Abstract
Background
Epidermal growth factor receptor (EGFR)-targeted agents have demonstrated clinical benefit in patients with cancer. Identifying tissue-of-origin-independent predictive biomarkers is important to optimally treat patients. We sought to identify a gene array profile that could predict responsiveness to panitumumab, a fully human EGFR-binding antibody, using preclinical models of human cancer.
Methods
Mice bearing 25 different xenograft models were treated twice weekly with panitumumab or immunoglobulin G2 control to determine their responsiveness to panitumumab. Samples from these xenografts and untreated xenografts were arrayed on the Affymetrix human U133A gene chip to identify gene sets predicting responsiveness to panitumumab using univariate and multivariate analyses. The predictive models were validated using the leave-one-group-out (LOO) method.
Results
Of the 25 xenograft models tested, 12 were responsive and 13 were resistant to panitumumab. Unsupervised analysis demonstrated that the xenograft models clustered by tissue type rather than responsiveness to panitumumab. After normalizing for tissue effects, samples clustered by responsiveness using an unsupervised multidimensional scaling. A multivariate selection algorithm was used to select 13 genes that could stratify xenograft models based on responsiveness after adjustment for tissue effects. The method was validated using the LOO method on a training set of 22 models and confirmed independently on three new models. In contrast, a univariate gene selection method resulted in higher misclassification rates.
Conclusion
A model was constructed from microarray data that prospectively predict responsiveness to panitumumab in xenograft models. This approach may help identify patients, independent of disease origin, likely to benefit from panitumumab.
Introduction
The epidermal growth factor receptor (EGFR) is a tyrosine kinase transmembrane receptor that mediates the mitogen-activated protein kinase (MAPK), phosphoinositide 3-kinase (PI3K), and STAT signaling pathways [1]. Activation of these pathways results in cellular proliferation, adhesion, migration, and survival [2–4]. EGFR is overexpressed in solid tumors, including colorectal, lung, head and neck, and breast carcinomas, and correlates with poorer prognosis in patients [5,6].
Panitumumab is a fully human monoclonal antibody that binds to the EGFR and prevents ligand-induced activation, resulting in arrest of tumor cell proliferation, production of angiogenic factors, and survival [7–10]. Panitumumab is approved as monotherapy for the treatment of metastatic colorectal cancer refractory to fluoropyrimidine-, oxaliplatin-, and irinotecan-based chemotherapy regimens, but it is not recommended for patients with mutations in KRAS codon 12 or 13 [11].
Currently, anti-EGFR therapies result in clinical benefit in approximately 32% to 44% of patients, with response rates of approximately 8% to 11% and median survival times ranging from approximately 6 to 7 months as monotherapy [12–16] and response rates of approximately 50% to 60% and median survival of approximately 20 to 24 months in combination with chemotherapy in the first line setting [12,17,18]. These relatively low response rates continue to challenge clinicians in determining the best treatment options for their patients, especially for those with metastatic late-stage disease, and underscore the need for better patient selection to maximize clinical benefit and the risk/benefit ratio. Although some progress has been made to help stratify patients using biomarkers such as gene amplification, mutations in genes including KRAS, BRAF, PI3K, and PTEN; and expression of EGFR ligands, many patients with normal expression of these markers progress on anti-EGFR therapy [19–25]. A further understanding of the underlying mechanisms of dependency on the EGFR signaling pathway may aid in identifying biomarkers that have a stronger predictive value than those that currently exist.
Microarray technology has been useful in identifying genes that can serve as potential biomarkers in a variety of settings. Numerous microarray studies have identified genes that can classify tumor pathologic subtype, metastatic potential, correlation to therapeutic outcome, and potential for disease recurrence [26]. More recently, efforts have focused on understanding EGFR pathway dependence [27] and responsiveness to EGFR inhibitors [25,28]. In colorectal tumors, transcript expression levels of single genes involved in the EGFR pathway (AREG and EREG) were significantly associated with progression-free survival [25,29] with cetuximab treatment; however, these results were not better predictors of response than using a simple genetic test for mutations in the KRAS gene [28]. Furthermore, many patients with wild-type KRAS do not benefit from anti-EGFR therapy [20]. Because pathways can have overlapping sets of transcriptional targets, univariate gene selection methods may not be sufficient to find the pathway(s) driving a particular tumor. Identification of a gene signature consisting of multiple genes using a multivariate selection methodology as described by Liu and Wu [30] that could predict responsiveness to targeted therapies, such as panitumumab, could ultimately improve the ability of clinicians to provide optimal treatment for their patients.
Microarray analysis on 25 different, untreated xenograft models was performed to determine a potential gene array profile that could predict responsiveness to panitumumab and to investigate any potential advantage of a multivariate selection methodology compared with a univariate selection for determining this predictive profile.
Materials and Methods
Xenograft Models
A total of 25 cell lines were selected for the xenograft models and for microarray analyses (Table 1). Female CD-1 nu/nu mice (Charles River Laboratories, Wilmington, MA) aged 5 to 6 weeks were received and housed in sterilized caging and acclimated. Xenograft models of each cell line were prepared by subcutaneous injection of 1 x 106 to 1x107 cells of a single cell line into the left flank of the mouse. The mice were observed daily, and tumors were allowed to grow to an average size of approximately 200 mm3 before treatment. Because archival tissue from the initial surgery/diagnosis is most commonly available for cancer patients, we sought to determine a predictive profile using tumors collected prior to panitumumab treatment. Therefore, untreated tumors from five animals from each xenograft model were subjected to microarray analysis.
Table 1.
