Abstract
Cancer progression is a complex series of events thought to incorporate the reversible developmental process of epithelial-to-mesenchymal transition (EMT). In vitro, the microRNA-200 family maintains the epithelial phenotype by posttranscriptionally inhibiting the E-cadherin repressors, ZEB1 and ZEB2. Here, we used in situ hybridization and immunohistochemistry to assess expression of miR-200 and EMT biomarkers in formalin-fixed paraffin-embedded human colorectal adenocarcinomas. In addition, laser capture microdissection and quantitative real-time polymerase chain reaction were employed to quantify levels of miR-200 in the normal epithelium, tumor core, invasive front, and stroma. We find that miR-200 is downregulated at the invasive front of colorectal adenocarcinomas that have destroyed and invaded beyond the basement membrane. However, regional lymph node metastases and vascular carcinoma deposits show strong expression of miR-200, suggesting this family of miRNAs is involved in the recapitulation of the primary tumor phenotype at metastatic sites. In contrast, adenomas and adenocarcinomas with intact basement membranes showed uniform miR-200 expression from the tumor core to the tumor-host interface. Taken together, these data support the involvement of EMT and mesenchymal-to-epithelial transition (MET) in the metastasis cascade and show that miR-200 is downregulated in the initial stages of stromal invasion but is restored at metastatic sites.
Introduction
Epithelial-to-mesenchymal transition (EMT) is recognized as an important step in invasion and metastasis, with mounting evidence suggesting metastasis is initiated by an EMT at the invasive front of primary carcinomas [1,2]. EMT is typified by down-regulation of E-cadherin, reorganization of the actin cytoskeleton from a cortical to a stress fiber distribution [3,4], and up-regulation of transcriptional repressors, including ZEB1 and ZEB2, which bind to E-boxes in the E-cadherin gene promoter to prevent its transcription [5]. A consequence of E-cadherin down-regulation is the dissociation of adherens junctions and the subsequent liberation of β-catenin from the plasma membrane. Accumulation of cytosolic β-catenin can lead to its translocation to the nucleus where it functions in a transcription complex to upregulate many other genes involved in migration, invasion, cell cycle progression, and differentiation. Immunohistologic examination and gene expression profiling of EMT biomarkers in colorectal cancer have revealed heterogeneous expression between the tumor core and the invasive front, indicating that a morphogenesis favoring the mesenchymal phenotype occurs in leading edge carcinoma cells [6–9]. Acquisition of the mesenchymal phenotype may well assist the migration of budding tumor cells, which are defined as single cells or clusters of up to five cells detached from the main tumor mass. Budding cells are positively correlated with tumor relapse, lymphatic and venous invasion, as well as lymph node, liver, and lung metastases [10–19]. Tumor budding is also associated with breakdown of the basement membrane, a thin layer of collagenous extracellular matrix that encapsulates the epithelium of normal colonic crypts and adenomatous glands. It has been shown that degradation of the basement membrane at the invasive front of colorectal adenocarcinomas positively correlates with metastasis and poor survival [20]. Moreover, metastases typically resemble the differentiation state of the primary epithelial tumor, suggesting that molecular changes acquired during the metastasis cascade are reversible [6]. The reversible process of EMT provides an attractive model for metastasis development because it describes tumor cell budding (dedifferentiation) and invasion at the primary site and redifferentiation (MET) at the metastatic site [6,21]. Taken together, these studies link EMT, budding cells, and loss of the basement membrane to invasion and metastasis.
EMT is regulated by members of the microRNA-200 family, which participate in a double-negative feedback loop with ZEB1 and ZEB2 [22–24]. The five members of the family are encoded in two genes on chromosomes 1 (miR-200b∼200a∼429) and 12 (miR-200c∼141). ZEB1 and ZEB2 repress transcription of both miR-200 genes to favor the mesenchymal phenotype, and conversely, miR-200 family members inhibit translation of ZEB1 and ZEB2, thereby promoting the epithelial phenotype by maintaining E-cadherin expression. In previous quantitative real-time polymerase chain reaction (qPCR) and immunohistochemical analyses on human ductal (epithelial) and metaplastic (mesenchymal) breast cancers, we found that ductal tumors have high levels of E-cadherin and miR-200, whereas invasive metaplastic tumors lack both, indicating that loss of miR-200 may contribute to tumor invasion [22]. Other groups have conducted immunohistochemistry and genetic profiling on colorectal adenocarcinomas, revealing that ZEB1 and ZEB2 are upregulated in leading edge malignant cells in which nuclear β-catenin and loss of E-cadherin are evident [7–9,20,25]. On the basis of these findings showing EMT at the invasive front of carcinomas and the fact that the pro-invasive properties of EMT are tightly regulated by the miR-200 family in vitro, we considered that loss of miR-200 could be an underlying mechanism of early invasion. We hypothesized that the miR-200 family would be downregulated at the tumor-host interface of colorectal adenocarcinomas where epithelial proteins are lost and mesenchymal proteins are gained. We show here that expression of miR-200 is down-regulated at the invasive front of colorectal adenocarcinomas with degraded basement membranes and budding cells, but miR-200 expression is strong in regional metastases. Taken together, these observations suggest that reversible EMT mediated by miR-200 contributes to colon cancer progression.
