Abstract
Polyploidy contributes to extensive intratumor genomic heterogeneity that characterizes advanced malignancies and is thought to limit the efficiency of current cancer therapies. It has been shown that telomere deprotection in p53-deficient mouse embryonic fibroblasts leads to high rates of polyploidization. We now show that tumor genome evolution through whole-genome duplication occurs in ∼15% of the karyotyped human neoplasms and correlates with disease progression. In a panel of human cancer and transformed cell lines representing the two known types of genomic instability (chromosomal and microsatellite), as well as the two known pathways of telomere maintenance in cancer (telomerase activity and alternative lengthening of telomeres), telomere dysfunction-driven polyploidization occurred independently of the mutational status of p53. Depending on the preexisting context of telomere maintenance, telomerase activity and its major components, human telomerase reverse transcriptase (hTERT) and human telomerase RNA component (hTERC), exert both reverse transcriptase-related (canonical) and noncanonical functions to affect tumor genome evolution through suppression or induction of polyploidization. These new findings provide a more complete mechanistic understanding of cancer progression that may, in the future, lead to novel therapeutic interventions.
Introduction
Chromosomal instability in neoplasia (CIN) is the most common form of genomic instability occurring in virtually all types and stages of cancer [1–3]. In contrast to microsatellite instability in neoplasia (MIN) that causes DNA mismatch repair errors [1], CIN massively affects the integrity and dosage of chromosomes through structural rearrangements and numerical aberrations such as aneuploidy and polyploidization [2]. Although most tumors are monoclonal in origin, chromosomal imbalances emerge in the early steps of carcinogenesis [4], are often distributed randomly among cancer cells [5], and may activate oncogenic pathways [6,7]. Such extensive intratumor genomic heterogeneity provides the grounds for a process of selection and adaptation that drives cancer cell populations into more malignant traits and is a major concern for all current and future oncotherapeutic strategies [8,9]. Radiotherapy and many anticancer drugs induce growth arrest in the G2/M phase of the cell cycle that frequently leads to polyploidization [10,11]. Drug- or irradiation-induced polyploidy usually leads to cell death by mitotic catastrophe [12]. However, it has been proposed that polyploidization may be associated with the emergence of cancer stem-like cells that confer therapy resistance to anticancer agents [13]. Therefore, a better understanding of the mechanisms regulating polyploidization is critical not only to decipher fundamental aspects of carcinogenesis but also for achieving efficient therapies against advanced malignancy.
Telomeres are specialized nucleoprotein complexes that protect the ends of eukaryotic chromosomes [14]. These highly repetitive entities are progressively depleted after each round of DNA replication in all dividing human somatic cells [15]. The loss of telomeric DNA is replenished by the action of the ribonucleoprotein telomerase, or by a rarer DNA recombination pathway, termed alternative lengthening of telomeres (ALT), that maintains telomere length in the absence of telomerase [16]. Because most normal human somatic tissues do not possess a constitutive means to fully maintain their telomeres, actively dividing cells demonstrate progressive telomeric length reductions with each cell division [17]. When a single, or a few, critically short telomeres occur, DNA damage responses are activated and cells undergo a growth arrest [15,18,19].
In normal cells, senescence or apoptosis acts as a biologic barrier to prevent neoplastic transformation [20–22]. To bypass these constraints, human malignancies sustain continuous growth by either activating telomerase [23,24] or engaging ALT [25,26].
Extreme telomere shortening is known to provoke terminal chromosome fusions and structural chromosome aberrations [18]. Such changes appear to occur early in neoplasia and coincide with chromosomal instability [2,27]. Telomere-driven genomic instability is characterized by frequent chromosomal break-fusion-bridge (B/F/B) cycles [28] that generate various types of oncogenic structural rearrangements and may affect numerical chromosomal constitution through whole chromosome losses because of anaphase lags [28–30].
Numerical chromosomal instability per se is also related to tumorigenesis: Cells and animals with reduced levels of centromere-associated protein-E (CENP-E) frequently become aneuploid because of random missegregation of one or a few chromosomes in the absence of DNA damage [31]. Depletion of CENP-E contributes to cellular transformation and causes a modest increase in spontaneous tumor formation [31]. In addition, patients with mosaic variegated aneuploidy syndrome, caused by mutations in the mitotic spindle checkpoint gene BUB1B, develop malignancies such as rhabdomyosarcomas, Wilms tumors, and leukemias [32]. However, induction of aneuploidy can also be an effective inhibitor of tumorigenesis [31]. While low levels of aneuploidy may provide the grounds for progressive oncogenic transformation, extremely accelerated rates of CIN may dramatically affect cancer cell homeostasis and may initially be a powerful anticancer protection mechanism [9,31].
A mechanistic link between telomere dysfunction, defective cell cycle checkpoints, and tetraploidization, through whole-genome endore-duplication, has recently been reported [33]. This model fits well with the hypothesis that polyploidy can lead to extended aneuploidy and heteroploidy in human solid tumors [34]. A combination of the two models predicts that, early in oncogenesis, telomere dysfunction and abrogated DNA damage responses allow the generation of somatic polyploid clones that can become highly aneuploid through subsequent divisions influenced by B/F/B-induced anaphase lags [8,28]. Tumor cell evolution will then be driven by selective pressure during continuous growth that leads to amplification of oncogenes and depletion of tumor suppressor pathways [35].
From earlier studies on the clonal evolution of tumor cell populations, we proposed that polyploidization through genome reduplication could reflect a process of genome evolution that is observed in vivo in tumor cells [36,37]. Genome reduplication occurs also in culture of immortalized human cancer cell lines and may be related to telomere dysfunction [38,39]. We now provide evidence that polyploidization is a common cause of neoplastic genome evolution in humans and correlates with progression of disease. We examined the effects of telomerase in a panel of cell lines representing the two known contexts of genomic instability in neoplasia, CIN and MIN, as well as the two known pathways of telomere maintenance (telomerase and ALT). We show that “telomere dysfunction-driven polyploidization” is a universal source of tumor evolution that occurs continuously during neoplastic cell growth in culture and can be triggered to be massive during telomeric crisis at the interface between different pathways of telomere maintenance. Interestingly, telomerase activity and the ectopic expression of its major components human telomerase reverse transcriptase (hTERT) or human telomerase RNA component (hTERC) exert both reverse transcriptase-related (canonical) and noncanonical functions to suppress or induce polyploidy. These new findings provide a more complete mechanistic understanding of cancer progression that may, in the future, lead to novel therapeutic interventions.
Materials and Methods
Data Mining from Cytogenetic Databases
On the basis of the International System for Human Cytogenetic Nomenclature 2009 [40], we examined neoplastic karyotypes that are included in two open access databases: 1) the National Cancer Institute, Mitelman Database of Chromosome Aberrations and Gene Fusions in Cancer (http://cgap.nci.nih.gov/Chromosomes/Mitelman) [41] and 2) the National Center for Biotechnology Information (NCBI) Spectral Karyotyping (SKY)/Multiplex Fluorescence In Situ Hybridization (M-FISH) and Comparative Genomic Hybridization database (http://www.ncbi.nlm.nih.gov/projects/sky) [42]. For the first database, we used the EXCEL software (Microsoft) to calculate frequencies of neoplastic genomes with documented evidence of polyploidization. Genome duplication or multiplication in tumor evolution has been directly recorded in a proportion of human neoplasms included in the Mitelman Database using the International System for Human Cytogenetic Nomenclature symbols idemx2, idemx3, etc., that symbolize whole-genome duplication (WGD), whole-genome triplication, etc. Taking into consideration the high rates of chromosome losses occurring in neoplastic genomes [34], we included cases with recorded numerical chromosome alterations ranging from near-triploidy (58–80 chromosomes) to extreme polyploidy. For the NCBI SKY/M-FISH and Comparative Genomic Hybridization database, as polyploidy deriving karyotypes, we considered those presenting an over diploid chromosome number (50–200 chromosomes) displaying in addition at least two individual pairs of identical structurally altered (marker) chromosomes or four individual homologous chromosomes in tetrasomy.
Cell Lines and Culture Conditions
HCT-116, HCT-116 p53-/- [43], and T-24 cells were provided by Drs C. Dimas and T. Vlahou (Biomedical Research Foundation of the Academy of Athens, Athens, Greece). Lisa-2, Ls-2, and HIO-118 were kindly provided by Dr D. Broccoli (Fox Chase Cancer Center, Philadelphia, PA). A-549 cell line was a gift from Dr G. V. Gorgoulis (School of Medicine, University of Athens, Athens, Greece). T-47D cell line was a gift from Dr A. Klinakis (Biomedical Research Foundation of the Academy of Athens). HeLa and breast cancer MCF-7 cells were a gift from Dr I. Irminger-Finger (Geneva Medical School, Geneva, Switzerland). The osteosarcoma cell line U2-OS was provided by Dr E. Gonos (Greek National Institute for Research, Athens, Greece). The colon cancer SW-480 and the osteosarcoma Saos2 cell lines were obtained from American Type Culture Collection (ATCC; Wesel, Germany). The ALT cell lines GM-847 and VA-13 were provided by Dr A. Londoño-Vallejo (Institute Curie, Paris, France). In addition, we used a VA-13 derivative cell line that stably expresses hTERC and hTERT and has reconstituted telomerase activity (VA-13TA)—described in Ford et al. [44]—and HCT-15 with four different derivative cell lines as described in Bechter et al. [45]. The HCT-15 cells express a dominant mutant version cDNA resulting in inhibition of wild-type endogenous hTERT. The sublines (SL1 and SL2) were Telomeric Repeat Amplification Protocol (TRAP; telomerase activity)-positive HCT-15 parental cells, and SL3 to SL8 were TRAP-negative sublines representing subsequent population doublings (PDs) of cells (PD27-PD107) after stable expression of the dominant negative cDNA against hTERT [45]. We also used a derivative subline of SL3 to SL8, in which telomerase activity was spontaneously restored after PD180 (SL10) [45]. To produce the G7-pssi10189-Krüppel-associated box (KRAB) cell lines, we infected ALT VA-13 cells with tetracycline-on-inducible (TRE) hTERT with puromycin-resistant M2 vector (rtTAsM2), which expresses a reverse tetracycline activator to switch on hTERT transcription and the KRAB transcriptional silencer with G418 resistance, which binds tetracycline-on and represses basal hTERT transcription. The VA-13InTAa, b, and c were G7-pssi10189-KRAB derivative polyclonal populations that were independently transduced with an additional lentiviral vector expressing wild-type hTERC (pssi6499) with blasticidin selection marker and red fluorescent protein for detection. The U2-OS and Saos2 cells were either transduced with green fluorescent protein (GFP) lentivirus pLOX-GFP-iresTK or hTERT lentivirus pLOX-GFP-TERT-iresTK (kindly donated by Dr D. Trono). All cell cultures were grown at 37°C and 5% CO2 in Dulbecco's modified Eagle's medium (Gibco, Grand Island, NY) supplemented with 10% FBS (Gibco), 25 units/ml penicillin (Sigma, St Louis, MO), and 25 pg/ml streptomycin (Invitrogen, Grand Island, NY).
