Abstract
A special class of biochemical reactions involves a set of enzymes that generate additional copies of themselves and transfer heritable information from parent to progeny molecules, thus providing the basis for genetics and Darwinian evolution. Such a process has been realized with a pair of self-replicating RNA enzymes that undergo exponential amplification at constant temperature. Exponential growth requires that the rate of production of new enzymes be directly proportional to the existing concentration of enzymes, which is the case for this system and provides a doubling time of ~20 min. However, the catalytic rate of the underlying enzymes is ~100-fold faster than the observed rate of replication. As in biological replication, other aspects of the system limit the generation time, chiefly the propensity of the substrate molecules to form non-productive complexes that limit their availability for replication. An analysis of this and other kinetic properties of the self-replicating RNA enzymes reveals how exponential amplification is achieved and how the rate of amplification might be increased.
The development of self-replicating chemical systems that undergo exponential amplification has been an important research goal since von Kiedrowski’s demonstration of a self-replicating hexadeoxynucleotide.1 Following that seminal work, other self-replicating systems have been devised, including ones based on nucleic acids,2–5 peptides,6–8 small organic molecules,9,10 and more complex architectures such as DNA nanoscale motifs11 and DNA crystals.12 These systems typically entail a template that directs the joining of two substrates to form a product that is identical to the template. Autocatalysis occurs when the product also can function as a template, directing the formation of additional products. Exponential growth, however, is usually not observed in these systems because amplification is limited by slow dissociation of the template-product complex.13,14
Recently, a self-replicating system was described that is based on an RNA enzyme with RNA ligase activity,15 and can undergo sustained exponential growth at constant temperature.16 The enzyme catalyzes the template-directed joining of two RNA substrates to form another copy of itself. The template-product complex dissociates in a non-rate-limiting manner, allowing amplification to proceed indefinitely so long as additional substrates are provided. The enzyme usually is made to operate as a cross-replicating pair, whereby two enzymes direct each other’s synthesis from a total of four component substrates (Figure 1).17 One enzyme, E, catalyzes the joining of two substrates, A´ and B´, to form the other enzyme, E´. Similarly, E´ catalyzes the joining of A and B to form E. The joining reaction catalyzed by either E or E´ results in formation of a 3´,5´-phosphodiester linkage between the two corresponding substrates, with concomitant release of inorganic pyrophosphate. No proteins or other informational macromolecules are required for replication, and in this sense the system is said to be self-sustaining.
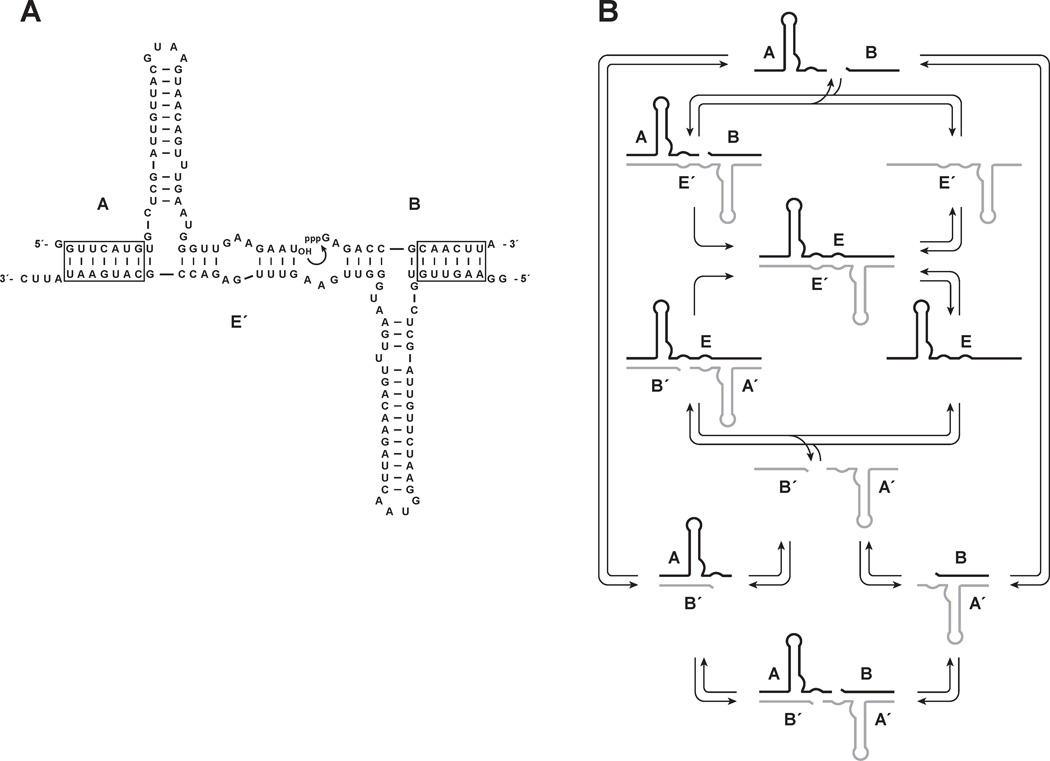
Figure 1.
Self-replicating RNA enzymes. (A) Sequence and secondary structure of the enzyme E´ and its substrates A and B. Curved arrow indicates the site of ligation to form the partner enzyme E, which similarly reacts with substrates A´ and B´ to form E´. Boxed regions indicate paired nucleotides that can vary in sequence. (B) Scheme for cross-replication of E (black) and E´ (gray), including equilibrium between free substrates and the A•B´, A´•B, and B´•A•A´•B complexes.
In the system of self-replicating RNA enzymes, heritable information can be transferred from parent to progeny molecules in the form of particular sequences located within two template regions of the enzyme and corresponding complementary regions of the substrates. Novel variation can arise due to recombination between these two regions, and different genetic variants can be made to encode corresponding functional variants of the enzyme. By combining exponential amplification of heritable information, the opportunity for mutation, and selection of advantageous phenotypes, the system is capable of undergoing Darwinian evolution in a self-sustained manner.16,18
The self-replicating system also has been used as a tool to measure the concentration of various target molecules.19,20 In this format, an aptamer domain is linked to the catalytic domain of the enzyme such that, upon binding its cognate ligand, the aptamer adopts a defined structure that results in activation of the enzyme. The exponential growth rate of the enzyme reflects the concentration of the ligand relative to the Kd of the aptamer-ligand interaction. This configuration enables the real-time, quantitative detection of various target molecules, analogous to quantitative PCR, but applicable to small-molecule and protein ligands.
The exponential growth rate of the self-replicating RNA enzymes is ~0.03 min−1, corresponding to a doubling time of ~20 min.16 It would be advantageous to increase this rate to allow more rapid self-sustained evolution and more rapid detection of target ligands. To guide these efforts, the present study sought to understand the kinetic properties of the RNA-catalyzed exponential amplification system. Such insights also may be useful in increasing the complexity of the synthetic evolving system, which presently is limited to a few thousand variants, and therefore has little capacity for inventive evolution. Two important constraints on the genetic complexity of the system are the modest fidelity of replication and the sequestration of reaction materials within non-productive complexes. The issue of replication fidelity has been addressed in another recent study.18 The present study provides a better understanding of the non-productive complexes and suggests how they might be overcome.
EXPERIMENTAL PROCEDURES
Materials
DNA templates were synthesized using an Applied Biosystems Expedite 8909 automated DNA/RNA synthesizer (Foster City, CA). Nucleoside phosphoramidites were purchased from Glen Research (Sterling, VA). Thermus aquaticus DNA polymerase and histidine-tagged T7 RNA polymerase were expressed and purified as previously described.21,22 Nucleoside and deoxynucleoside 5´-triphosphates were purchased from Sigma-Aldrich (St. Louis, MO), Antarctic phosphatase and T4 polynucleotide kinase were from New England Biolabs (Ipswich, MA), and [γ-32P]ATP (6 µCi/pmol) and [α-32P]ATP (3 µCi/pmol) were from Perkin Elmer (Waltham, MA).