Materials and Methods
Scratch Assay and Immunocytochemistry
Confluent SW480 cells were plated onto poly-L-lysine coated glass slides, and a scratch was made 24 hours later using a pipette tip. After 2 days, cells were fixed with paraformaldehyde (PFA) and permeabilized, and anti-E-cadherin (mouse anti-E-cadherin IgG2a, 1:500, 610182, BD Transduction Laboratories, North Ryde, Australia) or anti-ZEB1 (E:20X, 1:50, Santa Cruz Biotechnology, Santa Cruz, CA, sc-10572) antibodies [diluted in Triton/phosphate-buffered saline (PBS)] were applied for 1 hour at room temperature. After three PBS washes, secondary antibodies (Alexa Fluor goat anti-mouse IgG2a 488, donkey anti-goat 647; Life Technologies Australia Pty Ltd, Mulgrave, Australia), diluted in Triton/PBS, were applied to the cells for 1 hour at room temperature in the dark. 4′,6-Diamidino-2-phenylindole (DAPI) was added in the final 5 minutes, and then slides were washed thrice in PBS and mounted in Dako aqueous fluorescent mounting medium. Cells were photographed on an Olympus CellR fluorescence microscope.
miRNA In Situ Hybridization in Formalin-Fixed Paraffin-Embedded Human Colon Sections
Sections of 4 µm were fixed for 10 minutes in 4% PFA (pH 9–9.9) in PBS and then rinsed in three 5-minute PBS washes. Sections were deproteinated for 30 minutes in Proteinase K Buffer [50 mM Tris (pH 7.5), 5 mM EDTA, and 6.7 µg/ml Proteinase K] and then fixed in 4% PFA for 5 minutes and washed thrice in PBS. Acetylation was carried out for 10 minutes (30 ml of 0.1 M triethanolamine, 53.4 µl of concentrated HCl, and 76.3 µl of acetic anhydride) before three 5-minute PBS washes. Sections were prehybridized in hybridization buffer [50% formamide, 5x SSC (pH 4.5), 50 µg/ml yeast RNA, 1% sodium dodecyl sulfate, and 50 µg/ml heparin] in a humidified chamber (50% formamide, 5x SSC) for 2 hours at 30°C. 3′DIG-labeled miRCURY LNA miRNA probes [sn U6 hsa/mmu/rno (5′ cacgaatttgcgtgtcatcctt 3′, Tm = 75°C), scramble-miR (5′ ttcacaatgcgttatcggatgt 3′, Tm = 74°C), hsa-miR-200b (5′ gtcatcattaccaggcagtatta 3′, Tm = 71°C), and hsa-miR-200c (5′ ccatcattacccggcagtatta 3′, Tm = 74°C), Exiqon, Vedbaek, Denmark] were diluted to 20 nM in hybridization buffer and 50 µl was applied to each slide. Sections were coverslipped with parafilm and incubated overnight in a humidified chamber at 53°C. The next day, slides were placed in 5x SSC at 53°C and rocked for 30 minutes at room temperature to remove parafilm. Stringency washes were conducted at 53°C in 0.2x SSC for 3 hours, followed by 0.2x SSC at room temperature for 5 minutes. Sections were washed for 5 minutes in Buffer B1 [0.1 M Tris (pH 7.5), 0.15 M sodium chloride, and 0.24 mg/ml levamisole]. Sections were blocked for 1 hour in blocking solution [one-fifth fetal calf serum, one-fifth 10% blocking reagent (Roche Cat. No. 11096176001) in maleate buffer, and three-fifths maleate buffer] and placed in a humidified chamber at room temperature. For immunologic detection, anti-DIG antibody (Roche Diagnostics Australia Pty Ltd, Castle Hill, Australia) was diluted to 1:5000 in the blocking solution. Sections were incubated in a humidified chamber at room temperature, overnight in the dark. The next day, sections were washed thrice in Buffer B1 and then once in Buffer B3 [0.1 M