Cytogenetics
Logarithmically growing cell cultures were exposed to colcemid (0.1 µg/ml) (Gibco) for 1 to 3 hours, at 37°C in 5% CO2. Cells were harvested by trypsinization (Gibco), suspended in medium, and spun down (10 min, 1000 rpm). Supernatant was removed completely and 5 ml of 0.075 M KCl (Sigma) at room temperature was added drop by drop. The cells were incubated for 20 minutes at room temperature, and then 1 ml of fixative [3x methanol (Applichem GmbH, Darmstadt, Germany)/1x CH3COOH (Merck, Darmstadt, Germany)] was added. Cells were spun down (10 minutes at 1000 rpm), supernatant was removed, fixative was added, and the cells were recentrifuged for 10 minutes at 1000 rpm. Finally, cells were dropped onto wet microscope slides and left to air-dry. For the quantitation of chromosome number per metaphase, we combined inverted 4′,6′-diamidino-2-phenylindole (DAPI) staining, G-Banding, and molecular karyotyping b yM-FISH (MetaSystems GmbH, Altlussheim, Germany). G-Banding was performed after treatment with 0.25% trypsin (Gibco) and Giemsa (Carl Roth GmbH, Karlsruhe, Germany) staining. Multicolor FISH was performed according to the manufacturer's protocols (MetaSystems GmbH). For inverted DAPI banding, slides were counterstained and mounted with 0.1 µg/ml DAPI in VECTASHIELD antifade medium (Vector Laboratories, Burlingame, CA). Cytogenetic analyses were performed using a x63 magnification lens with a charge-coupled device (CCD) camera on a fluorescent Axio Imager Z1 Zeiss microscope and the MetaSystems Ikaros or Isis software.
Fluorescence In Situ Hybridization
For interphase or metaphase FISH, we used satellite probes specific for the centromeres of chromosomes 1, 2, 3, 4, 6, 7, 8, 9, 10, 11, 12, 16, 17, 18, 20, and X. Probes were purchased from Vysis (Abbott Park, IL) and Cytocell (Cambridge, United Kingdom). In brief, our protocol was based on pepsin (Invitrogen) pretreatment, formamide (Applichem) or NaOH (Sigma) target denaturation, overnight hybridization, and highstringency post-hybridization washes. Telomeric peptide nucleic acid (PNA) FISH was performed according to the manufacturer's instructions (Dako Cytomation, Glostrup, Denmark). Briefly slides were incubated at 3.7% formaldehyde (Carlo Erba Reagenti SpA, Milano, Italy), washed with 1x TBS (Dako Cytomation), immersed in pretreatment solution (Dako Cytomation), and dehydrated with cold ethanol series (VWR, Radnor, PA). Probe and target DNA were denatured at 80°C for 5 minutes and then slides were incubated for 1 hour at room temperature in the dark. For multicolor FISH, we used the 24XCyte Kit from MetaSystems GmbH. Staining was performed according to the manufacturer's instructions. All FISH preparations were mounted and counterstained with VECTASHIELD antifade medium containing 0.1 µg/ml DAPI (Vector). Digital images were captured and enhanced in a MetaSystems workstation as described above.
Telomeric Repeat Amplification Protocol
Telomerase activity of cell lysates was analyzed by the TRAP assay with a TRAPeze Telomerase Detection Kit (Chemicon, Billerica, MA) according to the manufacturer's instructions. Approximately 106 cells were harvested and lysed in 200 µl of 1x CHAPS lysis buffer [10 mM Tris-HCl (pH 7.5), 1 mM EGTA, 1 mM MgCl2, 0.5%CHAPS, 10% glycerol, 5 mM β-mercaptoethanol, and 0.1 mM benzamidine] on ice for 30 minutes. Polymerase chain reaction (PCR) products were electrophoresed in a 10% 19:1 acrylamide gel (Sigma) in 0.5x Tris/Borate/EDTA buffer (Sigma) using the Mini-PROTEAN II Gel System (Bio- Rad, Hercules, CA). Gels were stained either with 1:10,000 SYBR Green (Sigma) in 0.5x Tris/Borate/EDTA buffer for 15 minutes at room temperature and then exposed to UV light and visualized in a Kodak image acquisition station with Dolphin software or by radioisotopic detection using P32-labeled deoxycytidine triphosphates (dCTPs; Amersham, Uppsala, Sweden). Radiolabeled gels were dried in a Heto Lyo Pro3000 Dryer and visualized with a Typhoon 9200 Imager.
Telomerase Silencing
The telomerase-positive SW-480 cell line was transiently transfected using Lipofectamine 2000 (Invitrogen) according to themanufacturer's instructions, with ds small interfering RNA (siRNA) targeted against hTERT (Ambion, Grand Island, NY; sense, 5′-GGCUCUUUUUCU ACCGGAAtt-3′; antisense, 5′-UUCCGGUAGAAAAAGAGCCtg-3′). Cells were seeded in T25 flasks and/or six-well plates at appropriate cell densities 1 day before transfection to be subconfluent the first day of the experiment. The siRNA/Lipofectamine ratio was 20 pmol/µl. SW-480 cells were subjected to sequential transfections with siRNA, separated by 72- to 96-hour intervals. Cells were maintained in culture for 10, 30, or 60 days before harvest and collection of cell pellet. In addition, the SW-480 cell line was transduced with LentiLox 3.7 (pLL3.7), which contained a short hairpin RNA (shRNA) designed to silence hTERT. The shRNA for hTERT was synthesized at the FORTH Institute (Crete, Greece) and had the following sequence: sense, 5′-GATCCCCTTTCATCAGCAAGTTTGGATTCAAGAGATCCAAACTTGCTGATGAAATTTTTA-3′; antisense, 3′-GGGAAAGTAGTCGTTCAAA-CCC TAAGTTCTCTAGGTTTGAACGACTACTTTAAAAATTCGA-5′. Enrichment of transduced cells was performed with the aid of an ARIA cell sorter (Beckman Coulter, Nyon, Switzerland) using GFP as a selection marker. After selection, more than 90% of the cells were GFP positive. We also exposed the SW-480 cell line to the telomerase inhibitor N,N-1′,3′-phenylenebis-[2′,3′-dihydroxy-benzamide] (MST312) [46]. Briefly, 3 x 10-6 M of MST312 (kindly donated by Dr H. Seimiya) diluted in DMSO (Sigma) was added to the culture medium of subconfluent T25 flasks for 5 and 10 days. Medium was changed every 2 days. Similar procedure for telomerase inhibition through MST312 was followed for the telomerase-positive VA-13TA, A-549, and T-47D cell lines.
Reverse Transcription-PCR
Semiquantitative reverse transcription (RT)-PCR was performed as follows: 2 µg of total RNA was isolated from each cell line and incubated at 70°C for 5 minutes with 0.5 µg of oligo T17 (custom-made from the FORTH Institute). RT-PCR was followed with the mix containing 2 µl of 10x RT buffer (Promega, Madison, WI), 2.4 µl of 25 mM MgCl2 (Promega), 1 µl of 5 mM deoxynucleotide triphosphates (dNTPs; Promega), and 0.5 µl of reverse transcriptase (Promega) in H2O to a final volume of 20 µl. The transcription reaction was performed at 25°C for 5 minutes, 42°C for 1 hour, 70°C for 15 minutes, and then cooled down to 4°C. Amplification of the hTERT in 5 µl of cDNA was performed with 5 µl (20 µM) of each of the following primers: 5′-AGGCTGCAGAGTGCAGAGCAGCGTGGAGAGG-3′ and 5′-GCCTGAGCTGTACTTTGTACTTTGTCAA-3′; for hTERC, the primers were 5′-TTTCCCTAACCCTAA-3′ and 5′-AATCCGTCGAGCAGAGTT-3′, and for GAPDH, the primers were 5′-CTTACTCCTTGGAGGCCATG-3′ and 5′-TTAGCACCCCTGGCCAAGG-3′ (custom-made from the FORTH Institute). The PCR products were analyzed by electrophoresis in 8% nondenaturing polyacrylamide gel and stained with SYBR Green (1:10,000); images were acquired at a Kodak image acquisition station equipped with Dolphin software, and bands were quantified using ImageQuant (MD/APS software).