Xenograft Models of Human Cancer Cell Lines and In Vivo Response (as Observed by Tumor Growth Inhibition with Panitumumab Treatment)*.
| Tissue | Model | Response |
| Breast | MDA-MB468 | + |
| CNS | U118 | + |
| Colon | DLD-1 | + |
| Colon | HT-29 | + |
| Lung | A549 | + |
| Lung | NCI-H1650 | + |
| Lung | NCI-H1975 | + |
| Pancreas | BxPC-3 | + |
| Pancreas | MIAPaCa | + |
| Prostate | DU-145 | + |
| Prostate | PC-3 | + |
| Skin | A431 | + |
| Breast | BT-474 | - |
| Breast | MCF-7 | - |
| Breast | MDA-MB231 | - |
| Breast | ZR-75-1 | - |
| CNS | U-87 | - |
| Colon | Colo-205 | - |
| Lung | Calu-6 | - |
| Lung | NCI-H1299 | - |
| Lung | NCI-H460 | - |
| Lung | NCI-H82 | - |
| Lung | SK-MES-PD | - |
| Pancreas | CaPan-1bw | - |
| Pancreas | Panc-1 | - |
CNS indicates central nervous system.
Five animals per model were used, with the exception of CaPan-1bw and Panc-1, where 10 animals per model were used.
The mice were then treated with 5, 20, 100, 200, or 500 µg of panitumumab from a stock solution (20 mg/ml panitumumab in 50 mM acetate, 100 mM NaCl, pH 5.8) or immunoglobulin G2 (IgG2) control antibody twice weekly via intraperitoneal injection. Response was determined as a ≥40% inhibition of mean tumor volume in the treatment group compared with the control group at the last time point at the highest tested dose of panitumumab. Five to ten animals per dose group were tested to determine the response to panitumumab versus IgG2 control antibody treatment (see Legend, Table 1). Tumor volumes, calculated as length x width x height in millimeters cubed, and body weights were recorded at regular intervals. Statistical significance of observed differences between growth curves was evaluated by repeated measures analysis of variance followed by Scheffé post hoc testing for multiple comparisons.
All animal studies were performed under an internal Institutional Animal Care and Use Committee protocol and met all Association for Assessment and Accreditation of Laboratory Animal Care international specifications.
RNA Preparation for Microarray Assays
To minimize bias during the preparation of sample RNA from the tumors, we processed together samples from as many models as possible (between 6 and 12 models at a time). Any bias introduced from a particular batch would apply equally to all models processed in the same batch. Replicate runs from five different animals carrying the same xenograft model were processed in separate batches and hybridized on five different lots of microarray chips.
For each xenograft, 300-mm3 untreated tumors were collected. Total RNA was extracted from approximately 100 to 150 mg of tissue using Qiagen RNeasy Kit (Qiagen, Valencia, CA). Standard cRNA labeling and array processing were conducted per protocol (Affymetrix Technical Manual [31], Chapter 2: Eukaryotic Target Processing). First-strand synthesis was performed using 5 µg of total RNA, 10 pmol of T7-(dT)24 primer, and Superscript II RNase-H reverse transcriptase (Invitrogen, Carlsbad, CA). Double-stranded cDNA was purified using the MinElute Reaction Cleanup Kit (Qiagen). Biotinylated cRNA was synthesized using Bioarray High-Yield RNA Transcript Labeling Kit (Enzo Diagnostics, Farmingdale, NY) over 6 hours at 37°C, purified using the Qiagen RNeasy Mini Kit, and hybridized to the Affymetrix Human Genome U133A gene chip (Affymetrix, Santa Clara, CA). This chip contained approximately 22,000 probe sets.
Statistical Analysis
Data from Affymetrix .CEL files were imported into Matlab Version 7.0.1 (Mathworks, Inc, Natick, MA) and processed without background subtraction. Only the intensities from the perfect match probes were used. The minimum intensity from each chip was subtracted from all intensities on the chip and the value of 1 was added so that the subsequent minimum intensity on each chip equaled 1. The intensities were transformed using a natural log transformation. Finally, a chip-specific nonlinear normalization function that mapped intensities to a common distribution that was based on the entire data set was found. This was done so that the intensities from each chip had approximately the same minimum, range, and variance. This has a similar goal to the popular Robust Multichip Average method [32], but our method preserves the local structure seen in the original intensity distribution. Finally, the average of the transformed and normalized intensities for each probe in a probe set was computed. These averaged intensity values were used in all subsequent analysis. Some figures were prepared after reimporting data into Partek Pro Version 6.0 (Partek Incorporated, St Charles, MO). Tissue effects were estimated for each gene using the following model: where response is a nominal factor (R/NR). Least squares estimates for the tissue effect were subtracted from each intensity to produce tissue-adjusted intensities.
The number of genes was reduced from a total of approximately 22,000 on the microarray chip to approximately 9500 genes by selecting those that had the greatest variance in expression across all of the samples and those that had the greatest entropy (i.e., the union of the top 60% of entropy and variance). For the univariate selection method, a t test was used and the top genes were selected on the basis of the significance of the response effect. For the multivariate gene selection, an approach that could effectively search the most probable combinations was needed because too many gene combinations exist to search them all. For this, we used a genetic algorithm [33], which was designed using Matlab Genetic Algorithm and Direct Search Toolbox, Version 1.0.3 (R14SP2).
Ideally, the optimal search parameters that would not bias the results would be determined in a nested leave-one-group-out (LOO) method [34]. However, because we had a limited number of samples from certain tissues, we were unable to obtain these optimal parameters. Instead, we selected search parameters first before conducting a supervised examination of the data set to avoid biasing the results. Also, the best number of genes to include in the gene set was not initially known. Based on an unsupervised principal component analysis calculated using the covariance matrix, the first 13 principal components represented approximately 80% of the variability in the data. We decided to build the predictive model using 13 genes.
A genetic algorithm attempts to find the best gene sets, called individuals, in an iterative process. In each iteration, called a generation, the individuals are scored, and the best, most fit, individuals are increased in their likelihood of contributing their genes to the next generation. In our case, the individuals were scored using a linear discriminant method function, which is a measure of the within-group to between-group variance [35]. We started with the same number of individuals as there were candidate genes, and in each generation, the top 20% of the individuals were retained and their gene sets were passed along unaltered to the next search generation. The remaining 80% of the individuals in the new population were either a random combination of the genes from two individuals or the gene sets from one of the best individuals that were subject to mutation (substitute one gene for another one at random). In both cases, the parents were found using a fitness-weighted random selection method. The mutation rate was chosen such that each individual undergoing a mutation had, on average, one gene mutated. The search was conducted for up to 100 generations or until the best scoring individual no longer improved for 10 successive generations.
