Abstract
The setup of synthetic biological systems involving millions of bases is still limited by the required high quality of synthetic DNA. Important drivers to further open up the field are the accuracy and scale of chemical DNA synthesis and the downstream processing of longer DNA assembled from short fragments. We developed a new, highly parallel and miniaturized method for the preparation of high quality DNA termed “Megacloning” by using Next Generation Sequencing (NGS) technology in a preparative way. We demonstrate our method by processing both conventional and microarray-derived DNA oligonucleotides in combination with a bead-based high throughput pyrosequencing platform, gaining a 500-fold error reduction for microarray oligonucleotides in a first embodiment. We also show the assembly of synthetic genes as part of the Megacloning process. In principle, up to millions of DNA fragments can be sequenced, characterized and sorted in a single Megacloner run, enabling many new applications.
Current de novo gene construction1–4 rests on chemical oligonucleotide synthesis of the 1990s retaining error rates of 1 error in 300 base pairs and significant costs. Errors are typically avoided using manual selection of the best Sanger sequences using electrophoretic automation.
Recent innovations in programmable array technology5–8 offer the possibility to synthesize pools of thousands to millions of sequences per array with lengths comparable to conventional synthesis. The technology thus provides an extremely rich source of DNA oligonucleotides with great flexibility and superior efficiency regarding throughput and cost per basepair. However, the error rate of microarray-derived oligonucleotides is typically higher compared to conventional synthesis, making error avoidance or correction necessary. Furthermore it is challenging to divide the derived oligonucleotide pools, containing vast amounts of species, into sub pools -- necessary, for example, to guide the assembly of synthetic genes, chromosomal regions or whole pathways in synthetic biology.
The new method described here, termed “Megacloning”, turns NGS from a previously purely analytical method into a preparative tool, and represents a tremendous source of sequence-verified DNA where the yield from one NGS run is equivalent to that from hundreds to thousands of Sanger-sequence runs. It therefore addresses the challenge of error reduction for both conventional and microarray-derived DNA oligonucleotides. The output of the method are high quality DNA libraries containing “perfect parts” with desired and correct sequences in adjustable ratios useful for a wide range of (bio-)technological applications.
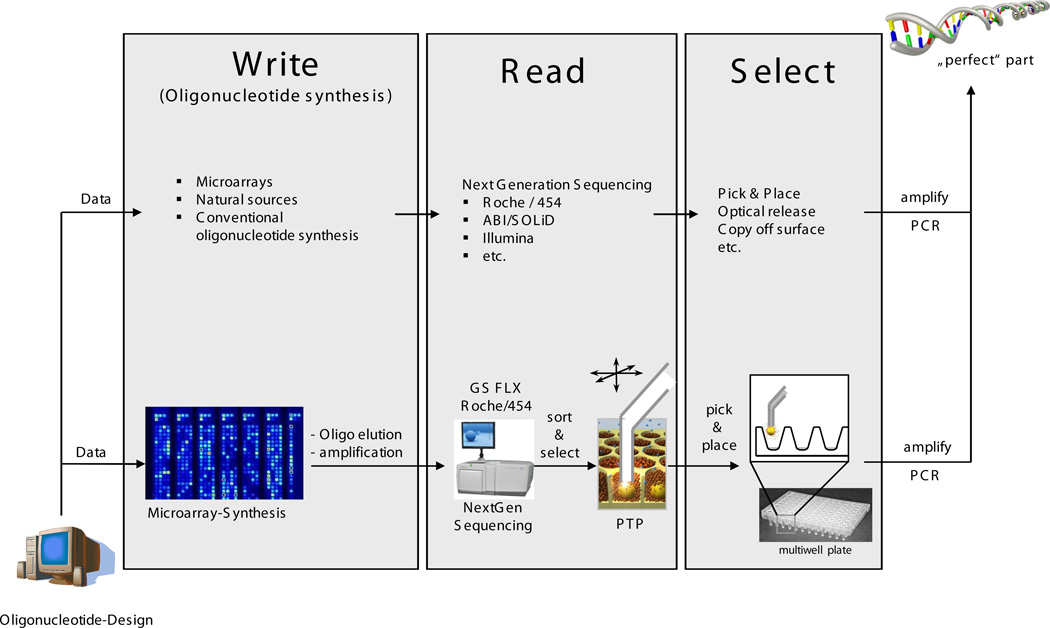
Here we present a proof-of-concept study aimed at the retrieval of clonal DNA with known sequence from an NGS platform post-sequencing (Fig.1). The workflow comprises input DNA of short length, an NGS run to generate sequence verified DNA clones, the localization of DNA with desired sequence on the sequencers substrate and finally the subsequent retrieval of the clones of choice. The sources for the input DNA are fairly independent of the Megacloning step. For the present work, input DNA was derived from conventional oligonucleotide synthesis and from DNA microarrays. The NGS platform used was the GS FLX device from Roche 454 Life Sciences9,10. Due to its open-top architecture, accessibility of the beads and the bead size, this platform is well suited for a pick-and-place approach using micropipettes to retrieve beads specifically from the 454-Picotiterplate (PTP) and transfer them into conventional multi-well plates for further processing.