Immunocytochemistry and Immuno-FISH
In situ dual-color immunofluorescent cytochemistry (IF) was performed on cells grown on coverslips. Cells were fixed in ice-cold methanol for 10 minutes at -20°C, permeabilized with 0.2% Triton for 12 minutes at 4°C, and blocked with 1% BSA for 1 hour at room temperature. Coverslips were incubated with appropriate antibodies in 1% BSA overnight at 4°C in humidity. For antibody detection, we used Alexa Fluor 488 and Alexa Fluor 568 (Molecular Probes, Grand Island, NY) and Cyanine 5 (BD Biosciences, Franklin Lakes, NJ) diluted 1:500 in 1% BSA for 1 hour at room temperature in humidity. For telomere-specific immuno-FISH, coverslips were post-fixed with ice-cold methanol for 10 minutes at -20°C and washed with phosphate-buffered saline, and then PNA FISH was applied as described above. Primary antibodies used for IF were specific for hTERT (Epitomics, Burlingame, CA), telomeric repeat factor 2 (TRF2; Santa Cruz Biotechnology, Santa Cruz, CA), γ-H2AX (clone JBW301; Millipore, Billerica, MA), γ-tubulin (Abcam, Cambridge, MA), and α-tubulin (Sigma; 1:500). Promyelocytic leukemia (PML; Santa Cruz Biotechnology), home-made 53BP1 and ataxia telangiectasia and Rad3 related (ATR) interacting protein (ATRIP; kindly donated by Dr T. Halazonetis), and replication protein A1 (RPA1; Calbiochem, Billerica, MA) antibodies (1:50–1:100) were also used. Coverslips were dehydrated and mounted onto microscope slides with VECTASHIELD containing DAPI as above. Digital images were captured in a MetaSystems workstation as described above.
Statistical Analysis
One-way analysis of variance, paired t test, and chi-square analyses were performed using the MINITAB software. Error bars represent SEM. In WGD graphs, error bars indicate SEM between two or more repetitive experiments.
Results
WGD Occurs in All Histopathologic Types of Human Neoplasia and Correlates with Disease Progression and Malignancy
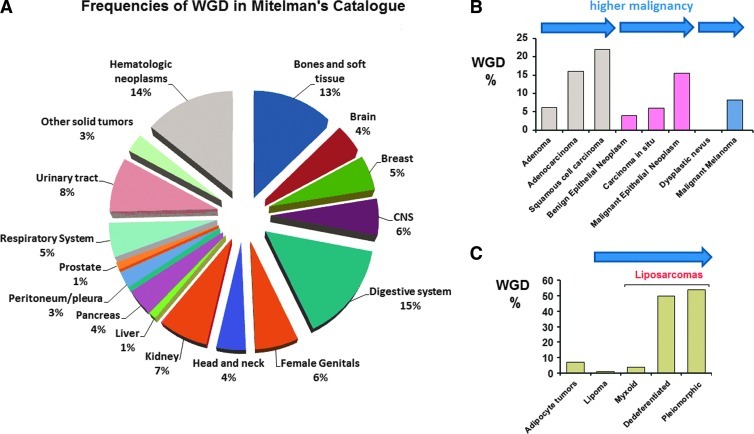
Extensive cytogenetic studies have shown that a significant number of cancer genomes evolve with polyploidy and widespread aneuploidy [10,47–49]. Such altered genomes exert hyperdiploid chromosomal contents with modal chromosome numbers varying from near-triploidy (58–80 chromosomes) to hyper-polyploidy (i.e., 200–400 chromosomes) [40]. To identify if polyploidization is a common process of tumor genome evolution and to estimate the frequencies, we applied cytogenetic criteria to examine neoplastic karyotypes of two National Cancer Institute databases [41,42]. On the basis of a broad and conservative analysis, we found that ∼15% of the total karyotyped neoplasias are evolutionary products of polyploidization through WGD (Figure 1A). In several tumor types, high frequencies of polyploidization were associated with disease progress and malignancy (Figure 1, B and C). Nevertheless, the proportions of recorded WGD may be substantially underestimated because of technical issues. To test this possibility, we examined the frequency of WGD in 290 human cancer specimens and tumor cell lines included in the public NCBI SKY/M-FISH Cancer Chromosome Database (http://www.ncbi.nlm.nih.gov/projects/sky) [42]. Approximately 60% of the molecular karyotypes included in the NCBI database, at the time of our analysis, could be considered by-products of WGD. Therefore, independently of tissue of origin, grade, or histopathologic type, a considerable proportion of karyotyped human cancer genomes have evolved toward malignancy through a process of polyploidization.
Figure 1.
Frequencies of polyploidization in Mitelman Catalog of Chromosome Abnormalities in Cancer: Data mining in a total of 59,772 karyotyped samples of human neoplasms included in Mitelman Database at the time of our analysis reveals that polyploidization through WGD occurs in virtually all types of human neoplasia, affecting about 15% of all cases. The percentages of WGD between different histopathologic entities varied between 1% and 15% (A). In several types of solid tumors, the frequencies of recorded WGD were found accelerated in cases representing disease progress or higher grades of malignancy: Note a three-fold to four-fold increase in the rates of WGD when adenomas are compared to adenocarcinomas, or squamous cell carcinomas, as well as between benign epithelial neoplasms or carcinomas in situ and malignant epithelial neoplasms. In melanocytic tumors, identification of WGD might be an indication of malignancy because benign dysplastic nevi do not exert WGD (B). Only 5% to 6% of total adipose tumors display evidence of WGD; however, the presence of WGD is strongly associated to disease progression from the benign lipomas and well-differentiated myxoid liposarcomas to the more malignant dedifferentiated or pleiomorphic liposarcomas (C).
Experimentally Immortalized and Cancer Human Cell Lines Using the ALT Display High Rates of WGD
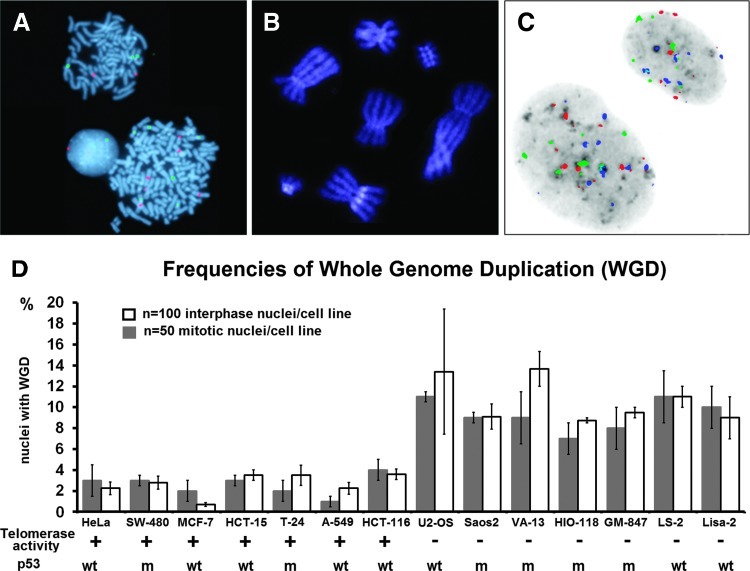
Compared to telomerase-positive examples, ALT tumors and experimentally immortalized cell lines demonstrate increased rates of endogenous telomere dysfunction and structural chromosome instability [50–52]. To investigate differences in the rates of WGD between cells using the ALT pathway and cells that require telomerase for continuous growth, we combined dual-color centromere-specific FISH with classic and molecular karyotyping (M-FISH/SKY) in seven telomerase-positive and seven ALT human cell lines (Figure 2). These duplicate data sets confirm each other with independent cytogenetic methods, providing, in parallel, higher throughput of information for percentages of polyploidy in the whole-cell population of a given harvest. Interphase and mitotic nuclei of the ALT cell lines of our comparative panel showed a significantly higher propensity for WGD (Figure 2D), linking polyploidization to increased rates of endogenous ALT telomere dysfunction. Consistent with previous results [53], p53 deficiency in the MIN HCT-116 cell line increased two-fold the rates of endogenous mitotic and interphase WGDs in the absence of telomere dysfunction, leading to stochastic selection of tetraploid subclones (Figure W1). However, further analysis did not reveal a correlation betweenWGD and mutations in p53 (Figure 2 and Table W1). These observations support the view that, independent of the status of p53, both telomerase-positive examples and ALT cell lines exert a proportion of mitotic or interphase WGD that is significantly more pronounced in the ALT cells.
Figure 2.
Mitotic and interphase polyploidies in a panel of human cancer and immortalized cell lines: WGD in cytogenetic preparations of SW-480 cells stained by DAPI (blue) and centromere-specific probes for chromosomes 3 (red, Texas Red) and 9 (green, fluorescein isothiocyanate) (A). Mitotic presence of diplochromosomes reveals endoreduplication-driven WGD in SW-480 cells lentivirally transduced with shRNA against hTERT (B). Interphase WGD in U2-OS nuclei labeled with inverted DAPI (gray) and centromeric probes specific for human centromeres 3 (red, Texas Red), 7 (green, fluorescein isothiocyanate), and 18 (blue, spectrum aqua) (630x) (C). Comparison of the rates of mitotic and interphase WGDs in 14 continuous human cell lines: Telomerase activity is indicated as (+) or (-) by TRAP. Status of p53 is indicated as wild type (WT) or mutated (M) (Table W1). HCT-15 and HCT-116 cell lines display MIN [45,53]. The remaining 12 cell lines can be categorized as CIN. The ALT cell lines that are characterized by extreme rates of telomere dysfunction display a significantly higher propensity for both interphase and mitotic WGDs (P < .0001 by analysis of variance) (D).