Validation was conducted using a full LOO analysis for both univariate and multivariate gene selection methods. In our study, a group corresponds to a tumor xenograft model (typically five animals per group). In each loop of the LOO, all data from one xenograft model were removed from the data set and to avoid biasing the results of the validation, gene selection was repeated during each loop of the LOO validation [36]. In each loop, a fully specified linear model was used to predict the responsiveness of the group that was left out. This provides a binary classification and a distance to the centroids of each class. The sum of squares error from the continuous distance to the correct class was calculated and further used to calculate the predicted error sums of squares, commonly called prediction sum of squares (PRESS) [37]. The classification was then compared to the actual response obtained from the original xenograft tumor growth inhibition results. The results of the LOO validation were compared with those obtained from a univariate selection method using the same size gene set. As a control, additional validation was achieved by randomly permuting the response labels within tissues and then repeating the entire LOO analysis.
To understand the minimal number of genes needed to predict and to avoid complications due to overfitting [34], we determined an optimal number of genes to use in a prediction set. This data set was used to predict the best gene signature for signature sizes from 1 to 13. These will not be considered internally validated until additional data become available.
Results
Identification of Panitumumab-Responsive Cell Lines in Xenograft Models
Panitumumab treatment of 25 established xenograft models demonstrated that 12 xenograft models, including three lung, two colon, two pancreas, two prostate, one breast, one glioblastoma, and one epidermoid cancer lines, were responsive to panitumumab as determined by tumor growth inhibition versus IgG2 control, whereas the remaining 13 models were not responsive to panitumumab (Table 1). In the 12 responsive models, tumor growth inhibition was statistically significant at the highest dose of panitumumab at 500 µg once every 2 weeks (P < .05). These results were used to assign the models to either responder or nonresponder groups.
Tissue Type Is a Major Factor in Clustering in the Principal Component Microarray Analysis
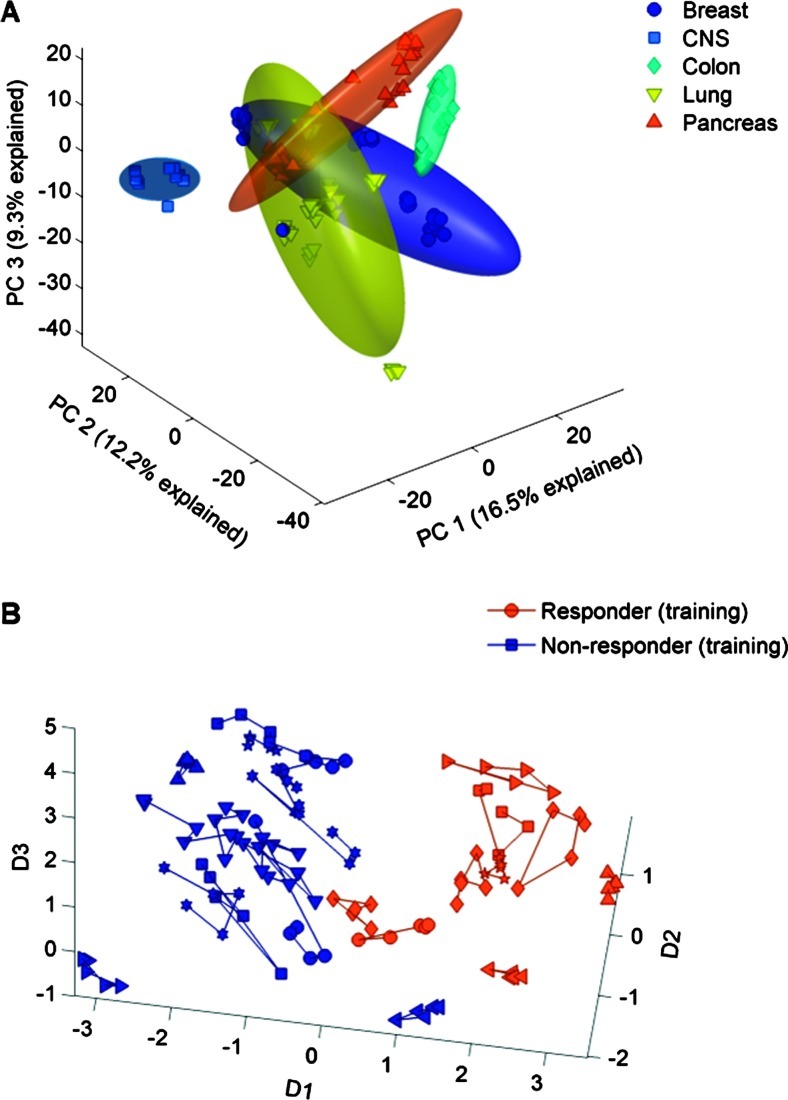
For each of the 25 untreated xenograft models, microarray analysis was performed on tumor samples from five different animals to determine the relative level of gene expression on both responsive and nonresponsive models. In an unsupervised principal component analysis clustering, the xenograft models separated on the basis of tissue type rather than responsiveness to panitumumab (Figure 1A). The principal component intensities for each model were used to calculate means and two SD radii for ellipsoids. In an analysis of variance, the average mean square error was 1.139 for tissue type and 0.819 for response to panitumumab, indicating that tissue type was the largest source of variability.
Figure 1.
(A) Tissue type is a major factor in clustering in the principal component analysis of 120 spots—each point is the average of five replicates. Data from the training set are shown. (B) Response to panitumumab is a major factor after subtracting tissue effects—each one is a separate model, connected by lines.
