathogenic microorganisms.
THE BEST-CHARACTERIZED PROBIOTICS
Many probiotics are culturable components of the microbiota that have been used for their beneficial functions since long before the term “probiotic” was coined. The most commonly used probiotic strains include the lactic acid bacteria (LAB), Gram-positive microbes that have been used for centuries in food production processes (yogurt, cheese, pickles). Members of the LAB such as Lactococcus and Streptococcus are also important components of the endogenous microbiota in the human ileum and jejunum and, at more moderate densities, in the colon (Hayashi et al. 2005). The approval for use in food combined with the absence of lipopolysaccharides (LPS) and lack of secreted proteases make many LAB ideal for use as probiotics. Similarly, as these organisms are Gram-positive, recombinant proteins can be secreted without getting trapped in the periplasm, making LAB species attractive vehicles for food-grade production of proteins and enzymes (Le Loir et al. 2005; Mierau and Kleerebezem 2005; Peterbauer et al. 2011).
The most widely used LAB species for cytokine delivery is Lactococcus lactis. This bacterium, unlike other Lactococcus species, is only transiently present in the human gut and is hence classified as a noncommensal (Nouaille et al. 2003). This transient colonization provides an advantage for the utilization of this probiotic strain as a delivery vehicle for protein vaccines and even, more recently, DNA vaccines. In these cases, a sustained colonization with a strain would be a disadvantage, as it may lead to overstimulation of the targeted pathway. The possibility to deliver proteins directly to the mucosa opens up new methods of therapeutic treatment in which traditional routes of medication fail as a result of, for example, low local availability of the therapeutic substance.
Regarding Gram-negative probiotics, the most commonly used strain is Escherichia coli Nissle 1917 (EcN) The army surgeon Alfred Nissle originally isolated this strain in 1917 from the feces of a soldier during the First World War who, in contrast to his comrades, did not develop infectious diarrhea during an outbreak of the highly contagious organism Shigella. Nissle’s observation suggested that EcN might provide colonization resistance to mucosal pathogens. Consistent with this hypothesis, the probiotic effect and biosafety of EcN has since been extensively shown in numerous trials and underlined by its long medical history in Central Europe as a microbial remedy (Kruis et al. 1997; Westendorf et al. 2005; Henker et al. 2007). At present, EcN is contained in a drug called Mutaflor, which is used for the treatment of both infectious diarrheal diseases and IBD (Schutz 1989). Furthermore, EcN has been administered to neonates to prevent the colonization of their digestive tract by multidrug-resistant pathogens (Lodinova-Zadnikova and Sonnenborn 1997; Boudeau et al. 2003; Kruis et al. 2004; Grabig et al. 2006; Henker et al. 2007).
LAB AND THEIR ROLE IN DELIVERING CYTOKINES AND OTHER MOLECULES TO RECONSTITUTE BARRIER DEFECTS
One of the recent advances in probiotics research involves the use of LAB to deliver cytokines directly to target sites within a host. LAB-delivered cytokines can be applied to treat diseases that weaken the mucosal barrier, such as IBD subtypes Crohn’s disease and ulcerative colitis. IBD is a major health concern in the Western world and manifests as chronic inflammation of the intestine that results in diarrhea, abdominal pain, and weight loss (Steidler et al. 2000). Although the etiology of IBD is unknown, it is clear that both genetic alterations in host pattern recognition receptors and proinflammatory genes as well as the microbiota play a role in causing the sustained intestinal inflammation seen in IBD patients (Nagalingam and Lynch 2012). Therefore, treatments aimed at reducing intestinal inflammation in these patients are highly desirable.
One way to reduce the chronic inflammation of IBD patients is through the administration of anti-inflammatory cytokines such as interleukin 10 (IL-10). This cytokine plays a central role in down-regulating inflammatory cascades and in the establishment of tolerance in the mucosa (Huibregtse et al. 2011). However, clinical trials of subcutaneous administration of IL-10 were disappointing because of the low efficacy and side effects (van Deventer et al. 1997; Colombel et al. 2001; Tilg et al. 2002). Owing to the acid sensitivity of IL-10, direct oral administration was not deemed a viable option. Instead, in an attempt to use the oral route of administration but protect IL-10 from degradation, Schotte et al. (2000) engineered an IL-10-producing L. lactis strain (LL-mIL10). They subsequently showed in a landmark study in 2000 that murine colitis significantly improved following treatment with this IL-10-producing strain (Fig. 1) (Steidler et al. 2000). Furthermore, the onset of colitis was inhibited in IL-10-deficient mice and the amount of IL-10 needed for the observed reduction in colitis was several orders of magnitudes lower than what was needed to reduce it by systemic administration (Steidler et al. 2000). Thus, delivery to the mucosa via LAB was shown to be a key element in the effectiveness of an IL-10-based treatment.
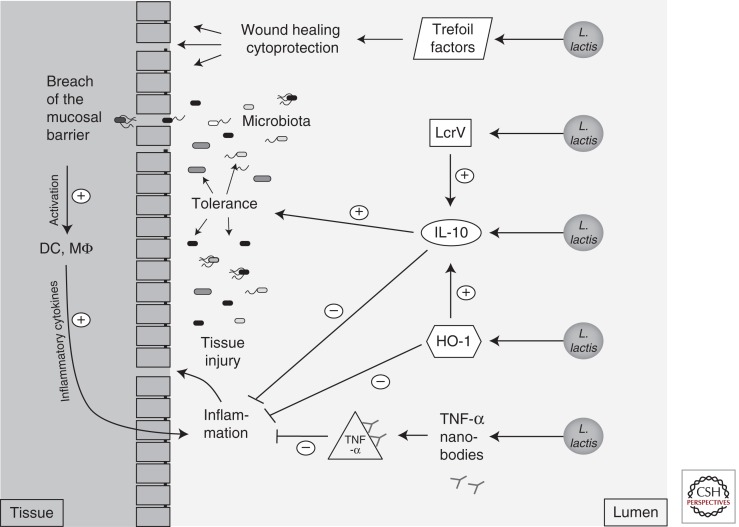
Figure 1.
Lactococcus lactis. The probiotic L. lactis was engineered to produce different cytokines: interleukin 10 (IL-10), heme oxygenase 1 (HO-1), the Yersinia protein LcrV, TNF-α nanobodies, or trefoil factors. After oral administration, L. lactis secretes these proteins in direct proximity to mucosal surfaces. These proteins can modulate the immune system in different ways to dampen the immune response and establish tolerance to the microbiota in the case of mucosal injury during IBD or cancer treatment.
A major issue to be resolved for clinical probiotic applications is the biological containment of genetically modified organisms. To achieve this in L. lactis, the thymidylate synthase gene (thyA) was replaced by the human IL-10 gene. Mutants in thyA require the presence of thymidine or thymine in the media to replicate, and therefore their growth is restricted to the human body and the accumulation of the strain in the environment is prevented (Steidler et al. 2003). The IL-10-producing L. lactis strain has since been administered to 10 Crohn’s disease patients during a phase 1 clinical trial. Out of 10 patients, eight had a clinical benefit and five went into complete clinical remission (Braat et al. 2006). These exciting findings have paved the way for subsequent clinical trials (Steidler et al. 2009).
Given the anti-inflammatory properties of IL-10, other strategies have been used to induce secretion of this cytokine in the gut mucosa. One such strategy involved a protein from a pathogen. Pathogenic yersiniae produce an anti-inflammatory protein called LcrV, which mediates evasion of the host’s immune response by enhancing IL-10 production (Fig. 1) (Foligne et al. 2007; Depaolo et al. 2008). Remarkably, an L. lactis strain producing LcrV was able to reduce inflammation in a trinitrobenzene sulfonic acid mouse model of colitis as efficiently as an L. lactis strain secreting IL-10 (Foligne et al. 2007).