Telomerase Depletion/Inhibition in CIN and MIN Tumor Cell Lines Is Related to Increased Rates of Telomere Dysfunction and Polyploidy
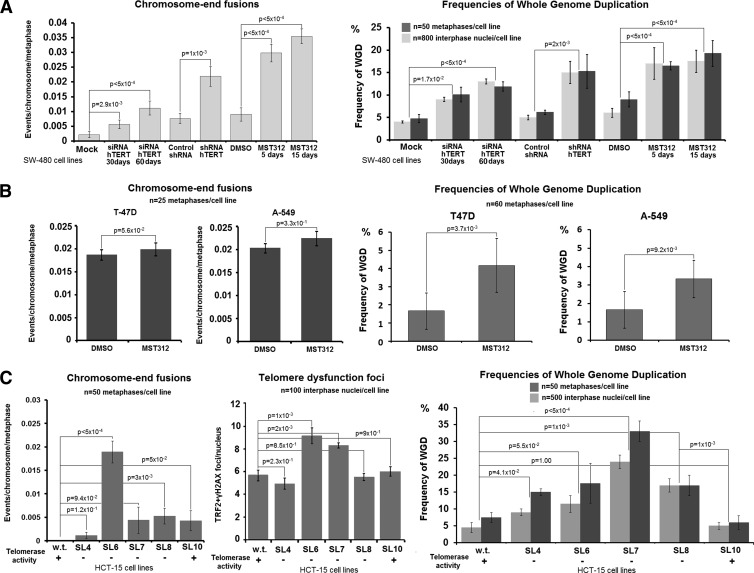
Mouse embryonic fibroblasts growing in the absence of telomerase activity show increased propensity to undergo WGD through endoreduplication and the formation of diplochromosomes [27]. Furthermore, knockdown of telomerase in the near-diploid MIN human colon cancer cell line HCT-116 is associated with increased rates of tetraploidization [53]. To confirm these findings in both MIN and CIN human cell lines, we tested depletion/inhibition of telomerase activity in another MIN cell line (HCT-15) and in the CIN cancer cell lines SW-480, T-47D, and A-549 (all express telomerase; Figure 3). Telomerase activity was suppressed in the SW-480 cells by transient hTERT siRNA transfection [50] and stable shRNA hTERT lentiviral transduction or treated with the telomerase inhibitor MST312 [46] (Figure 3A and Figure W2). The T47-D and A-549 cells were treated with MST312 (Figure 3B). We also examined multiple subclones of the stably transfected HCT-15 (MIN) cell line expressing a dominant negative cDNA against hTERT [45]. The HCT-15 (SL4-SL8) cells efficiently expressed the introduced dominant negative gene for more than 100 PDs and engaged ALT-like telomere elongation [45]. However, after 120 PDs, they spontaneously regained telomerase activity and the adverse effects of telomere dysfunction were rescued [45]. Telomere dysfunction in HCT-15 (SL4-SL8) cells was accompanied by increased rates of interphase and mitotic tetraploid nuclei. In contrast, spontaneous reconstitution of telomerase activity in the same cells suppressed the increased tendency toward tetraploidy (Figure 3C). These findings suggest that continuous growth after telomerase inhibition in telomerase-positive MIN and CIN human cancer cell lines is accompanied by telomere dysfunction and the spontaneous emergence of polyploidy.
Figure 3.
Inducible telomere dysfunction in CIN and MIN cell lines is associated with increased frequencies of WGD: Depletion of hTERT by serial transient siRNA transfections, lentiviral transduction with an shRNA against hTERT, or exposure to the telomerase inhibitor MST312 in the CIN SW-480 cells is associated with increased telomere dysfunction and significantly elevated levels of mitotic and interphase WGDs (A). Telomerase inhibition through MST312 for 10 days results to insignificant increase in end-to-end fusions but still leads to significant induction of WGD in two additional CIN cell lines with extremely low rates of endogenous polyploidization (A-549 and T47D) (B). Robust hTERT knockdown in different sublines of the MIN HCT-15 colon adenocarcinoma cell line, representing consequent PDs after retroviral introduction of an antimorph against hTERT. After PD180, telomerase activity is spontaneously restored (SL10) [45]. In this setting, telomerase knockdown leads to increase in telomere dysfunction and WGD, whereas restoration of telomerase activity suppresses WGD (C). Statistics by paired t test or chi-square test.
Stable Reconstitution of Telomerase Activity in ALT Cells Correlates with a High Prevalence of Polyploid Cells but Low Rates of Random Polyploidization
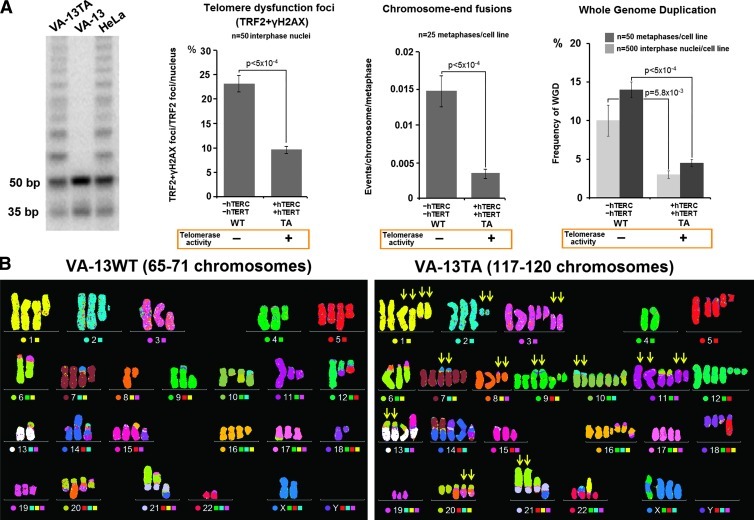
In view of the above results, we reasoned that reconstitution of telomerase activity in ALT cells that is known to repress endogenous telomere dysfunction [44] will also suppress the increased rates of endogenous WGD. To test this, we used the ALT VA-13 cells (T-antigen immortalized human embryonic WI38 fibroblasts) that were stably transfected with hTERT and hTERC and expressed telomerase activity for more than 200 PDs (VA-13TA) [44] (Figure 4A). Surprisingly, the VA-13TA cells showed an extremely high prevalence of polyploid nuclei (98% of total cells) with modal chromosome numbers ranging between 117 and 120 chromosomes (Figure 4B). The karyotypic characteristics of the near-pentaploid VA-13TA cells indicated that they were by-products of near-triploid progenitor VA-13 cells that had undergone one round of WGD. Nevertheless, the near-pentaploid VA-13TA exhibited only low rates of random mitotic and interphase WGDs similar to those of telomerase-positive cell lines (2–4%; Figures 2D and 4A). Such rare mitotic nuclei had undergone an additional round of WGD and displayed a hyperoctaploid genomic content of 220 to 240 chromosomes. Expectedly, the low frequencies of hyperoctaploid nuclei in VA-13TA cell line were significantly increased upon exposure of the cells to the telomerase inhibitor MST312 (Figure W4). Thus, in the ALT context, long-term reconstitution of telomerase activity reduces the rates of telomere dysfunction and WGD but can be related to high prevalence of polyploidization. This suggests that, in immortalized ALT cells, the initial steps of the simultaneous action of both known telomere maintenance pathways in mammals may be accompanied by massive telomere dysfunction.
Figure 4.
Constitutive telomerase activity in ALT cells reduces telomere dysfunction but is related to high prevalence of polyploidy: Long-term stable reconstitution of telomerase activity in the VA-13TA cell line through exogenous introduction of hTERC and hTERT (indicated by TRAP) reduces significantly endogenous telomere dysfunction, as indicated by the rates of chromosome end-to-end fusions and TIFs, and suppresses the rates of WGD (A). Prolonged exposure to telomerase activity in VA-13TA is accompanied by high prevalence of polyploid nuclei composed from 117 to 120 chromosomes (98%). Multicolor FISH indicates that the representative VA-13TA karyotype contains duplicated copies of several structurally altered chromosomes of the parental ALT VA-13 cells (arrows) (630x) (B). Statistics by paired t test or chi-square test.
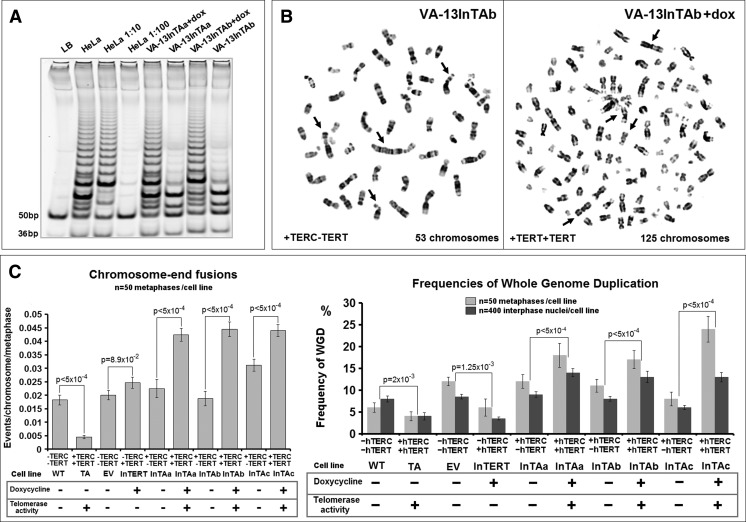
Inducible Telomerase Activity in ALT Cells Elicits Telomere Dysfunction and Polyploidization
To further investigate the unexpected predominance of polyploid nuclei in VA-13TA cells, we generated three stable cell line derivatives of the ALT VA-13 cells (InTAa, InTAb, and InTAc; Table W2). These multiclonal cell lines constitutively express hTERC and stably maintain a tetracycline-inducible hTERT cassette, capable of fully activating telomerase reverse transcriptase activity as indicated by TRAP assays (Figures 5A and W3B). In InTAa, b, and c cells, constitutive expression of hTERC was associated with increased rates of telomere dysfunction and polyploidization, as compared to parental VA-13, and cells were transduced with empty vectors (Figure 5, B and C). Consistent with our hypothesis, inducible reconstitution of telomerase activity accelerated preexisting rates of telomere dysfunction, as verified in mitotic cells by frequent end-to-end chromosome fusions, and in interphase nuclei by immunocytochemical co-localization of the telomere-specific shelterin component TRF2 and the DNA damage response (DDR) proteins γ-H2AX, PML, 53BP1, RPA1, and ATRIP (Figure 6A). As expected, immediate increase in telomere dysfunction was directly accompanied by high rates of mitotic and interphase polyploidizations (Figure 5C). Polyploid cells in neoplasia can form by several mechanisms including cell fusion, cytokinesis failure, or WGD due to an abortive cell cycle (endomitosis) [10]. To define the type of polyploidization process that is activated upon ectopic expression of TERC and TERT in ALT cells, we applied immunofluorescence assays using antibodies specific for α- and γ-tubulins and DAPI staining. This allows localization and enumeration of centrosomes per cell, as well as the identification of binucleated or multinucleated cells in a population [10,54]. The frequencies of amplified (more than two centrosome foci per cell) and of clustered amplified centrosomes were significantly increased upon induction of telomerase activity in VA-13InTAa, b, and c cells (Figure W5, A and B). However, the changes in the percentages of binucleated or multinucleated cells and of cells with scattered amplified centromeres were insignificant (Figure W5C). Therefore, most of the observed telomere dysfunction-driven polyploidy resulted from WGD through endomitosis and not due to generalized cytokinesis failure that generates multinucleated cells with scattered centrosomes [10]. These results reveal that the introduction of telomerase activity in cells using the ALT pathway is triggering immense telomere dysfunction responses that are capable of conferring further tumor evolution through whole-genome endoreduplication. Hence, telomerase activity may facilitate induction of polyploidy in certain contexts.