Response Is a Major Factor in Gene Regulation after Subtracting Tissue Effects
A mixed-effects nested model with tissue, response, and tumor model was applied to each gene, where model was treated as a random effect. Tissue and response were contained in the tumor model. Variance components were estimated using Restricted Estimated Maximum Likelihood. Adjusting for multiple tests using Bonferroni led to no differentially expressed genes between responders and non-responders. At a false discovery rate of 0.05 [38], nearly 500 genes that were statistically different between responders and nonresponders were found.
Because tissue was the major contributing source of variability, we sought to reduce the effects of tissue on the prediction of response, which would interfere with our multivariate gene selection procedure. For each of the 9500 genes evaluated, we adjusted the observed intensities by subtracting the marginal mean intensity for each tissue. Thus, for a given gene, the average adjusted intensities for each tissue were the same. After subtracting the tissue effect, a multidimensional scaling analysis showed that the samples clustered by responsiveness (Figure 1B).
A Set of 13 Genes Can Predict Responsiveness
Twenty-two xenograft models were selected to be the training set because these were derived from tissues that included examples of both responder and nonresponder (i.e., the tissues were balanced). The remaining three models were responders from completely different tissue types than those used to build the model and were used as an independent test set.
We used a genetic algorithm combined with a linear discriminant to select 13 genes that could separate responsive and nonresponsive models. Validation runs were conducted with the LOO method. In this analysis, all data from one model were removed from the data set. The remaining data were used to select a set of genes and to build a predictive model, which was then applied on the xenograft model left out. The prediction was then compared to the actual response obtained from the original xenograft tumor growth inhibition results. Twenty-two independent tests were performed and response prediction was 100%. As a control, the same analyses were conducted on xenograft models that were randomly assigned to the responsive or nonresponsive group regardless of actual responsiveness. This random assignment resulted in an overall predictive rate of 55% (Table 2).
Table 2.
Prediction of Responsiveness via Univariate or Multivariate Methodology and Comparison with Xenograft Results*.
| Method | Correct Prediction | Overall Prediction (%) | |
| Response (+, %) | Nonresponse (-, %) | ||
| Thirteen gene set—multivariate | 100 | 100 | 100 |
| Five gene set—multivariate | 100 | 100 | 100 |
| Four gene set—multivariate | 100 | 100 | 100 |
| Thirteen gene set—univariate | 64 | 81 | 75 |
| Five gene set—univariate | 64 | 81 | 75 |
| Four gene set—univariate | 62 | 69 | 67 |
| Random assignment | 44 | 61 | 55 |
N = 13, randomized control.
Four, 5, or 13 genes were selected by either methodology. A control experiment denoted as “Random assignment,” which randomly assigned models as either positive or negative for response, was included (n = 13 genes).
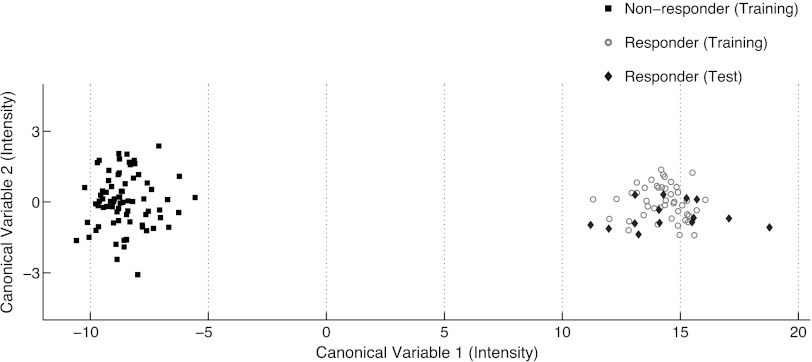
After LOO validation of the multivariate algorithm, a final set of 13 genes was selected from all 22 models. This predictive model was used to prospectively predict responsiveness in the three independent xenograft models (Figure 2).
Figure 2.
LOO validation using the top 13 genes selected by multivariate methodology.
Multivariate Compared with Univariate Gene Selection
To compare the gene selection methods, we chose an additional set of 13 genes using a univariate gene selection methodology. A t test was used to select the top 13 genes with the most significant difference in expression between responders from nonresponders, and a classification model was built using a linear discriminant analysis. The first two canonical variables are plotted in Figure 2. Genes selected using the multivariate method had a Mahalanobis distance between group means of 22.8, whereas genes selected using the t test had a distance of only 6.9. These results indicate that separation of groups is greater using a multivariate methodology versus univariate methodology.
Optimization of the Number of Genes
The intention of the previous analysis was to obtain an unbiased estimate of the performance of the algorithm. In addition, we used this same data to determine the optimal number of genes to prospectively predict outcomes. This is usually a balance between predictive power (reduction in bias) of the gene set and its ability to generalize (minimum variance). Too few genes will tend to be lacking in predictive power and too many genes will tend to overfit the data and simply add variance without aiding, and perhaps harming, predictive power [39].
To determine the optimal number of genes, we varied the number from 1 to 13 genes. For each specified gene set size, the LOO analysis was repeated. We found that as few as four genes resulted in 100% correct prediction (Table 2). Interestingly, both 5 and 13 genes also resulted in 100% correct prediction. After testing both methods, the univariate gene selection methodology resulted in lower prediction rates (67% to 75%) than the multivariate gene set selection methodology (100%) when compared using 4, 5, and 13 genes. These results further confirm that multivariate methodology provided the best predictive power.