Opposite to IL-10 on the cytokine spectrum is tumor necrosis factor α (TNF-α), a cytokine that mediates many of the clinical symptoms of IBD. Although systemic administration of anti-TNF-α antibodies has become an established therapy for Crohn’s disease and ulcerative colitis, it has drawbacks, similar to those seen with the systemic administration of IL-10. The therapy is costly and has poor patient compliance and several adverse side effects (Vandenbroucke et al. 2010). It was believed that oral administration and local production of the antibody at the site of inflammation in the gut could alleviate these problems and partially restore normal mucosal function. Thus, L. lactis was engineered to produce anti-TNF-α nanobodies (Fig. 1), small and stable single-domain antibody fragments that were derived from heavy-chain camelid antibodies. As expected, daily administration of L. lactis producing these nanobodies reduced colonic inflammation in both dextran sodium sulfate (DSS)-treated and IL-10-deficient mice. Remarkably, the effect of the anti-TNF-α nanobodies produced by L. lactis was restricted to the gut and did not interfere with systemic function of TNF-α (Vandenbroucke et al. 2010). Therefore, LAB-mediated delivery of IL-10 and anti-TNF-α antibodies appears to be more beneficial than systemic delivery to control mucosal inflammation.
Another host factor that improves the integrity of the gut mucosal barrier is the enzyme heme oxygenase 1 (HO-1), which provides protection against oxidative stress and has anti-inflammatory and other immunomodulatory functions (Fig. 1) (Vijayan et al. 2010). Notably, an L. lactis strain secreting this enzyme proved to be effective in reducing morbidity and mortality in a model of LPS-induced mucosal injury (Pang et al. 2008) and in a model of hemorrhagic shock (Pang et al. 2009) in rats. Possible mechanisms for the protective effects in the gut mucosa of the L. lactis strain secreting HO-1 are the observed reductions in TNF-α and myeloperoxidase levels accompanied by increased IL-10 production.
Other molecules that have a broad protective effect on the mucosa are peptides of the trefoil factor family (TFF): Human TFF-1 and -2 are produced by mucus-producing cells in the stomach and duodenum, whereas TFF-3 is highly expressed in goblet cells in the small and large intestine (Kjellev 2009). TFFs are cytoprotective and promote epithelial wound healing and reconstitution of the gastrointestinal tract (Vandenbroucke et al. 2004) and are therefore excellent candidates for mucosa restoration (Fig. 1). However, one major drawback to the use of trefoil factors as therapeutic agents is that they do not reach the colon when administered orally. Because TFFs bind to mucus and are absorbed in the cecum, intrarectal administration has proven effective (Tran et al. 1999; Vandenbroucke et al. 2004). Intragastric administration of TFF-secreting L. lactis to mice has also been proven to be effective in the prevention and healing of acute DSS-induced colitis and chronic colitis in IL-10-deficient mice (Vandenbroucke et al. 2004).
Because TFF-1 and -3 are also secreted by human salivary glands and thus present in saliva, an L. lactis strain secreting TFF-1 was recently formulated into a mouthwash for treatment of oral mucositis, a very common and painful complication of radio- or chemotherapy in cancer patients (Caluwaerts et al. 2010). Cytotoxic anticancer drugs that affect fast-growing cancer cells also affect mucosal cells with their rapid mitotic rate, leading to atrophy, swelling, erythema, and ulceration (Raber-Durlacher et al. 2010). The TFF-1-secreting L. lactis strain was highly efficacious in alleviating oral mucositis in a hamster model (Caluwaerts et al. 2010) and in patients in a phase 1b clinical trial, leading to a clinical phase 2/3 trial to begin in 2013 (http://www.actogenix.com).
Taken together, these studies underline the potential of using probiotics to deliver molecules directly at the target site in order to restore the mucosal barrier function without interfering with systemic immunity.
THE PROBIOTIC E. coli NISSLE 1917
Although engineering Gram-positive probiotic strains like LAB to deliver molecules of interest has been a relatively uncomplicated affair because of the comparatively simple nature of their cell walls, Gram-negative probiotics like EcN have also been engineered to secrete molecules (Rao et al. 2005; Choi et al. 2012). In an attempt to block infection with the human immunodeficiency virus (HIV), EcN was engineered to secrete a hybrid peptide comprising the HIV protein gp41 (which catalyzes receptor-mediated membrane fusion) and the EcN hemolysin A (Hly), which allows direct export from the EcN cytoplasm into the extracellular medium. Remarkably, the secreted peptide inhibited HIV fusion and entry into the host cells. Moreover, the engineered EcN colonized mice for weeks to months, indicating that secretion of microbicides by this commensal may be a way to prevent HIV entry (Rao et al. 2005). In this scenario, the persistent colonization of EcN constitutes an advantage over LAB, which only transiently colonize the gut mucosa. In another recent study, EcN was engineered to deliver epidermal growth factor as a means to enhance wound healing (Choi et al. 2012). Therefore, although LAB have been the probiotics of choice for targeted delivery of molecules, EcN may provide distinct advantages when persistent colonization is desirable. For this reason, understanding the mechanisms by which EcN colonizes and persists in the gut is critical to enhancing its efficacy and potentially developing other Gram-negative bacteria for use as probiotics.
Since its serendipitous discovery, EcN has been widely used to shorten the duration of diarrhea in children and to alleviate intestinal inflammation in patients with IBD, and in particular ulcerative colitis. Although the molecular mechanisms by which EcN exerts its beneficial effects are largely unknown (Schultz and Lindstrom 2008), several studies have tried to understand what makes EcN a probiotic. Because EcN administration alleviates gastrointestinal tract inflammatory disorders and is highly protective against pathogenic bacteria and fungi including Listeria monocytogenes, Candida albicans, and Salmonella enterica serovar Typhimurium (Hockertz 1991, 1997; Mandel et al. 1995), one might expect that this probiotic strain would down-regulate inflammation. However, there is strong evidence that this bacterium instead enhances the host cell-mediated response, leading to a modulation of the balance between both pro- and anti-inflammatory local cytokines (Fig. 2) (Cross et al. 2004; Ukena et al. 2005).
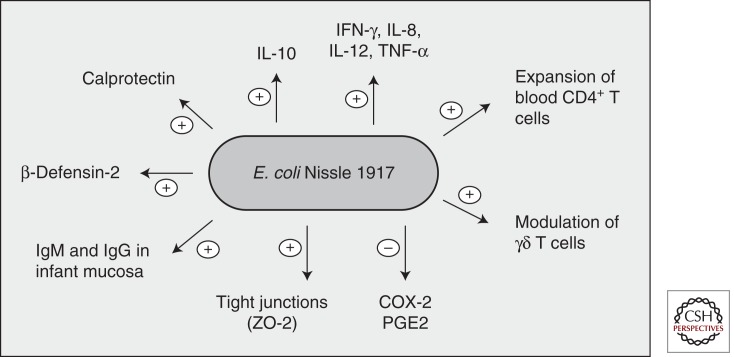
Figure 2.
Immune modulation by E. coli Nissle 1917. The probiotic E. coli Nissle 1917 modulates the host immune system in multiple ways (summary of data from in vitro and in vivo experiments).
One of the mechanisms by which EcN limits intestinal inflammation while maintaining intestinal immunological homeostasis is through the induction of naïve and memory peripheral blood CD4+-T-cell clonal expansion without the activation of mucosal and lamina propria T cells (Fig. 2) (Sturm et al. 2005). EcN also increases the activation, cell cycle progression, and cytokine secretion of γδ T cells, which play an important role in the immune response to bacterial antigens (Fig. 2). Remarkably, γδ-T-cell activation is followed by apoptosis of these cells, probably as a way to limit intestinal inflammation (Guzy et al. 2008). EcN has also been shown to mitigate experimental colitis in mice while also reducing expression of the proinflammatory cytokines and interferon γ (IFN-γ), which are activated through Toll-like receptor 4 (TLR4) and TLR5 (Fig. 2) (Grabig et al. 2006). EcN administration can also induce systemic humoral immunity in infants as well as induce specific IgA and IgM antibodies in the mucosa (Fig. 2) (Cukrowska et al. 2002; Ouwehand et al. 2002).