Tris (pH 9.5), 0.1 M sodium chloride, 0.05 M magnesium chloride, 0.24 mg/ml levamisole, and 1% Tween-20]. Buffer B4 [Buffer B3 plus 20 µl/ml nitro-blue tetrazolium/5-bromo-4-chloro-3′-indolyphosphate (NBT/BCIP)] was applied to the sections, which were then coverslipped with parafilm. Sections were incubated in a humidified chamber, in the dark, overnight at room temperature. Slides were placed in tris-ethylenediaminetetraacetic acid (TE; pH 8) for 5 minutes, with rocking, then dehydrated on a heat block at 50°C. Slides were dipped in histolene or xylene and then mounted in a xylene-based mounting medium. Images were captured on the Olympus CellR or Hamamatsu Nanozoomer and analyzed using AnalySIS LS Research software (Olympus, Australia Pty Ltd, Mt Waverley, Australia), NDP Hamamatsu software, Microsoft Excel, and Graphpad Prism.
Immunofluorescence
Formalin-fixed paraffin-embedded (FFPE) sections were deparaffinized in three 5-minute washes of histolene and then rehydrated in two 10-minute washes of 100% ethanol, two 10-minute washes of 95% ethanol, and two 5-minute washes of MQ-H2O. Sections were placed in PBS for 5 minutes before being immersed in 10 mM sodium citrate buffer (pH 6.0) for 10 minutes at 95 to 99°C. Following antigen retrieval, sections were removed from heat and allowed to cool in the citrate buffer for 30 minutes. Sections were rinsed in three 5-minute washes of MQ-H2O and once in PBS for 5 minutes. Sections were blocked in 1% BSA in 0.3% Triton X-100/PBS for 1 hour at room temperature in a humidified chamber. Primary antibodies were diluted in the blocking solution (mouse anti-β-catenin IgG1, 1:200, BD Transduction Laboratories, 610154; mouse anti-E-cadherin IgG2a, 1:100, BD Transduction Laboratories, 610182; mouse anti-CK20 IgG2a, 1:50, Dako, M7019; mouse anti-AE1/AE3 IgG1, 1:400, Dako Australia Pty Ltd, Campbellfield, Australia, M3515; goat anti-ZEB1 E:20X, 1:50, Santa Cruz Biotechnology, sc-10572). Sections were incubated overnight at 4°C and then rinsed in three 5-minute washes of PBS. Secondary antibodies (Alexa Fluor goat anti-mouse IgG2a 488, goat anti-mouse IgG1 555, and donkey anti-goat 647; Life Technologies Australia Pty Ltd) were diluted in 0.3% Triton/PBS and applied to the sections for 2 hours in a humidified chamber in the dark. DAPI was added in the last 5 minutes. Sections were rinsed thrice in PBS before being mounted in Dako aqueous fluorescent mounting medium. For triplicate staining, primary antibodies were applied together, but secondary antibodies were staggered to prevent cross-reaction. Donkey anti-goat 647 was applied for 2 hours and rinsed off, and then goat anti-mouse 488 and 555 were applied together.
Immunohistochemistry
ZEB1 was detected with either goat anti-ZEB1 (E:20X, 1:50, Santa Cruz Biotechnology, sc-10572) or rabbit anti-ZEB1 antiserum (1:400 from Darling et al. [26]). Laminin was detected with mouse anti-laminin (LAM-89; Sigma Immunochemicals, Sigma-Aldrich, Castle Hill, Australia). Biotin-conjugated secondary antibodies (biotinylated anti-goat IgG, anti-rabbit IgG, or anti-mouse IgG; Vector Laboratories, Burlingame, CA, BA-9500) were applied (1:400), and ABC and DAB kits (Vector Laboratories) were used to visualize expression. Sections were counterstained with 1% methyl green.