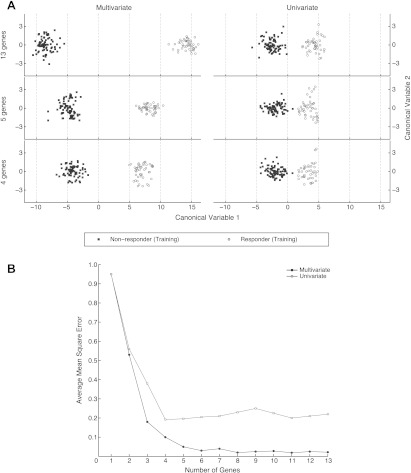
The separation achieved using the best gene set of a given size, which were identified using the whole data set, is shown in Figure 3A. This Figure shows the first two canonical variables using the given number of genes selected by either the univariate or multivariate method. From these plots, the intergroup variability compared to the intragroup variability is clearly greater in the multivariate analysis for all three gene sets than in any group using the univariate selection method. The distance between groups using four genes selected together was 10.9, whereas the distance using 13 genes selected independently was 6.9. The separation distance between groups also increased as the number of genes in the set increased; however, because of previously described overfitting concerns, and because adding more than five genes did not significantly reduce the PRESS, we maintained the optimal gene set at five genes.
Figure 3.
(A) Multivariate versus univariate separation of groups (responders versus nonresponders) based on gene sets of 4, 5, and 13 genes. The top 4, 5, and 13 genes in each set were selected. The axes are canonical variables 1 and 2. (B) A set of five genes resulted in the best separation and the least prediction error in this data set. This plot illustrates residual error from a linear model using a different number of genes to predict (PRESS method), using univariate selection (red) and multivariate (blue) methods.
To quantify the separation from each of the runs, we used the PRESS model to estimate the error of each prediction in the context of an LOO analysis. The average mean square error over all results for each number of genes using both multivariate and univariate analyses is shown in Figure 3B. In the multivariate selection method, there was a significant reduction in error (P < .05) for each new gene (up to five genes) that was added to the model. However, increasing the number of genes beyond 5, that is, from 6 to 13 genes, did not improve the error significantly. Therefore, five genes in the gene set appeared to be the optimal number of genes. Using univariate analysis, the average mean square error plateaued with four genes, suggesting that a four-gene set is optimal in the univariate selection model. The PRESS was always equal or lower for the multivariate method than for the univariate, although we did not try to use the univariate or multivariate selection method for more than 13 genes.
A Final Search Identified a Set of Five Genes that Can Predict Responsiveness
We conducted a final search for the best set of five genes. In this search, the genetic algorithm was run 500 times, compared with 100 times in the previous searches, and the initial population was increased to 1 million individual gene sets. The simulation was stopped after 50 generations or five generations with no change in the best score. The top 5 genes and biologic function are listed (Table 3).
Table 3.
Gene Signature of the Top 5 Genes (Multivariate Method).
| Number | Gene Symbol | Probe ID | Gene/Biologic Function |
| 1 | TP73L | 209863_S_AT | Tumor protein p73-like |
| 2 | POLG | 213008_AT | Polymerase (DNA directed), gamma |
| 3 | MRPS10 | 218106_S_AT | Mitochondrial ribosomal protein S10 |
| 4 | YWHAH | 201020_AT | Tyrosine 3/tryptophan 5-monooxygenase activation protein, eta polypeptide |
| 5 | ROR1 | 205805_S_AT | Receptor tyrosine kinase-like orphan receptor 1 |
Discussion
The advancements in anti-EGFR therapy for many cancers, including colorectal, lung, and head and neck, are promising and further validate the role of the EGFR pathway in tumor growth [40]. Although efficacy across tumor types with small molecule or antibody monotherapies remains relatively low in unselected patient populations [12], it is improved in a selected population [19,20,41]. To understand the mechanism of action of panitumumab, we used animal models, microarray, and multivariate methodology to identify a gene signature that would predict responsiveness to panitumumab in xenograft models. These findings indicate that additional pathways and genes that are not inhibited with anti-EGFR therapies are involved in tumor growth and metastases. Gene expression profiling can identify these genes, and, as biomarkers, these genes can potentially be useful in defining a patient population that will respond and/or have a good prognosis with certain therapies.
Public DNA and microarray databases, such as the Stanford Micro-array Database, have been useful in developing tumor classification models in breast tumors [42] and lymphoma [43,44]. The usefulness of multivariate subset selection in this context has been demonstrated [30,45]. Here, we show the advantage of using a genetic algorithm compared with a univariate method for predicting responsiveness to panitumumab in animal models of human disease.
Multidimensional scaling of data normalized for tissue shows responsive animals cluster together (Figure 1B), suggesting that a tissueindependent and EGFR mutation-independent signature may exist. We also observed that in the multidimensional scaling, the responders were clustered more tightly than nonresponders. Given that panitumumab specifically inhibits ligand-induced autophosphorylation of EGFR, the gene signature may be expected to be similar among responders, whereas nonresponsiveness can arise from many other pathways being activated. This would result in a wide array of gene transcriptional variations in a broad, loosely clustered gene space.
These data may also suggest that it is possible to find a signature in the entire gene set (Figure 1B). However, for clinical usefulness, selecting a small number of genes that could be assayed from a formalin-fixed, paraffin-embedded section is more practical. Therefore, we decided to use a subset selection method to find a small set of genes that would adequately separate responders and nonresponders.
Our multivariate method was built on 22 heterogeneous tumor xenografts representing five different tissue types, and we successfully identified an optimal gene signature of five genes that could predict responsiveness to panitumumab in tumor models. Our predictive model was internally validated using the LOO method with test models, each resulting in 100% correct predictive power. Although the gene signatures (from 4 to 13 genes) from the multivariate analysis all had 100% correct prediction rate, we determined that signatures with more than five genes did not significantly reduce the marginal error rate.
Most of the genes identified have been implicated in tumorigenesis or in cell proliferation. Tumor-protein 73-like plays a role in apoptosis and cell cycle regulation and has also been implicated in increasing the transcription rate of vascular endothelial growth factor [46]. ROR1 is an orphan receptor tyrosine kinase. Although its function is unclear, knock-out mice studies suggest a role in development of various tissues. YWHAH is a 14-3-3 family member that interacts with p53 and may regulate apoptosis. Tumor models with high levels of expression of these genes tend to respond to treatment with anti-EGFR antibody. The last two genes in the signature (MRPS10 and PolG) are related to mitochondrial function. High levels of these gene products correlate with nonresponsiveness in this study. MRPS10 is a nuclear encoded mitochondrial ribosomal protein S10. It is a structural component of the mitochondrial ribosome and is necessary for protein production within the mitochondria. PolG is the only DNA polymerase for mitochondrial DNA replication. The role of these genes in predicting responsiveness is unclear and warrants further investigation.