Figure 5.
Inducible reconstitution of telomerase activity in ALT cells increases telomere dysfunction and polyploidy through WGD: TRAP assay shows inducible telomerase activity in the VA-13 derivative multiclonal cell lines InTAa and InTAb, after 5 days in doxycycline (A). Reconstitution of telomerase activity in InTAb cells increases frequencies of chromosome end-to-end fusions (arrows) and WGD (inverted DAPI, 630x) (B). Constitutive expression of hTERC in the three InTA (a, b, and c) cell lines is related to a significant increase in telomere dysfunction-driven chromosome terminal fusions as compared to parental WT cells and the TA cell line that stably expresses telomerase activity. The rates of telomere dysfunction after 5 days in doxycycline and activation of the telomerase holoenzyme are highly accelerated. Elevated frequencies of WGD in mitotic and interphase nuclei of different VA-13 cell lines correspond to the rates of telomere dysfunction (C). Statistics by paired t test or chi-square test.
Figure 6.
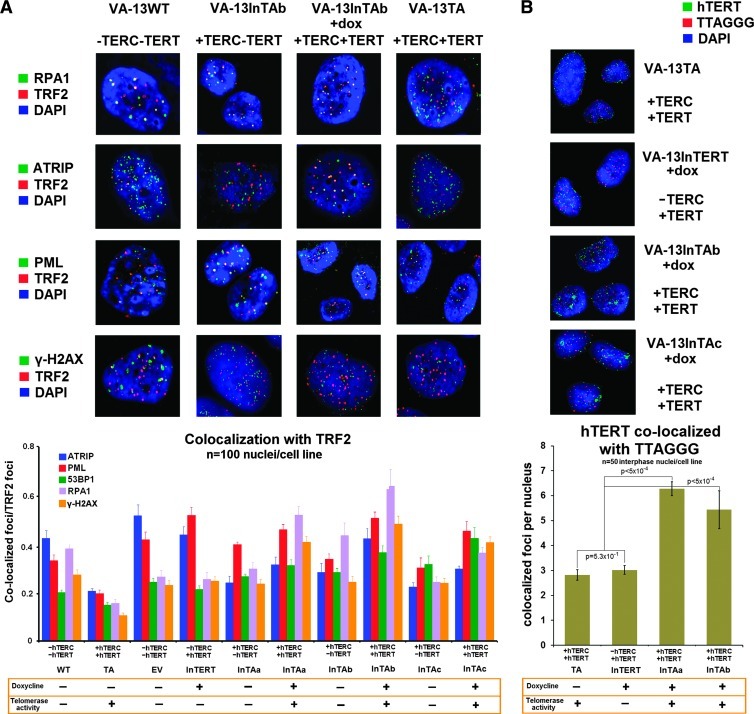
Nuclear association of telomeric binding TRF2 with DDR proteins and of hTERT with telomeric repeats in VA-13 cells. Examples of nuclear co-localization of TRF2-specific antibodies, with the DDR proteins RPA1, ATRIP, PML, and γ-H2AX, by immunofluorescence, in parental ALT VA-13 (WT), in InTAb (constitutive expression of hTERC), in InTAb + doxycycline cells (inducible expression of hTERT and telomerase activity after 5 days of doxycycline in culture), and in VA-13TA cells that constitutively express both holoenzyme components and display telomerase activity for more than 200 PDs (630x). The graph depicts frequencies of spatial interaction of TRF2 with ATRIP, PML, 53BP1, RPA1, and γ-H2AX by dual-color IF in 100 nuclei/cell line. Constitutive overexpression of hTERC in the three independent InTA (a, b, and c) cell lines is associated with significantly reduced co-localization rates of TRF2 with PML (P = .001, P = .002, and P = .001) and ATRIP (P = .007, P = .003, and P = .002), whereas rates of classic TIFs (co-localization of TRF2 with γ-H2AX) remain unaffected. Doxycycline-induced reconstitution of telomerase activity in these cell lines restored levels of co-localization between TRF2 and PML (P = .012, P = .000, and P = .012, respectively) or ATRIP (P = .007, P = .001, and P = .001, respectively), increasing in parallel frequencies of classic TIFs and the rates of spatial association of TRF2 with 53BP1 (P = .045, P = .018, and P = .034, respectively) and RPA1 (P < .0001, P = .001, and P < .0001). Inducible expression of hTERT alone, in the InTERT cells, did not exert any effects in spatial association of TRF2 with DDR components (ATRIP, P = .192; PML, P = .060; 53BP1, P = .139; RPA1, P = .786; γ-H2AX, P = .123) (A). Immuno-FISH depicts nuclear localization of an antibody specific for hTERT and TTAGGG telomeric repeats (630x). Compared to the stable +hTERC+hTERT TA cells, the rates of spatial association of telomerase with the telomeres are significantly elevated after 5 days of the interphase between the sole action of ALT and the introduction of telomerase activity (InTAc + doxycycline, P = .529; InTAa and InTAb, P < .0001) (B) (all statistics by paired t test).
Ectopic Expression of hTERC and Reconstitution of Telomerase Activity Antagonize ALT Introducing Telomere Dysfunction and Polyploidization
To examine the effects of ectopic expression of hTERC, and both hTERC and hTERT, in telomere dysfunction-driven polyploidization in the ALT pathway, we generated an additional VA-13 derivative cell line that conditionally expresses hTERT but does not display telomerase activity due to endogenous absence of hTERC (InTERT; Figure W3) [44]. We then examined spatial association of the DDR elements γ-H2AX, PML, 53BP1, RPA1, and ATRIP with TRF2 in our panel of VA-13 derivative cell lines (Figure 6A). Suppression of the ALT pathway has been associated with diminished spatial interaction of shelterin components and single-strand DNA binding DDR proteins such as RPA1 with the ALT-associated promyelocytic leukemia body (APB) bodies [55–59]. Our results indicate that polyploidy-inducing constitutive overexpression of hTERC, in ALT cells that lack hTERC RNA, is associated with significantly reduced rates of telomeric localization of PML, RPA1, and ATRIP but substantially increased rates of γ-H2AX and 53BP1, as compared to parental and control VA-13 cells. Reconstitution of telomerase activity restored suppression of telomeric association of PML, RPA1, and ATRIP with TRF2, but the rates of γ-H2AX and 53BP1 assembly at telomeres were notably increased (Figure 6A). γ-H2AX recruits 53BP1 and both bind at double-stranded DNA breaks [60]. Escalated telomere dysfunction was verified by increased frequency of chromosomal end fusions (Figure 5, B and C). To further investigate the role of telomerase activity in triggering immediate DDR at ALT telomeres, we used immuno-FISH to examine nuclear localization of exogenously expressed hTERT, relative to the chromosome termini. In the absence of hTERC, inducible expression of telomerase activity resulted in diffuse nuclear fluorescence of an hTERT-specific antibody, while spatial association with telomeres was low. Inducible telomerase activity in VA-13InTERT cells resulted in accumulation of hTERT in nuclear foci that appear to be Cajal bodies [61]. Co-localization of hTERT with telomeric repeats was substantially higher as compared to the stably transfected telomerase-positive VA-13TA cells (Figure 6B). Overall, these results indicate that overexpression of hTERC alone, as well as the combined action of hTERC and hTERT, antagonizes the ALT pathway of telomere maintenance, eliciting, respectively, constitutive or transient telomeric DDR that triggers WGD.
A Noncanonical Function of hTERT Suppresses Polyploidization in the ALT Pathway
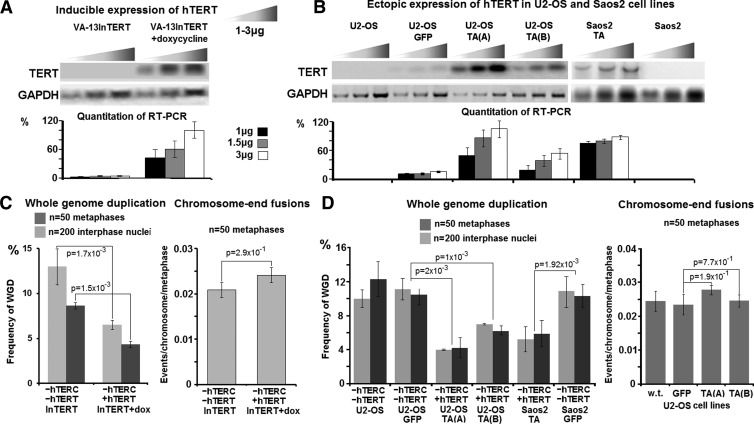
The introduction of hTERT in VA-13 cells, in the absence of hTERC, did not affect association of PML, RPA1, and ATRIP, with TRF2 and APBs and was not related to significant increase in the frequencies of classic telomere dysfunction-induced foci (TIFs; i.e., co-localized TRF2 and γ-H2AX nuclear foci; Figure 6A). InTERT, a VA-13 derivative cell line, was grown in the presence of doxycycline for 5 and 30 days. RT-PCR of the catalytic subunit of telomerase showed that hTERT expression was restored and remained as such during the duration of the experiment (Figure 7A). Semiquantitative RT-PCR showed that the levels of inducible hTERT expression were similar to those observed in telomerase-positive cancer cell lines such as HeLa or SW-480 (Figure W3A). At 5 and 30 days after inducible ectopic expression of hTERT, in the absence of hTERC and telomerase activity, a significant decrease in the endogenous rates of WGD was noted in VA-13InTERT + doxycycline cells (Table W2). Interestingly, suppression of WGD was independent of the rates of telomere dysfunction (Figures 5C and 7C). To rule out cell line-specific effects, we examined the hTERC/hTERT null human ALT osteosarcoma cell lines U2-OS and Saos2 [62], transduced either with a GFP-expressing lentivirus or the same construct expressing hTERT (Figure 7B). Again, ectopic expression of hTERT alone, in the absence of telomerase activity, did not affect rates of telomere dysfunction but was accompanied by significant suppression of endogenous WGD (Figure 7D). Collectively, these results demonstrate that, in human ALT cells not expressing hTERC, hTERT exerts a noncanonical telomerase activity-independent function, acting as a negative regulator of spontaneous polyploidization during continuous neoplastic cell growth.
