Abstract
The cystic fibrosis transmembrane conductance regulator (CFTR) requires dynamic fluctuations between states in its gating cycle for proper channel function, including changes in the interactions between the nucleotide-binding domains (NBDs) and between the intracellular domain (ICD) coupling helices and NBDs. Such motions are also linked with fluctuating phosphorylation-dependent binding of CFTR’s disordered regulatory (R) region to the NBDs and partners. Folding of CFTR is highly inefficient, with the marginally stable NBD1 sampling excited states or folding intermediates that are aggregation-prone. The severe CF–causing F508del mutation exacerbates the folding inefficiency of CFTR and leads to impaired channel regulation and function, partly as a result of perturbed NBD1–ICD interactions and enhanced sampling of these NBD1 excited states. Increased knowledge of the dynamics within CFTR will expand our understanding of the regulated channel gating of the protein as well as of the F508del defects in folding and function.
Improved knowledge of the dynamics within CFTR—including the R region and NBD1—will expand our understanding of its function, as well as the implications of the F508del defect that is present in many cystic fibrosis patients.
Understanding how proteins function often requires knowledge of structure and conformational stability and dynamics. Unlike mechanical parts, proteins are self-assembling dynamic components that require flexibility for function. Conformational changes are required during the folding process and for progression through functional cycles. Aberrations in stability and/or dynamics can lead to protein misfolding or dysfunction, primary causes of many diseases including cystic fibrosis (CF). Changes in stability can be independent of significant structural changes to a protein’s ground state, as is the case for the cystic fibrosis transmembrane conductance regulator (CFTR). As discussed in previous articles, deletion of F508 in CFTR (F508del), widely observed in CF patients, results in a significant decrease in folding and processing efficiency as well as in gating defects (Riordan et al. 1989; Cheng et al. 1990; Dalemans et al. 1991; Drumm et al. 1991). An early indication that the mutation does not sterically inhibit proper folding or remove a critical structural component was the observation that the processing efficiency could be significantly improved by expressing the protein at 26°C instead of 37°C (Denning et al. 1992). The mutation decreases CFTR’s thermostability, but a small amount of properly processed mutant protein does arrive at the cell membrane and has residual activity. Although the mutation does not prohibit the conformations required for proper gating, the activity of F508del is rapidly lost with increase in temperature, indicating that the mutation affects CFTR stability and dynamics. The pathology is characterized by the inefficiency of folding and processing and a reduction in the lifetime of the open conformation during the gating cycle.
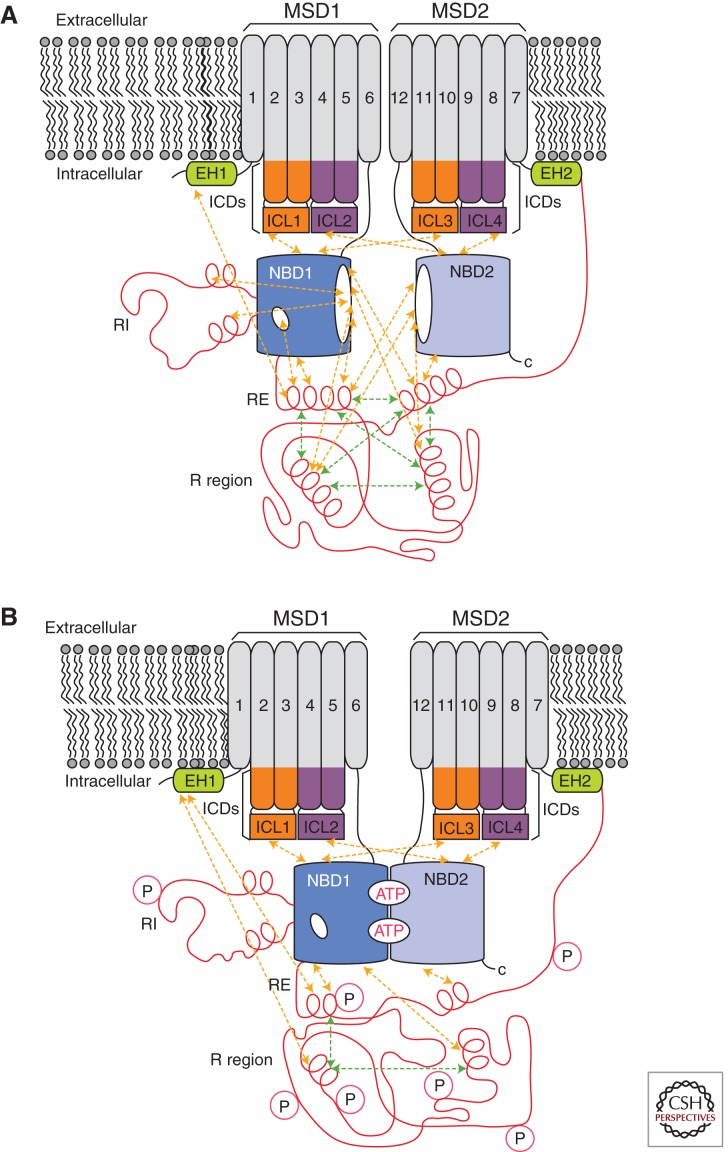
CFTR comprises two repeated units, each containing a membrane-spanning domain (MSD1 and MSD2) and a cytoplasmic nucleotide-binding domain (NBD1 and NBD2) (Riordan et al. 1989). The carboxyl terminus of NBD1 is connected to the amino terminus of MSD2 via a 200-residue intrinsically disordered regulatory (R) region (Fig. 1) (Riordan et al. 1989; Dulhanty and Riordan 1994; Ostedgaard et al. 2000). Folding of wild-type (WT) CFTR is generally found to be inefficient, although efficiency varies with cell type (Lukacs et al. 1994; Ward and Kopito 1994; Varga et al. 2004). The dominant F508del mutation exacerbates the inefficiency. Improperly folded CFTR is retained by endoplasmic reticulum (ER) quality control mechanisms, a process that is readily measured by monitoring the degree of glycosylation by SDS polyacrylamide gel electrophoresis (Cheng et al. 1990). Although some reports indicate that CFTR domains initiate their folding cotranslationally in an independent fashion (Kleizen et al. 2005; Hoelen et al. 2010), interdomain interactions are required for final processing and escape from the ER quality control machinery (Cui et al. 2007). Notably, acquisition of a protease-resistant NBD2 fold has been shown to require appropriate interactions with the preceding domains (Du et al. 2005). Deletion of F508 in NBD1 negatively affects proper folding of NBD2, possibly by introducing a nonphysiological tight interaction between NBD1 and NBD2 (Du et al. 2005). Interestingly, NBD2 is not required for transportation out of the ER (Cui et al. 2007) or CFTR gating (Wang et al. 2010b). Misfolding of NBD2 in F508del may result in CFTR being retained in the ER by the quality control machinery, unable to proceed to the cell surface.
Figure 1.
Schematic representation of nonphosphorylated (A) and phosphorylated (B) CFTR. The membrane-spanning domains (MSDs) and nucleotide-binding domains (NBDs) are shown in gray and blue, respectively. The R region, regulatory insertion (RI), and regulatory extension (RE) are shown in red. The intracellular domains (ICDs) are shown in orange or purple with the coupling helices or intracellular loops (ICLs) identified. The elbow helices (EHs) are shown in green. Intramolecular interactions are indicated by dashed arrows.
At the cell surface, MSD1 and MSD2 of CFTR form a regulated anion channel that cycles through a closed state, an open-ready state, and an open state. The cytoplasmic extensions of the transmembrane helices form helical bundle structural regions, which collectively constitute the intracellular domains (ICDs) believed to transmit information from the NBDs to the transmembrane pore. Gating of the channel is known to require phosphorylation of the R region (Cheng et al. 1991; Berger et al. 1993), binding of ATP at the two sites at the NBD heterodimer interface, and hydrolysis of ATP at the site formed by the ATP-binding core of NBD2 (Cheung et al. 2008). Phosphorylation is believed to relieve R-region inhibition of channel opening, but the conformational change effected by the phosphorylation is poorly understood. NBD1 and -2 form a functional dimer (reviewed by Cheung et al. 2008), but the changes in the dimer conformation that open and close the channel are also poorly understood.
Here we will discuss gating cycle dynamics, the dynamics of the R region and NBD1, and changes in CFTR’s dynamic behavior resulting from deletion of F508.