Figure 1.
Strategy overview. The general approach includes DNA from a variety of sources. After Next Generation Sequencing the DNA will be sorted and retrieved selectively whereas the technologies used depend on the NGS platform. The particular approach described here includes microarrays as well as conventional sources of oligonucleotides. For sequencing prior sorting and selection the GS FLX platform (454/Roche) was used.
First we established a technical setup for the controlled extraction of beads. The PTP at this stage contained a "natural" sample, and extraction was done using a micropipette controlled by a micro actuator device (supplementary Fig.1 and 2 online). To assess the fidelity of our setup we compared the reads coming from the GS FLX platform with Sanger-derived sequences of DNA amplified from extracted beads. The alignment of Sanger sequences to the NGS reads matched 99.9%. Only two mismatches were obtained in 2,410 basepairs. Both were putative insertions in the GS FLX reads occurring at homopolymer stretches and therefore have a high likelihood of being platform-specific base calling artefacts9 (supplementary note online).
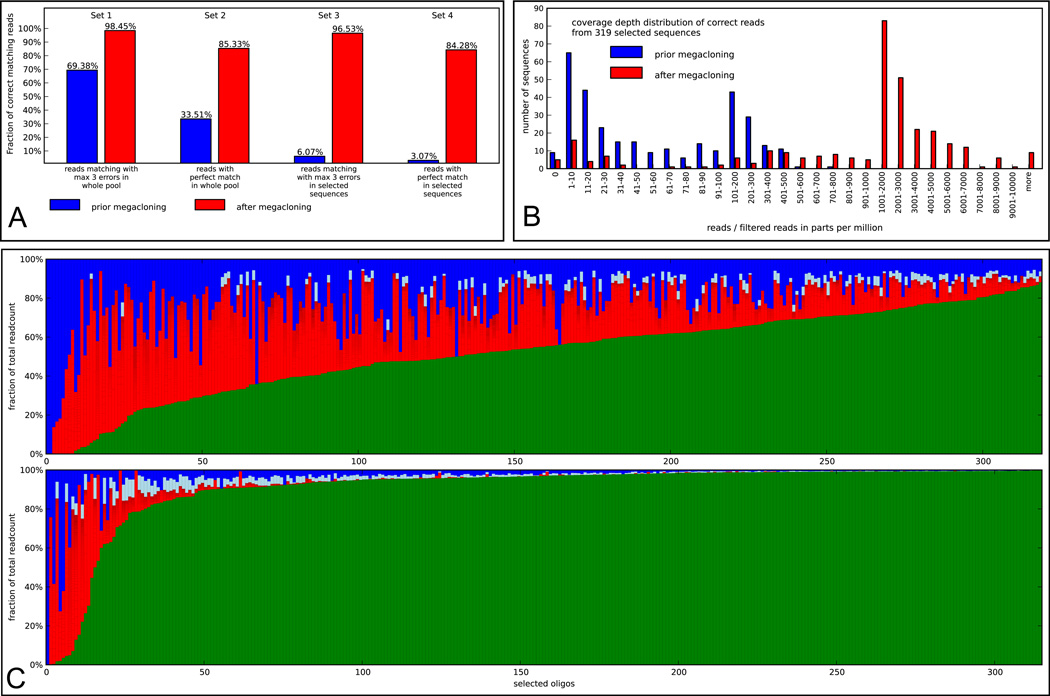
Next we collected a set of 319 beads with DNA clones from a microarray derived pool containing initially 3918 sequences. The beads for extraction were selected to ensure that their GS FLX reads matched perfectly to sequences in our starting pool. The obtained DNA and the untreated pool were compared after being sequenced independently on a Genome Analyzer II (Illumina GAII). 3.1 % of reads from the initial (non-enriched) DNA pool mapped without errors to the set of 319 selected sequences. In the enriched pool the fraction of reads mapping perfectly to the target sequences was 84.3 %. The increase by factor 27.2 shows clearly a successful enrichment of selected and correct sequences (Fig.2A and 2B). Also the analysis of reads on the level of single target sequences shows that for 94 % of the sequences in the selected pool 50% or more of the reads were correct (Fig. 2C). Error prone sequences contained a high number of different species likely to be caused by known sequence variations on the GAII as reported previously11.