The immunomodulatory effects of EcN have also been observed in colonic epithelial cells, where both cell debris and cell extract fractions of EcN induced the secretion of the proinflammatory cytokine IL-8 (Lammers et al. 2002) while decreasing the expression of cyclooxygenase-2 (COX-2) and the secretion of prostaglandin E2 (PGE2), two molecules that have been implicated in colorectal carcinogenesis (Fig. 2) (Otte et al. 2009). Furthermore, EcN has been shown to be a potent activator of the antimicrobial peptide human β-defensin-2 through flagellin stimulation of NF-κB- and AP-1-mediated signaling, thereby enhancing the colonic epithelial chemical defense system (Fig. 2) (Fellermann and Stange 2001; Ganz 2003; Wehkamp et al. 2004; Splichal et al. 2005; Schlee et al. 2007). Another mechanism by which EcN enhances the mucosal barrier is through up-regulation of the tight junction-associated protein zonula occludens 2 (ZO-2) in intestinal epithelial cells (Fig. 2) (Schulze and Downward 2001; Ukena et al. 2007). Remarkably, EcN counteracted the reduced expression of ZO-2 resulting from enteropathogenic E. coli infection (Zyrek et al. 2007). Furthermore, oral administration of EcN to DSS-treated mice reduced loss of body weight and colon shortening and also conferred protection from the DSS colitis-associated increase in mucosal permeability to lumenal substances (Ukena et al. 2007).
Taken together, these studies point to an immunomodulatory role for EcN, with a balanced activation of the immune response counteracted by inactivation mechanisms. However, these studies do not explain the molecular mechanisms by which EcN modulates the immune response in vivo and ameliorates intestinal inflammation.
Important insight into the mechanisms of probiotic activity exhibited by EcN may be found by comparing its genome to those of other E. coli strains. Although all E. coli strains colonize the gut, differences in the genome content explain why some E. coli strains do not cause disease while others are pathogenic (Hacker and Kaper 2000; Hentschel and Hacker 2001). Many differentiating genetic determinants are acquired by horizontal gene transfer and oftentimes cluster on the chromosome in genomic islands that carry factors that enhance the fitness of a strain in a given environment (Kaper and Hacker 1999; Hacker and Carniel 2001). Therefore, horizontal gene transfer plays an important role in the adaptation of bacteria to specialized niches and may explain why EcN is a probiotic (Koonin et al. 2001). Indeed, EcN represents an excellent example of bacterial genome evolution within the pathogenic E. coli serotype O6 lineage. Although EcN serotype O6:K5:H1 is typical of E. coli strains associated with urinary tract infections, EcN is completely nonpathogenic (Gunzer et al. 2002). Although EcN lacks prominent virulence genes, it exhibits several fitness factors that contribute to its colonization efficiency and survival within the host (Fig. 3) (Reid et al. 2001; Sanders 2003). However, the contribution of many of the putative fitness factors that promote EcN colonization and survival in the intestine is not well understood.
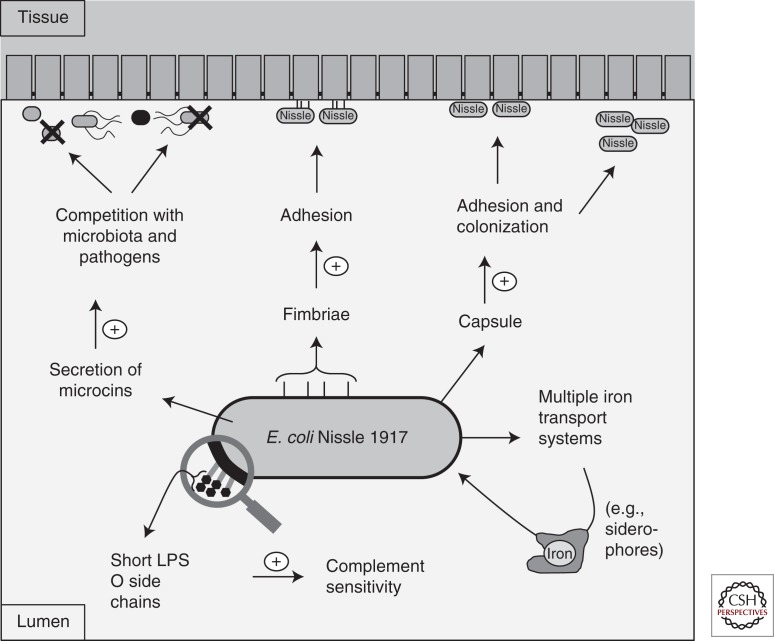
Figure 3.
Fitness factors of E. coli Nissle 1917. The probiotic E. coli Nissle 1917 possesses multiple fitness factors that enable it to colonize the gut and compete with the resident microbiota and pathogenic bacteria.
Several putative fitness determinants of EcN are localized on four genomic islands that have been partially sequenced and analyzed. Comparative genomic hybridization studies with the available genomes of E. coli K-12 strain MG1655 and uropathogenic E. coli O6 strains CFT073 and 536 showed structural similarities on the genomic level and established that EcN is strongly related to the highly virulent uropathogenic strain CFT073 (Grozdanov et al. 2004; Vejborg et al. 2010). Some fitness factors encoded by both CFT073 and EcN are curli and both type 1 and F1C fimbriae (Fig. 3). In particular, F1C fimbrie of EcN are required for biofilm formation, adherence to epithelial cells, intestinal colonization, and persistence in the gut of infant mice (Blum et al. 1995; Stentebjerg-Olesen et al. 1999; Lasaro et al. 2009). An important difference between EcN and CFT073 is in the structure of LPS (Fig. 3). A point mutation introducing a stop codon in the gene for the O6 antigen polymerase makes the O6 polysaccharide side chain very short, consisting of only a single “repeating unit” of the oligosaccharide building block typical of the O6 antigen. This peculiar characteristic is believed to contribute to EcN serum complement sensitivity and to be responsible for the semirough phenotypic aspect of the colonies grown on solid nutrient media (Fig. 3). This change could also play a role in the special immunomodulating properties exhibited by EcN, which is free of immunotoxic side effects in patients (Grozdanov et al. 2002). Another characteristic of EcN is the presence of an extracellular capsule of the K5 serotype, which is found in only 1% of E. coli isolates and is important for adhesion and colonization (Fig. 3) (Herias et al. 1997; Burns and Hull 1998). Despite this, and in contrast to other extraintestinal pathogenic capsule-forming E. coli, K5 does not contribute to serum resistance, and EcN is rapidly killed in the classic serum resistance test (Hughes et al. 1982). Moreover, the EcN K5 capsule was shown to stimulate TLR5 and to increase the induction of chemokines in both intestinal epithelial cells and ex vivo mouse small intestine (Hafez et al. 2009, 2010). Although the K5 capsule seems to play an important role in vitro, to date it is not known whether it contributes to the probiotic effect of EcN in vivo.
Probably the most striking feature of EcN’s genome is the multiple mechanisms present to acquire the essential metal nutrient iron (Fig. 3) (Crosa et al. 2004). Most bacteria display an absolute requirement for iron, and its acquisition is generally difficult because of its low solubility and potential toxicity (Andrews and Schmidt 2007). Bacteria can survive in iron-limiting conditions using specialized iron transport mechanisms, which are usually induced by low iron availability and provide specificity for alternative sources of this metal (Skaar 2010). One of the mechanisms used by Enterobacteriaceae to acquire iron is to secrete small chelators termed siderophores. EcN produces several siderophores, including enterobactin, salmochelin, aerobactin, and yersiniabactin (Bäumler et al. 1998; Hantke et al. 2003; Valdebenito et al. 2006). Moreover, EcN has the hemin- and citrate-dependent iron acquisition systems, as well as the Iha siderophore receptor, which was initially identified as a putative nonfimbrial adherence-conferring molecule in the uropathogenic strain CFT073 (Tarr et al. 2000; Torres et al. 2001; Welch et al. 2002; Léveillé et al. 2006; Hancock et al. 2010). Besides the ferric iron uptake transporters, EcN also produces EfeU, an elemental ferrous iron uptake system of the oxidase-dependent iron transporters (OFeT) family, which is a homolog of the yeast iron permease Ftr1p. Notably, the efeU gene has been shown to be inactivated in E. coli K-12 by a frameshift mutation (Grosse et al. 2006). When the mutation is repaired, EfeU is functional and alleviates iron starvation in a strain defective in all other iron transporters (Cao et al. 2007).
Overall, it appears that EcN has maintained the same redundancy in iron uptake systems as its closest relative, the uropathogenic strain CFT073. Although iron uptake promotes CFT073’s colonization of the bladder and the kidney (Garcia et al. 2011), it is probable that the many specialized iron uptake systems in EcN contribute to its colonization of the intestine and promote competition for a niche with the resident microbiota (Crosa et al. 2004), particularly if the intestine is inflamed.