Quantification of miRNA Staining Intensity
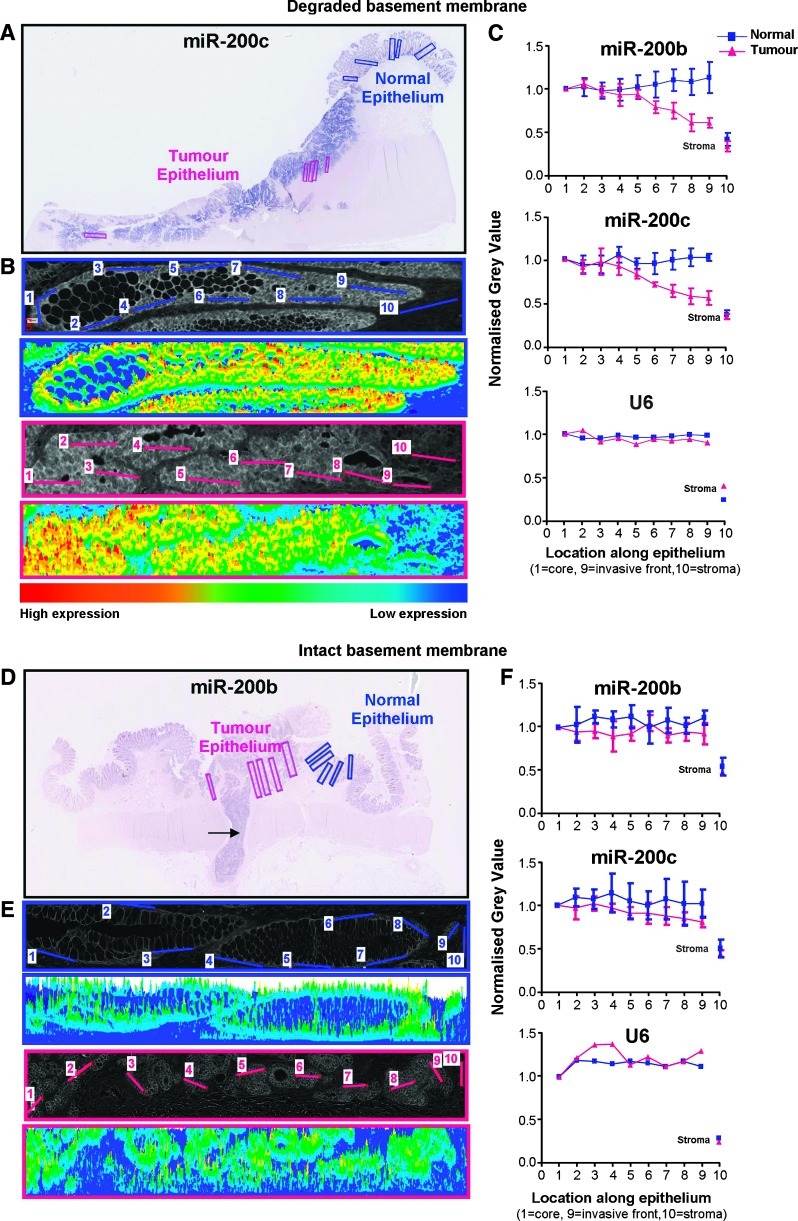
In situ hybridization (ISH) slides were scanned on a Hamamatsu Nanozoomer using the x40 objective. For each specimen, five regions of tumor and five regions of normal epithelium were selected and analyzed using Olympus AnalySIS software. Each region was white-balanced, converted to grayscale, and then inverted. For each region, 10 lines were drawn from the inner to the outer part of the tumor (or from the lumen to the muscularis mucosa in normal epithelium; see Figure 4 for examples), and the average staining intensity along each line was calculated and normalized to the value for the first line in the region. The final line measured in each region was within the stroma, where miR-200 expression was absent. For each line, values from the five regions were then averaged and graphs of miRNA signal across the tumor and normal epithelia were plotted in Prism, showing SEM.
Figure 4.
Quantification of ISH intensity. Representative results from adenocarcinomas with degraded (A–C) and intact (D–F) basement membranes. (A) ISH for miR-200c of a moderately differentiated pT3N0 adenocarcinoma with budding cells and degradation of the basement membrane at the tumor-host interface. (B, E) Representative examples of analyzed regions (inverted images and heat maps) of normal (blue) and tumor (pink) epithelia. Each region was white-balanced, converted to grayscale, and then inverted before the intensity of the miR-200 signal was measured across five regions (one region is shown) of normal and tumor epithelia. In each region, 10 lines were drawn to produce histograms of gray values, which were then averaged across the five regions (±SD). (C, F) Quantification of the miR-200 stain. Intensity values were measured from the lumen to the base of the crypt in normal epithelium and from the core to the invasive front of tumor epithelium. The final measurement was taken from the stroma. Data were normalized to the first point in normal and tumor regions (±SD). (D) Quantification of miR-200b staining across a well/moderately differentiated pT1N1 adenocarcinoma. This tumor was confined to the submucosa, displayed strong borders at the tumor-host interface, and had invaded into an adjacent blood vessel (arrow).