Preparation of Oligonucleotides
The enzymes E and E´ were prepared by in vitro transcription of corresponding synthetic DNA templates. Substrates A and A´ were obtained by site-specific cleavage of E and E´, respectively, using E. coli M1 RNA and an external guide RNA to generate products terminating in a 2´- and 3´-hydroxyl.23 The external guide RNAs had the sequence 5´-CGUAAGUUGCGGUCUCACCA-3´ and 5´-AUAUUCAUGCGGUCUCACCA-3´ for A and A´, respectively (region of hybridization underlined). Substrates B and B´ were prepared by in vitro transcription, with a hammerhead ribozyme appended to their 3´ terminus to provide a homogeneous 3´ end upon self-cleavage during transcription. The transcribed RNAs had the sequence 5´-GAGACCGCAACUUA•UACGGAAACGUACUGAUGAGGCCGAAAG-GCCGAAAAGUUG-3´ and 5´-GAGACCGCAUGAAUAUUC•UACGGAAACGUACUGAU-GAGGCCGAAAGGCCGAAAAUAUUC-3´ for B and B´, respectively (hammerhead pairing underlined; cleavage site indicated by a dot). The A and A´ substrates were [5´-32P]-labeled by dephosphorylating using Antarctic phosphatase, incubating at 65 °C for 5 min to inactivate the phosphatase, and phosphorylating using T4 polynucleotide kinase and [γ-32P]ATP. The B and B´ substrates were internally labeled by carrying out in vitro transcription in the presence of [α-32P]ATP.
Determination of Reaction Rates and Equilibrium Constants
All reactions were carried out in the presence of 25 mM MgCl2 and 50 mM EPPS (pH 8.5) at 44 °C. Aliquots were removed at various times and quenched by adding a solution containing 8 M urea and Na2EDTA in three-fold excess over Mg2+. Substrates and products were separated by denaturing polyacrylamide gel electrophoresis (PAGE) and quantitated using a PharosFX molecular imager (Bio-Rad, Hercules, CA).
Determination of Michaelis-Menten Parameters
The component reactions, A + B → E and A´ + B´ → E´, were carried out under enzyme-excess, single-turnover conditions using trace radiolabeled amounts of the substrate being evaluated (<5 nM for A or A´; 20 nM for B or B´), saturating concentrations of the other substrate (15 µM), and various concentrations of the enzyme that spanned KM. The reactions were initiated by mixing a solution containing the radiolabeled substrate with a solution containing the other substrate and enzyme, both at 44 °C. First-order rate constants, kobs, were obtained from a plot of fraction reacted vs. time, fitting the data to a single-exponential equation:
| (1) |
where F(t) is the fraction reacted at time t, and Fmax is the maximum extent of the reaction. Values for kcat and KM were obtained from a plot of kobs vs. [E] (or [E´]), fitting the data to the Michaelis-Menten equation:
| (2) |
Determination of Dissociation Rates by Pulse-Chase Experiments
A mixture containing 0.5 µM [5´-32P]-labeled E, 1 µM E´, 25 mM MgCl2, and 50 mM EPPS (pH 8.5) was equilibrated first at 23 °C for 20 min, then at 44 °C for 30 min. A chase solution was added that also had been equilibrated at 44 °C for 30 min and contained a large excess of unlabeled E (30 µM final concentration), 25 mM MgCl2, and 50 mM EPPS (pH 8.5). Aliquots were withdrawn at various times and subjected to gel-shift analysis in a 10% non-denaturing polyacrylamide gel containing 12 mM MgCl2, 2 mM Na2EDTA, and 90 mM Tris-borate (pH 8.3). The fraction of shifted material vs. time was fit to a single-exponential equation, and kdissoc was calculated as the exponential decay rate. The same procedure was applied to a mixture containing 0.5 µM [α-32P]-labeled B (or B´) and 1 µM A´ (or A), chasing with 30 µM unlabeled B (or B´). In that case the labeled material was fully shifted prior to the chase, but >90% was displaced within 30 s.
Determination of Kd of the E•E´ Complex
Gel-shift analysis was carried out employing trace amounts (<1 nM) of [5´-32P]-labeled E and various concentrations of unlabeled E´, which were equilibrated at 23 °C for 20 min, then at 44 °C for 1 h. The material was analyzed by non-denaturing polyacrylamide gel electrophoresis, as above. The fraction of labeled E that bound to E´ was determined as a function of [E´] and the data were fit to a saturation plot, from which Kd was determined.
Determination of Kd of the A•B´ and A´•B Complexes
The component reactions were monitored using the same protocol as for determining the Michaelis-Menten parameters, employing trace amounts of [5´-32P]-labeled A (or A´) and various concentrations of unlabeled B´ (or B), which first were allowed to reach equilibrium, then were mixed with B and E´ (or B´ and E). Values for kobs were determined as above, and a plot of kobs vs. [E´] (or [E]) was used to determine a value for KMapp in each instance. The KMapp values were related to kcat and KM that had been determined in the absence of competing substrate to provide an estimate of Kd:
| (3) |
Kd was determined separately for two different concentrations of competing substrate for each of the two component reactions.
Self-Replication Reactions
Reactions were initiated by combining an equilibrated solution containing [5´-32P]-labeled A, [5´-32P]-labeled A´, E, and E´ with a second equilibrated solution containing B and B´, both solutions also containing 25 mM MgCl2 and 50 mM EPPS (pH 8.5). Unless otherwise stated, the reactions employed 5 µM each of A, A´, B, and B´ and 0.1 µM each of E and E´, and were carried out at 44 °C. The reactions were sampled at various times and the products were separated by PAGE to determine the yield of radiolabeled E and E´. The data were fit to the logistic growth equation:
| (4) |
where [E]t is the concentration of E (or E´) at time t, a is the maximum extent of growth, b is the degree of sigmoidicity, and c is the exponential growth rate. The pseudo-first-order rate constant for the initial phase of the reaction was obtained from the derivative of the logistic growth equation, which at t = 0 is:
| (5) |
The initial rate of reaction also was measured directly for various starting concentrations of both E and E´, collecting data over the first 10% of the reaction. These data were fit to a linear plot, rejecting any data for which the linear regression coefficient was <0.98.
Kinetic Modeling
A model was constructed based on Figure 1B, incorporating the exponential amplification profiles for E and E´ shown in Figure 2A and the experimentally determined Kd values for the E•E´, A•B´, and A´•B complexes. The starting concentrations of the B and B´ substrates were adjusted for their maximum extent of reaction, which were 3.9 and 2.9 µM, respectively. The total concentration of each substrate over time was calculated by subtracting the concentration of product from the initial concentration of substrate. The concentrations of complexes E•E´, A•B´, and A´•B were calculated from the experimentally determined Kd values, considering the relevant equilibrium and solving the quadratic equation, for example, for A•B´:
| (6) |
where [A]tot and [B´]tot are the total concentrations of A and B´, respectively.