One feature of the inflammatory response is that it releases antimicrobial proteins that sequester metal ions to further limit their availability to pathogens, a process known as nutritional immunity (Kehl-Fie and Skaar 2010). One mechanism is the release of the antimicrobial proteins lipocalin-2 and calprotectin by both epithelial cells and the neutrophils recruited to the site of infection. Lipocalin-2 binds to a subset of catecholate siderophores including enterochelin, the siderophore secreted by all Enterobacteriaceae to acquire iron, thereby limiting the growth of strains that rely on enterochelin as the sole scavenger of iron (Smith 2007). Calprotectin sequesters zinc and manganese, thereby limiting their availability and thus limiting the growth of sensitive bacteria (Kehl-Fie and Skaar 2010). Remarkably, S. enterica serovar Typhimurium (and likely other intestinal pathogens) is resistant to both lipocalin-2 and calprotectin. Resistance to lipocalin-2 is mediated by the siderophore salmochelin (Müller et al. 2009), a glycosylated enterochelin that is too big to fit in the lipocalin-2 binding pocket (Fischbach et al. 2006). A high-affinity zinc transporter, ZnuABC, is essential for zinc uptake when this element is limited and contributes to the low sensitivity to calprotectin exhibited by S. Typhimurium (Liu et al. 2012). It is thus not surprising that both lipocalin-2 and calprotectin provide this pathogen with an advantage in growing in the inflamed gut and competing with the microbiota (Raffatellu et al. 2009; Liu et al. 2012).
Although the mechanisms by which EcN ameliorates diarrhea are not completely understood, fitness factors enhancing its survival in the inflamed gut similar to those used by S. Typhimurium will likely play an important role. Notably, EcN was shown to induce a significant increase of calprotectin in the small intestine of gnotobiotic piglets. Contrary to this, calprotectin did not increase in the gut after infection with the nonpathogenic E. coli strain O86 or with the enteropathogenic E. coli strain O55 (Splichal et al. 2005), suggesting that calprotectin may play a role in the probiotic activity of EcN. Building on this, further studies are necessary to assess whether EcN colonization and its probiotic function are enhanced in the inflamed gut when antimicrobials like lipocalin-2 and calprotectin are highly expressed. In this scenario, high-affinity metal transporters may boost EcN colonization of the inflamed gut and may provide a means to compete for metals with other organisms, including pathogens.
Other factors that may help EcN to compete with bacteria in the gastrointestinal tract are the microcins MccH47 and MccM (Fig. 3). Microcins are low-molecular-weight antimicrobial peptides that, similar to bacteriocins of Gram-positive strains, display potent bactericidal activity against phylogenetically related bacteria that lack complementary immunity proteins (Baquero and Moreno 2006). MccH47 and MccM bind to the siderophore salmochelin and are taken up by catecholate siderophore receptors, thus exhibiting a “Trojan horse” mechanism of entry into strains (Patzer et al. 2003; Duquesne et al. 2007; Vassiliadis et al. 2010). In light of these observations, the MccH47 and MccM microcins may enhance EcN’s competition with the microbiota and bacterial pathogens. EcN was shown to inhibit human intestinal epithelial cell invasion of adherent and invasive E. coli, S. Typhimurium, Yersinia enterocolitica, Shigella flexneri, Legionella pneumophila, and L. monocytogenes (Boudeau et al. 2003; Altenhoefer et al. 2004). The mechanism of this inhibition is not known, but it is possible that at least in some conditions EcN secretes anti-invasive components that counteract pathogens, which likely include the MccH47 and MccM microcins (Baquero and Moreno 2006). Studies in animal models are needed to determine the contribution of metal transporters, microcins, and other factors to the probiotic effect of EcN.
CONCLUDING REMARKS
Our understanding of the intestinal mucosal barrier function and its alteration in IBD patients has dramatically improved in recent years. In light of this, production of cytokines, enzymes, and other molecules by gut commensals in order to strengthen the mucosa has become a rapidly expanding field of research. Engineered probiotics like LAB are an excellent tool to deliver molecules directly to the mucosa without the side effects and shortcomings of systemic delivery. Notably, they are safe to use in humans and have been closely associated with humans for centuries. Moreover, those in use do not permanently colonize the gut and have been engineered to be environmentally safe, which ensures a tightly controlled administration of the desired molecules. Because of this, there has been an expansion in clinical applications of LAB to deliver molecules. Some products, for example, the TFF-1-producing L. lactis mouthwash, are now in phase 2/3 clinical trials, and more will hopefully follow soon.
With the advent of deep sequencing, we have also gained new knowledge on the complexity of the microbiota that colonizes our intestine and its alterations in a variety of pathologies including IBD and infections. We now appreciate that inflammation shapes the microbial communities of the gut and that only the fittest survive in an inflamed environment. Intestinal pathogens like S. Typhimurium have acquired several mechanisms to thrive in the inflamed gut and compete with the microbiota. By understanding how pathogens survive in the inflamed gut, we can isolate or engineer probiotic strains that share similar traits to colonize the inflamed gut and compete with pathogens for a niche; EcN, which shares many fitness factors of intestinal pathogens, is a natural example of such probiotics. Moreover, the persistent residence of Gram-negative bacteria like EcN in the inflamed gut mucosa may make these organisms attractive candidates for molecule delivery when long-term colonization of relatively harsh host environments is desirable.
ACKNOWLEDGMENTS
We acknowledge Sean-Paul Nuccio for help with editing this article. Work in M.R.’s lab is supported by National Institutes of Health Public Health Service grant AI083619 and by the Pacific Southwest Regional Center of Excellence. J.B. is supported by a postdoctoral fellowship of the American Heart Association (11POST7090006).
Footnotes
Editors: Pascale Cossart and Stanley Maloy
Additional Perspectives on Bacterial Pathogenesis available at www.perspectivesinmedicine.org
REFERENCES
- Altenhoefer A, Oswald S, Sonnenborn U, Enders C, Schulze J, Hacker J, Oelschlaeger TA 2004. The probiotic Escherichia coli strain Nissle 1917 interferes with invasion of human intestinal epithelial cells by different enteroinvasive bacterial pathogens. FEMS Immunol Med Microbiol 40: 223–229 [DOI] [PubMed] [Google Scholar]
- Andrews NC, Schmidt PJ 2007. Iron homeostasis. Annu Rev Physiol 69: 69–85 [DOI] [PubMed] [Google Scholar]
- Anukam KC, Reid G 2008. Probiotics: 100 years (1907–2007) after Elie Metchnikoff's observations. In Communicating current research and educational topics and trends in applied microbiology, 2007 ed. (ed. Mendez-vilas A), pp. 466–474 Formatex.org, Spain [Google Scholar]
- Artis D 2008. Epithelial-cell recognition of commensal bacteria and maintenance of immune homeostasis in the gut. Nat Rev Immunol 8: 411–420 [DOI] [PubMed] [Google Scholar]