Figure 7.
Ectopic expression of hTERT in the absence of telomerase activity suppresses endogenous polyploidization in ALT cells: Semiquantitative RT-PCR indicates inducible and stable expression of hTERT, in three ALT human cell lines (A, B). In the absence of telomerase activity, the inducible introduction of hTERT in the VA-13 derivative InTERT cell line for 30 days did not affect telomeric integrity, as indicated by the frequencies of terminal chromosome fusions, but reduced significantly the rates of endogenous mitotic and interphase WGDs (C). Similarly, the lentiviral transduction of U2-OS cells with particles carrying GFP or hTERT + GFP had insignificant effects on telomere functionality but significantly suppressed WGD. Suppression of WGD upon ectopic expression of hTERT was also observed in Saos2 cells. Transduction rates for U2-OS TA(A) and U2-OS GFP reached 90% to 95%, 50% to 60% for U2-OS TA(B), and 60% to 75% for Saos2TA and Saos2 GFP (D). Statistics by paired t test or chi-square test.
Discussion
Polyploidization through WGD occurs frequently in all types and stages of human neoplasia and may be associated with disease progression. About 15% of the karyotyped human malignancies show evidence of polyploidization. However, only 49% of ∼60,000 cases of Mitelman Catalogue (at the time of our analysis) displayed a diploid or pseudodiploid karyotype of 46 chromosomes, suggesting that well over 50% of all human cancers exhibit major chromosome imbalances [41]. The tendency of cancer cells to undergo extensive chromosome losses [29,30,34], combined with previous technical difficulties in precise chromosome recognition and interpretation of cytogenetic aberrations [63], as well as insufficient karyotypic recording of WGD, may have grossly underestimated the frequencies of polyploidization as an almost universal process of malignant genome evolution. Indeed, in the substantially smaller but more detailed NCBI M-FISH/SKY database, about 60% of the solid tumor cases displayed evidence of clonal evolution through WGD or polyploidy. Consistent with these results, a recent meta-analysis, of allelic copy-ratio profiles derived from single nucleotide polymorphism (SNP) arrays from 3155 cancer samples, showed that the frequencies of WGD varied between 25% and 50% across tumor types, reflecting differences in disease-specific biology and clinical progression status [64].
From the pivotal work of Barbara McClintock [65], telomere dysfunction is known to produce structural chromosomal instability and can cause chromosome losses through B/F/B cycles and anaphase lagging [28,66]. There are frequent reports of high rates of polyploidization in tissues and cells suffering from telomere dysfunction [27,67]. Moreover, the association of tetraploidization with dysfunctional telomeres in POT1a/b and p53-deficient mouse embryonic fibroblasts suggested a common mechanism for the induction of polyploidization in the early stages of tumorigenesis, when telomere dysfunction can result from excessive telomere shortening [33]. Inhibition of telomerase activity is currently under clinical trials as a novel method to treat human cancers [68,69]. Our data show that ablation of telomerase in both CIN and MIN cell lines is associated with the induction of telomere crisis and increased tendency for polyploidy. Despite some limited clonal structural or numerical chromosome anomalies, MIN tumors exert a remarkable propensity to maintain diploidy [1,70]. Indeed, in the presence of functional p53, spontaneous restoration of telomerase activity in the MIN context dropped telomere dysfunction-driven WGD substantially, indicating negative selection for tetraploidy. However, in the absence of p53, several subclones of the HCT-116 cells were composed solely of near-tetraploid cells. Therefore, both CIN and MIN telomerase-positive cell lines respond to extreme telomere dysfunction through WGD. Clonal expansion of tetraploid or polyploid cells appears to occur independently of p53 in CIN telomerase-positive or ALT cell lines. In contrast, MIN cells may depend on defective p53 to maintain polyploidy.
As predicted from previous studies [44], prolonged reconstitution of telomerase activity in ALT cells repressed a proportion of ongoing telomere dysfunction events. Telomerase reconstitution also suppressed the propensity for polyploidization to rates similar to those observed in telomerase-positive cell lines. However, compared to parental ALT VA-13, virtually all of the telomerase-positive VA-13TA cells became polyploid. Hence, during the process of ectopic restoration of telomerase activity, massive polyploidization may occur. Indeed, when we isolated or combined the effects of hTERC and hTERT, in constitutive or inducible systems, we observed that the expression of hTERC in hTERT/hTERC-null ALT cells decreased spatial interaction of telomeres with APB bodies. This effect suppressed recombinatorial ALT telomere maintenance [56,58,71] and raised endogenous telomere dysfunction, increasing the incidence of WGD. Interestingly, the inducible concerted action of hTERC and hTERT, to elongate telomeres, dramatically affected telomere integrity of the ALT cells resulting in massive local DDR that triggered terminal chromosome fusions and increased rates of polyploidization. These results indicate that the initial steps of reconstitution of telomerase activity in the ALT pathway provoke a transient telomeric crisis that is restored upon prolonged action of telomerase. Thus, telomerase activity despite antagonistic effects to the ALT machinery eventually reduces telomere dysfunction and ongoing polyploidization. However, initial telomeric insults can be significant enough to allow emergence and clonal expansion of cells with duplicated polyploid genomic content.
The introduction of hTERT, in the presence of constitutive expression of hTERC, activated the telomerase holoenzyme to initiate telomere maintenance but also restored spatial association of the single-stranded DNA-binding proteins ATRIP and RPA1 with telomeres at levels similar to the parental and mock ALT cells. Combined with the increased rates of telomere dysfunction and WGD during constitutive expression of hTERC, the previous observations support the hypothesis that, in the absence of hTERT, excessive hTERC may interfere with the machinery of ALT telomere maintenance through the suppression of terminal DNA recombinatorial replication. Apparently, the inducible assembly of telomerase holoenzyme occupied a proportion of ALT-interfering hTERC, so the low frequencies of the association of RPA1/ATRIP with single-stranded telomeric DNA were rescued. However, the simultaneous activation of both pathways of telomere maintenance in ALT cells was largely ineffective because it was associated with immediate escalation of telomere dysfunction and further increase in the rates of WGD. Since these effects are not detrimental for continuous growth, after a number of PDs in culture, a proportion of ALT telomeres will be replenished by telomerase, resulting in less frequent DDR responses at telomeres. Thus, most of the ALT machinery will be suppressed, and high rates of endogenous WGD will be reduced to similar levels with those observed in several telomerase-positive cell lines. Interestingly, hTERT, the major component of telomerase, might be involved in a negative feedback loop, acting as a direct suppressor of polyploidy through a reverse transcriptase-independent, noncanonical function [72,73] that remains to be clarified (Figure W6).
In summary, depending on the preexisting pathway of telomere maintenance, both telomerase inhibition and the introduction of telomerase activity are permissive for clonal evolution in neoplasia. This allows increased genome plasticity by the generation of novel structural chromosome anomalies and also through polyploidization.
Supplementary Material
Acknowledgments
We thank T. Halazonetis, J. Karlseder, H. Seimiya, D. Thanos, D. Trono, and DAKO, for reagents. We also thank all cell line donors, as indicated in the Materials and Methods section, as well as Drs A. Efstratiadis and G. Grigoriadis for continuous support and valuable discussions.
Abbreviations
- CIN
chromosomal instability in neoplasia
- ALT
alternative lengthening of telomeres
Footnotes
This work was supported by the Biomedical Research Foundation of the Academy of Athens (Athens, Greece), in an intramural funding for S.G., grant 05NON-EU-449 of the Greek Secretariat of Research of the Greek Ministry of Development, and the EU COST Action BM0703 “Cangenin.” The authors declare no competing financial interests.
This article refers to supplementary materials, which are designated by Tables W1 and W2 and Figures W1 to W6 and are available online at www.neoplasia.com.