Figure 2.
(A) Comparison of the initial microarray oligonucleotide pool (blue) and the pool enriched with the Megacloner technology (red) based on the results of the Illumina GAII runs. The bars in Set 1 represent the fraction of reads that could be mapped allowing up to three errors, bars in Set 2 show the fractions of perfectly matching reads to the sequence set of the initial pool (3918 sequences). Differences between the blue and the red bar in Set 2 represent the enrichment of correct sequences by Megacloning. The bars in Set 3 and Set 4 show the fractions of reads mapping to sequences from the selected pool (319 sequences). Differences between blue and red bars in Set 3 show the enrichment of selected 319 sequences prior vs. after Megacloning, blue and red bars in Set 4 represent the enrichment of sequences which are in the set of 319 selected sequences and which are correct. (B) Histogram of read counts in the Illumina GAII data of the initial pool (blue) and the enriched Megacloned sample (red). Only reads mapping without errors to one of the 319 selected target sequences have been taken into account. To compare the two NGS runs on the basis of read counts the numbers have been converted into parts-per-million-units (ppm) taking the number of filtered reads as basis. (C) Composition of reads from the Illumina GAII data including 319 selected sequences in the initial pool (top) and the enriched pool (bottom). The oligonucleotides are sorted by the fraction of correct reads. Green: correct reads, red: error prone reads (compartments in the red bars represent single sequences with a readcount of 0.1% or more of total reads for the particular sequence). Light blue: sum of non-unique error prone reads where each sequence represents less than 0.1% of total reads for the particular sequence. Blue: unique reads. In the Illumina GAII dataset from the enriched sample just 315 out of 319 selected sequences could be detected.
To test the assembly of gene fragments based on Megacloned oligonucleotides stemming from a microarray we assembled two gene fragments of approximately 220 bp in length combining either nine or ten Megacloned, bead-derived amplicons in a PCR-based gene assembly reaction12,13. The obtained assemblies were cloned and Sanger sequenced. Seven out of eight clones matched the target sequence perfectly. Interestingly, one clone showed insertions and deletions all located within a 23 bp wide region. Errors in assemblies originated from inaccuracies in the starting material could be expected to be distributed evenly over the entire constructs. Since this sequence was otherwise free of errors these defects were likely caused by miss-assembly rather than errors in the building blocks used (supplementary note online).
To further evaluate the capabilities of the Megacloning approach towards generation of biologically functional genes we applied DNA fragments of 274–394 bp in length to the process and extracted 32 beads from the PTP carrying putatively "correct" sequences. These DNA fragments were the product of gene assembly reactions12 using overlapping 40mer oligonucleotides synthesized using conventional phosphoramidite chemistry and could be assembled into a model gene encoding β-D-Glucuronidase (UidA)14 (2,080 bp).
Three obtained Sanger sequences from the bead DNA were totally unrelated to the expected sequence and were likely caused by wrong bead extraction or contamination. The remaining 29 sequences covered 7,195 basepairs and matched without errors to the expected target sequences (supplementary note online).
We then assembled the model gene out of nine DNA fragments from the set of 29 matching beads. Absence of errors in the full length gene construct was again checked by Sanger sequencing and the biological functionality of the gene was tested in an enzymatic assay based on the conversion of X-Glc substrate into blue dye15 (supplementary note online). Besides the proof of feasibility of generating biological functional genes this experiment further mimics other applications of our technology such as the use of sheared natural DNA and their subsequent sorting and re-ordering.