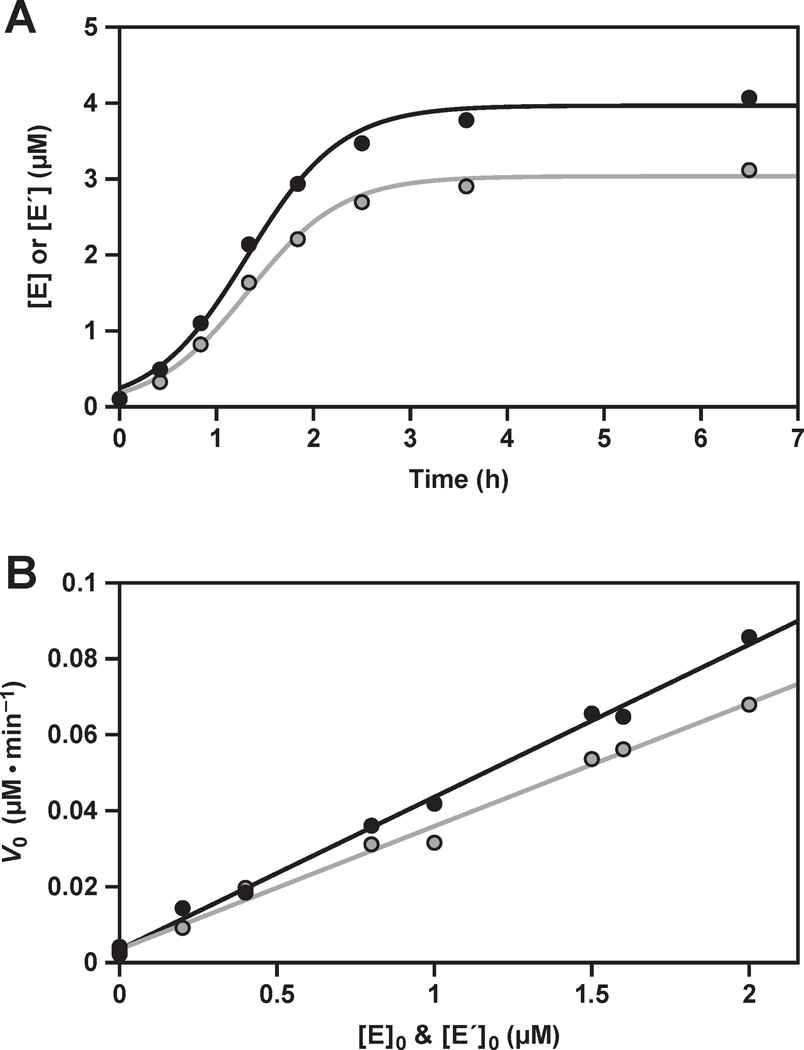
Figure 2.
Exponential amplification of the replicating RNA enzymes. (A) Time course of amplification, starting with 0.1 µM each of E and E´, 5 µM each of A, A´, B, and B´, and 25 mM MgCl2, carried out at pH 8.5 and 44 °C. The data were fit to the logistic growth equation, which gave an exponential growth rate of 0.034 min−1 for the production of both E (black) and E´ (gray). The pseudo-first order rate constant was calculated from the derivative of the logistic growth equation, applied to the first 15% of the reaction, which gave observed rates of 0.037 and 0.025 min−1 for the production of E and E´, respectively. (B) Initial rate of reaction (V0) for the production of both E and E´, under the same conditions as above, but with various starting concentrations of the two enzymes. A linear fit to the data gave exponential growth rates of 0.040 and 0.032 min−1 for the production of E and E´, respectively, and rates for the spontaneous reaction in the absence of enzymes of 3.5 × 10−9 M • min−1 for both E and E´.
RESULTS
The system of self-replicating RNA enzymes involves two component reactions: A + B → E and A´ + B´ → E´ (Figure 1). These reactions first were studied in isolation to determine the relevant underlying catalytic parameters. Each component reaction involves binding of two substrates to the enzyme, followed by a chemical step that is thermodynamically highly favorable and essentially irreversible, then dissociation of the E•E´ complex to complete the catalytic cycle. Non-rate-limiting product release is essential for exponential amplification.
The A and A´ substrates bind to the E´ and E enzymes, respectively, through three discontinuous regions of Watson-Crick pairing (Figure 1A). Two of these regions involve four base pairs of defined sequence and the third involves 6–7 base pairs of variable sequence. The B and B´ substrates bind to their respective enzymes through two discontinuous regions of Watson-Crick pairing, one involving four base pairs of defined sequence and the other involving 6–7 base pairs of variable sequence. The bound substrates also may engage in tertiary interactions with the enzyme, for example, through interactions involving the central stem-loop of A and A´. The ligated product binds to the enzyme through five discontinuous regions of Watson-Crick pairing, as well as potential tertiary interactions.
When all four substrates are present in the same mixture, there is the opportunity to form A•B´ and A´•B complexes due to the partial complementarity of these molecules (Figure 1B). Such complexes are expected to reduce the effective concentration of the substrates and therefore reduce the efficiency of the RNA-catalyzed reaction. The four substrates also can form a B´•A•A´•B complex in which the A and A´ components, likely pre-associated as A•B´ and A´•B complexes, bind to each other through a central region of four base pairs. This tetramolecular complex has been shown to have a low level of catalytic activity, resulting in the spontaneous formation of E and E´ molecules that then undergo autocatalytic amplification.19
The complete self-replication reaction involves reciprocal synthesis of E and E´ from the four component substrates. A plot of enzyme concentration vs. time has a sigmoidal shape (Figure 2A), indicative of exponential growth limited by the fixed concentration of substrates.16 The reactions are typically performed in the presence of 25 mM MgCl2 at pH 8.5 and 44 °C, resulting in an exponential growth rate of 0.03 min−1. The maximum extent of the reaction is typically 80% for substrates that are prepared by in vitro transcription. However, another study has shown that a maximum extent of >90% can be achieved when both the B and B´ substrates are prepared synthetically,24 which may be indicative of sequence heterogeneity at the 5´ end of the transcribed materials.25,26 There is some degradation of the RNA during the course of the reaction, which occurs at a constant rate of ~5% h−1. The degradation rate is similar for the unreacted substrates and ligated products, and thus has little effect on the determination of the fraction reacted.
A formal demonstration of exponential growth is obtained by measuring the initial rate of reaction as a function of the starting concentration of enzyme, fitting the data to the equation:
| (7) |
where the initial rate, (d[E]/dt)0, is proportional to the starting enzyme concentration, [E]0, raised to the reaction order p. A plot of (d[E]/dt)0 vs. [E]0p has a slope equal to the autocatalytic rate constant, kauto, and a y-intercept equal to the rate of reaction in the absence of enzyme, kspont. Exponential growth occurs when the reaction order is 1.0 (ref. 13). Data were obtained for starting concentrations of both E and E´ of 0–2 µM, and fit well to a plot with reaction order 1.0 (r = 0.998, 0.996), confirming exponential growth (Figure 2B). The autocatalytic rate constants are 0.040 and 0.032 min−1 for the production of E and E´, respectively, coinciding with the overall exponential growth rate of 0.03 min−1. The efficiency of replication, kauto/kspont, is ~107 M−1.