- Arumugam M, Raes J, Pelletier E, Le Paslier D, Yamada T, Mende DR, Fernandes GR, Tap J, Bruls T, Batto JM, et al. 2011. Enterotypes of the human gut microbiome. Nature 473: 174–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atarashi K, Tanoue T, Shima T, Imaoka A, Kuwahara T, Momose Y, Cheng G, Yamasaki S, Saito T, Ohba Y, et al. 2011. Induction of colonic regulatory T cells by indigenous Clostridium species. Science 331: 337–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aureli P, Capurso L, Castellazzi AM, Clerici M, Giovannini M, Morelli L, Poli A, Pregliasco F, Salvini F, Zuccotti GV 2011. Probiotics and health: An evidence-based review. Pharmacol Res 63: 366–376 [DOI] [PubMed] [Google Scholar]
- Baquero F, Moreno F 2006. The microcins. FEMS Microbiol Lett 23: 117–124 [Google Scholar]
- Bäumler AJ, Norris TL, Lasco T, Voight W, Reissbrodt R, Rabsch W, Heffron F 1998. IroN, a novel outer membrane siderophore receptor characteristic of Salmonella enterica. J Bacteriol 180: 1446–1453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bevins CL, Salzman NH 2011. Paneth cells, antimicrobial peptides and maintenance of intestinal homeostasis. Nat Rev Microbiol 9: 356–368 [DOI] [PubMed] [Google Scholar]
- Blum G, Marre R, Hacker J 1995. Properties of Escherichia coli strains of serotype O6. Infection 23: 234–236 [DOI] [PubMed] [Google Scholar]
- Boudeau J, Glasser AL, Julien S, Colombel JF, Darfeuille-Michaud A 2003. Inhibitory effect of probiotic Escherichia coli strain Nissle 1917 on adhesion to and invasion of intestinal epithelial cells by adherent-invasive E. coli strains isolated from patients with Crohn’s disease. Aliment Pharmacol Ther 18: 45–56 [DOI] [PubMed] [Google Scholar]
- Braat H, Rottiers P, Hommes DW, Huyghebaert N, Remaut E, Remon JP, van Deventer SJ, Neirynck S, Peppelenbosch MP, Steidler L 2006. A phase I trial with transgenic bacteria expressing interleukin-10 in Crohn’s disease. Clin Gastroenterol Hepatol 4: 754–759 [DOI] [PubMed] [Google Scholar]
- Burns SM, Hull SI 1998. Comparison of loss of serum resistance by defined lipopolysaccharide mutants and an acapsular mutant of uropathogenic Escherichia coli O75:K5. Infect Immun 66: 4244–4253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caluwaerts S, Vandenbroucke K, Steidler L, Neirynck S, Vanhoenacker P, Corveleyn S, Watkins B, Sonis S, Coulie B, Rottiers P 2010. AG013, a mouth rinse formulation of Lactococcus lactis secreting human Trefoil Factor 1, provides a safe and efficacious therapeutic tool for treating oral mucositis. Oral Oncol 46: 564–570 [DOI] [PubMed] [Google Scholar]
- Cao J, Woodhall MR, Alvarez J, Cartron ML, Andrews SC 2007. EfeUOB (YcdNOB) is a tripartite, acid-induced and CpxAR-regulated, low-pH Fe2+ transporter that is cryptic in Escherichia coli K-12 but functional in E. coli O157:H7. Mol Microbiol 65: 857–875 [DOI] [PubMed] [Google Scholar]
- Choi HJ, Ahn JH, Park SH, Do KH, Kim J, Moon Y 2012. Enhanced wound healing by recombinant Escherichia coli Nissle 1917 via human epidermal growth factor receptor in human intestinal epithelial cells: Therapeutic implication using recombinant probiotics. Infect Immun 80: 1079–1087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobb BA, Wang Q, Tzianabos AO, Kasper DL 2004. Polysaccharide processing and presentation by the MHCII pathway. Cell 117: 677–687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombel JF, Rutgeerts P, Malchow H, Jacyna M, Nielsen OH, Rask-Madsen J, Van Deventer S, Ferguson A, Desreumaux P, Forbes A, et al. 2001. Interleukin 10 (Tenovil) in the prevention of postoperative recurrence of Crohn’s disease. Gut 49: 42–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotter PD, Hill C, Ross RP 2005. Bacteriocins: Developing innate immunity for food. Nat Rev Microbiol 3: 777–788 [DOI] [PubMed] [Google Scholar]
- Coyne MJ, Reinap B, Lee MM, Comstock LE 2005. Human symbionts use a host-like pathway for surface fucosylation. Science 307: 1778–1781 [DOI] [PubMed] [Google Scholar]
- Crosa JH, Mey AR, Payne SM 2004. Iron transport in bacteria. ASM Press, Washington, DC [Google Scholar]
- Cross ML, Ganner A, Teilab D, Fray LM 2004. Patterns of cytokine induction by gram-positive and gram-negative probiotic bacteria. FEMS Immunol Med Microbiol 42: 173–180 [DOI] [PubMed] [Google Scholar]
- Cukrowska B, Lodinova-Zadnikova R, Enders C, Sonnenborn U, Schulze J, Tlaskalova-Hogenova H 2002. Specific proliferative and antibody responses of premature infants to intestinal colonization with nonpathogenic probiotic E. coli strain Nissle 1917. Scand J Immunol 55: 204–209 [DOI] [PubMed] [Google Scholar]
- Depaolo RW, Tang F, Kim I, Han M, Levin N, Ciletti N, Lin A, Anderson D, Schneewind O, Jabri B 2008. Toll-like receptor 6 drives differentiation of tolerogenic dendritic cells and contributes to LcrV-mediated plague pathogenesis. Cell Host Microbe 4: 350–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiBaise JK, Zhang H, Crowell MD, Krajmalnik-Brown R, Decker GA, Rittmann BE 2008. Gut microbiota and its possible relationship with obesity. Mayo Clin Proc 83: 460–469 [DOI] [PubMed] [Google Scholar]
- Duquesne S, Destoumieux-Garzon D, Peduzzi J, Rebuffat S 2007. Microcins, gene-encoded antibacterial peptides from enterobacteria. Nat Prod Rep 24: 708–734 [DOI] [PubMed] [Google Scholar]
- FAO/WHO 2001. Health and nutritional properties of probiotics in food including powder milk with live lactic acid bacteria. Cordoba, Argentina, October 1–4 [Google Scholar]
- Fava F, Danese S 2011. Intestinal microbiota in inflammatory bowel disease: Friend of foe? World J Gastroenterol 17: 557–566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fellermann K, Stange EF 2001. Defensins—Innate immunity at the epithelial frontier. Eur J Gastroenterol Hepatol 13: 771–776 [DOI] [PubMed] [Google Scholar]
- Feng T, Wang L, Schoeb TR, Elson CO, Cong Y 2010. Microbiota innate stimulation is a prerequisite for T cell spontaneous proliferation and induction of experimental colitis. J Exp Med 207: 1321–1332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischbach MA, Lin H, Zhou L, Yu Y, Abergel RJ, Liu DR, Raymond KN, Wanner BL, Strong RK, Walsh CT, et al. 2006. The pathogen-associated iroA gene cluster mediates bacterial evasion of lipocalin 2. Proc Natl Acad Sci 103: 16502–16507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Floch MH, Walker WA, Madsen K, Sanders ME, Macfarlane GT, Flint HJ, Dieleman LA, Ringel Y, Guandalini S, Kelly CP, et al. 2011. Recommendations for probiotic use—2011 update. J Clin Gastroenterol 4 (Suppl): S168–S171 [DOI] [PubMed] [Google Scholar]
- Foligne B, Dessein R, Marceau M, Poiret S, Chamaillard M, Pot B, Simonet M, Daniel C 2007. Prevention and treatment of colitis with Lactococcus lactis secreting the immunomodulatory Yersinia LcrV protein. Gastroenterology 133: 862–874 [DOI] [PubMed] [Google Scholar]
- Ganz T 2003. Defensins: Antimicrobial peptides of innate immunity. Nat Rev Immunol 3: 710–720 [DOI] [PubMed] [Google Scholar]
- Garcia EC, Brumbaugh AR, Mobley HL 2011. Redundancy and specificity of Escherichia coli iron acquisition systems during urinary tract infection. Infect Immun 79: 1225–1235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gophna U, Sommerfeld K, Gophna S, Doolittle WF, Veldhuyzen van Zanten SJ 2006. Differences between tissue-associated intestinal microfloras of patients with Crohn’s disease and ulcerative colitis. J Clin Microbiol 44: 4136–4141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabig A, Paclik D, Guzy C, Dankof A, Baumgart DC, Erckenbrecht J, Raupach B, Sonnenborn U, Eckert J, Schumann RR, et al. 2006. Escherichia coli strain Nissle 1917 ameliorates experimental colitis via Toll-like receptor 2- and Toll-like receptor 4-dependent pathways. Infect Immun 74: 4075–4082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosse C, Scherer J, Koch D, Otto M, Taudte N, Grass G 2006. A new ferrous iron-uptake transporter, EfeU (YcdN), from Escherichia coli. Mol Microbiol 62: 120–131 [DOI] [PubMed] [Google Scholar]