FULL-LENGTH CFTR
Unlike the other ATP-binding cassette (ABC) transporters to which it is related, CFTR functions as a channel rather than as an active transporter (Anderson et al. 1991). Despite this difference, CFTR shows significant sequence similarity with ABC transporters (Riordan et al. 1989) and likely undergoes similar conformational rearrangements. As a channel, CFTR exists in a closed conformation, which does not permit chloride transport, and an open conformation, which permits rapid chloride flow through the pore formed by the MSDs. Single-channel recordings of CFTR indicate that the channel has two phases, a burst interval, during which the channel is open but flickers rapidly through a closed state, and an interburst interval, during which the channel is closed (Winter et al. 1994), indicating that CFTR has at least three conformations: a closed state, an open state, and an open-ready state. The open-ready state does not allow chloride flow but, unlike the closed state, is poised to rapidly transition to the open state. As a large polytopic membrane protein containing multiple domains and disordered segments, full-length CFTR has resisted efforts to directly resolve the structure of any of these conformations by X-ray crystallography. Insight into the structure and dynamics of full-length CFTR has come from crystal structures of related ABC transporters, molecular modeling, cross-linking studies, electrophysiology experiments, and electron microscopy. In this section we review what is known about the dynamics of full-length CFTR as it passes through the gating cycle.
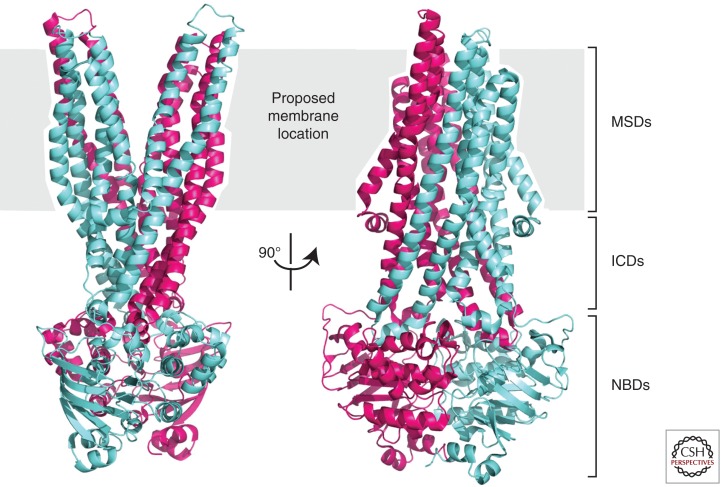
CFTR-related ABC transporters representing different stages of the transport cycle have been crystallized (Dawson and Locher 2006; Hollenstein et al. 2007; Pinkett et al. 2007; Ward et al. 2007). The structures suggest that the MSDs collectively form two discrete helical bundles that move with respect to each other, forming alternately outward- and inward-facing conformations. In the outward-facing conformation (as exemplified by the Sav1866 structure [Dawson and Locher 2006]), the helical bundles point away from each other on the extracellular side of the protein, diverging near the midpoint of the membrane, and the NBDs form tight dimers with both composite ATP-binding sites tightly juxtaposed and nucleotide-filled (Fig. 2). In the inward-facing conformation (as exemplified by the MsbA nucleotide-free structures [Ward et al. 2007]), the helical bundles point away from each other on the intracellular side of the membrane and the NBDs are at least partially separated and nucleotide-free (Hollenstein et al. 2007; Ward et al. 2007). A range of separation is observed for the inward-facing conformation, suggesting a great deal of flexibility in the inward-facing conformation (Ward et al. 2007). In the MsbA “open apo” structure, the NBDs are completely separated. The outward-facing conformation appears to be promoted by nucleotide-induced NBD tight dimer formation transmitted through an interaction between the NBDs and the ICDs. The inward-facing conformation may be triggered by nucleotide hydrolysis and release.
Figure 2.
Ribbon diagram representation of the outward-facing conformation of Sav1866 in two orthogonal views. The subunits of the dimer are shown in blue and purple. Note that the two transmembrane helical bundles move apart as they near the extracellular side of the membrane (PDB 2HYD [Dawson and Locher 2006]).
The relationship between the inward- and outward-facing conformations observed in these crystal structures of related transporters and the range of channel motions in CFTR has not been conclusively established. The outward-facing conformation has been assumed to be representative of the open conformation of the CFTR channel in part because it is the nucleotide-bound state in the observed structures (Mornon et al. 2009). By analogy, the closed conformation may be represented by the inward-facing conformation. ABC transporters operate by an “alternating access” mechanism (Mitchell 1957; Jardetzky 1966). They bind substrate in one conformation and then undergo a conformational change that allows release of the substrate on the opposite side of the membrane. The substrate is never able to freely pass through the transporter. Because CFTR is closely related to these ABC transporters, it is envisioned that it undergoes a similar conformational change. However, as it is a channel and not an active transporter, it is not clear that the analogy holds and that CFTR maintains the same dynamic modes. In particular, CFTR is believed to have three different conformations rather than two, although one or more of these conformations may be very similar. As with other ABC transporters, channel gating has been shown to be tightly coupled to ATP hydrolysis (Csanady et al. 2009), but the effect of nucleotide binding on the position of the NBDs with respect to each other has not been resolved. Channel opening may require induced dimerization or a more moderate conformational change in preexisting dimers (Smith et al. 2002; Dawson and Locher 2006; Jones and George 2007, 2009).
Electron microscopy images obtained from single-particle analysis of noncrystalline as well as two-dimensional crystals of CFTR in a nucleotide-free state and a nucleotide-bound phosphorylated state lend some support to CFTR having inward- and outward-facing conformations (Rosenberg et al. 2004; Awayn et al. 2005; Zhang et al. 2009). The images suggest that in the absence of nucleotide the helical bundles have a greater separation on the cytoplasmic side, whereas in the nucleotide-bound form the helical bundles are closer to the outward-facing model. The role of nucleotide binding and conformational changes at the NBDs is not discernible from these low-resolution images. However, the images do suggest that the NBDs do not completely separate as they do in one of the MsbA structures (open apo) (Ward et al. 2007).
The crystal structures of Sav1866 and MsbA, bacterial ABC transporters, were used to construct structural models of the opened and closed states of full-length CFTR that have been partially validated experimentally (Mornon et al. 2008, 2009; Serohijos et al. 2008a; Alexander et al. 2009; Huang et al. 2009; Kanelis et al. 2010; Moran 2010). The channel pore architecture has been experimentally probed using cysteine mutants and thiol-reactive reagents to determine the accessibility of particular sites in the pore. Taken together, these studies are strongly supportive of the Sav1866-based homology models (Zhang et al. 2005; Fatehi and Linsdell 2008; Alexander et al. 2009). They show a pore that is wide on the extracellular side and becomes more inaccessible to molecular probes, especially anionic probes, moving toward the cytoplasmic surface. There is also evidence that the pore architecture undergoes significant conformational change during the gating cycle (Zhang et al. 2005; Fatehi and Linsdell 2008). Activation of the channel (by forskolin and 3-isobutyl-1-methylxantine [IBMX]) appears to significantly enhance access of anions to the pore (Fatehi and Linsdell 2008). Accessibility for cations is less stringent, and even large organic cations can reach cysteines introduced into the putative inner vestibule of the pore in the inactive state. Fatehi and Linsdell further propose that there are two functionally distinct closed states: the inactivated channel and the “activated but still closed” channel conformation. They suggest that the latter conformation is activated by protein kinase A (PKA) but still requires ATP-dependent conformational changes at the NBDs to open the channel to chloride flow. The activated but still closed conformation is likely related to the open-ready conformation observed in single-channel gating studies.
The homology models also identify specific interactions between the ICDs and the NBDs, which have been tested experimentally by chemical cross-linking studies. At a gross level, these studies confirm close interactions between the NBDs and the ICD “coupling helices” that lie at the NBD interface (called ICLs or CLs), which supports the notion that the positioning of the MSDs is controlled by interactions between the NBDs and ICLs (Serohijos et al. 2008a). At a specific level, cross-linking studies confirm interactions between ICL1, ICL3, and ICL4 and NBD1 (He et al. 2008, 2010). These studies also show that cross-linking NBD1 to ICL3 or -4, but not ICL1, strongly inhibits channel gating (He et al. 2008), a result that suggests that the NBD–ICL interface must change during the gating cycle. Similar results were noted for NBD2–ICL interfaces (He et al. 2008). Significant flexibility in NBD–ICL interactions is also indicated by the large distribution of linker lengths that can cross-link particular NBD–ICL cysteine pairs introduced into a Cys-less CFTR (Serohijos et al. 2008a). Although cross-linking can inhibit channel gating, suggesting a requirement for flexibility, NBD1–ICL4 and NBD2–ICL2 cross-linking are not strongly influenced by PKA phosphorylation or adenosine 5′-(β, γ-imido)triphosphate (AMPPNP) binding. These findings imply that the interfaces between NBD1 and ICL4 and between NBD2 and ICL2, respectively, remain relatively tight during the gating cycle (He et al. 2008). On the other hand, nuclear magnetic resonance (NMR) evidence indicates that ICL1 only binds to the PKA-phosphorylated state of mouse NBD1 (mNBD1) (phosphorylated on the RI and RE; see below) (Kanelis et al. 2010), suggesting that activation of CFTR by PKA does alter the NBD1–ICL1 interaction.