The absence of errors in 7,195 bp of DNA yielded from 29 extracted beads raised the question of achievable error rates from the Megacloner process. Therefore we explored the potential of this approach using a statistical model. This model considers two main error sources, referring to wrong sequencing calls and polymerase errors during DNA amplification16. The calculations estimated the probability of finding one error in our extracted sequence space of approximately 7,200 bp with 29 % which is in line with our findings. The theoretical error rate of bead amplicons after Megacloning using the setup employed in this study was estimated to be 1 error in 21 kbp (supplementary note online). Compared with the error rate in the starting material of 1 error in 40 basepairs (determined from GAII data of the initial microarray pool) this equals a 500-fold error reduction.
We furthermore calculated the expectable amount of reads from NGS that match the target sequences of a given pool without errors. These numbers are crucial to estimate the complexity of pools that can be processed in one Megacloner run and hence the resulting efficiency and cost structure and are influenced mainly by three parameters: the error rate of the starting pool, the sequencing accuracy, and the length of the variable sequence (supplementary Fig.14 online). With an error rate of 1 error in 40 bp and an average sequencing accuracy of 99.9% in the GS FLX we expect a 5 to 10-fold cost reduction on the level of DNA fragments (compared to conventional oligonucleotide synthesis) that can be achieved already with the presented prototype device (supplementary note online). Since these fragments are widely free of errors further savings can be expected in gene synthesis due to reduction of subsequent sequencing cost for final quality control.
In this work we demonstrated the targeted retrieval of bead-bound DNA from a high throughput sequencer without major modifications to the sequencing process. Previous methods for error correction in DNA pools7,17–21 do not adequately handle collections of closely related sequences that occur in repetitive sequences or multi-gene family libraries. They also do not enable hierarchical assembly strategies the way that ordered selection and physical separation of clonal DNA as described here can.
The new Megacloner process has been proven to be useful for retrieval and sorting of correct and functional sequences and increased the portion of error free sequences in a sample significantly. This technology allows the processing of DNA from microarrays but also from a variety of other sources such as conventional oligonucleotide synthesis or natural DNA fragments.
The extrapolation of the statistical model was fully supported by the experimental data and suggested that DNA with error rates of one error in 21 kbp could be obtained with the used setup from input DNA having an error rate of 1 in 40 bp. Such raw material could be easily obtained by state-of-the-art microarray technologies that are a fundamental part of our entire concept.
Further increase of DNA quality could be achieved primarily by addressing the amplification step of bead bound DNA for example with more powerful polymerases since this contribution to errors is 4.7-fold higher than the expected error rate of the Megacloner itself (supplementary note online). Another accessible parameter for optimizing the overall process in terms of error rates is improvement of DNA starting material quality. Also optimization of sequencing accuracy could be a way to improve the ability to select correct parts after NGS. This is, however, subject of ongoing optimization in the scope of NGS development including ligase based methods with improved accuracy22.
The pool used in our conceptual study contained ~4,000 sequences. According to our results and extrapolations this can be increased to approximately 30,000 sequences per pool with the described setup. Since the bead extraction is generally independent from the pool complexity, it is mainly limited by the used NGS platform and the starting material quality (supplementary note online). Unpublished data from Kosuri and colleagues23 demonstrate that more advanced microarray formats are able to deliver libraries with even higher complexity and of sufficient quality to fit into a gene assembly process. Therefore, with an appropriate degree of automation gaining an extraction frequency of two or three beads per minute, achievable with state-of-the-art robotics, the work up of one PTP becomes possible within days resulting in over one million basepairs per plate. Hence, the downstream process (amplification, cleanup, assembly), will represent the next bottleneck.
Our next focus in the present context is improvement and automation of physical bead extraction. The workflow employed in this study still involved a considerable degree of manual steps and human intervention which was identified as the most important source of error in terms of extraction of unwanted beads. The success rate of approximately 90% (29 beads out of 32) has therefore to be increased for the bead localization and retrieval process.
The method described here holds the potential to decrease production cost for synthetic DNA by one or more orders of magnitude. This source of high quality DNA could aid the field of synthetic biology as well as the production of libraries for antibodies or enzyme variants. In addition to synthetic sources the sorting of natural DNA could enable the quick reconstruction or combination of DNA fragments to assemble genes, chromosomes or genomes while simultaneously including synthetic parts of DNA.