Interestingly, this signature predicts responsiveness well when used prospectively on tumors arising from different tissue types. The model was built using five different tissue types, and we successfully prospectively predicted outcomes on two new tissue types that were not used to build the predictive model. This supports the hypothesis that one mechanism of action for EGFR in tumor growth exists and that this mechanism is consistent across all tissue types. Further, our data show that this tissue-independent molecular signature can be identified, and, eventually, may be used on patient samples to potentially predict outcomes.
We noted during our LOO and subsequent analysis that it is possible and likely that any given run of the genetic algorithm may produce a different set of five genes. Our experience has been consistent with other published results [30] such that the top set from any run performs well in predicting an outcome. We have also shown that it is possible to substitute highly correlated genes for any or all of the five genes in the set that can be used to build highly predictive models (data not shown). We suggest that the gene in the correlation group may represent a biological phenomenon useful in some aspect of the prediction.
In light of numerous attempts to predict response or other clinical outcomes using univariate gene selection methods, our study indicates that better results may be obtained using multivariate gene selection methods. Multivariate methods have been shown to offer improvements in other settings, such as in tumor classification [30], but are not widely used to stratify patients. Our results indicate that for any number of genes in the model to predict responsiveness, multivariate modeling results in smaller error than univariate modeling (Figure 3A).
Recently, EREG and AREG expression levels have been shown to be predictive of disease control [25]. In our data set, EREG and AREG expression levels were well correlated (P < .001 for Pearson correlation coefficient of 0.86). However, a linear discriminant classifier based on the expression of these genes gave a relatively high error rate of 0.36.
In summary, the goal of clinicians is to treat patients with the best therapy that will provide the greatest probability of success in terms of response and survival. With anti-EGFR therapies, more than 40% of patients have a partial response or stable disease, whereas another 50% have progressive disease in the monotherapy setting. If clinicians had biomarkers that would help define tumors that were dependent on EGFR signaling, the success rate in treating patients would increase significantly. In this study, we identified a gene signature that was predictive in animal models. However, because there are fundamental differences between the animal models and human subjects, this model will not likely perform as well in humans. Instead, we suggest that multivariate gene selection techniques may be important to identify signatures driving tumor growth. If this holds, we could then use this algorithm on patient samples to identify biomarkers and aid in future trial designs.
Acknowledgments
The authors thank Ali Hassan, PhD (Complete Healthcare Communications, Inc), whose work was funded by Amgen Inc, for editorial assistance.
Abbreviations
- EGFR
epidermal growth factor receptor
- IgG2
immunoglobulin G2
- LOO
leave-one-group-out
- PRESS
prediction sum of squares
Footnotes
This study was funded by Amgen Inc. All authors are employees of and stockholders in Amgen Inc.
References
- 1.Yarden Y, Sliwkowski MX. Untangling the ErbB signalling network. Nat Rev Mol Cell Biol. 2001;2:127–137. doi: 10.1038/35052073. [DOI] [PubMed] [Google Scholar]
- 2.Cohenuram M, Saif MW. Panitumumab the first fully human monoclonal antibody: from the bench to the clinic. Anticancer Drugs. 2007;18:7–15. doi: 10.1097/CAD.0b013e32800feecb. [DOI] [PubMed] [Google Scholar]
- 3.Herbst RS, Shin DM. Monoclonal antibodies to target epidermal growth factor receptor-positive tumors: a new paradigm for cancer therapy. Cancer. 2002;94:1593–1611. doi: 10.1002/cncr.10372. [DOI] [PubMed] [Google Scholar]
- 4.Ritter CA, Arteaga CL. The epidermal growth factor receptor-tyrosine kinase: a promising therapeutic target in solid tumors. Semin Oncol. 2003;30:3–11. doi: 10.1053/sonc.2003.50027. [DOI] [PubMed] [Google Scholar]
- 5.Gullick WJ, Hughes CM, Mellon K, Neal DE, Lemoine NR. Immunohistochemical detection of the epidermal growth factor receptor in paraffin-embedded human tissues. J Pathol. 1991;164:285–289. doi: 10.1002/path.1711640403. [DOI] [PubMed] [Google Scholar]
- 6.Nicholson RI, Gee JM, Harper ME. EGFR and cancer prognosis. Eur J Cancer. 2001;37:S9–S15. doi: 10.1016/s0959-8049(01)00231-3. [DOI] [PubMed] [Google Scholar]
- 7.Foon KA, Yang XD, Weiner LM, Belldegrun AS, Figlin RA, Crawford J, Rowinsky EK, Dutcher JP, Vogelzang NJ, Gollub J, et al. Preclinical and clinical evaluations of ABX-EGF, a fully human anti-epidermal growth factor receptor antibody. Int J Radiat Oncol Biol Phys. 2004;58:984–990. doi: 10.1016/j.ijrobp.2003.09.098. [DOI] [PubMed] [Google Scholar]