A critical point made by these models is that F508 forms part of the interface between NBD1 and ICL4. As discussed below, deletion of F508 has a limited effect on the ground state structure of NBD1 itself. The structure of the mutant NBD1 is the same as that of the WT, with the exception of altered surface properties in the vicinity of F508 and structural rearrangement of the F494–W496 loop. Deletion of F508 and the resulting change in NBD1 surface properties were hypothesized to disrupt the interaction with the ICL4 coupling helix that contains R1070. In F508del CFTR, mutation of V510, which is near the position of F508 in the WT, to aspartic acid partially corrects the CF defect and promotes CFTR maturation (Loo et al. 2010). The V510D mutation likely introduces a salt bridge with R1070, strengthening this interface. These data support the hypothesis that F508 disrupts this critical ICL4 interaction. Similar results are seen with an R1070W mutation, which might be expected to enhance the hydrophobic contacts at this interface or fill the void introduced by deletion of F508, or with a combined R1070D/V510R mutation, which would reverse the proposed salt bridge (Farinha et al. 2010; Loo et al. 2010). These additional mutations support the idea that the F508del phenotype can be attributed at least partially to a disruption of this interface, although as we discuss below, the V510D mutation also thermodynamically stabilizes NBD1 itself. The data on the V510D mutation indicate that interdomain interactions between NBD1 and ICL4 are required for proper folding and maturation of CFTR. Thus, restoring this interface via therapeutic compounds designed to enhance this interaction might be expected to partially correct the F508del defect. Examining the effects of stabilizing and destabilizing mutations at this interface may be a useful approach for exploring the dynamics of this interface through the gating cycle.
Molecular modeling of CFTR based on related ABC transporters, electron microscopy, cross-linking, and electrophysiology experiments have yielded significant insight into the overall organization and dynamics of full-length CFTR. Disruption of the NBD1–ICL4 interaction by F508del provides a partial answer for how the mutation disrupts processing and gating. Further explanation of how F508del disrupts CFTR processing arose from a closer look at the dynamics and thermodynamics of the isolated NBDs. CFTR’s processing efficiency has been shown to be related to the instability and aggregation tendencies of the NBDs. Deletion of F508, which is found in NBD1, further decreases the stability of NBD1. The R region, which is an important site of phosphodependent regulation, is also highly dynamic. The highly dynamic natures of the R region and NBD1 are evidence of a shallow energetic landscape, with an equilibrium between multiple accessible conformations that is sensitive to subtle sequence changes. As a result of this thermodynamic sensitivity, NBD1 dynamics and interactions are highly responsive to destabilizing and stabilizing mutations. The next section focuses on the complex dynamics of the R region and NBD1 and how these impact CFTR processing and function.
CFTR REGIONS AND DOMAINS
R Region
The R region is the principal physiological regulator of CFTR function, and its mechanism of action has been the focus of intense study. A dominant view is that the nonphosphorylated R region inhibits channel opening. This view is supported by evidence indicating that removal of portions of the R region encompassing residues 760–783 or 817–838 produces channels that open without activation by phosphorylation (Baldursson et al. 2001; Xie et al. 2002). Activation of the channel requires PKA phosphorylation of multiple serines (there are >10 sites), most of which are in the R region (Picciotto et al. 1992; Chappe et al. 2004), although there are also protein kinase C (PKC) (Picciotto et al. 1992; Chappe et al. 2004) and inhibitory adenosine monophosphate-stimulated kinase (AMPK) sites (King et al. 2009; Kongsuphol et al. 2009). Critically, there is no requirement for phosphorylation at any one specific site (Cheng et al. 1991; Rich et al. 1993). Phosphorylation at an increasing number of sites enhances the level of channel activation in a roughly additive manner (Chang et al. 1993; Rich et al. 1993; Wilkinson et al. 1997). The R region is disordered, as shown by NMR and circular dichroism experiments (Dulhanty and Riordan 1994; Ostedgaard et al. 2000; Baker et al. 2007), providing a key component in understanding the plasticity in its behavior.
As a disordered protein segment, the R region is highly dynamic and does not assume a single ground state conformation whether or not it is phosphorylated. Rather, NMR experiments indicate that the isolated R region rapidly exchanges between many different conformations (Baker et al. 2007). Individual segments of the R region can transiently populate secondary structural elements such as α-helices (Baker et al. 2007). Phosphorylation reduces the global α-helical content of the R region, which likely alters R-region binding preferences. R-region structural ensembles generated using discrete molecular dynamics (MD) simulations suggest that phosphorylation increases the R-region radius of gyration (Hegedus et al. 2008); however, NMR-based diffusion measurements indicate that phosphorylation actually decreases the hydrodynamic radius of the R region (Bozoky et al. 2010). Electron microscopy images provide evidence of a high-variability region of density adjacent to the ICDs that is not accounted for by the Sav1866 protein structure and suggests one possible location for a dynamic R region (Zhang et al. 2009). Electron microscopy images of nanogold-labeled CFTR also support a position of the R region near the ICDs.
R-region flexibility makes phosphorylation sites highly accessible and is likely to play an important role in its regulatory function. Disordered proteins often function as protein interaction hubs, which integrate protein inputs (Dunker et al. 2005). Besides interactions with PKA, PKC, and AMPK, the R region has been reported to interact with NBD1, NBD2, the disordered carboxy-terminal portion of CFTR, and the STAS domain of SLC26A3 in a dynamic fashion (Bozoky et al. 2010). NMR evidence for the interactions indicates that they are transient (lifetimes of less than milliseconds) and that multiple sections of the R region are involved. The R region may serve to integrate various cellular control parameters to enable generation of an appropriate level of channel response. A particularly important set of R interactions may be with the NBDs. The R region interacts with the NBDs and causes changes on multiple NBD1 surfaces, including some that overlap with the proposed NBD2 interaction interface. R-region phosphorylation was shown to decrease binding to NBD1 (Baker et al. 2007), possibly by reducing the population of NBD1-interacting α-helices. It is important to note that the data suggest that multiple portions of the R region interact with the NBDs in a dynamic fashion. No specific segments of the R region have been identified as critical for NBD1 interaction. This points to a model in which various segments of the nonphosphorylated R region bind to the NBD dimer interface in a transient manner and sterically inhibit formation of a productive heterodimer, thus preventing channel opening. Phosphorylation reduces R region–NBD interactions, facilitating dimer formation.
Another hypothesis is that the R region directly controls the position of the MSDs in the gating cycle (Hegedus et al. 2008), rather than exerting its regulatory effect through the NBDs. In this distance constraint model, the size of the R-region ensemble directly controls the physical separation of the MSDs through its amino-terminal tether to NBD1 and its carboxy-terminal tether to MSD2. Consistent with this, channel gating may not require NBD dimerization in and of itself (Cui et al. 2007; Wang et al. 2007, 2010b). Channels with NBD2 removed have been found to have a low open probability (Cui et al. 2007; Wang et al. 2007; Zhang et al. 2011). Electron microscopy positioning of the R region near to the ICDs is also consistent with this idea. Computer models of phosphorylated and nonphosphorylated R regions and their interactions with various binding partners based on NMR and small-angle X-ray scattering data are in progress to more clearly define the R-region conformational ensemble within the context of CFTR.