Laser Capture Microdissection
FFPE sections were deparaffinized, rehydrated, and then stained with hematoxylin and eosin (H&E). Following staining, sections were dipped briefly in 100% ethanol, allowed to dry and then scanned on a Hamamatsu Nanozoomer. Laser capture microdissection (LCM) was conducted within 24 hours of H&E staining using a Zeiss PALM LCM microscope; 105 µm2 of tissue was captured in duplicate from areas of normal epithelium, tumor core, invasive front, and stroma tissue. Total RNA was purified using the Qiagen miRNeasy FFPE kit and eluted in 30 µl. Slides were coverslipped and scanned after LCM.
Multiplex Reverse Transcription, Multiplex Preamplification, and qPCR Multiplex Primer Pool for Reverse Transcription Reaction
Twenty-five microliters of each 5x TaqMan Reverse Transcription (RT) primer [from Applied Biosystems kits: miR-200a (#4427975000502), miR-200b (#4427975-001800), miR-200c (#4427975-000505), miR-203 (#4427975-000507), miR-205 (#4427975-000509), miR-16 (#4427975-000391), miR-24 (#4427975-000402), miR-93 (#4427975-001090), let-7a (#4427975-000377), and RNU6 (#4427975-001093)] was pooled and then dehydrated in a SpeedVac at 50°C. The primer pool was resuspended in 100 µl of RNase-free H2O to give a 5x multiplex RT pool containing 62.5 nM of each primer, which was sufficient for 50 multiplex reactions.
Reverse transcription. RT was carried out in a total volume of 10 µl with reagents from the TaqMan MicroRNA RT Kit (Applied Biosystems, p/n 4366597, Life Technologies Australia Pty Ltd). Briefly, 5 µl of neat total RNA was added to 5 µl of Mastermix (2 µl of 5x Multiplex RT primer pool, 0.2 µl of 100 mM deoxynucleotide triphosphates (dNTPs), 1 µl of 50 U/µl Multiscribe reverse transcriptase, 1 µl of 10x RT buffer, 0.125 µl of RNase inhibitor, and 0.675 µl of RNase-free water). The reaction was mixed then iced for 5 minutes. Conditions for the RT reaction were 30 minutes at 16°C, 30 minutes at 42°C, and 5 minutes at 85°C, followed by a 4°C soak.
Preamplification PCR of cDNA. Preamplification was conducted according to the Applied Biosystems protocol with the exception that all recommended reaction volumes were halved. One microliter of each 20x TaqMan qPCR primer was combined, and the pool was diluted by the addition of 90 µl of TE (pH 7.5) to create a 0.2x pooled assay mix. cDNA was diluted slightly from 10 to 13 µl by the addition of 3 µl of RNase-free water, which allowed for two reactions, if required. Preamplification was carried out in a total volume of 25 µl, where 6.25 µl of the diluted cDNA was combined with 12.5 µl of 2x TaqMan PreAmp Mastermix (Applied Biosystems, p/n 4391128) and 6.25 µl of 0.2x pooled assay mix. Cycling parameters were 95°C for 10 minutes, followed by 14 cycles of 95°C for 15 seconds and 60°C for 4 minutes. The preamplification products were placed on ice upon completion of the reaction. Preamplified products were diluted to 1:20 in TE before qPCR.
Quantitative real-time RT-PCR of miRNA. Real-time PCRs of individual miRNAs were conducted in a total volume of 10 µl [0.5 µl of 20x TaqMan primer, 5 µl of 2x TaqMan Mastermix (Applied Biosystems p/n 4364343), 2 µl of RNase-free water, and 2.5 µl of diluted preamplified cDNA]. Real-time PCR was performed in triplicate on a Rotor-Gene 6000 PCR machine (Corbett Research/Qiagen, Chadstone Centre, Australia) under the conditions of 95°C for 10 minutes, followed by 50 cycles at 95°C for 15 seconds and 60°C for 1 minute. Data were collected and analyzed using comparative quantification Rotor-Gene software. MicroRNA levels for each sample were normalized to a pool of four unchanging control miRNAs (let-7a, miR-16, miR-24, and miR-93).