- 8.Yang XD, Jia XC, Corvalan JR, Wang P, Davis CG. Development of ABX-EGF, a fully human anti-EGF receptor monoclonal antibody, for cancer therapy. Crit Rev Oncol Hematol. 2001;38:17–23. doi: 10.1016/s1040-8428(00)00134-7. [DOI] [PubMed] [Google Scholar]
- 9.Yang XD, Jia XC, Corvalan JR, Wang P, Davis CG, Jakobovits A. Eradication of established tumors by a fully human monoclonal antibody to the epidermal growth factor receptor without concomitant chemotherapy. Cancer Res. 1999;59:1236–1243. [PubMed] [Google Scholar]
- 10.Jakobovits A, Amado RG, Yang X, Roskos L, Schwab G. From XenoMouse technology to panitumumab, the first fully human antibody product from transgenic mice. Nat Biotechnol. 2007;25:1134–1143. doi: 10.1038/nbt1337. [DOI] [PubMed] [Google Scholar]
- 11.Vectibix® (Panitumumab), author Full Prescribing Information. Thousand Oaks, CA: Amgen Inc; 2011. [Google Scholar]
- 12.Cunningham D, Humblet Y, Siena S, Khayat D, Bleiberg H, Santoro A, Bets D, Mueser M, Harstrick A, Verslype C, et al. Cetuximab monotherapy and cetuximab plus irinotecan in irinotecan-refractory metastatic colorectal cancer. N Engl J Med. 2004;351:337–345. doi: 10.1056/NEJMoa033025. [DOI] [PubMed] [Google Scholar]
- 13.Saltz LB, Meropol NJ, Loehrer PJ, Sr, Needle MN, Kopit J, Mayer RJ. Phase II trial of cetuximab in patients with refractory colorectal cancer that expresses the epidermal growth factor receptor. J Clin Oncol. 2004;22:1201–1208. doi: 10.1200/JCO.2004.10.182. [DOI] [PubMed] [Google Scholar]
- 14.Van Cutsem E, Peeters M, Siena S, Humblet Y, Hendlisz A, Neyns B, Canon JL, Van Laethem JL, Maurel J, Richardson G, et al. Open-label phase III trial of panitumumab plus best supportive care compared with best supportive care alone in patients with chemotherapy-refractory metastatic colorectal cancer. J Clin Oncol. 2007;25:1658–1664. doi: 10.1200/JCO.2006.08.1620. [DOI] [PubMed] [Google Scholar]
- 15.Van Cutsem E, Siena S, Humblet Y, Canon JL, Maurel J, Bajetta E, Neyns B, Kotasek D, Santoro A, Scheithauer W, et al. An open-label, single-arm study assessing safety and efficacy of panitumumab in patients with metastatic colorectal cancer refractory to standard chemotherapy. Ann Oncol. 2008;19:92–98. doi: 10.1093/annonc/mdm399. [DOI] [PubMed] [Google Scholar]
- 16.Jonker DJ, O'Callaghan CJ, Karapetis CS, Zalcberg JR, Tu D, Au HJ, Berry SR, Krahn M, Price T, Simes RJ, et al. Cetuximab for the treatment of colorectal cancer. N Engl J Med. 2007;357:2040–2048. doi: 10.1056/NEJMoa071834. [DOI] [PubMed] [Google Scholar]
- 17.Douillard J, Cassidy J, Jassem J, Rivera F, Kocáková I, Rogowski W, Canon JR, Yanez EP, Xu F, Gansert JL. Randomized, open-label, phase III study of panitumumab (pmab) with FOLFOX4 versus FOLFOX4 alone as first-line treatment (tx) for metastatic colorectal cancer (mCRC): efficacy by skin toxicity (ST) [abstract] J Clin Oncol. 2010;28:3528. [Google Scholar]
- 18.Van Cutsem E, Köhne CH, Hitre E, Zaluski J, Chang Chien CR, Makhson A, D'Haens G, Pinter T, Lim R, Bodoky G, et al. Cetuximab and chemotherapy as initial treatment for metastatic colorectal cancer. N Engl J Med. 2009;360:1408–1417. doi: 10.1056/NEJMoa0805019. [DOI] [PubMed] [Google Scholar]
- 19.Tsao MS, Sakurada A, Cutz JC, Zhu CQ, Kamel-Reid S, Squire J, Lorimer I, Zhang T, Liu N, Daneshmand M, et al. Erlotinib in lung cancer—molecular and clinical predictors of outcome. N Engl J Med. 2005;353:133–144. doi: 10.1056/NEJMoa050736. [DOI] [PubMed] [Google Scholar]
- 20.Amado RG, Wolf M, Peeters M, Van Cutsem E, Siena S, Freeman DJ, Juan T, Sikorski R, Suggs S, Radinsky R, et al. Wild-type KRAS is required for panitumumab efficacy in patients with metastatic colorectal cancer. J Clin Oncol. 2008;26:1626–1634. doi: 10.1200/JCO.2007.14.7116. [DOI] [PubMed] [Google Scholar]
- 21.Lievre A, Bachet JB, Le Corre D, Boige V, Landi B, Emile JF, Cote JF, Tomasic G, Penna C, Ducreux M, et al. KRAS mutation status is predictive of response to cetuximab therapy in colorectal cancer. Cancer Res. 2006;66:3992–3995. doi: 10.1158/0008-5472.CAN-06-0191. [DOI] [PubMed] [Google Scholar]
- 22.Benvenuti S, Sartore-Bianchi A, Di Nicolantonio F, Zanon C, Moroni M, Veronese S, Siena S, Bardelli A. Oncogenic activation of the RAS/RAF signaling pathway impairs the response of metastatic colorectal cancers to anti-epidermal growth factor receptor antibody therapies. Cancer Res. 2007;67:2643–2648. doi: 10.1158/0008-5472.CAN-06-4158. [DOI] [PubMed] [Google Scholar]
- 23.Frattini M, Saletti P, Romagnani E, Martin V, Molinari F, Ghisletta M, Camponovo A, Etienne LL, Cavalli F, Mazzucchelli L. PTEN loss of expression predicts cetuximab efficacy in metastatic colorectal cancer patients. Br J Cancer. 2007;97:1139–1145. doi: 10.1038/sj.bjc.6604009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Freeman DJ, Juan T, Reiner M, Hecht JR, Meropol NJ, Berlin J, Mitchell E, Sarosi I, Radinsky R, Amado RG. Association of K-ras mutational status and clinical outcomes in patients with metastatic colorectal cancer receiving panitumumab alone. Clin Colorectal Cancer. 2008;7:184–190. doi: 10.3816/CCC.2008.n.024. [DOI] [PubMed] [Google Scholar]