The principle that we applied here using the GS FLX platform should be generally applicable also to other available NGS platforms such as Illumina’s GAII, SOLiD, the Polonator or others. In the present context, the advantage of the GS FLX platform is the robot-accessible platform architecture and the comparably large size of the beads. Due to different architectures of the other platforms such as partially closed systems and significantly smaller DNA carriers, the harvest from those will require a different mechanism such as optical approaches including photo-sensitive and cleavable linker-molecules. Advantage of these platforms is a considerable higher number of DNA clones which potentially could increase the capacity and throughput of the technology up to the gigabase level.
Methods
Oligo synthesis, Sequence design, adaptors etc
Oligonucleotides used for this work were synthesized on programmable microarray synthesizers using light directed synthesis methods5. Conventional oligonucleotides used for gene assembly were obtained from Sigma Aldrich. Harvesting of oligonucleotides from microarray surfaces was performed by chemical cleavage of succinate-ester bonds using ammonia hydrochloride solution.
Amplification of microarray derived oligonucleotide pools by emPCR
Microarray derived oligonucleotide pools were amplified prior to NGS using emulsion PCR24. Therefore universal terminal sequences were attached during synthesis and served as primer regions. Amplification primers contained adaptors for sequencing on the Illumina GAII platform and/or the 454 GS FLX (supplementary Fig. 11 online)
Sequencing on the 454 GS FLX
The sample preparation for the PCR amplified oligonucleotides was done according to the manufacturer’s protocols (Roche/454). In order to keep the DNA intact after sequencing we exchanged the bleaching cleaning buffer with TE-Buffer prior to the sequencing run to avoid degradation of DNA during the final cleaning steps of the Roche sequencer.
Data analysis of 454 data and image conversion
NGS reads obtained from the GS FLX sequencer were aligned to the target sequences in the oligonucleotide pool in order to find the best matching sequence for every read and to perform further analysis such as error rate estimation etc. Perfect matching sequences have been selected and localized in the sequencer image by using the coordinates attached to every read sequence. For sequence data analysis we used various Python scripts using the BioPython package. The images from the GS FLX sequencer were converted into the TIFF standard format using the Python Imaging Library (PIL).
Bead localization and extraction
After aligning the GS FLX reads to the set of target sequences we selected reads that perfectly matched one of the desired oligonucleotide sequences in the pool. For localization of beads we located the corresponding chemiluminescent signals in the converted raw image from the GS FLX platform using the x- and y-coordinates that were included in the NGS raw data. To locate beads in the PTP we identified reference points in the raw image and their corresponding positions in the PTP using suitable patterns of light signals. Based on these reference points the bead positions on the PTP were calculated using an algorithm for scaling and rotation (supplementary Fig.8 online). The extraction was performed with a micropipette with an outer diameter of 28 µm. For pipette handling we used a 3-axis micro actuator (supplementary Fig.1 and 2 online). Prior to extraction of beads the PTP was stored under a water layer to prevent desiccation and shrinking of beads. After picking, the beads were transferred immediately into a PCR vial and stored under water until further processing.
Amplification of DNA from beads
Amplification of bead bound DNA was performed with the primers “454-A” and “454-B”, targeting the Roche/454 adaptors, or “slx-fw-long” and “slx-rev-long” for Illumina adaptors (supplementary note online). For amplification of fragments with 40mer variable region primers were 5’-biotinylated to facilitate subsequent removal of primer regions on a streptavidin matrix. PCR conditions: 20 mM Tris-HCl (pH 8.8), 10 mM ammonium-sulfate, 10 mM potassium chloride, 2 mM magnesium-sulfate, 0.1 % Triton X-100, 200 µM each dNTP, 2% (v/v) DMSO, 1 µM each primer, 50 U/ml native pfu polymerase (Fermentas). Cycling: initial denaturation 96° (2 min); then 30 cycles of 96° (30 s), 63° (30 s), 72° (30 s), and final elongation 72° (3 min). After amplification, all PCR products were analyzed on PAGE (supplementary Fig. 9 online) to check specificity and yield. For generation of the subpool containing 319 sequences we estimated the concentration on the basis of the gel analysis and mixed the amplicons in equimolar concentrations.
Illumina sequencing and data analysis
Since the sample contained suitable adaptors all steps regarding adaptor ligation have been omitted. All other steps were done according to the protocols from Illumina.