NBD1
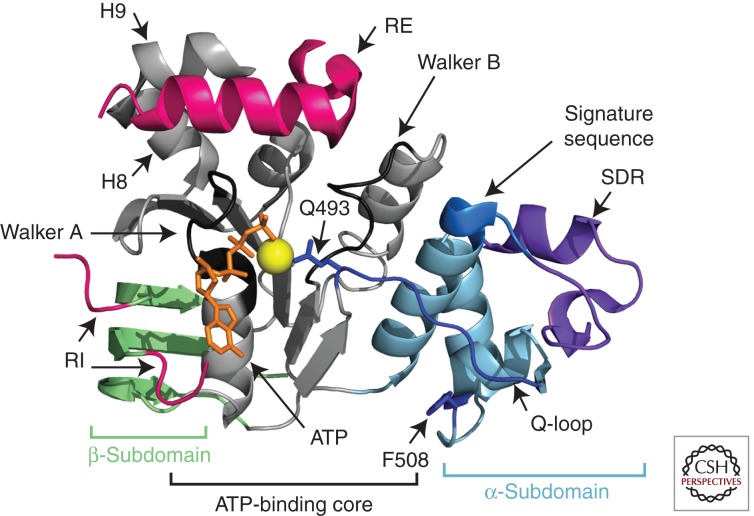
As NBD1 is the site of the dominant F508del mutation, this domain has been the subject of intensive structural, dynamic, and thermodynamic analyses. Crystal structures of NBD1 show that it is divided into the three basic regions seen in nucleotide-binding domains of other ABC transporters (Lewis et al. 2004, 2005; Thibodeau et al. 2005; Atwell et al. 2010): the β-sheet subdomain, the ATP-binding core, and the α-helical subdomain (Fig. 3). The ATP-binding core contains the Walker A and Walker B motifs and forms the primary contacts with ATP in the monomeric form. Two additional elements containing phosphorylation sites could be resolved in some structures: the regulatory extension (RE), which is formed by the first 30 residues of the R region and is alternatively classified as the carboxyl terminus of NBD1, and the regulatory insertion (RI), a 30-residue, largely disordered insertion between the first and second β-strands unique to CFTR’s NBD1. Analysis of several published NBD1 crystal structures reveals that F508del does not grossly perturb the structure of the ground state of the core NBD1 domain. It only causes significant structural changes near the location of the mutation (Lewis et al. 2004, 2005, 2010; Thibodeau et al. 2005; Atwell et al. 2010). Nevertheless, several lines of evidence suggest that the mutation does alter NBD1 dynamic modes or excited states.
Figure 3.
Ribbon diagram representation of CFTR NBD1. The ATP-binding core, α-subdomain, and β-subdomain are shown in gray, blue, and green, respectively. The regulatory extension (RE) and the amino and carboxyl termini of the regulatory insertion (RI) are shown in pink. The central portion of the RI is disordered and is not shown. ATP and magnesium are shown in orange and yellow, respectively. Other features, including the structurally diverse region (SDR), the Q-loop within the α-subdomain, and helices 8 and 9 (H8 and H9), are indicated by the labeled arrows. (PDB 2BBO [Lewis et al. 2010].)
The RI and RE show the most pronounced conformational variability in comparisons of the different crystal structures (Lewis et al. 2010), strongly suggesting that they are mobile. Residues 654–669 in the RE consistently form an α-helix, but the helix packs in different positions in the various crystal structures. One dominant interaction site is near the Walker A motif, an interaction that would inhibit NBD heterodimerization. Hydrogen/deuterium (H/D) exchange experiments show that the amide protons in this α-helix readily exchange in solution, indicating that the helix is unstable (Lewis et al. 2010). NMR-derived chemical shifts provide evidence that the preceding two helices (H8 and H9) also exchange with a coil conformation (Hudson et al. 2012). Crystal structures show short helices at both the amino and carboxyl termini of the RI; however, electron density is absent for the central portion, implying that this region of the RI is disordered. The two short helices at either end of the RI are also found in different orientations in the crystal structures, again implying mobility. The RI affects the geometry of ATP binding in some structures (Lewis et al. 2004). H/D exchange experiments confirm that amide protons in the RI are highly solvent-exposed, indicating that the terminal helices and interaction with ATP are not stable.
The RI and RE contain phosphoregulatory sites at residues 422, 660, and 670 (Townsend et al. 1996; Csanady et al. 2005). NMR spectral changes caused by phosphorylation of mNBD1 were shown to be similar to changes caused by removal of the RE, indicating that phosphorylation disrupts interactions between the NBD1 core and the RE (Kanelis et al. 2010). Phosphorylation also seems to disrupt the RI interaction with the NBD1 core, so that the RI appears to sample more disordered conformations. Kanelis et al. proposed a model in which the RE and RI form transient inhibitory interactions with the NBD1 core, much like the R region does. These inhibitory interactions reduce dimerization probability and are in turn inhibited by phosphorylation. Phosphorylation of the RI may also enhance ATP affinity by disrupting the RI’s interaction with the ATP-binding core, a possible regulatory mechanism. Deletion of F508 reduces the magnitude of the changes resulting from phosphorylation, suggesting that the phosphorylated RI and RE interact more strongly with the NBD1 core in the mutant (Kanelis et al. 2010). This strengthened interaction was hypothesized to reduce NBD heterodimerization propensity and channel open probability. It was also observed that F508del inhibits the interaction between ICL1 and the phosphorylated mNBD1. Collectively, these data suggest that F508del alters interactions between the regulatory regions and NBD1 in the absence of large structural changes to the ground state of NBD1, implying that properties other than the altered surface near F508 have been changed. These additional altered properties must result from changes in dynamic modes or excited states that are not observed in the static crystal structures.
NMR evidence also indicates that NBD1 is a highly dynamic protein with extensive motion in the millisecond-to-picosecond timescales. Even the ATP-binding core of the protein experiences millisecond-to-microsecond timescale dynamics, as shown by loss of NMR signal intensities (Chong and Forman-Kay 2010; Kanelis et al. 2010). The exchanging population in the ATP-binding core is likely to be small, as H/D exchange reveals that exchange in this region is relatively slow (Lewis et al. 2010). Analysis of crystallographic data indicates that the α-subdomain has variable orientations with respect to the ATP-binding core (up to 7° rotation), strongly suggesting a flexible linkage (Lewis et al. 2010). This was not unexpected, because the orientation of the α-subdomain with respect to the ATP-binding core has been shown to be nucleotide-dependent in other ABC transporters (Smith et al. 2002; Jones and George 2007, 2009). This rotation is postulated to play a critical role in the power stroke of ABC transporters (Hopfner et al. 2000; Smith et al. 2002), but its role in CFTR is unknown. Whether the CFTR α-subdomain reorients in response to nucleotide changes is also unknown. Conformational variability in the Q-loop, which links the ATP-binding core subdomain with the amino terminus of the α-subdomain, is not observed in the crystal structures, possibly because most of the structures have bound ATP or a nonhydrolyzable analog. One MD study indicates that deletion of F508 destabilizes the Q-loop, causing it to dissociate from the body of the domain, and leads to α-subdomain orientational freedom (Wieczorek and Zielenkiewicz 2008). Crystallographic conformational variability and/or high B-factors are observed for the 509–511 loop, a portion of the structurally diverse region (SDR residues 541–547) that likely interacts with NBD2, the Walker B loop (572–579), the α-helix following the H-loop, and the loop preceding the Walker B β-strand. High B-factors are observed for the final β-strand of the β-subdomain, which likely binds to the ICDs. Comparison of H/D exchange for WT and F508del indicates that the mutation results in a significant increase in H/D exchange rate in the sequence vicinity of the 508 mutation, extending from before residue 504 to beyond residue 517 (Lewis et al. 2010). Changes in the H/D exchange rate elsewhere in the domain are minimal, at least when Mg-ATP is bound to the domain. This is consistent with the loose packing of the F508 residue and the variable conformations for this region observed in the crystal structures. It is also consistent with the minimal structural changes observed in the ground state structure on deletion of F508.
Thus, two mechanisms explain how F508del negatively affects processing and gating. First, the mutation directly changes the ICL-interacting surface of NBD1, negatively affecting these interactions that are critical for processing and likely for gating as well. Second, F508del changes the dynamic and thermodynamic properties of NBD1, including enhanced interactions with the RI, the RE, and the R region. This second mechanism likely involves a change in the overall energetic landscape of NBD1 on deletion of F508, resulting in perturbed excited states and differential sampling of these excited states. Because of the changes in the energetic landscape, this second mechanism also relates to the folding and unfolding properties of NBD1. Several lines of evidence indicate that the change in the NBD1 energetic landscape is a key part of the explanation for the negative effects of F508del.
The nature of a subset of the suppressor mutations for the F508del defect, in particular, suggests that NBD1 stability is central to the defect. F508del causes a substantial, ∼6°C–7°C drop in thermal melting temperature of purified NBD1 (Protasevich et al. 2010). Many of the suppressor mutations, like G550E, R553Q, and R555K (Teem et al. 1993, 1996), which are relatively distant from the F508 position, increase NBD1 thermal stability and reverse some of the processing and functional defects, presumably without reverting the surface changes caused by deletion of F508. V510D, which may rescue F508del by facilitating the NBD1–ICL4 interaction, also stabilizes NBD1 (Protasevich et al. 2010), suggesting a dual mechanism of action. Recently, substitution of proline residues at key positions in the Q-loop, the SDR, and the RI in context of the I539T suppressor mutation has been shown to restore channel function and thermostability to full-length F508del CFTR. MD studies support a role for these proline mutations in reducing flexibility and increasing the temperatures of unfolding transitions for F508del NBD1 (Aleksandrov et al. 2012). Most convincingly, for a subset of suppressor mutants, a good correlation is observed between the improvement in NBD1 stability and CFTR processing efficiency (Thibodeau et al. 2010; Rabeh et al. 2011).