References
- 1.Rajagopalan H, Jallepalli PV, Rago C, Velculescu VE, Kinzler KW, Vogelstein B, Lengauer C. Inactivation of hCDC4 can cause chromosomal instability. Nature. 2004;428:77–81. doi: 10.1038/nature02313. [DOI] [PubMed] [Google Scholar]
- 2.Rajagopalan H, Lengauer C. Aneuploidy and cancer. Nature. 2004;432:338–341. doi: 10.1038/nature03099. [DOI] [PubMed] [Google Scholar]
- 3.Bayani J, Selvarajah S, Maire G, Vukovic B, Al-Romaih K, Zielenska M, Squire JA. Genomic mechanisms and measurement of structural and numerical instability in cancer cells. Semin Cancer Biol. 2007;17:5–18. doi: 10.1016/j.semcancer.2006.10.006. [DOI] [PubMed] [Google Scholar]
- 4.Gorgoulis VG, Vassiliou LV, Karakaidos P, Zacharatos P, Kotsinas A, Liloglou T, Venere M, Ditullio RA, Jr, Kastrinakis NG, Levy B, et al. Activation of the DNA damage checkpoint and genomic instability in human precancerous lesions. Nature. 2005;434:907–913. doi: 10.1038/nature03485. [DOI] [PubMed] [Google Scholar]
- 5.Nowell PC. The clonal evolution of tumor cell populations. Science. 1976;4260:23–28. doi: 10.1126/science.959840. [DOI] [PubMed] [Google Scholar]
- 6.Weinberg RA. The integration of molecular genetics into cancer management. Cancer. 1992;70:1653–1658. doi: 10.1002/1097-0142(19920915)70:4+<1653::aid-cncr2820701603>3.0.co;2-1. [DOI] [PubMed] [Google Scholar]
- 7.Vogelstein B, Kinzler KW. Cancer genes and the pathways they control. Nat Med. 2004;10:789–799. doi: 10.1038/nm1087. [DOI] [PubMed] [Google Scholar]
- 8.Gagos S, Irminger-Finger I. Chromosome instability in neoplasia: chaotic roots to continuous growth. Int J Biochem Cell Biol. 2005;37:1014–1033. doi: 10.1016/j.biocel.2005.01.003. [DOI] [PubMed] [Google Scholar]
- 9.Greaves M, Maley CC. Clonal evolution in cancer. Nature. 2012;481:306–313. doi: 10.1038/nature10762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Storchova Z, Pellman D. From polyploidy to aneuploidy, genome instability and cancer. Nat Rev Mol Cell Biol. 2004;5:45–54. doi: 10.1038/nrm1276. [DOI] [PubMed] [Google Scholar]
- 11.Sabisz M, Skladanowski A. Cancer stem cells and escape from drug-induced premature senescence in human lung tumor cells: implications for drug resistance and in vitro drug screening models. Cell Cycle. 2009;8:3208–3217. doi: 10.4161/cc.8.19.9758. [DOI] [PubMed] [Google Scholar]
- 12.Vakifahmetoglu H, Olsson M, Zhivotovsky B. Death through a tragedy: mitotic catastrophe. Cell Death Differ. 2008;15:1153–1162. doi: 10.1038/cdd.2008.47. [DOI] [PubMed] [Google Scholar]
- 13.Erenpreisa J, Cragg SM. MOS, aneuploidy and the ploidy cycle of cancer cells. Review. Oncogene. 2010;29:5447–5451. doi: 10.1038/onc.2010.310. [DOI] [PubMed] [Google Scholar]
- 14.Blackburn EH. Telomeres. Trends Biochem Sci. 1991;16:378–381. doi: 10.1016/0968-0004(91)90155-o. [DOI] [PubMed] [Google Scholar]
- 15.Shay JW, Wright WE. Senescence and immortalization: role of telomeres and telomerase. Carcinogenesis. 2005;26:867–874. doi: 10.1093/carcin/bgh296. [DOI] [PubMed] [Google Scholar]
- 16.Rizki A, Lundblad V. Defects in mismatch repair promote telomerase-independent proliferation. Nature. 2001;411:713–716. doi: 10.1038/35079641. [DOI] [PubMed] [Google Scholar]
- 17.Huffman EK, Levene DS, Tesmer MV, Shay WJ, Wright EW. Telomere shortening is proportional to the size of the G-rich telomeric 3′-overhang. J Biol Chem. 2000;275:19719–19722. doi: 10.1074/jbc.M002843200. [DOI] [PubMed] [Google Scholar]
- 18.Zou Y, Sfeir A, Gryaznov SM, Shay JW, Wright WE. Does a sentinel or a subset of short telomeres determine replicative senescence? Mol Biol Cell. 2004;15:3709–3718. doi: 10.1091/mbc.E04-03-0207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sabatier L, Ricoul M, Pottier G, Murnane JP. The loss of a single telomere can result in instability of multiple chromosomes in a human tumor cell line. Mol Cancer Res. 2005;3:139–150. doi: 10.1158/1541-7786.MCR-04-0194. [DOI] [PubMed] [Google Scholar]
- 20.Wright WE, Shay JW. Time, telomeres and tumours: is cellular senescence more than an anticancer mechanism? Trends Cell Biol. 1995;5:293–297. doi: 10.1016/s0962-8924(00)89044-3. [DOI] [PubMed] [Google Scholar]
- 21.Wright WE, Shay JW. Cellular senescence as a tumor-protection mechanism: the essential role of counting. Curr Opin Genet Dev. 2001;11:98–103. doi: 10.1016/s0959-437x(00)00163-5. [DOI] [PubMed] [Google Scholar]
- 22.Halazonetis TD, Gorgoulis VG, Bartek J. An oncogene-induced DNA damage model for cancer development. Science. 2008;319:1352–1355. doi: 10.1126/science.1140735. [DOI] [PubMed] [Google Scholar]
- 23.Kim NW, Piatyszek MA, Prowse KR, Harley CB, West MD, Ho PL, Coviello GM, Wright WE, Weinrich SL, Shay JW. Specific association of human telomerase activity with immortal cells and cancer. Science. 1994;266:2011–2015. doi: 10.1126/science.7605428. [DOI] [PubMed] [Google Scholar]
- 24.Shay JW, Bacchetti S. A survey of telomerase activity in human cancer. Eur J Cancer. 1997;33:787–791. doi: 10.1016/S0959-8049(97)00062-2. [DOI] [PubMed] [Google Scholar]
- 25.Muntoni A, Reddel RR. The first molecular details of ALT in human tumor cells. Hum Mol Genet. 2005;14:191–196. doi: 10.1093/hmg/ddi266. [DOI] [PubMed] [Google Scholar]
- 26.Nabetani A, Ishikawa F. Alternative lengthening of telomeres pathway: recombination-mediated telomere maintenance mechanism in human cells. J Biochem. 2011;149:5–14. doi: 10.1093/jb/mvq119. [DOI] [PubMed] [Google Scholar]
- 27.Blasco MA, Lee HW, Hande MP, Samper E, Lansdorp PM, DePinho RA, Greider CW. Telomere shortening and tumor formation by mouse cells lacking telomerase RNA. Cell. 1997;91:25–34. doi: 10.1016/s0092-8674(01)80006-4. [DOI] [PubMed] [Google Scholar]
- 28.Gisselsson D, Pettersson L, Höglund M, Heidenblad M, Gorunova L, Wiegant J, Mertens F, Dal Cin P, Mitelman F, Mandahl N. Chromosomal breakage-fusion-bridge events cause genetic intratumor heterogeneity. Proc Natl Acad Sci USA. 2000;97:5357–5362. doi: 10.1073/pnas.090013497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Grady WM. Genomic instability and colon cancer. Cancer Metastasis Rev. 2004;23:11–27. doi: 10.1023/a:1025861527711. [DOI] [PubMed] [Google Scholar]
- 30.Gorringe LK, Chin S, Pharoah P, Staines MJ, Oliveira C, Edwards PA, Caldas C. Evidence that both genetic instability and selection contribute to the accumulation of chromosome alterations in cancer. Carcinogenesis. 2005;26:923–930. doi: 10.1093/carcin/bgi032. [DOI] [PubMed] [Google Scholar]
- 31.Weaver BA, Silk AD, Montagna C, Verdier-Pinard P, Cleveland DW. Aneuploidy acts both oncogenically and as a tumor suppressor. Cancer Cell. 2007;11:25–36. doi: 10.1016/j.ccr.2006.12.003. [DOI] [PubMed] [Google Scholar]
- 32.Hanks S, Coleman K, Reid S, Plaja A, Firth H, Fitzpatrick D, Kidd A, Mehes K, Nash R, Robin N, et al. Constitutional aneuploidy and cancer predisposition caused by biallelic mutations in BUB1B. Nat Genet. 2004;36:1159–1161. doi: 10.1038/ng1449. [DOI] [PubMed] [Google Scholar]
- 33.Davoli T, Denchi EL, de Lange T. Persistent telomere damage induces bypass of mitosis and tetraploidy. Cell. 2010;141:81–93. doi: 10.1016/j.cell.2010.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Duesberg P, Rausch C, Rasnick D, Hehlmann R. Genetic instability of cancer cells is proportional to their degree of aneuploidy. Proc Natl Acad Sci USA. 1998;95:13692–13697. doi: 10.1073/pnas.95.23.13692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cahill DP, Kinzler KW, Vogelstein B, Lengauer C. Genetic instability and Darwinian selection in tumours. Trends Cell Biol. 1999;9:57–60. [PubMed] [Google Scholar]
- 36.Gagos S, Iatridou-Kyrkou K, Liosi A, Karakitsos P, Papageorgaki P, Kyroudi A, Pathak S. Clonal evolution of an immunoblastic type non-Hodgkin's lymphoma with der(6)t(1;6)(q11;p11) as its primary cytogenetic abnormality. Cancer Genet Cytogenet. 1995;79:59–63. doi: 10.1016/0165-4608(94)00112-o. [DOI] [PubMed] [Google Scholar]
- 37.Antachopoulos CT, Gagos S, Iliopoulos DC, Karayannacos PE, Tseleni-Balafouta S, Alevras P, Koundouris C, Skalkeas GD. Low-dose heparin treatment does not inhibit SW480 human colon cancer growth and metastasis in vivo. In Vivo. 1996;10:527–531. [PubMed] [Google Scholar]
- 38.Gagos S, Papaioannou G, Chiourea M, Merk-Loretti S, Jefford CE, Mikou P, Irminger-Finger I, Liossi A, Blouin JL, Dahoun S. Unusually stable abnormal karyotype in a highly aggressive melanoma negative for telomerase activity. Mol Cytogenet. 2008;1:20. doi: 10.1186/1755-8166-1-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gagos S, Iliopoulos D, Tseleni-Balafouta S, Agapitos M, Antachopoulos C, Kostakis A, Karayannakos P, Skalkeas G. Cell senescence and a mechanism of clonal evolution leading to continuous cell proliferation, loss of heterozygosity, and tumor heterogeneity: studies on two immortal colon cancer cell lines. Cancer Genet Cytogenet. 1996;90:157–165. doi: 10.1016/s0165-4608(96)00049-0. [DOI] [PubMed] [Google Scholar]
- 40.Shaffer GL, Slovak LM, Campbell JL, editors. ISCN: An International System for Human Cytogenetic Nomenclature. Basel: S. Karger; 2009. [Google Scholar]
- 41.Mitelman F, Johansson B, Mertens F. Mitelman Database of Chromosome Aberrations and Gene Fusions in Cancer. 2007. Available at: http://cgap.nci.nih.gov/Chromosomes/Mitelman.