- Grozdanov L, Zahringer U, Blum-Oehler G, Brade L, Henne A, Knirel YA, Schombel U, Schulze J, Sonnenborn U, Gottschalk G, et al. 2002. A single nucleotide exchange in the wzy gene is responsible for the semirough O6 lipopolysaccharide phenotype and serum sensitivity of Escherichia coli strain Nissle 1917. J Bacteriol 184: 5912–5925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grozdanov L, Raasch C, Schulze J, Sonnenborn U, Gottschalk G, Hacker J, Dobrindt U 2004. Analysis of the genome structure of the nonpathogenic probiotic Escherichia coli strain Nissle 1917. J Bacteriol 186: 5432–5441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunzer F, Hennig-Pauka I, Waldmann KH, Sandhoff R, Grone HJ, Kreipe HH, Matussek A, Mengel M 2002. Gnotobiotic piglets develop thrombotic microangiopathy after oral infection with enterohemorrhagic Escherichia coli. Am J Clin Pathol 118: 364–375 [DOI] [PubMed] [Google Scholar]
- Guzy C, Paclik D, Schirbel A, Sonnenborn U, Wiedenmann B, Sturm A 2008. The probiotic Escherichia coli strain Nissle 1917 induces γδ T cell apoptosis via caspase- and FasL-dependent pathways. Int Immunol 20: 829–840 [DOI] [PubMed] [Google Scholar]
- Hacker J, Carniel E 2001. Ecological fitness, genomic islands and bacterial pathogenicity. A Darwinian view of the evolution of microbes. EMBO Rep 2: 376–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hacker J, Kaper JB 2000. Pathogenicity islands and the evolution of microbes. Annu Rev Microbiol 54: 641–679 [DOI] [PubMed] [Google Scholar]
- Hafez M, Hayes K, Goldrick M, Warhurst G, Grencis R, Roberts IS 2009. The K5 capsule of Escherichia coli strain Nissle 1917 is important in mediating interactions with intestinal epithelial cells and chemokine induction. Infect Immun 77: 2995–3003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafez M, Hayes K, Goldrick M, Grencis RK, Roberts IS 2010. The K5 capsule of Escherichia coli strain Nissle 1917 is important in stimulating expression of Toll-like receptor 5, CD14, MyD88, and TRIF together with the induction of interleukin-8 expression via the mitogen-activated protein kinase pathway in epithelial cells. Infect Immun 78: 2153–2162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock V, Vejborg RM, Klemm P 2010. Functional genomics of probiotic Escherichia coli Nissle 1917 and 83972, and UPEC strain CFT073: Comparison of transcriptomes, growth and biofilm formation. Mol Genet Genomics 284: 437–454 [DOI] [PubMed] [Google Scholar]
- Hantke K, Nicholson G, Rabsch W, Winkelmann G 2003. Salmochelins, siderophores of Salmonella enterica and uropathogenic Escherichia coli strains, are recognized by the outer membrane receptor IroN. Proc Natl Acad Sci 100: 3677–3682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi H, Takahashi R, Nishi T, Sakamoto M, Benno Y 2005. Molecular analysis of jejunal, ileal, caecal and recto-sigmoidal human colonic microbiota using 16S rRNA gene libraries and terminal restriction fragment length polymorphism. J Med Microbiol 54 (Pt 11): 1093–1101 [DOI] [PubMed] [Google Scholar]
- Henker J, Laass M, Blokhin BM, Bolbot YK, Maydannik VG, Elze M, Wolff C, Schulze J 2007. The probiotic Escherichia coli strain Nissle 1917 (EcN) stops acute diarrhoea in infants and toddlers. Eur J Pediatr 166: 311–318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hentschel U, Hacker J 2001. Pathogenicity islands: The tip of the iceberg. Microbes Infect 3: 545–548 [DOI] [PubMed] [Google Scholar]
- Herias MV, Midtvedt T, Hanson LA, Wold AE 1997. Escherichia coli K5 capsule expression enhances colonization of the large intestine in the gnotobiotic rat. Infect Immun 65: 531–536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hockertz S 1991. Immunomodulating effect of killed, apathogenic Escherichia coli, strain Nissle 1917, on the macrophage system. Arzneimittelforschung 41: 1108–1112 [PubMed] [Google Scholar]
- Hockertz S 1997. Augmentation of host defence against bacterial and fungal infections of mice pretreated with the non-pathogenic Escherichia coli strain Nissle 1917. Arzneimittelforschung 47: 793–796 [PubMed] [Google Scholar]
- Hogenauer C, Hammer HF, Krejs GJ, Reisinger EC 1998. Mechanisms and management of antibiotic-associated diarrhea. Clin Infect Dis 27: 702–710 [DOI] [PubMed] [Google Scholar]
- Hooper LV, Gordon JI 2001. Commensal host-bacterial relationships in the gut. Science 292: 1115–1118 [DOI] [PubMed] [Google Scholar]
- Høverstad T, Midtvedt T 1986. Short-chain fatty acids in germfree mice and rats. J Nutr 116: 1772–1776 [DOI] [PubMed] [Google Scholar]
- Hughes C, Phillips R, Roberts AP 1982. Serum resistance among Escherichia coli strains causing urinary tract infection in relation to O type and the carriage of hemolysin, colicin, and antibiotic resistance determinants. Infect Immun 35: 270–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huibregtse IL, Zaat SA, Kapsenberg ML, Sartori da Silva MA, Peppelenbosch MP, van Deventer SJ, Braat H 2011. Genetically modified Lactococcus lactis for delivery of human interleukin-10 to dendritic cells. Gastroenterol Res Pract 2012: 639291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov II, Atarashi K, Manel N, Brodie EL, Shima T, Karaoz U, Wei D, Goldfarb KC, Santee CA, Lynch SV, et al. 2009. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell 139: 485–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jervis HR, Biggers DC 1964. Mucosal enzymes in the cecum of conventional and germfree mice. Anat Rec 148: 591–597 [DOI] [PubMed] [Google Scholar]
- Kaper JB, Hacker J 1999. Pathogenicity islands and other mobile virulence elements. ASM Press, Washington, DC [Google Scholar]
- Karin M, Lawrence T, Nizet V 2006. Innate immunity gone awry: Linking microbial infections to chronic inflammation and cancer. Cell 124: 823–835 [DOI] [PubMed] [Google Scholar]
- Kehl-Fie TE, Skaar EP 2010. Nutritional immunity beyond iron: A role for manganese and zinc. Curr Opin Chem Biol 14: 218–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kjellev S 2009. The trefoil factor family—Small peptides with multiple functionalities. Cell Mol Life Sci 66: 1350– 1369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin EV, Makarova KS, Aravind L 2001. Horizontal gene transfer in prokaryotes: Quantification and classification. Annu Rev Microbiol 55: 709–742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruis W, Schutz E, Fric P, Fixa B, Judmaier G, Stolte M 1997. Double-blind comparison of an oral Escherichia coli preparation and mesalazine in maintaining remission of ulcerative colitis. Aliment Pharmacol Ther 11: 853–858 [DOI] [PubMed] [Google Scholar]
- Kruis W, Fric P, Pokrotnieks J, Lukas M, Fixa B, Kascak M, Kamm MA, Weismueller J, Beglinger C, Stolte M, et al. 2004. Maintaining remission of ulcerative colitis with the probiotic Escherichia coli Nissle 1917 is as effective as with standard mesalazine. Gut 53: 1617–1623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lammers KM, Helwig U, Swennen E, Rizzello F, Venturi A, Caramelli E, Kamm MA, Brigidi P, Gionchetti P, Campieri M 2002. Effect of probiotic strains on interleukin 8 production by HT29/19A cells. Am J Gastroenterol 97: 1182–1186 [DOI] [PubMed] [Google Scholar]