The NGS raw data obtained from Illumina GAII was processed by the following steps.
Truncation of reads to the length of the variable regions (40 bp)
filtering out reads containing ambiguities (filtered reads)
group reads with similar sequences (bins)
Subsequently for each read we identified the best matching target sequence from the oligonucleotide pool by mapping all reads to a pseudo-genome using razerS (www.seqan.de/projects). The pseudo-genome was generated by concatenation of the variable parts of pool sequences separated by 40mer poly-T stretches. The corresponding target sequence could then be determined by the matching position in the pseudo-genome. Alignments from the razerS output was used to determine insertions, deletions and substitutions. To compare the two GAII runs based on the number of correct reads we converted the read counts into parts-per-million units (ppm) taking the number of filtered reads prior the matching procedure (after step 2) as a basis.
Assembly of gene fragments from conventional oligonucleotides
Gene fragments >200 bp included were assembled from conventionally synthesized 40mer oligonucleotides having a constant overlap region of 20 nucleotides to the adjacent oligomer. Primer regions for 454 sequencing and restriction sites for primer removal were included during assembly. The assembly reaction contained 5 nM of each construction oligonucleotide and 200 nM of terminal primers. PCR conditions: 1× KOD polymerase buffer (Novagen), 1.25 mM MgSO4, 40 µM each dNTP, 5 U/ml KOD Hot Start Polymerase (Novagen). Cycling for gene assembly: initial denaturation 96° (4 min); then 30 cycles of 96° (10 s), 55–40°C touchdown (30 s), 72° (10 s). For subsequent amplification: 96° (10 s), 55°C (30 s), 72° (30 s), final elongation 72° (3 min).
Assembly of genes from >200 bp fragments
Gene assembly up to 2 kbp were performed according to the protocol used for assembly of >200 bp from oligonucleotides.
Primer removal & cleanup of bead amplicons prior gene assembly
For removal of primer regions amplicons were incubated with LguI in 1× Tango buffer (Fermentas) for 3 h at 37°. For >200 bp fragments small restriction fragments containing primer regions were removed by PCR purification columns (GenElute PCR Clean-Up, Sigma Aldrich). For cleanup of microarray derived fragments with 40mer variable region biotinylated primers were used during bead DNA amplification and restriction products containing biotin residues were removed using streptavidin matrix. The 40mer fragments were ethanol precipitated and dissolved in water prior further processing.
Assembly of genes from 40mer dsDNA fragments
For the assembly of genes from 40mer dsDNA we used a two stage assembly protocol including a primerless PCR followed by a PCR for amplification of the resulting products described previously13.
Supplementary Material
Acknowledgements
We thank B. A. Roe, F. Z. Najar and D. D. White for sequencing support. J. Jäger for technical consulting and D. Summerer, T. Brefort, S. Kosuri and D. Levner for discussions and comments.
Footnotes
Declaration of competing financial interests
M. Matzas, N. Kefer, N. Siebelt, V. Boisguérin, J.T. Leonard, P.F. Stähler, A. Keller, C.F. Stähler and P. Häberle have potentially competing financial interests in companies affiliated to the febit group.