- 25.Khambata-Ford S, Garrett CR, Meropol NJ, Basik M, Harbison CT, Wu S, Wong TW, Huang X, Takimoto CH, Godwin AK, et al. Expression of epiregulin and amphiregulin and K-ras mutation status predict disease control in metastatic colorectal cancer patients treated with cetuximab. J Clin Oncol. 2007;25:3230–3237. doi: 10.1200/JCO.2006.10.5437. [DOI] [PubMed] [Google Scholar]
- 26.Virtanen C, Woodgett J. Clinical uses of microarrays in cancer research. Methods Mol Med. 2008;141:87–113. doi: 10.1007/978-1-60327-148-6_6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chung CH, Seeley EH, Roder H, Grigorieva J, Tsypin M, Roder J, Burtness BA, Argiris A, Forastiere AA, Gilbert J, et al. Detection of tumor epidermal growth factor receptor pathway dependence by serum mass spectrometry in cancer patients. Cancer Epidemiol Biomarkers Prev. 2010;19:358–365. doi: 10.1158/1055-9965.EPI-09-0937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.de Reynies A, Boige V, Milano G, Faivre J, Laurent-Puig P. KRAS mutation signature in colorectal tumors significantly overlaps with the cetuximab response signature. J Clin Oncol. 2008;26:2228–2230. doi: 10.1200/JCO.2007.15.9186. author reply 2230–2221. [DOI] [PubMed] [Google Scholar]
- 29.Jacobs B, De Roock W, Piessevaux H, Van Oirbeek R, Biesmans B, De Schutter J, Fieuws S, Vandesompele J, Peeters M, Van Laethem JL, et al. Amphiregulin and epiregulin mRNA expression in primary tumors predicts outcome in metastatic colorectal cancer treated with cetuximab. J Clin Oncol. 2009;27:5068–5074. doi: 10.1200/JCO.2008.21.3744. [DOI] [PubMed] [Google Scholar]
- 30.Liu F, Wu B. Multi-group cancer outlier differential gene expression detection. Comput Biol Chem. 2007;31:65–71. doi: 10.1016/j.compbiolchem.2007.02.004. [DOI] [PubMed] [Google Scholar]
- 31.GeneChip® Expression Analysis Technical Manual P/N 701024 Rev. 1. Santa Clara, CA: Affymetrix Inc; 2009. [Google Scholar]
- 32.Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- 33.Goldberg DE, editor. Genetic Algorithms in Search, Optimization and Machine Learning. Boston, MA: Addison-Wesley Longman Publishing; 1989. [Google Scholar]
- 34.Aliferis CF, Tsamardinos I, Massion P, Statnikov A, Hardin D. Why classification models using array gene expression data perform so well: a preliminary investigation of explanatory factors; International Conference on Mathematics and Engineering Techniques in Medicine and Biological Sciences; June 23–26, 2003; Las Vegas, NV. 2003. [Google Scholar]
- 35.Hastie T, Tibshirani R, Friedman J, editors. The Elements of Statistical Learning: Data Mining, Inference, and Prediction. New York, NY: Springer; 2001. [Google Scholar]
- 36.Ambroise C, McLachlan GJ. Selection bias in gene extraction on the basis of microarray gene-expression data. Proc Natl Acad Sci USA. 2002;99:6562–6566. doi: 10.1073/pnas.102102699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kutner MH, editor. Applied Linear Statistical Models. Boston, MA: McGraw-Hill Irwin; 2005. [Google Scholar]
- 38.Efron B, Tibshirani R. Empirical bayes methods and false discovery rates for microarrays. Genet Epidemiol. 2002;23:70–86. doi: 10.1002/gepi.1124. [DOI] [PubMed] [Google Scholar]
- 39.Miller AJ, editor. Subset Selection in Regression. London, UK: Chapman and Hall; 1990. [Google Scholar]
- 40.Rocha-Lima CM, Soares HP, Raez LE, Singal R. EGFR targeting of solid tumors. Cancer Control. 2007;14:295–304. doi: 10.1177/107327480701400313. [DOI] [PubMed] [Google Scholar]
- 41.Siena S, Tabernero J, Cunningham D, Koralewski P, Ruff P, Rother M, Johnson CW, Zhang A, Gansert JL, Douillard J. Randomized phase III study of panitumumab (pmab) with FOLFOX4 compared to FOLFOX4 alone as first-line treatment (tx) for metastatic colorectal cancer (mCRC): PRIME trial analysis by epidermal growth factor receptor (EGFR) tumor staining [abstract] J Clin Oncol. 2010;28:3566. [Google Scholar]
- 42.Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, Pollack JR, Ross DT, Johnsen H, Akslen LA, et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 43.Golub TR, Slonim DK, Tamayo P, Huard C, Gaasenbeek M, Mesirov JP, Coller H, Loh ML, Downing JR, Caligiuri MA, et al. Molecular classification of cancer: class discovery and class prediction by gene expression monitoring. Science. 1999;286:531–537. doi: 10.1126/science.286.5439.531. [DOI] [PubMed] [Google Scholar]
- 44.Alizadeh AA, Eisen MB, Davis RE, Ma C, Lossos IS, Rosenwald A, Boldrick JC, Sabet H, Tran T, Yu X, et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. 2000;403:503–511. doi: 10.1038/35000501. [DOI] [PubMed] [Google Scholar]
- 45.Nguyen DV, Rocke DM. Partial least squares proportional hazard regression for application to DNA microarray survival data. Bioinformatics. 2002;18:1625–1632. doi: 10.1093/bioinformatics/18.12.1625. [DOI] [PubMed] [Google Scholar]
- 46.Vikhanskaya F, Bani MR, Borsotti P, Ghilardi C, Ceruti R, Ghisleni G, Marabese M, Giavazzi R, Broggini M, Taraboletti G. p73 Overexpression increases VEGF and reduces thrombospondin-1 production: implications for tumor angiogenesis. Oncogene. 2001;20:7293–7300. doi: 10.1038/sj.onc.1204896. [DOI] [PubMed] [Google Scholar]