Steady-State Catalytic Parameters
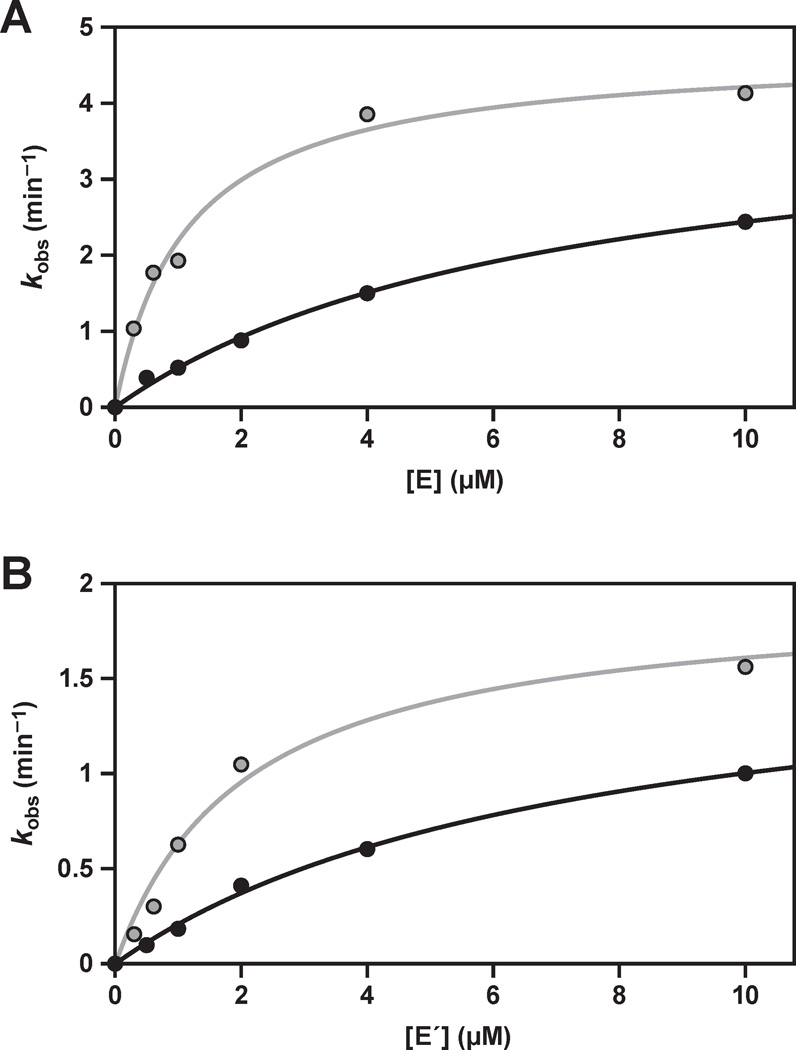
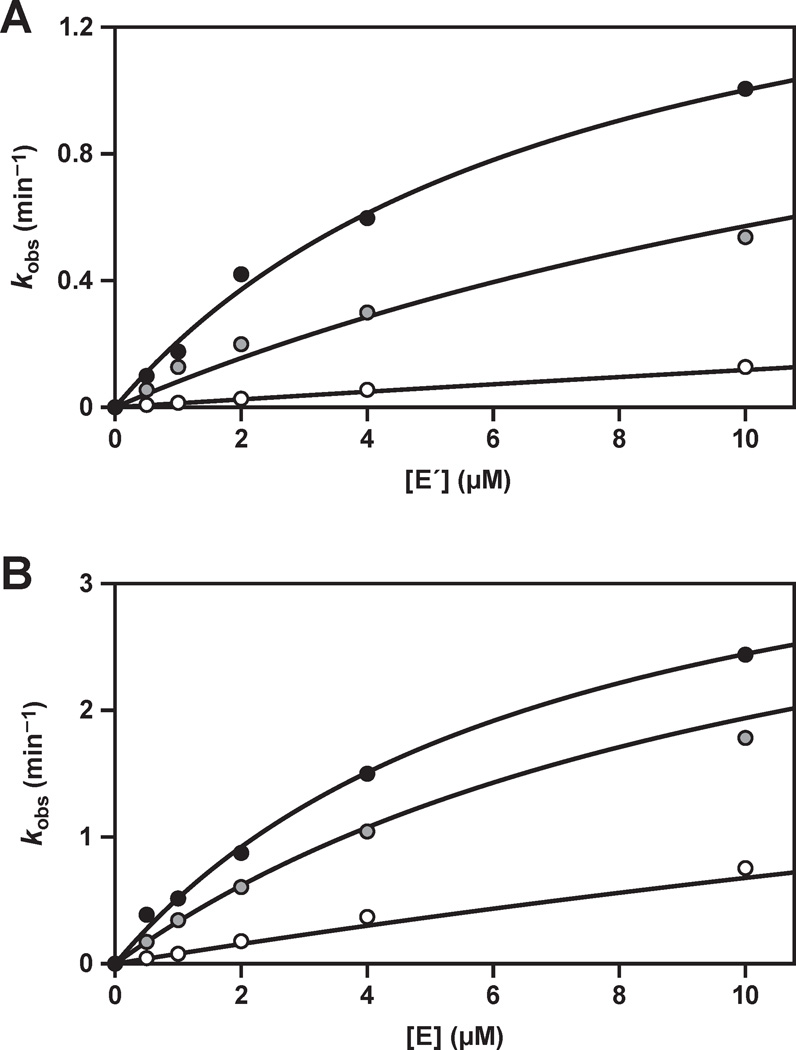
Michaelis-Menten parameters were determined for each of the two component reactions, measured under enzyme-excess, single-turnover conditions for each of the two substrates (Figure 3). These reactions employed trace concentration of the substrate being evaluated, saturating concentration of the other substrate, and various concentrations of the enzyme that spanned KM. The values for kcat all are in the range of 2–5 min−1, with the E-catalyzed reaction being about two-fold faster compared to the E´-catalyzed reaction (Table 1). These rates are about 100-fold faster than the observed rate of reaction for the complete self-replication system (Figure 2A), suggesting that other steps in the replication cycle are rate-limiting.
Figure 3.
Catalytic activity of the replicating RNA enzymes in the component ligation reactions, carried out under enzyme-excess, single-turnover conditions. (A) E-catalyzed ligation of A´ (black) or B´ (gray); or (B) E´-catalyzed ligation of A (black) or B (gray); with saturating concentrations of the other substrate and various concentrations of enzyme. The data were fit to the Michaelis-Menten equation: kobs = kcat [E] / (KM + [E]).
Table 1.
Michaelis-Menten Parameters for the Component Ligation Reactions
| enzyme | substrate | kcat (min−1) | KM (µM) |
|---|---|---|---|
| E | A´ | 4.1 ± 0.3 | 7.0 ± 0.9 |
| E | B´ | 4.7 ± 0.2 | 1.1 ± 0.2 |
| E´ | A | 1.7 ± 0.1 | 7.4 ± 1.1 |
| E´ | B | 1.9 ± 0.2 | 2.1 ± 0.5 |
The KM values for the four substrates, also determined under enzyme-excess, single-turnover conditions, are in the range of 1–8 µM, with the KM values for A and A´ being several-fold higher compared to those for B and B´ (Table 1). This disparity has been noted previously,4,16 and may reflect the more complex folded structure of the A and A´ substrates, which results in a smaller fraction of these molecules adopting the properly folded state.