- Lasaro MA, Salinger N, Zhang J, Wang Y, Zhong Z, Goulian M, Zhu J 2009. F1C fimbriae play an important role in biofilm formation and intestinal colonization by the Escherichia coli commensal strain Nissle 1917. Appl Environ Microbiol 75: 246–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Loir Y, Azevedo V, Oliveira SC, Freitas DA, Miyoshi A, Bermudez-Humaran LG, Nouaille S, Ribeiro LA, Leclercq S, Gabriel JE, et al. 2005. Protein secretion in Lactococcus lactis: An efficient way to increase the overall heterologous protein production. Microb Cell Fact 4: 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Léveillé S, Caza M, Johnson JR, Clabots C, Sabri M, Dozois CM 2006. Iha from an Escherichia coli urinary tract infection outbreak clonal group A strain is expressed in vivo in the mouse urinary tract and functions as a catecholate siderophore receptor. Infect Immun 74: 3427–3436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu CH, Lee SM, Vanlare JM, Kasper DL, Mazmanian SK 2008. Regulation of surface architecture by symbiotic bacteria mediates host colonization. Proc Natl Acad Sci 105: 3951–3956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JZ, Jellbauer S, Poe AJ, Ton V, Pesciaroli M, Kehl-Fie TE, Restrepo NA, Hosking MP, Edwards RA, Battistoni A, et al. 2012. Zinc sequestration by the neutrophil protein calprotectin enhances Salmonella growth in the inflamed gut. Cell Host Microbe 11: 227–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodinova-Zadnikova R, Sonnenborn U 1997. Effect of preventive administration of a nonpathogenic Escherichia coli strain on the colonization of the intestine with microbial pathogens in newborn infants. Biol Neonate 71: 224–232 [DOI] [PubMed] [Google Scholar]
- Mahowald MA, Rey FE, Seedorf H, Turnbaugh PJ, Fulton RS, Wollam A, Shah N, Wang C, Magrini V, Wilson RK, et al. 2009. Characterizing a model human gut microbiota composed of members of its two dominant bacterial phyla. Proc Natl Acad Sci 106: 5859–5864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandel L, Trebichavsky I, Splichal I, Schulze J 1995. Stimulation of intestinal immune cells by E. coli in gnotobiotic piglets. Adv Exp Med Biol 371A: 463–464 [DOI] [PubMed] [Google Scholar]
- Marco ML, Pavan S, Kleerebezem M 2006. Towards understanding molecular modes of probiotic action. Curr Opin Biotechnol 17: 204–210 [DOI] [PubMed] [Google Scholar]
- Maslowski KM, Vieira AT, Ng A, Kranich J, Sierro F, Yu D, Schilter HC, Rolph MS, Mackay F, Artis D, et al. 2009. Regulation of inflammatory responses by gut microbiota and chemoattractant receptor GPR43. Nature 461: 1282–1286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazmanian SK, Liu CH, Tzianabos AO, Kasper DL 2005. An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell 122: 107–118 [DOI] [PubMed] [Google Scholar]
- Mierau I, Kleerebezem M 2005. 10 years of the nisin-controlled gene expression system (NICE) in Lactococcus lactis. Appl Microbiol Biotechnol 68: 705–717 [DOI] [PubMed] [Google Scholar]
- Mollenbrink M, Bruckschen E 1994. Treatment of chronic constipation with physiologic Escherichia coli bacteria. Results of a clinical study of the effectiveness and tolerance of microbiological therapy with the E. coli Nissle 1917 strain (Mutaflor). Med Klin (Munich) 89: 587–593 [PubMed] [Google Scholar]
- Mukhopadhya I, Hansen R, El-Omar EM, Hold GL 2012. IBD—What role do Proteobacteria play? Nat Rev Gastroenterol Hepatol 9: 219–230 [DOI] [PubMed] [Google Scholar]
- Müller SI, Valdebenito M, Hantke K 2009. Salmochelin, the long-overlooked catecholate siderophore of Salmonella. Biometals 22: 691–695 [DOI] [PubMed] [Google Scholar]
- Nagalingam NA, Lynch SV 2012. Role of the microbiota in inflammatory bowel diseases. Inflamm Bowel Dis 18: 968–984 [DOI] [PubMed] [Google Scholar]
- Nissle A 1959. Explanations of the significance of colonic dysbacteria & the mechanism of action of E. coli therapy (mutaflor). Medizinische 4: 1017–1022 [PubMed] [Google Scholar]
- Nouaille S, Ribeiro LA, Miyoshi A, Pontes D, Le Loir Y, Oliveira SC, Langella P, Azevedo V 2003. Heterologous protein production and delivery systems for Lactococcus lactis. Genet Mol Res 2: 102–111 [PubMed] [Google Scholar]
- Otte JM, Mahjurian-Namari R, Brand S, Werner I, Schmidt WE, Schmitz F 2009. Probiotics regulate the expression of COX-2 in intestinal epithelial cells. Nutr Cancer 61: 103–113 [DOI] [PubMed] [Google Scholar]
- Ouwehand AC, Salminen S, Isolauri E 2002. Probiotics: An overview of beneficial effects. Antonie Van Leeuwenhoek 82: 279–289 [PubMed] [Google Scholar]
- Pang Q, Ji Y, Li Y, Bermudez-Humaran LG, Hu G, Zeng Y 2008. Intragastric administration with recombinant Lactococcus lactis producing heme oxygenase-1 prevents lipopolysaccharide-induced endotoxemia in rats. FEMS Microbiol Lett 283: 62–68 [DOI] [PubMed] [Google Scholar]
- Pang QF, Ji Y, Bermudez-Humaran LG, Zhou QM, Hu G, Zeng Y 2009. Protective effects of a heme oxygenase-1-secreting Lactococcus lactis on mucosal injury induced by hemorrhagic shock in rats. J Surg Res 153: 39–45 [DOI] [PubMed] [Google Scholar]
- Patzer SI, Baquero MR, Bravo D, Moreno F, Hantke K 2003. The colicin G, H and X determinants encode microcins M and H47, which might utilize the catecholate siderophore receptors FepA, Cir, Fiu and IroN. Microbiology 149: 2557–2570 [DOI] [PubMed] [Google Scholar]
- Peterbauer C, Maischberger T, Haltrich D 2011. Food-grade gene expression in lactic acid bacteria. Biotechnol J 6: 1147–1161 [DOI] [PubMed] [Google Scholar]
- Raber-Durlacher JE, Elad S, Barasch A 2010. Oral mucositis. Oral Oncol 46: 452–456 [DOI] [PubMed] [Google Scholar]
- Raffatellu M, George MD, Akiyama Y, Hornsby MJ, Nuccio SP, Paixao TA, Butler BP, Chu H, Santos RL, Berger T, et al. 2009. Lipocalin-2 resistance confers an advantage to Salmonella enterica serotype Typhimurium for growth and survival in the inflamed intestine. Cell Host Microbe 5: 476–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao S, Hu S, McHugh L, Lueders K, Henry K, Zhao Q, Fekete RA, Kar S, Adhya S, Hamer DH 2005. Toward a live microbial microbicide for HIV: Commensal bacteria secreting an HIV fusion inhibitor peptide. Proc Natl Acad Sci 102: 11993–11998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid G, Howard J, Gan BS 2001. Can bacterial interference prevent infection? Trends Microbiol 9: 424–428 [DOI] [PubMed] [Google Scholar]
- Sanders ME 2003. Probiotics: Considerations for human health. Nutr Rev 61: 91–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlee M, Wehkamp J, Altenhoefer A, Oelschlaeger TA, Stange EF, Fellermann K 2007. Induction of human β-defensin 2 by the probiotic Escherichia coli Nissle 1917 is mediated through flagellin. Infect Immun 75: 2399–2407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schotte L, Steidler L, Vandekerckhove J, Remaut E 2000. Secretion of biologically active murine interleukin-10 by Lactococcus lactis. Enzyme Microb Technol 27: 761–765 [DOI] [PubMed] [Google Scholar]
- Schultz M, Lindstrom AL 2008. Rationale for probiotic treatment strategies in inflammatory bowel disease. Expert Rev Gastroenterol Hepatol 2: 337–355 [DOI] [PubMed] [Google Scholar]
- Schulze A, Downward J 2001. Navigating gene expression using microarrays—A technology review. Nat Cell Biol 3: E190–E195 [DOI] [PubMed] [Google Scholar]
- Schutz E 1989. The treatment of intestinal diseases with Mutaflor. A multicenter retrospective study. Fortschr Med 107: 599–602 [PubMed] [Google Scholar]
- Servin AL 2004. Antagonistic activities of lactobacilli and bifidobacteria against microbial pathogens. FEMS Microbiol Rev 28: 405–440 [DOI] [PubMed] [Google Scholar]