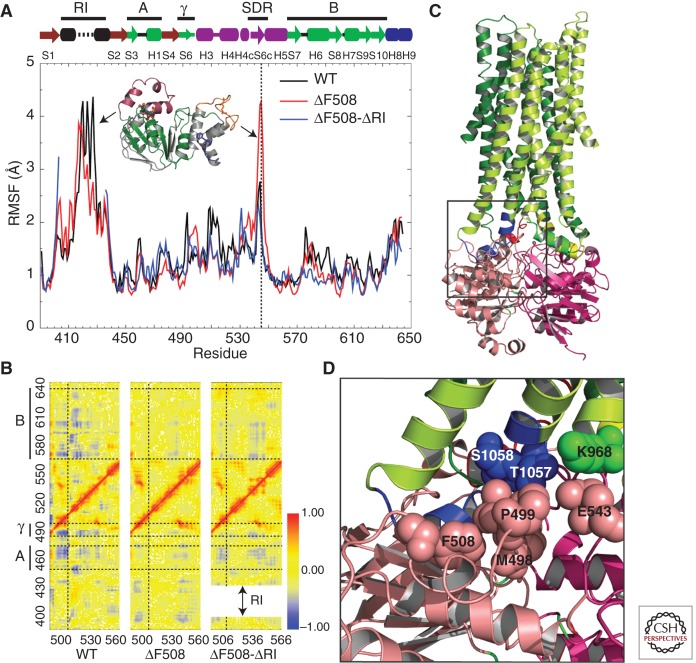
In addition to stabilizing point mutations, complete removal of the RI, which is relatively distant from the α-subdomain, also dramatically enhances the solution stability of NBD1 (Atwell et al. 2010). Furthermore, removal of the RI has been shown to suppress trafficking and gating defects associated with F508del (Aleksandrov et al. 2010). Not only does removal of RI improve solution stability of NBD1, it also restores function to F508del CFTR at physiologically relevant temperatures, as shown by iodide efflux and electrophysiological measurements (Aleksandrov et al. 2010). Partial removal of RI did not recapitulate this effect on NBD1 stability and restoration of function, indicating that the dynamics of RI (residues 401–436) plays an inhibitory role in CFTR function. As mentioned earlier, NBD1 crystal structures present only portions of the RI, and these in multiple conformations. The complete RI could not be resolved because of its intrinsic flexibility. Discrete MD simulations of NBD1 with a structural model for the RI revealed that the RI dynamically couples different regions of NBD1 (Fig. 4A,B) (Aleksandrov et al. 2010). Residue-wise root mean square fluctuation (RMSF) during the simulation reiterates that the RI is intrinsically flexible and dynamic (Fig. 4A). Most importantly, dynamic coupling exists between the F508-containing loop and the ATP-binding core subdomain of NBD1, which is lost on deletion of F508 (Fig. 4B). Such coupling is partially restored on deletion of the RI, indicating that the RI influences the dynamics of the F508-containing loop (Fig. 4B). Therefore, deletion of F508 could cause dynamic effects that influence the NBD1–ICL3–ICL4 interfaces and hence perturb the overall gating characteristics of the channel (Fig. 4C,D). Given that the structure of NBD1 with and without F508 is not significantly different, NBD1 dynamics likely play a crucial role in trafficking, maturation, and function of CFTR. Based on these results, one might hypothesize that conformational restriction of the RI region in NBD1 would improve maturation, trafficking, and function of F508del CFTR.
Figure 4.
CFTR NBD1 dynamics. (A) Root mean square fluctuations (RMSFs) of every residue in NBD1 as a function of time, obtained from MD simulations. The vertical dotted line indicates fluctuations of the SDR loop. Black arrows represent RMSF peaks for the corresponding structural regions in the plot. The ATP-binding core subdomain (A, G451–L475; B, D565–Q637) and γ-switch (Q493–P499) are colored green and labeled by the bold lines at the top for reference. (B) Pairwise Cα correlation map for residues 490–560 in comparison with those of all other regions in NBD1. Horizontal dotted lines separate the ATP-binding core subdomain (A and B) and γ-switch, as indicated by the bold lines on the left of the y-axis. The vertical dotted line corresponds to position F508. The shift in the y-axis of the F508del–ΔRI correlation map is a consequence of RI deletion. Red dots (correlation coefficient = 1) indicate residue pairs that move in concert in the same direction, and blue dots (correlation coefficient = −1) indicate residue pairs that move with opposite velocities all the time. (C) Structural model of full-length CFTR (http://dokhlab.unc.edu/research/CFTR/home.html). The area of the NBD1–ICL3–ICL4 interface is shown in the inset. (D) A detailed view of the location and orientation of the residues in the NBD1–ICL3–ICL4 interface from C. Residues F508, P499, M498, and E543 from NBD1 (salmon), S1058 and T1057 from ICL4 (blue), and K968 from ICL3 (cyan) are shown as spheres. (From Aleksandrov et al. 2010; reproduced, with permission.)
As we have suggested, changes to the NBD1 energetic landscape not only alter its interaction with regulatory regions, but they also affect its self-association. Extensive chemical and thermal denaturation studies monitored by a variety of biophysical techniques have identified an aggregation-prone, partially unfolded intermediate state on the unfolding pathway that is enhanced by F508del (Richardson et al. 2007; Protasevich et al. 2010; Wang et al. 2010a). F508del does not seem to affect unfolding of this partially unfolded intermediate, suggesting that the F508 region is already unfolded in the intermediate. F508 and many suppressor mutations are located in the α-helical subdomain, implying that the partially unfolded state involves this subdomain (Wang et al. 2010a). The RI is not located in or near the α-helical subdomain; however, it still may affect the conformation of this subdomain. One possible mechanism is that ATP affinity is enhanced on removal of the RI. ATP binding in turn may affect the α-helical subdomain orientation and stability via its interaction with the Q-loop. A second possibility is dynamic coupling between the RI and the SDR region as observed in MD simulations (Aleksandrov et al. 2010). Deletion of the RI stabilizes the SDR region in these MD studies and may impart an effect on NBD1 stability similar to α-helical subdomain suppressor mutations (Aleksandrov et al. 2010). The partially unfolded intermediate identified by the Hunt and Brouillette groups appears to be aggregation-prone and thus likely reduces both processing and gating efficiency. A partially unfolded configuration for the α-subdomain, which may be similar to that identified by Wang et al. (2010a), can be observed in MD simulations, suggesting that the dynamics of NBD1 can be accurately captured using simulations. In addition, MD folding simulations led to the conclusion that F508 deletion alters the kinetics of NBD1 folding (Serohijos et al. 2008b). These studies provide atomistic explanations for the experimentally observed decrease in NBD1 folding yields on deletion of F508 (Thibodeau et al. 2005). Different folding intermediates are observed and different transient contact patterns are formed for WT and F508del NBD1 in these simulations (Serohijos et al. 2008b). In this study, the H5–S6 loop and the S7–H5 loop near the α-subdomain–ATP-binding core interface were found to represent significant structural differences between WT and F508del NBD1. NMR dispersion experiments have also identified a weakly-populated excited state for WT NBD1 protein that is enhanced by F508del (Chong and Forman-Kay 2010). The relationship between the excited states observed by these different methods and groups is unclear, but collectively they support the notion that F508del profoundly perturbs the NBD1 energetic landscape.
The nature of some of the NBD1 excited states might be investigated by analysis of NBD1 fragments that have been found to be competent to bind ATP. An early report identified a 67-residue fragment comprising residues 450 through 516 of NBD1 that could bind to ATP with submillimolar affinity (Thomas et al. 1991). Subsequently, a fragment comprising residues 404–589 was shown to be capable of folding and binding to ATP with an affinity of ∼90 µM (Qu et al. 1997). This fragment is missing the first β-strand of NBD1 and the carboxy-terminal portion of the ATP-binding core. More recently, in vitro translation experiments monitored by fluorescence resonance energy transfer confirm that NBD1 residues 389 through 500, which comprise the β-subdomain and parts of the ATP-binding core, can fold autonomously into a subdomain that can bind to ATP (Khushoo et al. 2011). This study further showed that ATP stimulates proper folding of this fragment. The ATP-binding capacity of these NBD1 fragments suggests that these fragments can form folded structures. However, they are unlikely to assume the conformation observed in the ground state structures, as critical portions are missing in each instance. Thus, these fragments may form nonnative conformations that may be in equilibrium with the ground state conformation in the context of the full-length NBD1, providing insight into the accessible energy landscape.