Dissociation of the E•E´ Complex
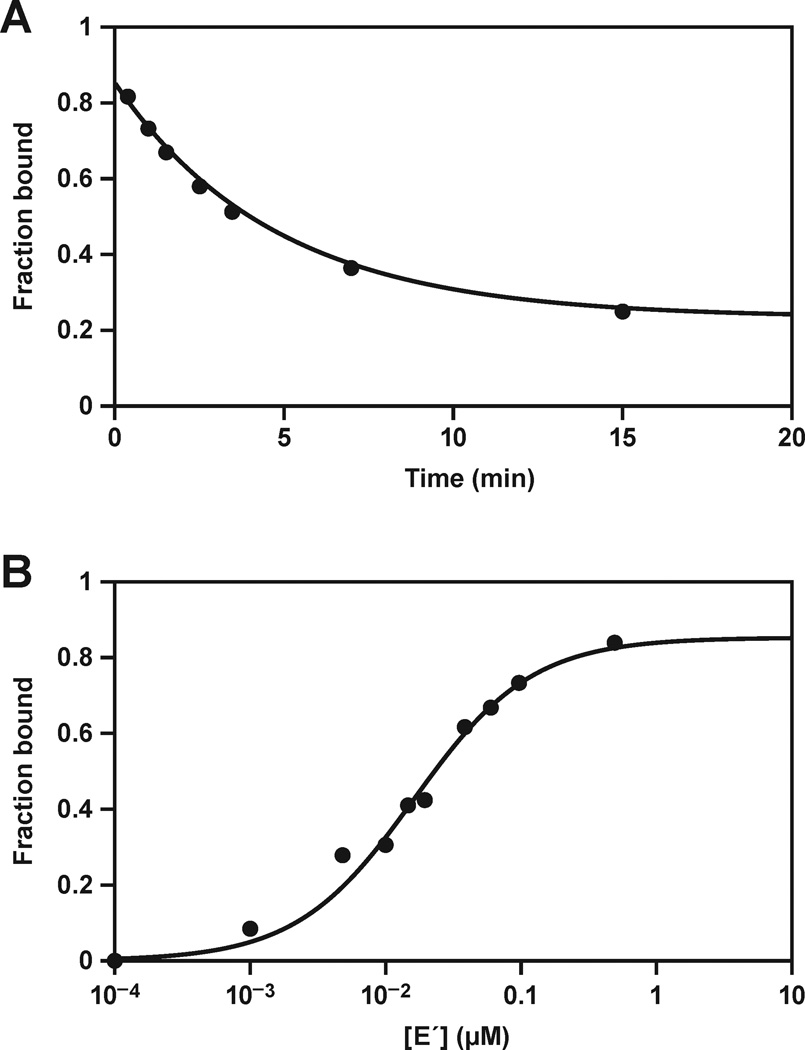
The rate of dissociation of the E•E´ complex was determined by pulse-chase experiments in which radiolabeled E and unlabeled E´ first were allowed to bind at equilibrium, then a large excess of unlabeled E was added, and aliquots were withdrawn at various times to determine the fraction of radiolabeled E that remained bound to E´. The data fit well to a single exponential decay, giving a value for kdissoc of 0.21 min−1 at 44 °C (Figure 4A). This value is similar to that reported previously for a less optimized form of the enzyme,17 and is about 10-fold slower compared to the value for kcat, as described above. Thus multiple turnover of the two component reactions is limited by the rate of product release, even though this is not the case for the complete self-replication system, which has an even slower observed rate.
Figure 4.
Stability of the E•E´ complex. (A) The rate of dissociation of E•E´ was determined by pre-equilibrating radiolabeled E and unlabeled E´, then chasing with a large excess of unlabeled E. The fraction of labeled E that remained bound to E´ at various times was determined by gel-shift analysis, and the data were fit to an exponential decay equation. (B) The Kd of the E•E´ complex was determined by gel-shift analysis, measuring the fraction of labeled E that bound to various concentrations of unlabeled E´. The data were fit to a saturation plot: Fbound = [E´] / (Kd + [E´]).
The Kd of the E•E´ complex was determined by gel-shift analysis, giving a value of 16 nM at 44 °C (Figure 4B). The self-replication reaction typically employs a starting concentration of 5 µM each of the four substrates and 0.01–0.1 µM each of the two enzymes, and results in the production of ~4 µM each of new E and E´ molecules at maximum extent. Thus, throughout most of the course of the reaction, the concentrations of the two enzymes are substantially in excess of the Kd of the E•E´ complex, suggesting that the majority of enzyme molecules are engaged in this complex rather than being free in solution. However, the dissociation rate of this complex is sufficiently fast that free E and E´ molecules are continually made available to bind additional substrates. The calculated rate of association of the E•E´ complex is given by: kassoc = kdissoc / Kd = 1.3 × 107 M−1 min−1. This rate is only about 10-fold slower than the rate of helical nucleation of complementary RNA molecules under similar reaction conditions.27,28
Dissociation of the A•B´ and A´•B Complexes
The rates of dissociation of the A•B´ and A´•B complexes are too fast to measure reliably using pulse-chase methods, with >90% of the labeled material being displaced within 30 s, corresponding to a dissociation rate of >4 min−1. Gel-shift analyses also proved unreliable due to the difficulty in preventing dissociation of these complexes during non-denaturing gel electrophoresis, even when performed at low temperature. Instead, a strategy was devised to measure the rate of ligation in the presence of varying amounts of one of the competing substrates. The reaction A + B → E was carried out in the presence of varying amounts of B´, and the reaction A´ + B´ → E´ was carried out in the presence of varying amounts of B. The Kd values for A•B´ and A´•B were determined by quantitative analysis of the competitive inhibition (see Materials and Methods).
The calculated Kd values for the A•B´ and A´•B complexes are 0.058 and 0.16 µM, respectively (Figure 5). These values indicate that in the complete self-replication system most of the substrate molecules are present as non-productive complexes, rather than being free in solution. If the association rates of the A•B´ and A´•B complexes are similar to the association rate of the E•E´ complex, then the dissociation rates of A•B´ and A´•B would be ~1 min−1. The attempted pulse-chase experiments indicate that these rates are several fold faster, close to the rate of helical nucleation of RNA duplexes. In any case, dissociation of the A•B´ and A´•B complexes would not be rate limiting for the complete self-replication system. Rather, it appears that the observed rate of reaction in the complete system is limited by the concentrations of free substrates. Based on total substrate concentrations of 5 µM each and the Kd values of the non-productive complexes, the concentrations of free substrates would be only 0.5 µM for A and B´ and 0.8 µM for A´ and B, which are substantially below the KM of the corresponding enzyme-substrate complexes.
Figure 5.
Stability of the A•B´ and A´•B complexes. (A) E´-catalyzed ligation of A and B in the presence of no B´ (black), 0.1 µM B´ (gray), or 1 µM B´ (white); or (B) E-catalyzed ligation of A´ and B´ in the presence of no B (black), 0.1 µM B (gray), or 1 µM B (white); employing various concentrations of enzyme. The data were fit to Eq. 3 (see Experimental Procedures) to obtain the Kd of the A•B´ and A´•B complexes.
Considering the free rather than the total concentrations of substrates, the calculated rates of the two component reactions are given by:
| (8a) |
| (8b) |
where kobsE and kobsE´ are the rates of the E- and E´-catalyzed reactions, respectively. The calculated rate of the slower E´-catalyzed reaction closely matches the observed rate of the complete self-replication system, which is ~0.03 min−1 (Figure 2A).