- 42.NCI and NCBI's SKY/M-FISH and CGH Database, author. 2001. Available at: http://www.ncbi.nlm.nih.gov/sky/skyweb.cgi.
- 43.Bunz F, Dutriaux A, Lengauer C, Waldman T, Zhou S, Brown JP, Sedivy JM, Kinzler KW, Vogelstein B. Requirement for p53 and p21 to sustain G2 arrest after DNA damage. Science. 1998;282:1497–1501. doi: 10.1126/science.282.5393.1497. [DOI] [PubMed] [Google Scholar]
- 44.Ford LP, Zou Y, Pongracz K, Gryaznov SM, Shay JW, Wright WE. Telomerase can inhibit the recombination-based pathway of telomere maintenance in human cells. J Biol Chem. 2001;276:32198–32203. doi: 10.1074/jbc.M104469200. [DOI] [PubMed] [Google Scholar]
- 45.Bechter OE, Zou Y, Walker W, Wright WE, Shay JW. Telomeric recombination in mismatch repair deficient human colon cancer cells after telomerase inhibition. Cancer Res. 2004;64:3444–3451. doi: 10.1158/0008-5472.CAN-04-0323. [DOI] [PubMed] [Google Scholar]
- 46.Seimiya H, Oh-hara T, Suzuki T, Naasani I, Shimazaki T, Tsuchiya K, Tsuruo T. Telomere shortening and growth inhibition of human cancer cells by novel synthetic telomerase inhibitors MST-312, MST-295, and MST-1991. Mol Cancer Ther. 2002;1:657–665. [PubMed] [Google Scholar]
- 47.Fabarius A, Hehlmann R, Duesberg PH. Instability of chromosome structure in cancer cells increases exponentially with degrees of aneuploidy. Cancer Genet Cytogenet. 2003;143:59–72. doi: 10.1016/s0165-4608(03)00003-7. [DOI] [PubMed] [Google Scholar]
- 48.Mitelman F, Johansson B, Mertens F. The impact of translocations and gene fusions on cancer causation. Nat Rev Cancer. 2007;7:233–245. doi: 10.1038/nrc2091. [DOI] [PubMed] [Google Scholar]
- 49.Ozery-Flato M, Linhart C, Trakhtenbrot L, Izraeli S, Shamir R. Large-scale analysis of chromosomal aberrations in cancer karyotypes reveals two distinct paths to aneuploidy. Genome Biol. 2011;12:R61. doi: 10.1186/gb-2011-12-6-r61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gagos S, Chiourea M, Christodoulidou A, Apostolou E, Raftopoulou C, Deustch S, Jefford CE, Irminger-Finger I, Shay JW, Antonarakis SE. Pericentromeric instability and spontaneous emergence of human neoacrocentric and minute chromosomes in the alternative pathway of telomere lengthening. Cancer Res. 2008);68:8146–8155. doi: 10.1158/0008-5472.CAN-08-0945. [DOI] [PubMed] [Google Scholar]
- 51.Mitchell MA, Johnson JE, Pascarelli K, Beeharry N, Chiourea M, Gagos S, Lev D, von Mehren M, Kipling D, Broccoli D. Doxorubicin resistance in a novel in vitro model of human pleomorphic liposarcoma associated with alternative lengthening of telomeres. Mol Cancer Ther. 2010;9:682–692. doi: 10.1158/1535-7163.MCT-09-0705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lundberg G, Sehic D, Lansberg JK, Õra I, Frigyesi A, Castel V, Navarro S, Piqueras M, Martinsson T, Noguera R, et al. Alternative lengthening of telomeres—an enhanced chromosomal instability in aggressive non-MYCN amplified and telomere elongated neuroblastomas. Genes Chromosomes Cancer. 2011;50:250–262. doi: 10.1002/gcc.20850. [DOI] [PubMed] [Google Scholar]
- 53.Pantic M, Zimmermann S, El Daly H, Opitz OG, Popp S, Boukamp P, Martens UM. Telomere dysfunction and loss of p53 cooperate in defective mitotic segregation of chromosomes in cancer cells. Oncogene. 2006;25:4413–4420. doi: 10.1038/sj.onc.1209486. [DOI] [PubMed] [Google Scholar]
- 54.Pihan GA, Wallace J, Zhou Y, Doxsey SJ. Centrosome abnormalities and chromosome instability occur together in pre-invasive carcinomas. Cancer Res. 2003;63:1398–1404. [PubMed] [Google Scholar]
- 55.Nakamura JA, Redon EC, Bonner MW, Sedelnikova AO. Telomere-dependent and telomere-independent origins of endogenous DNA damage in tumor cells. Aging (Albany NY) 2009;1:212–219. doi: 10.18632/aging.100019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Grudic A, Jul-Larsen A, Haring SJ, Wold MS, Lønning PE, Bjerkvig R, Bøe SO. Replication protein A prevents accumulation of single-stranded telomeric DNA in cells that use alternative lengthening of telomeres. Nucleic Acids Res. 2007;35:7267–7278. doi: 10.1093/nar/gkm738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hass SC, Gakhar L, Wold SM. Functional characterization of a cancer causing mutation in human replication protein A. Mol Cancer Res. 2010;8:1017–1026. doi: 10.1158/1541-7786.MCR-10-0161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mason CA, Roy R, Simmons TD, Wold SM. Functions of alternative replication protein A in initiation and elongation. Biochemistry. 2010;49:5919–5928. doi: 10.1021/bi100380n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Misri S, Pandita S, Kumar R, Pandite KT. Telomeres, histone code, and DNA damage response. Cytogenet Genome Res. 2008;122:297–307. doi: 10.1159/000167816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Goodarzi AA, Jeggo AP. ‘A mover and a shaker’: 53BP1 allows DNA doublestrand breaks a chance to dance and unite. F1000 Biol Rep. 2009;1:21. doi: 10.3410/B1-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhao Y, Abreu E, Kim J, Stadler G, Eskiocak U, Terns MP, Shay JW, Wright WE. Processive and distributive extension of human telomeres by telomerase under homeostatic and nonequilibrium conditions. Mol Cell. 2011;42:297–307. doi: 10.1016/j.molcel.2011.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang Y, Cai L, Wei RX, Hu H, Jin W, Zhu XB. Different expression of alternative lengthening of telomere (ALT)-associated proteins/mRNAs in osteosarcoma cell lines. Oncol Lett. 2011;6:1327–1332. doi: 10.3892/ol.2011.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Albertson DG, Collins C, McCormick F, Gray JW. Chromosome aberrations in solid tumors. Nat Genet. 2003;34:369–376. doi: 10.1038/ng1215. [DOI] [PubMed] [Google Scholar]
- 64.Carter LS, Cibulskis K, Helman E, McKenna A, Shen H, Zack T, Laird PW, Onofrio RC, Winckler W, Weir BA, et al. Absolute quantification of somatic DNA alterations in human cancer. Nat Biotechnol. 2012;30:413–421. doi: 10.1038/nbt.2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jones RN. McClintock's controlling elements: the full story. Cytogenet Genome Res. 2005;109:90–103. doi: 10.1159/000082387. [DOI] [PubMed] [Google Scholar]
- 66.Martinez AC, van Wely KH. Are aneuploidy and chromosome breakage caused by a CINgle mechanism? Cell Cycle. 2010;12:2275–2280. doi: 10.4161/cc.9.12.11865. [DOI] [PubMed] [Google Scholar]
- 67.Lazzerini Denchi E, Celli G, de Lange T. Hepatocytes with extensive telomere deprotection and fusion remain viable and regenerate liver mass through endoreduplication. Genes Dev. 2006;20:2648–2653. doi: 10.1101/gad.1453606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Shay JW, Wright WE. Hallmarks of telomeres in ageing research. J Pathol. 2007;211:114–123. doi: 10.1002/path.2090. [DOI] [PubMed] [Google Scholar]
- 69.Shay JW, Reddel RR, Wright WE. Cancer and telomeres—an ALTernative to telomerase. Science. 2012;336:1388–1390. doi: 10.1126/science.1222394. [DOI] [PubMed] [Google Scholar]
- 70.Lengauer C, Kinzler KW, Vogelstein B. Genetic instabilities in human cancers. Nature. 1998;396:643–649. doi: 10.1038/25292. [DOI] [PubMed] [Google Scholar]
- 71.Draskovic I, Arnoult N, Steiner V, Bacchetti S, Lomonte P, Londoño-Vallejo A. Probing PML body function in ALT cells reveals spatio-temporal requirements for telomere recombination. Proc Natl Acad Sci USA. 2009;106:15726–15731. doi: 10.1073/pnas.0907689106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sarin KY, Cheung P, Gilison D, Lee E, Tennen RI, Wang E, Artandi MK, Oro AE, Artandi SE. Conditional telomerase induction causes proliferation of hair follicle stem cells. Nature. 2005;436:1048–1052. doi: 10.1038/nature03836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Imamura S, Uchiyama J, Koshimizu E, Hanai J, Raftopoulou C, Murphey RD, Bayliss PE, Imai Y, Burns CE, Masutomi K, et al. A non-canonical function of zebrafish telomerase reverse transcriptase is required for developmental hematopoiesis. PLoS One. 2008;3:e3364. doi: 10.1371/journal.pone.0003364. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.