In summary, NBD1 stability and changes in the energetic landscape of NBD1 on deletion of F508 are likely a critical part of the F508 defect. Suppression of the processing defect by stabilizing mutations and low-temperature rescue support the key role these changes in NBD1 dynamics play in the F508 defect. CFTR channel opening and closing has been shown to be tightly coupled to ATP binding and hydrolysis at the NBDs (Csanady et al. 2009). The challenge for the future will be to understand at a molecular level how the nucleotide-binding status of the NBDs, dimer conformation, and dynamics are allosterically linked to MSD conformation and channel gating. It will also be interesting to see how the absence of NBD2 will affect these linkages.
CONCLUDING REMARKS
CFTR shows a wide range of dynamics that enable proper folding and gating. The MSDs and ICDs likely rearrange in response to activation by PKA phosphorylation and in response to nucleotide binding by the NBDs, forming a closed state, an open-ready state, and an open state. The precise nature of these conformational changes awaits higher-resolution electron microscopy images and more channel-gating experiments building on already extensive studies. MSD interactions with NBD1 via the ICDs are required for proper folding and gating. Disruption of ICD and NBD1 interactions is one mechanism by which F508del disrupts folding and gating. F508del also appears to alter the transient and complex dynamic interactions between the NBD1 and the highly flexible, disordered phosphoregulatory elements of CFTR that include the R region, RI, and RE, further impairing channel gating. NMR data-based computational models of these regulatory interactions will be useful for understanding exactly how the phosphoregulatory elements and their regulatory interactions are changed by phosphorylation and how they are affected by deletion of F508. NBD1 itself is highly dynamic. Although F508del causes only minor changes to the NBD1 ground state structure, it alters the stability, folding pathway, and binding properties of this marginally stable domain. A variety of data indicate that F508del changes the energy landscape for this domain and alters the frequency with which higher energy states are accessed. The dynamics and thermodynamics of NBD1 remain exciting areas of study, both because changes in NBD1 dynamics have been directly linked to misprocessing, instability, and channel-gating defects and because some of these higher energy states may have functional relevance. As such, information about dynamics will be key for building a model to describe how the R region and NBDs allosterically control the MSDs and channel activity. In addition, knowledge of the dynamics involved in domain and full-length CFTR folding and interactions with cellular processing machinery will be critical for mechanistic understanding of the F508del defects.
ACKNOWLEDGMENTS
The authors thank Drs. John R. Riordan, Adrian W.R. Serohijos, Tamas Hegedus, Lihua He, Andrei Aleksandrov, Rhea Hudson, Jennifer Dawson, Robert Vernon, and Zoltán Bozóky for valuable discussions. In addition, funding from the Cystic Fibrosis Foundation Therapeutics and Cystic Fibrosis Canada to J.D.F.-K. and from the National Institutes of Health (R01GM080742) to N.V.D. is gratefully acknowledged.
Footnotes
Editors: John R. Riordan, Richard C. Boucher, and Paul M. Quinton
Additional Perspectives on Cystic Fibrosis available at www.perspectivesinmedicine.org
REFERENCES
- Aleksandrov AA, Kota P, Aleksandrov LA, He L, Jensen T, Cui L, Gentzsch M, Dokholyan NV, Riordan JR 2010. Regulatory insertion removal restores maturation, stability and function of ΔF508 CFTR. J Mol Biol 401: 194–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aleksandrov AA, Kota P, Cui L, Jensen T, Alekseev AE, Reyes S, He L, Gentzsch M, Aleksandrov LA, Dokholyan NV, et al. 2012. Allosteric modulation balances stability and restores function of ΔF508 CFTR. J Mol Biol 419: 41–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander C, Ivetac A, Liu X, Norimatsu Y, Serrano JR, Landstrom A, Sansom M, Dawson DC 2009. Cystic fibrosis transmembrane conductance regulator: Using differential reactivity toward channel-permeant and channel-impermeant thiol-reactive probes to test a molecular model for the pore. Biochemistry 48: 10078–10088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson MP, Gregory RJ, Thompson S, Souza DW, Paul S, Mulligan RC, Smith AE, Welsh MJ 1991. Demonstration that CFTR is a chloride channel by alteration of its anion selectivity. Science 253: 202–205 [DOI] [PubMed] [Google Scholar]
- Atwell S, Brouillette CG, Conners K, Emtage S, Gheyi T, Guggino WB, Hendle J, Hunt JF, Lewis HA, Lu F, et al. 2010. Structures of a minimal human CFTR first nucleotide-binding domain as a monomer, head-to-tail homodimer, and pathogenic mutant. Protein Eng Des Sel 23: 375–384 [DOI] [PubMed] [Google Scholar]
- Awayn NH, Rosenberg MF, Kamis AB, Aleksandrov LA, Riordan JR, Ford RC 2005. Crystallographic and single-particle analyses of native- and nucleotide-bound forms of the cystic fibrosis transmembrane conductance regulator (CFTR) protein. Biochem Soc Trans 33: 996–999 [DOI] [PubMed] [Google Scholar]
- Baker JM, Hudson RP, Kanelis V, Choy WY, Thibodeau PH, Thomas PJ, Forman-Kay JD 2007. CFTR regulatory region interacts with NBD1 predominantly via multiple transient helices. Nat Struct Mol Biol 14: 738–745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldursson O, Ostedgaard LS, Rokhlina T, Cotten JF, Welsh MJ 2001. Cystic fibrosis transmembrane conductance regulator Cl– channels with R domain deletions and translocations show phosphorylation-dependent and -independent activity. J Biol Chem 276: 1904–1910 [DOI] [PubMed] [Google Scholar]
- Berger HA, Travis SM, Welsh MJ 1993. Regulation of the cystic fibrosis transmembrane conductance regulator Cl– channel by specific protein kinases and protein phosphatases. J Biol Chem 268: 2037–2047 [PubMed] [Google Scholar]
- Bozoky Z, Baker JM, Thomas PJ, Bear CE, Forman-Kay JD 2010. The regulatory R region of CFTR serves as a dynamic integrator. Pediatr Pulmonol 45 (Suppl): 220 [Google Scholar]
- Chang XB, Tabcharani JA, Hou YX, Jensen TJ, Kartner N, Alon N, Hanrahan JW, Riordan JR 1993. Protein kinase A (PKA) still activates CFTR chloride channel after mutagenesis of all 10 PKA consensus phosphorylation sites. J Biol Chem 268: 11304–11311 [PubMed] [Google Scholar]
- Chappe V, Hinkson DA, Howell LD, Evagelidis A, Liao J, Chang XB, Riordan JR, Hanrahan JW 2004. Stimulatory and inhibitory protein kinase C consensus sequences regulate the cystic fibrosis transmembrane conductance regulator. Proc Natl Acad Sci 101: 390–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng SH, Gregory RJ, Marshall J, Paul S, Souza DW, White GA, O’Riordan CR, Smith AE 1990. Defective intracellular transport and processing of CFTR is the molecular basis of most cystic fibrosis. Cell 63: 827–834 [DOI] [PubMed] [Google Scholar]
- Cheng SH, Rich DP, Marshall J, Gregory RJ, Welsh MJ, Smith AE 1991. Phosphorylation of the R domain by cAMP-dependent protein kinase regulates the CFTR chloride channel. Cell 66: 1027–1036 [DOI] [PubMed] [Google Scholar]
- Cheung JC, Kim Chiaw P, Pasyk S, Bear CE 2008. Molecular basis for the ATPase activity of CFTR. Arch Biochem Biophys 476: 95–100 [DOI] [PubMed] [Google Scholar]
- Chong PA, Forman-Kay JD 2010. Alteration of CFTR NBD1 dynamics upon deletion of F508 underlying the basic defect. Pediatr Pulmonol 45 (Suppl): 240 [Google Scholar]
- Csanady L, Chan KW, Nairn AC, Gadsby DC 2005. Functional roles of nonconserved structural segments in CFTR’s NH2-terminal nucleotide binding domain. J Gen Physiol 125: 43–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csanady L, Vergani P, Gadsby DC 2009. Strict coupling between CFTR’s catalytic cycle and gating of its Cl– ion pore revealed by distributions of open channel burst durations. Proc Natl Acad Sci 107: 1241–1246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui L, Aleksandrov L, Chang XB, Hou YX, He L, Hegedus T, Gentzsch M, Aleksandrov A, Balch WE, Riordan JR 2007. Domain interdependence in the biosynthetic assembly of CFTR. J Mol Biol 365: 981–994 [DOI] [PubMed] [Google Scholar]