Replication with Varying Initial Substrate Concentrations
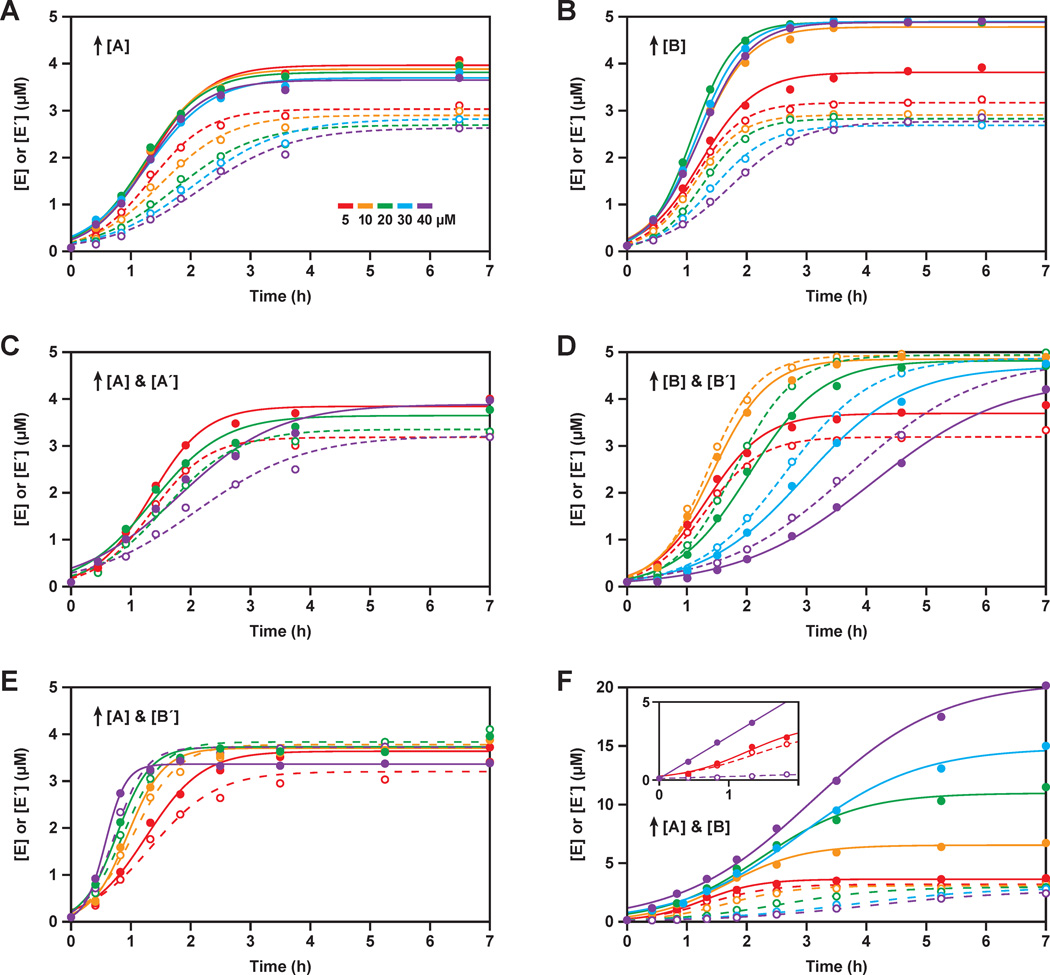
To explore the role of the non-productive complexes in reducing the concentrations of free substrates and thereby limiting the rate of self-replication, reactions were carried out using various initial concentrations of one or two substrates, while maintaining 5 µM initial concentration of each of the other substrates. When the concentration of A was progressively increased from 5 to 40 µM, the rate of the reaction A´ + B´ → E´ progressively declined (Figure 6A). This is explained by the higher concentrations of A favoring formation of the A•B´ complex, thereby reducing the concentration of free B´ that is available to react with A´. In contrast, the rate of the reaction A + B → E was nearly unchanged as the concentration of A was increased. The higher concentrations of A, in excess of the concentration of B´, might be expected to increase the rate of production of E. However, the concentration of free B remains limiting, and the decreased rate of production of E´ reduces the amount of catalyst available to drive the production of E.
Figure 6.
Exponential amplification carried out in the presence of unbalanced concentrations of the four substrates. Unless otherwise stated, all reactions employed 0.1 µM each of E and E´, 5 µM each of A, A´, B, and B´, and 25 mM MgCl2, and were carried out at pH 8.5 and 44 °C. The starting concentrations of one or two substrates were increased from 5 µM (red), to 10 µM (orange), 20 µM (green), 30 µM (blue), or 40 µM (violet). The data were fit to the logistic growth equation, with solid and dashed lines indicating the production of E and E´, respectively. (A) Increasing [A]; (B) increasing [B]; (C) increasing both [A] and [A´]; (D) increasing both [B] and [B´]; (E) increasing both [A] and [B´]; (F) increasing both [A] and [B]. For clarity, not all concentrations are plotted in (C) and (E). Inset in (F) shows a linear fit to the data for the reaction employing 40 µM A and B (violet), in comparison to the reaction with 5 µM A and B (red).
The results were analogous when the concentration of B was increased from 5 to 40 µM, while maintaining 5 µM concentration of each of the other three substrates. This manipulation caused the rate of the reaction A´ + B´ → E´ to be reduced, while the rate of the reaction A + B → E was unchanged (Figure 6B). However, increasing the concentration of B to ≥10 µM resulted in an increased maximum extent of reaction for the production of E, reaching >95%. This is consistent with the hypothesis that a fraction of the in vitro transcribed B molecules are unreactive, with the large excess of B compensating for this deficiency.
Next, the concentrations of both A and A´ were increased from 5 to 40 µM, while maintaining the concentrations of B and B´ at 5 µM (Figure 6C). This resulted in slightly increased rates of production of both E and E´ when the concentrations of A and A´ were increased to 10 µM, but then decreased rates of production when the concentrations of A and A´ were further increased to 20–40 µM. There are two competing effects at play. Raising the concentrations of A and A´ results in increased saturation of the corresponding enzymes, but also results in increased formation of the non-productive A•B´ and A´•B complexes. The KM values for A and A´ are 7.7 and 6.7 µM, respectively. Concentrations of A and A´ in excess of B´ and B, respectively, are available to bind to the corresponding enzymes, but when these excess concentrations reach saturation they no longer provide benefit with regard to the observed rate of reaction. They do, however, continue to enhance the rate of formation of the non-productive complexes, thus reducing the availability of both the B and B´ substrates.
Similar results were obtained when the concentrations of both B and B´ were increased from 5 to 40 µM, while maintaining the concentrations of A and A´ at 5 µM (Figure 6D). There was a modest increase in the rates of production of both E and E´ when the concentrations of B and B´ were increased to 10 µM, but then decreased rates of production when the concentrations of B and B´ were further increased to 20–40 µM.
When the concentrations of both A and B´ were increased from 5 to 40 µM, while maintaining the concentrations of A´ and B at 5 µM, the rates of production of both E and E´ increased progressively (Figure 6E). At 40 µM concentrations of A and B´, the exponential growth rate was 0.07 min−1, which is the fastest ever observed with this system, corresponding to a doubling time of ~10 min. The elevated concentrations of both A and B´ result in increased concentration of the non-productive A•B´ complex, but also increased concentrations of free A and B´ substrates, the latter contributing to a faster rate of reaction. Because of the equilibrium: A + B´ ⇆ A•B´, there is a square-root relationship between the increase in the total concentrations of A and B´ and the resulting increase in the concentrations of free A and B´ that are available to react.
Finally, the concentrations of A and B were increased simultaneously from 5 to 40 µM, while maintaining the concentrations of A´ and B´ at 5 µM (Figure 6F). This resulted in progressively decreased rates of production of E´, which can be attributed to reduced concentrations of free A´ and B´ due to their increased sequestration within the non-productive complexes. At the same time, the concentrations of free A and B progressively increased, as evidenced by the increased rate of production of E during the initial phase of the reaction (Figure 6F, inset). However, this is a linear rate of reaction because it is not supported by increased production of the autocatalytic partner E´. At 40 µM concentrations of both A and B, the linear rate of production of E was 0.4 min−1, which is close to the rate of dissociation of the E•E´ complex. This is in contrast to the behavior observed with 5 µM concentrations of substrates, where the reaction already exhibits sigmoidicity within the first two hours.