- Shaw MH, Kamada N, Kim YG, Nunez G 2012. Microbiota-induced IL-1β, but not IL-6, is critical for the development of steady-state TH17 cells in the intestine. J Exp Med 209: 251–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skaar EP 2010. The battle for iron between bacterial pathogens and their vertebrate hosts. PLoS Pathog 6: e1000949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith KD 2007. Iron metabolism at the host pathogen interface: Lipocalin 2 and the pathogen-associated iroA gene cluster. Int J Biochem Cell Biol 39: 1776–1780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Splichal I, Fagerhol MK, Trebichavsky I, Splichalova A, Schulze J 2005. The effect of intestinal colonization of germ-free pigs with Escherichia coli on calprotectin levels in plasma, intestinal and bronchoalveolar lavages. Immunobiology 209: 681–687 [DOI] [PubMed] [Google Scholar]
- Spor A, Koren O, Ley R 2011. Unravelling the effects of the environment and host genotype on the gut microbiome. Nat Rev Microbiol 9: 279–290 [DOI] [PubMed] [Google Scholar]
- Steidler L, Hans W, Schotte L, Neirynck S, Obermeier F, Falk W, Fiers W, Remaut E 2000. Treatment of murine colitis by Lactococcus lactis secreting interleukin-10. Science 289: 1352–1355 [DOI] [PubMed] [Google Scholar]
- Steidler L, Neirynck S, Huyghebaert N, Snoeck V, Vermeire A, Goddeeris B, Cox E, Remon JP, Remaut E 2003. Biological containment of genetically modified Lactococcus lactis for intestinal delivery of human interleukin 10. Nat Biotechnol 21: 785–789 [DOI] [PubMed] [Google Scholar]
- Steidler L, Rottiers P, Coulie B 2009. Actobiotics as a novel method for cytokine delivery. Ann NY Acad Sci 1182: 135–145 [DOI] [PubMed] [Google Scholar]
- Stentebjerg-Olesen B, Chakraborty T, Klemm P 1999. Type 1 fimbriation and phase switching in a natural Escherichia coli fimB null strain, Nissle 1917. J Bacteriol 181: 7470–7478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturm A, Rilling K, Baumgart DC, Gargas K, Abou-Ghazale T, Raupach B, Eckert J, Schumann RR, Enders C, Sonnenborn U, et al. 2005. Escherichia coli Nissle 1917 distinctively modulates T-cell cycling and expansion via Toll-like receptor 2 signaling. Infect Immun 73: 1452–1465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarr PI, Bilge SS, Vary JC Jr, Jelacic S, Habeeb RL, Ward TR, Baylor MR, Besser TE 2000. Iha: A novel Escherichia coli O157:H7 adherence-conferring molecule encoded on a recently acquired chromosomal island of conserved structure. Infect Immun 68: 1400–1407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilg H, van Montfrans C, van den Ende A, Kaser A, van Deventer SJ, Schreiber S, Gregor M, Ludwiczek O, Rutgeerts P, Gasche C, et al. 2002. Treatment of Crohn’s disease with recombinant human interleukin 10 induces the proinflammatory cytokine interferon γ. Gut 50: 191–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tlaskalova-Hogenova H, Stepankova R, Kozakova H, Hudcovic T, Vannucci L, Tuckova L, Rossmann P, Hrncir T, Kverka M, Zakostelska Z, et al. 2011. The role of gut microbiota (commensal bacteria) and the mucosal barrier in the pathogenesis of inflammatory and autoimmune diseases and cancer: Contribution of germ-free and gnotobiotic animal models of human diseases. Cell Mol Immunol 8: 110–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres AG, Redford P, Welch RA, Payne SM 2001. TonB-dependent systems of uropathogenic Escherichia coli: Aerobactin and heme transport and TonB are required for virulence in the mouse. Infect Immun 69: 6179–6185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran CP, Cook GA, Yeomans ND, Thim L, Giraud AS 1999. Trefoil peptide TFF2 (spasmolytic polypeptide) potently accelerates healing and reduces inflammation in a rat model of colitis. Gut 44: 636–642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ukena SN, Westendorf AM, Hansen W, Rohde M, Geffers R, Coldewey S, Suerbaum S, Buer J, Gunzer F 2005. The host response to the probiotic Escherichia coli strain Nissle 1917: Specific up-regulation of the proinflammatory chemokine MCP-1. BMC Med Genet 6: 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ukena SN, Singh A, Dringenberg U, Engelhardt R, Seidler U, Hansen W, Bleich A, Bruder D, Franzke A, Rogler G, et al. 2007. Probiotic Escherichia coli Nissle 1917 inhibits leaky gut by enhancing mucosal integrity. PLoS ONE 2: e1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdebenito M, Crumbliss AL, Winkelmann G, Hantke K 2006. Environmental factors influence the production of enterobactin, salmochelin, aerobactin, and yersiniabactin in Escherichia coli strain Nissle 1917. Int J Med Microbiol 296: 513–520 [DOI] [PubMed] [Google Scholar]
- Vandenbroucke K, Hans W, Van Huysse J, Neirynck S, Demetter P, Remaut E, Rottiers P, Steidler L 2004. Active delivery of trefoil factors by genetically modified Lactococcus lactis prevents and heals acute colitis in mice. Gastroenterology 127: 502–513 [DOI] [PubMed] [Google Scholar]
- Vandenbroucke K, de Haard H, Beirnaert E, Dreier T, Lauwereys M, Huyck L, Van Huysse J, Demetter P, Steidler L, Remaut E, et al. 2010. Orally administered L. lactis secreting an anti-TNF Nanobody demonstrate efficacy in chronic colitis. Mucosal Immunol 3: 49–56 [DOI] [PubMed] [Google Scholar]
- van Deventer SJ, Elson CO, Fedorak RN 1997. Multiple doses of intravenous interleukin 10 in steroid-refractory Crohn’s disease. Crohn’s Disease Study Group. Gastroenterology 113: 383–389 [DOI] [PubMed] [Google Scholar]
- Vassiliadis G, Destoumieux-Garzon D, Lombard C, Rebuffat S, Peduzzi J 2010. Isolation and characterization of two members of the siderophore-microcin family, microcins M and H47. Antimicrob Agents Chemother 54: 288–297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vejborg RM, Friis C, Hancock V, Schembri MA, Klemm P 2010. A virulent parent with probiotic progeny: Comparative genomics of Escherichia coli strains CFT073, Nissle 1917 and ABU 83972. Mol Genet Genomics 283: 469–484 [DOI] [PubMed] [Google Scholar]
- Vijayan V, Mueller S, Baumgart-Vogt E, Immenschuh S 2010. Heme oxygenase-1 as a therapeutic target in inflammatory disorders of the gastrointestinal tract. World J Gastroenterol 16: 3112–3119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinolo MA, Rodrigues HG, Nachbar RT, Curi R 2011. Regulation of inflammation by short chain fatty acids. Nutrients 3: 858–876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wehkamp J, Harder J, Wehkamp K, Wehkamp-von Meissner B, Schlee M, Enders C, Sonnenborn U, Nuding S, Bengmark S, Fellermann K, et al. 2004. NF-κB- and AP-1-mediated induction of human beta defensin-2 in intestinal epithelial cells by Escherichia coli Nissle 1917: A novel effect of a probiotic bacterium. Infect Immun 72: 5750–5758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch RA, Burland V, Plunkett G III, Redford P, Roesch P, Rasko D, Buckles EL, Liou SR, Boutin A, Hackett J, et al. 2002. Extensive mosaic structure revealed by the complete genome sequence of uropathogenic Escherichia coli. Proc Natl Acad Sci 99: 17020–17024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westendorf AM, Gunzer F, Deppenmeier S, Tapadar D, Hunger JK, Schmidt MA, Buer J, Bruder D 2005. Intestinal immunity of Escherichia coli NISSLE 1917: A safe carrier for therapeutic molecules. FEMS Immunol Med Microbiol 43: 373–384 [DOI] [PubMed] [Google Scholar]
- Wu GD, Chen J, Hoffmann C, Bittinger K, Chen YY, Keilbaugh SA, Bewtra M, Knights D, Walters WA, Knight R, et al. 2011. Linking long-term dietary patterns with gut microbial enterotypes. Science 334: 105–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zyrek AA, Cichon C, Helms S, Enders C, Sonnenborn U, Schmidt MA 2007. Molecular mechanisms underlying the probiotic effects of Escherichia coli Nissle 1917 involve ZO-2 and PKCζ redistribution resulting in tight junction and epithelial barrier repair. Cell Microbiol 9: 804–816 [DOI] [PubMed] [Google Scholar]