- Dalemans W, Barbry P, Champigny G, Jallat S, Dott K, Dreyer D, Crystal RG, Pavirani A, Lecocq JP, Lazdunski M 1991. Altered chloride ion channel kinetics associated with the ΔF508 cystic fibrosis mutation. Nature 354: 526–528 [DOI] [PubMed] [Google Scholar]
- Dawson RJ, Locher KP 2006. Structure of a bacterial multidrug ABC transporter. Nature 443: 180–185 [DOI] [PubMed] [Google Scholar]
- Denning GM, Anderson MP, Amara JF, Marshall J, Smith AE, Welsh MJ 1992. Processing of mutant cystic fibrosis transmembrane conductance regulator is temperature-sensitive. Nature 358: 761–764 [DOI] [PubMed] [Google Scholar]
- Drumm ML, Wilkinson DJ, Smit LS, Worrell RT, Strong TV, Frizzell RA, Dawson DC, Collins FS 1991. Chloride conductance expressed by ΔF508 and other mutant CFTRs in Xenopus oocytes. Science 254: 1797–1799 [DOI] [PubMed] [Google Scholar]
- Du K, Sharma M, Lukacs GL 2005. The ΔF508 cystic fibrosis mutation impairs domain-domain interactions and arrests post-translational folding of CFTR. Nat Struct Mol Biol 12: 17–25 [DOI] [PubMed] [Google Scholar]
- Dulhanty AM, Riordan JR 1994. Phosphorylation by cAMP-dependent protein kinase causes a conformational change in the R domain of the cystic fibrosis transmembrane conductance regulator. Biochemistry 33: 4072–4079 [DOI] [PubMed] [Google Scholar]
- Dunker AK, Cortese MS, Romero P, Iakoucheva LM, Uversky VN 2005. Flexible nets. The roles of intrinsic disorder in protein interaction networks. FEBS J 272: 5129–5148 [DOI] [PubMed] [Google Scholar]
- Farinha CM, Da Paula A, Amaral MD 2010. Rescuing F508del-CFTR by genetic revertants, low temperature and small molecules. Pediatr Pulmonol 45 (Suppl): 23020146367 [Google Scholar]
- Fatehi M, Linsdell P 2008. State-dependent access of anions to the cystic fibrosis transmembrane conductance regulator chloride channel pore. J Biol Chem 283: 6102–6109 [DOI] [PubMed] [Google Scholar]
- He L, Aleksandrov AA, Serohijos AW, Hegedus T, Aleksandrov LA, Cui L, Dokholyan NV, Riordan JR 2008. Multiple membrane-cytoplasmic domain contacts in the cystic fibrosis transmembrane conductance regulator (CFTR) mediate regulation of channel gating. J Biol Chem 283: 26383–26390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L, Aleksandrov LA, Cui L, Jensen TJ, Nesbitt KL, Riordan JR 2010. Restoration of domain folding and interdomain assembly by second-site suppressors of the ΔF508 mutation in CFTR. FASEB J 24: 3103–3112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegedus T, Serohijos AW, Dokholyan NV, He L, Riordan JR 2008. Computational studies reveal phosphorylation-dependent changes in the unstructured R domain of CFTR. J Mol Biol 378: 1052–1063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoelen H, Kleizen B, Schmidt A, Richardson J, Charitou P, Thomas PJ, Braakman I 2010. The primary folding defect and rescue of ΔF508 CFTR emerge during translation of the mutant domain. PLoS ONE 5: e15458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollenstein K, Frei DC, Locher KP 2007. Structure of an ABC transporter in complex with its binding protein. Nature 446: 213–216 [DOI] [PubMed] [Google Scholar]
- Hopfner KP, Karcher A, Shin DS, Craig L, Arthur LM, Carney JP, Tainer JA 2000. Structural biology of Rad50 ATPase: ATP-driven conformational control in DNA double-strand break repair and the ABC-ATPase superfamily. Cell 101: 789–800 [DOI] [PubMed] [Google Scholar]
- Huang SY, Bolser D, Liu HY, Hwang TC, Zou X 2009. Molecular modeling of the heterodimer of human CFTR’s nucleotide-binding domains using a protein–protein docking approach. J Mol Graph Model 27: 822–828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson RP, Chong PA, Protasevich II, Vernon R, Noy E, Bihler H, Li AJ, Kalid O, Sela-Culang I, Mense M, et al. 2012. Conformational changes relevant to channel activity and folding within the first nucleotide binding domain of the cystic fibrosis transmembrane conductance regulator. J Biol Chem 287: 28480–28494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jardetzky O 1966. Simple allosteric model for membrane pumps. Nature 211: 969–970 [DOI] [PubMed] [Google Scholar]
- Jones PM, George AM 2007. Nucleotide-dependent allostery within the ABC transporter ATP-binding cassette: A computational study of the MJ0796 dimer. J Biol Chem 282: 22793–22803 [DOI] [PubMed] [Google Scholar]
- Jones PM, George AM 2009. Opening of the ADP-bound active site in the ABC transporter ATPase dimer: Evidence for a constant contact, alternating sites model for the catalytic cycle. Proteins 75: 387–396 [DOI] [PubMed] [Google Scholar]
- Kanelis V, Hudson RP, Thibodeau PH, Thomas PJ, Forman-Kay JD 2010. NMR evidence for differential phosphorylation-dependent interactions in WT and ΔF508 CFTR. EMBO J 29: 263–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khushoo A, Yang Z, Johnson AE, Skach WR 2011. Ligand-driven vectorial folding of ribosome-bound human CFTR NBD1. Mol Cell 41: 682–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King JD Jr, Fitch AC, Lee JK, McCane JE, Mak DO, Foskett JK, Hallows KR 2009. AMP-activated protein kinase phosphorylation of the R domain inhibits PKA stimulation of CFTR. Am J Physiol Cell Physiol 297: C94–C101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleizen B, van Vlijmen T, de Jonge HR, Braakman I 2005. Folding of CFTR is predominantly cotranslational. Mol Cell 20: 277–287 [DOI] [PubMed] [Google Scholar]
- Kongsuphol P, Cassidy D, Hieke B, Treharne KJ, Schreiber R, Mehta A, Kunzelmann K 2009. Mechanistic insight into control of CFTR by AMPK. J Biol Chem 284: 5645–5653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis HA, Buchanan SG, Burley SK, Conners K, Dickey M, Dorwart M, Fowler R, Gao X, Guggino WB, Hendrickson WA, et al. 2004. Structure of nucleotide-binding domain 1 of the cystic fibrosis transmembrane conductance regulator. EMBO J 23: 282–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis HA, Zhao X, Wang C, Sauder JM, Rooney I, Noland BW, Lorimer D, Kearins MC, Conners K, Condon B, et al. 2005. Impact of the ΔF508 mutation in first nucleotide-binding domain of human cystic fibrosis transmembrane conductance regulator on domain folding and structure. J Biol Chem 280: 1346–1353 [DOI] [PubMed] [Google Scholar]
- Lewis HA, Wang C, Zhao X, Hamuro Y, Conners K, Kearins MC, Lu F, Sauder JM, Molnar KS, Coales SJ, et al. 2010. Structure and dynamics of NBD1 from CFTR characterized using crystallography and hydrogen/deuterium exchange mass spectrometry. J Mol Biol 396: 406–430 [DOI] [PubMed] [Google Scholar]
- Loo TW, Bartlett MC, Clarke DM 2010. The V510D suppressor mutation stabilizes ΔF508-CFTR at the cell surface. Biochemistry 49: 6352–6357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukacs GL, Mohamed A, Kartner N, Chang XB, Riordan JR, Grinstein S 1994. Conformational maturation of CFTR but not its mutant counterpart (ΔF508) occurs in the endoplasmic reticulum and requires ATP. EMBO J 13: 6076–6086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell P 1957. A general theory of membrane transport from studies of bacteria. Nature 180: 134–136 [DOI] [PubMed] [Google Scholar]
- Moran O 2010. Model of the cAMP activation of chloride transport by CFTR channel and the mechanism of potentiators. J Theor Biol 262: 73–79 [DOI] [PubMed] [Google Scholar]