DISCUSSION
A kinetic framework for the self-replicating RNA enzymes can be established from the results obtained in this study. A central conclusion is that the rate of RNA-catalyzed RNA ligation is not rate limiting for the complete replication system. The kcat values for the two component ligation reactions are in the range of 2–5 min−1. If these were rate-limiting, exponential amplification would occur with a doubling time of ~20 s and would reach maximum extent in just a few minutes. Multiple turnover for the two component reactions is limited by the rate of dissociation of the E•E´ complex, which is about 10-fold slower than kcat. However, this too is not rate limiting for the complete replication system. If it were, the system would not exhibit sigmoidal growth and would not proceed with reaction order 1.0, indicative of exponential amplification, and the reaction rate would be about 10-fold faster than the observed rate of ~0.03 min−1.
The observed rate for the complete replication system is restricted by the availability of free substrates, which are present at concentrations well below the KM of the corresponding enzyme-substrate complexes. Due to the inherent complementarity of the system, substrates A and B bind to E´, but also bind to opposing substrates B´ and A´, respectively. Similarly, A´ and B´ bind to E, but also to B and A, respectively. It is the formation of the non-productive A•B´ and A´•B complexes that reduces the availability of free substrates. The KM values of the enzymes for their corresponding substrates are in the range of 1–8 µM, but the Kd values of the non-productive complexes are ~0.1 µM. Thus, when operating at substrate concentrations of 5 µM each, only ~10% of the substrate molecules are free in solution. As a result, the enzymes are poorly saturated and the observed rate is only ~0.03 min−1. Nonetheless, exponential growth is observed because the non-productive complexes have a fast dissociation rate (>1 min−1), which continually makes substrate molecules available to bind to the enzymes.
By increasing the concentration of one of the four substrates, in excess of its partner in the non-productive complex, it is possible to increase the free concentration of that substrate. However, this does not increase the rate of the corresponding component reaction because the other substrate for that reaction remains limiting (Figure 6A,B). Instead the rate of the reciprocal component reaction decreases because the elevated substrate concentration results in more complete sequestration of that substrate’s partner within the non-productive complex. If the concentrations of both substrates for the same component reaction are increased, then the rate of that reaction is increased due to enhanced saturation of the enzyme with both substrates (Figure 6F). However, the rate of the reciprocal reaction is decreased due to enhanced sequestration of both of the reciprocal substrates.
The rates of the two component reactions can be increased by increasing the concentrations of both A and A´ (or both B and B´), but once saturation is reached for the enzyme-substrate complexes, there is no further benefit for the rate of exponential amplification (Figure 6C,D). Substrate concentrations substantially in excess of the KM only contribute to the formation of non-productive complexes. A special case arises when the concentrations are increased for both components of the same non-productive complex (Figure 6E). This results in increased sequestration of both excess substrates within the same non-productive complex, but also increased free concentrations of those substrates due to the equilibrium between bound and free components. The concentration of free components increases as the square root of the increased concentration of the bound components, still contributing to enhanced saturation of both enzymes, and therefore increasing the rate of exponential amplification.
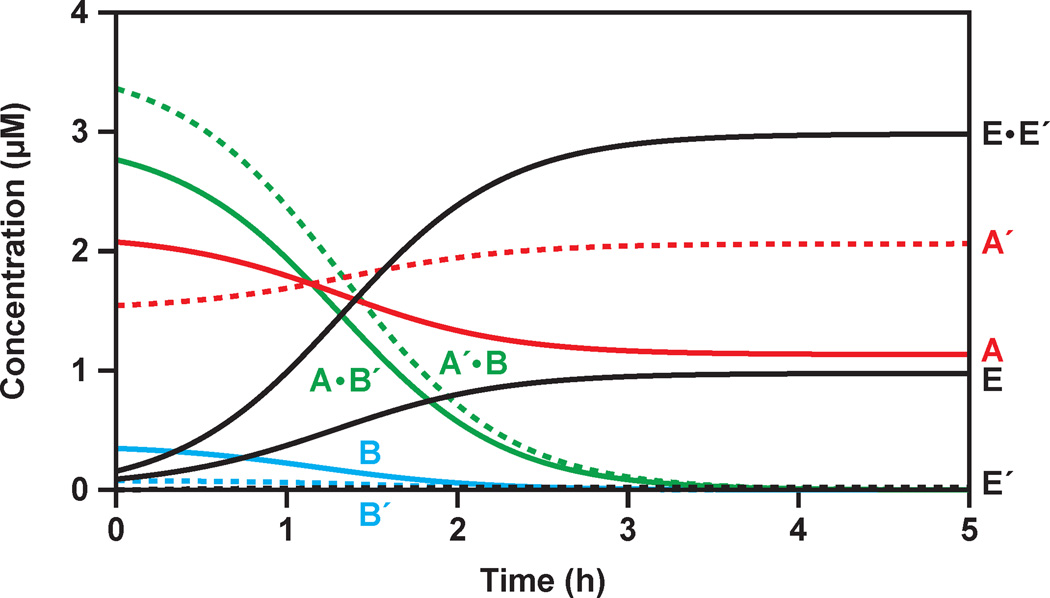
Kinetic modeling was carried out to better understand the overall reaction system and to seek ways to improve the rate of exponential amplification. The exponential amplification profiles (Figure 2A) were fit to a model based on Figure 1B, including the non-productive A•B´ and A´•B complexes, but not the spontaneous reaction pathway initiated by the B´•A•A´•B complex. The starting concentrations of the B and B´ substrates were adjusted for their maximum extent of reaction (see Results). The experimentally determined Kd values for the E•E´, A•B´, and A´•B complexes were used to calculate the concentrations of free and bound reactants throughout the course of amplification (see Experimental Procedures).
Figure 7 depicts the estimated concentrations of the various components over time, starting with 0.1 µM each of E and E´. As expected, the majority of substrate molecules exist as A•B´ and A´•B complexes and the majority of enzyme molecules exist as the E•E´ complex. The overall kinetic profile is controlled by the relative equilibrium constants of the various complexes, with exponential amplification occurring at a rate of 0.034 min−1.
Figure 7.
Estimation of the concentrations of various reaction components during the course of exponential amplification, based on the data shown in Figure 2A and the experimentally determined Kd values for the E•E´, A•B´, and A´•B complexes. The starting concentrations of A and A´ (red) were 5 µM each; the starting concentrations of B and B´ (blue), adjusted for their maximum extent of reaction, were 3.5 and 3 µM, respectively; and the starting concentrations of E and E´ (black) were 0.1 µM each. At the outset of the reaction most of the substrates existed as A•B´ and A´•B complexes (green), with the excess A and A´ molecules remaining free in solution.
There are several ways in which faster rates of exponential amplification might be achieved. A modest increase in kcat of the two component reactions would result in a proportional increase in the observed rates of these reactions (Eq. 8) and in the overall rate of amplification. However, if kcat were to increase by more than 7-fold, then dissociation of the E•E´ complex would become rate limiting and the system would no longer exhibit exponential growth. The best that could achieved would be an exponential growth rate of ~0.2 min−1, corresponding to a doubling time of 3 min. Faster exponential growth also would result if the Km of the enzyme-substrate complexes were reduced relative to the Kd of the non-productive A•B´ and A´•B complexes. This would provide more complete saturation of the enzymes for a given concentration of free substrates, contributing to faster observed rates of reaction. Again, however, the maximum rate of exponential growth would be limited by the rate of dissociation of the E•E´ complex. Faster product dissociation would allow a higher maximum rate of exponential amplification. It also would benefit the amplification rate, even without an improved kcat or Km, because it would provide higher concentrations of free E and E´ to carry out the ligation reaction.