Literature
- 1.Endy D. Foundations for engineering biology. Nature. 2005;438:449–453. doi: 10.1038/nature04342. [DOI] [PubMed] [Google Scholar]
- 2.Menzella HG, et al. Combinatorial polyketide biosynthesis by de novo design and rearrangement of modular polyketide synthase genes. Nat. Biotechnol. 2005;23:1171–1176. doi: 10.1038/nbt1128. [DOI] [PubMed] [Google Scholar]
- 3.Gibson DG, et al. Creation of a Bacterial Cell Controlled by a Chemically Synthesized Genome. Science. 2010 doi: 10.1126/science.1190719. [DOI] [PubMed] [Google Scholar]
- 4.Carr PA, Church GM. Genome engineering. Nat. Biotechnol. 2009;27:1151–1162. doi: 10.1038/nbt.1590. [DOI] [PubMed] [Google Scholar]
- 5.Gao X, et al. A flexible light-directed DNA chip synthesis gated by deprotection using solution photogenerated acids. Nucleic Acids Res. 2001;29:4744–4750. doi: 10.1093/nar/29.22.4744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Singh-Gasson S, et al. Maskless fabrication of light-directed oligonucleotide microarrays using a digital micromirror array. Nat. Biotechnol. 1999;17:974–978. doi: 10.1038/13664. [DOI] [PubMed] [Google Scholar]
- 7.Tian J, et al. Accurate multiplex gene synthesis from programmable DNA microchips. Nature. 2004;432:1050–1054. doi: 10.1038/nature03151. [DOI] [PubMed] [Google Scholar]
- 8.Porreca GJ, et al. Multiplex amplification of large sets of human exons. Nat. Methods. 2007;4:931–936. doi: 10.1038/nmeth1110. [DOI] [PubMed] [Google Scholar]
- 9.Margulies M, et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wicker T, Schlagenhauf E, Graner A, Close TJ, Keller B, Stein N. 454 sequencing put to the test using the complex genome of barley. BMC Genomics. 2006;7:275. doi: 10.1186/1471-2164-7-275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Willenbrock H, et al. Quantitative miRNA expression analysis: comparing microarrays with next-generation sequencing. RNA. 2009;15:2028–2034. doi: 10.1261/rna.1699809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stemmer WP, Crameri A, Ha KD, Brennan TM, Heyneker HL. Single-step assembly of a gene and entire plasmid from large numbers of oligodeoxyribonucleotides. Gene. 1995;164:49–53. doi: 10.1016/0378-1119(95)00511-4. [DOI] [PubMed] [Google Scholar]
- 13.Richmond KE, et al. Amplification and assembly of chip-eluted DNA (AACED): a method for high-throughput gene synthesis. Nucleic Acids Res. 2004;32:5011–5018. doi: 10.1093/nar/gkh793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jefferson RA, Burgess SM, Hirsh D. beta-Glucuronidase from Escherichia coli as a gene-fusion marker. Proc. Natl. Acad. Sci. USA. 1986;83:8447–8451. doi: 10.1073/pnas.83.22.8447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Couteaudier Y, Daboussi MJ, Eparvier A, Langin T, Orcival J. The GUS gene fusion system (Escherichia coli beta-D-glucuronidase gene), a useful tool in studies of root colonization by Fusarium oxysporum. Appl Environ Microbiol. 1993;59:1767–1773. doi: 10.1128/aem.59.6.1767-1773.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cline J, Braman JC, Hogrefe HH. PCR fidelity of pfu DNA polymerase and other thermostable DNA polymerases. Nucleic Acids Res. 1996;24:3546–3551. doi: 10.1093/nar/24.18.3546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carr PA, Park JS, Lee Y-J, Yu T, Zhang S, Jacobson JM. Protein-mediated error correction for de novo DNA synthesis. Nucleic Acids Res. 2004;32:e162. doi: 10.1093/nar/gnh160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Smith J, Modrich P. Removal of polymerase-produced mutant sequences from PCR products. Proc. Natl. Acad. Sci. USA. 1997;94:6847–6850. doi: 10.1073/pnas.94.13.6847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bang D, Church GM. Gene synthesis by circular assembly amplification. Nat. Methods. 2008;5:37–39. doi: 10.1038/nmeth1136. [DOI] [PubMed] [Google Scholar]
- 20.Fuhrmann M, Oertel W, Berthold P, Hegemann P. Removal of mismatched bases from synthetic genes by enzymatic mismatch cleavage. Nucleic Acids Res. 2005;33:e58. doi: 10.1093/nar/gni058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Binkowski BF, Richmond KE, Kaysen J, Sussman MR, Belshaw PJ. Correcting errors in synthetic DNA through consensus shuffling. Nucleic Acids Res. 2005;33:e55. doi: 10.1093/nar/gni053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McKernan KJ, et al. Sequence and structural variation in a human genome uncovered by short-read, massively parallel ligation sequencing using two-base encoding. Genome Res. 2009;19:1527–1541. doi: 10.1101/gr.091868.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kosuri S, et al. A scalable gene synthesis platform using high-fidelity DNA microchips. Manuscript submitted for publication. Wyss Institute for Biologically Inspired Engineering; [Google Scholar]
- 24.Williams R, Peisajovich SG, Miller OJ, Magdassi S, Tawfik DS, Griffiths AD. Amplification of complex gene libraries by emulsion PCR. Nat. Methods. 2006;3:545–550. doi: 10.1038/nmeth896. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.