- Mornon JP, Lehn P, Callebaut I 2008. Atomic model of human cystic fibrosis transmembrane conductance regulator: Membrane-spanning domains and coupling interfaces. Cell Mol Life Sci 65: 2594–2612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mornon JP, Lehn P, Callebaut I 2009. Molecular models of the open and closed states of the whole human CFTR protein. Cell Mol Life Sci 66: 3469–3486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostedgaard LS, Baldursson O, Vermeer DW, Welsh MJ, Robertson AD 2000. A functional R domain from cystic fibrosis transmembrane conductance regulator is predominantly unstructured in solution. Proc Natl Acad Sci 97: 5657–5662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picciotto MR, Cohn JA, Bertuzzi G, Greengard P, Nairn AC 1992. Phosphorylation of the cystic fibrosis transmembrane conductance regulator. J Biol Chem 267: 12742–12752 [PubMed] [Google Scholar]
- Pinkett HW, Lee AT, Lum P, Locher KP, Rees DC 2007. An inward-facing conformation of a putative metal-chelate-type ABC transporter. Science 315: 373–377 [DOI] [PubMed] [Google Scholar]
- Protasevich I, Yang Z, Wang C, Atwell S, Zhao X, Emtage S, Wetmore D, Hunt JF, Brouillette CG 2010. Thermal unfolding studies show the disease causing F508del mutation in CFTR thermodynamically destabilizes nucleotide-binding domain 1. Protein Sci 19: 1917–1931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu BH, Strickland EH, Thomas PJ 1997. Localization and suppression of a kinetic defect in cystic fibrosis transmembrane conductance regulator folding. J Biol Chem 272: 15739–15744 [DOI] [PubMed] [Google Scholar]
- Rabeh WM, Di Bernardo S, Okiyoneda T, Liu Y, Mulvihill CM, Du K, Xu H, Konermann L, Lukacs G, Bossard F 2011. Conformational stabilization of the NBD1 is necessary, but not sufficient for CFTR biogenesis. Pediatr Pulmonol 45 (Suppl): 220 [Google Scholar]
- Rich DP, Berger HA, Cheng SH, Travis SM, Saxena M, Smith AE, Welsh MJ 1993. Regulation of the cystic fibrosis transmembrane conductance regulator Cl– channel by negative charge in the R domain. J Biol Chem 268: 20259–20267 [PubMed] [Google Scholar]
- Richardson JM, Thibodeau PH, Watson J, Thomas PJ 2007. Identification of a non-native state of NBD1 that is affected by ΔF508. Pediatr Pulmonol 45 (Suppl): 203 [Google Scholar]
- Riordan JR, Rommens JM, Kerem B, Alon N, Rozmahel R, Grzelczak Z, Zielenski J, Lok S, Plavsic N, Chou J-L, et al. 1989. Identification of the cystic fibrosis gene: Cloning and characterization of complementary DNA. Science 245: 1066–1073 [DOI] [PubMed] [Google Scholar]
- Rosenberg MF, Kamis AB, Aleksandrov LA, Ford RC, Riordan JR 2004. Purification and crystallization of the cystic fibrosis transmembrane conductance regulator (CFTR). J Biol Chem 279: 39051–39057 [DOI] [PubMed] [Google Scholar]
- Serohijos AW, Hegedus T, Aleksandrov AA, He L, Cui L, Dokholyan NV, Riordan JR 2008a. Phenylalanine-508 mediates a cytoplasmic-membrane domain contact in the CFTR 3D structure crucial to assembly and channel function. Proc Natl Acad Sci 105: 3256–3261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serohijos AW, Hegedus T, Riordan JR, Dokholyan NV 2008b. Diminished self-chaperoning activity of the ΔF508 mutant of CFTR results in protein misfolding. PLoS Comput Biol 4: e1000008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith PC, Karpowich N, Millen L, Moody JE, Rosen J, Thomas PJ, Hunt JF 2002. ATP binding to the motor domain from an ABC transporter drives formation of a nucleotide sandwich dimer. Mol Cell 10: 139–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teem JL, Berger HA, Ostedgaard LS, Rich DP, Tsui LC, Welsh MJ 1993. Identification of revertants for the cystic fibrosis ΔF508 mutation using STE6-CFTR chimeras in yeast. Cell 73: 335–346 [DOI] [PubMed] [Google Scholar]
- Teem JL, Carson MR, Welsh MJ 1996. Mutation of R555 in CFTR-ΔF508 enhances function and partially corrects defective processing. Receptors Channels 4: 63–72 [PubMed] [Google Scholar]
- Thibodeau PH, Brautigam CA, Machius M, Thomas PJ 2005. Side chain and backbone contributions of Phe508 to CFTR folding. Nat Struct Mol Biol 12: 10–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thibodeau PH, Richardson JM III, Wang W, Millen L, Watson J, Mendoza JL, Du K, Fischman S, Senderowitz H, Lukacs GL, et al. 2010. The cystic fibrosis-causing mutation ΔF508 affects multiple steps in cystic fibrosis transmembrane conductance regulator biogenesis. J Biol Chem 285: 35825–35835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas PJ, Shenbagamurthi P, Ysern X, Pedersen PL 1991. Cystic fibrosis transmembrane conductance regulator: Nucleotide binding to a synthetic peptide. Science 251: 555–557 [DOI] [PubMed] [Google Scholar]
- Townsend RR, Lipniunas PH, Tulk BM, Verkman AS 1996. Identification of protein kinase A phosphorylation sites on NBD1 and R domains of CFTR using electrospray mass spectrometry with selective phosphate ion monitoring. Protein Sci 5: 1865–1873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varga K, Jurkuvenaite A, Wakefield J, Hong JS, Guimbellot JS, Venglarik CJ, Niraj A, Mazur M, Sorscher EJ, Collawn JF, et al. 2004. Efficient intracellular processing of the endogenous cystic fibrosis transmembrane conductance regulator in epithelial cell lines. J Biol Chem 279: 22578–22584 [DOI] [PubMed] [Google Scholar]
- Wang W, Bernard K, Li G, Kirk KL 2007. Curcumin opens cystic fibrosis transmembrane conductance regulator channels by a novel mechanism that requires neither ATP binding nor dimerization of the nucleotide-binding domains. J Biol Chem 282: 4533–4544 [DOI] [PubMed] [Google Scholar]
- Wang C, Protasevich I, Yang Z, Seehausen D, Skalak T, Zhao X, Atwell S, Spencer Emtage J, Wetmore DR, Brouillette CG, et al. 2010a. Integrated biophysical studies implicate partial unfolding of NBD1 of CFTR in the molecular pathogenesis of F508del cystic fibrosis. Protein Sci 19: 1932–1947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Wu J, Bernard K, Li G, Wang G, Bevensee MO, Kirk KL 2010b. ATP-independent CFTR channel gating and allosteric modulation by phosphorylation. Proc Natl Acad Sci 107: 3888–3893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward CL, Kopito RR 1994. Intracellular turnover of cystic fibrosis transmembrane conductance regulator. Inefficient processing and rapid degradation of wild-type and mutant proteins. J Biol Chem 269: 25710–25718 [PubMed] [Google Scholar]
- Ward A, Reyes CL, Yu J, Roth CB, Chang G 2007. Flexibility in the ABC transporter MsbA: Alternating access with a twist. Proc Natl Acad Sci 104: 19005–19010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieczorek G, Zielenkiewicz P 2008. ΔF508 mutation increases conformational flexibility of CFTR protein. J Cyst Fibros 7: 295–300 [DOI] [PubMed] [Google Scholar]
- Wilkinson DJ, Strong TV, Mansoura MK, Wood DL, Smith SS, Collins FS, Dawson DC 1997. CFTR activation: Additive effects of stimulatory and inhibitory phosphorylation sites in the R domain. Am J Physiol 273: L127–L133 [DOI] [PubMed] [Google Scholar]
- Winter MC, Sheppard DN, Carson MR, Welsh MJ 1994. Effect of ATP concentration on CFTR Cl– channels: A kinetic analysis of channel regulation. Biophys J 66: 1398–1403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie J, Adams LM, Zhao J, Gerken TA, Davis PB, Ma J 2002. A short segment of the R domain of cystic fibrosis transmembrane conductance regulator contains channel stimulatory and inhibitory activities that are separable by sequence modification. J Biol Chem 277: 23019–23027 [DOI] [PubMed] [Google Scholar]
- Zhang ZR, Song B, McCarty NA 2005. State-dependent chemical reactivity of an engineered cysteine reveals conformational changes in the outer vestibule of the cystic fibrosis transmembrane conductance regulator. J Biol Chem 280: 41997–42003 [DOI] [PubMed] [Google Scholar]
- Zhang L, Aleksandrov LA, Zhao Z, Birtley JR, Riordan JR, Ford RC 2009. Architecture of the cystic fibrosis transmembrane conductance regulator protein and structural changes associated with phosphorylation and nucleotide binding. J Struct Biol 167: 242–251 [DOI] [PubMed] [Google Scholar]
- Zhang L, Aleksandrov LA, Riordan JR, Ford RC 2011. Domain location within the cystic fibrosis transmembrane conductance regulator protein investigated by electron microscopy and gold labelling. Biochim Biophys Acta 1808: 399–404 [DOI] [PubMed] [Google Scholar]