Improved replicator designs should aim to improve the catalytic efficiency, kcat/KM, of the RNA-catalyzed reactions, within the limit set by the rate of dissociation of the E•E´ complex. This might be achieved by increasing the stability of enzyme-substrate complexes, so long as that did not result in comparable stabilization of the A•B´ and A´•B complexes, which rely on the same base-pairing interactions. It would be desirable, for example, to improve KM through enhanced tertiary interactions in the region of the ligation junction, which would not pertain to the non-productive complexes. In vitro evolution experiments are currently underway to develop self-replicating RNA enzymes with an improved kcat/KM. Such enzymes would have the added advantage of being able to operate efficiently at lower concentrations of substrates, thus enabling implementation of more complex populations during self-sustained evolution and more sensitive detection of target molecules during ligand-dependent exponential amplification.
The RNA-catalyzed exponential amplification system is the only known genetic system outside of terrestrial biology. It provides a working model of biological replication and has some capacity for self-sustained Darwinian evolution. In both natural and synthetic genetic systems, replication is the primary determinant of fitness. Replication entails not only the literal duplication of the genetic material, but also ensuring an adequate supply of the components that support the replicative process. As the present study demonstrates, when the replication machinery is starved for fuel, its performance is restricted. Genetic variants that are less subject to these restrictions would enjoy a selective advantage, exhibiting faster exponential growth rates that would allow them to outcompete others in an evolving population.
ACKNOWLEDGMENTS
This work was supported by grant NNX07AJ23G from NASA and grant R01GM065130 from the NIH. A.C.F. was supported by a postdoctoral cross-disciplinary fellowship from the Human Frontier Science Organization.
REFERENCES
- 1.von Kiedrowski G. A self-replicating hexadeoxynucleotide. Angew. Chemie. 1986;25:932–935. [Google Scholar]
- 2.Zielinski WS, Orgel LE. Autocatalytic synthesis of a tetranucleotide analogue. Nature. 1987;327:346–347. doi: 10.1038/327346a0. [DOI] [PubMed] [Google Scholar]
- 3.von Kiedrowski G, Wlotzka B, Helbing J, Mazten M, Jordan S. Parabolic growth of a self-replicating hexadeoxynucleotide bearing a 3´–5´-phosphoamidate linkage. Angew. Chemie. 1991;30:423–426. [Google Scholar]
- 4.Paul N, Joyce GF. A self-replicating ligase ribozyme. Proc. Natl. Acad. Sci. USA. 2002;99:12733–12740. doi: 10.1073/pnas.202471099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vaidya N, Manapat ML, Chen IA, Xulvi-Brunet R, Hayden EJ, Lehman N. Spontaneous network formation among cooperative RNA replicators. Nature. 2012;491:72–77. doi: 10.1038/nature11549. [DOI] [PubMed] [Google Scholar]
- 6.Lee DH, Granja JR, Martinez JA, Severin K, Ghadiri MR. A self-replicating peptide. Nature. 1996;382:525–528. doi: 10.1038/382525a0. [DOI] [PubMed] [Google Scholar]
- 7.Yao S, Ghosh I, Zutshi R, Chmielewski J. A pH-modulated, self-replicating peptide. J. Am. Chem. Soc. 1997;119:10559–10560. [Google Scholar]
- 8.Rubinov B, Wagner N, Rapaport H, Ashkenasy G. Self-replicating amphiphilic β-sheet peptides. Angew. Chemie. 2009;48:6683–6686. doi: 10.1002/anie.200902790. [DOI] [PubMed] [Google Scholar]
- 9.Tjivikua T, Ballester P, Rebek J. A self-replicating system. J. Am. Chem. Soc. 1990;112:1249–1250. [Google Scholar]
- 10.Terfort A, von Kiedrowski G. Self-replication by condensation of 3-amino-benzamidines and 2-formylphenoxyacetic acids. Angew. Chemie. 1992;5:654–656. [Google Scholar]
- 11.Wang T, Sha RJ, Dreyfus R, Leunissen ME, Maass C, Pine DJ, Chaikin PM, Seeman NC. Self-replication of information-bearing nanoscale patterns. Nature. 2011;478:225–228. doi: 10.1038/nature10500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schulman R, Yurke B, Winfree E. Robust self-replication of combinatorial information via crystal growth and scission. Proc. Natl. Acad. Sci. USA. 2012;109:6405–6410. doi: 10.1073/pnas.1117813109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.von Kiedrowski G. Minimal Replicator Theory I: Parabolic versus Exponential Growth. In: Dugas H, editor. Bioorganic Chemistry. vol. 3. New York: Springer-Verlag; 1993. pp. 115–146. [Google Scholar]
- 14.Issac R, Chmielewski J. Approaching exponential growth with a self-replicating peptide. J. Am. Chem. Soc. 2002;124:6808–6809. doi: 10.1021/ja026024i. [DOI] [PubMed] [Google Scholar]
- 15.Rogers J, Joyce GF. The effect of cytidine on the structure and function of an RNA ligase ribozyme. RNA. 2001;7:395–404. doi: 10.1017/s135583820100228x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lincoln TA, Joyce GF. Self-sustained replication of an RNA enzyme. Science. 2009;323:1229–1232. doi: 10.1126/science.1167856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim DE, Joyce GF. Cross-catalytic replication of an RNA ligase ribozyme. Chem. Biol. 2004;11:1505–1512. doi: 10.1016/j.chembiol.2004.08.021. [DOI] [PubMed] [Google Scholar]
- 18.Sczepanski JT, Joyce GF. Synthetic evolving systems that implement a user-specified genetic code of arbitrary design. Chem. Biol. 2012;19:1324–1332. doi: 10.1016/j.chembiol.2012.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lam BJ, Joyce GF. Autocatalytic aptazymes enable ligand-dependent exponential amplification of RNA. Nat. Biotechnol. 2009;27:288–292. doi: 10.1038/nbt.1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lam BJ, Joyce GF. An isothermal system that couples ligand-dependent catalysis to ligand-independent exponential amplification. J. Am. Chem. Soc. 2011;133:3191–3197. doi: 10.1021/ja111136d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pluthero FG. Rapid purification of high-activity Taq DNA polymerase. Nucleic Acids Res. 1993;21:4850–4851. doi: 10.1093/nar/21.20.4850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ellinger T, Ehricht R. Single-step purification of T7 RNA polymerase with a 6-histidine tag. BioTechniques. 1998;24:718–720. doi: 10.2144/98245bm03. [DOI] [PubMed] [Google Scholar]
- 23.Forster AC, Altman S. External guide sequences for an RNA enzyme. Science. 1990;249:783–786. doi: 10.1126/science.1697102. [DOI] [PubMed] [Google Scholar]
- 24.Olea C, Jr, Horning DP, Joyce GF. Ligand-dependent exponential amplification of a self-replicating L-RNA enzyme. J. Am. Chem. Soc. 2012;134:8050–8053. doi: 10.1021/ja302197x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pleiss JA, Derrick ML, Uhlenbeck OC. T7 RNA polymerase produces 5´ end heterogeneity during in vitro transcription from certain templates. RNA. 1998;4:1313–1317. doi: 10.1017/s135583829800106x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Helm M, Brulé H, Giegé R, Florentz C. More mistakes by T7 RNA polymerase at the 5´ ends of in vitro-transcribed RNAs. RNA. 1999;5:618–621. doi: 10.1017/s1355838299982328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nelson JW, Tinoco I. Comparison of the kinetics of ribo-, deoxyribo- and hybrid oligonucleotide double-strand formation by temperature-jump kinetics. Biochemistry. 1982;21:5289–5295. doi: 10.1021/bi00264a026. [DOI] [PubMed] [Google Scholar]
- 28.Santoro SW, Joyce GF. Mechanism and utility of an RNA-cleaving DNA enzyme. Biochemistry. 1998;37:13330–13342. doi: 10.1021/bi9812221. [DOI] [PubMed] [Google Scholar]