Results
An In Vitro Model Suggests Detaching Colon Cancer Cells Undergo EMT
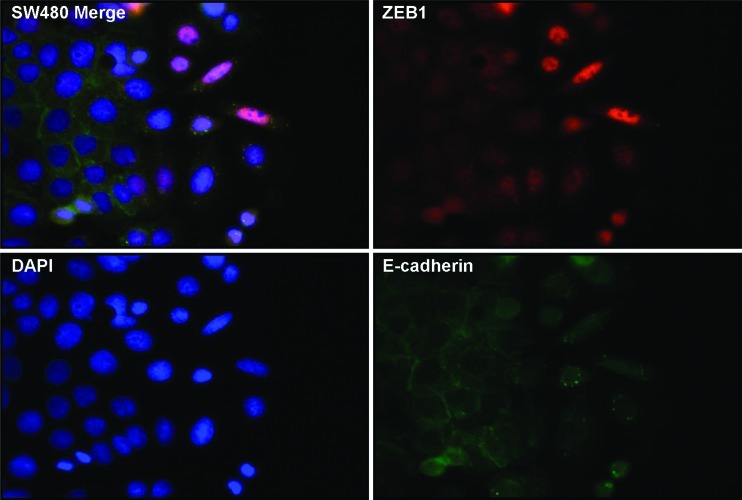
Up-regulation of mesenchymal proteins and repression of epithelial proteins are proposed to be important molecular features for the progression of tumor cells to an invasive phenotype. Here, we sought to investigate in vitro whether colon cancer cells, given the opportunity to detach from a confluent epithelial layer, exhibit typical characteristics of EMT. We scratched away a strip of cells from a confluent monolayer of SW480 colon cancer cells to provide a space into which the cells could migrate and monitored EMT changes in cells at the edge of the monolayer by staining for the epithelial marker E-cadherin and the mesenchymal marker ZEB1. At the leading edge of the scratch, epithelial cells transformed from compact structures with round nuclei into large, elongated, migrating cells with oval-shaped nuclei (Figure 1). Consistent with an EMT, migrating cells displayed internalization of E-cadherin and nuclear expression of ZEB1, whereas ZEB1 was absent in confluent epithelial cells expressing membrane-bound E-cadherin. Because ZEB1 expression is usually mutually exclusive with miR-200 expression, this suggests that miR-200 is downregulated in migrating cells. Noteworthy, SW480 cells are highly plastic and on the cusp of EMT; thus, no cytokine stimulation (other than factors already present in the medium) was required to exert these EMT effects. Furthermore, SW480s are regarded as E-cadherin- defective; however, this clone showed weak membranous E-cadherin, which was internalized in cells at the leading edge. Variations in SW480 cells have been previously reported [27,28] and this clone is likely to be the SW480-ADH variant described by Palmer et al. [29] that expresses E-cadherin. Taken together, the morphologic transition, loss of membrane-bound E-cadherin, and gain of nuclear ZEB1 are indicative of EMT contributing to the migratory and invasive characteristics of colon cancer cells.
Figure 1.
E-cadherin and ZEB1 expression in an in vitro model of EMT. Cultured SW480 colon adenocarcinoma cells migrating into a scratched area were immunofluorescently stained for ZEB1 (red) and E-cadherin (green). DAPI staining is shown in blue.
ISH Shows miR-200 Expression Is Lost at the Invasive Front of Colorectal Adenocarcinomas with Degraded Basement Membranes
If EMT contributes to cancer progression, then the miR-200 family of microRNAs might be downregulated in invasive front regions of adenocarcinomas. To assess this, we examined the levels of miR-200b and miR-200c by ISH in a panel of 28 human colorectal adenocarcinomas that exhibited budding cells and/or other invasive properties. ISH was conducted on miR-200b and miR-200c to represent the two miR-200 gene transcripts on chromosomes 1 (miR-200b ∼200a∼429) and 12 (miR-200c∼141). To indicate the extent to which tumor cells had undergone EMT, we immunostained adjacent sections for the following epithelial and mesenchymal markers: E-cadherin, cytokeratins, and ZEB1. Because nuclear localization of β-catenin and destruction of the basement membrane are further evidence of EMT, we also immunostained sections for β-catenin and laminin, a component of the basement membrane.
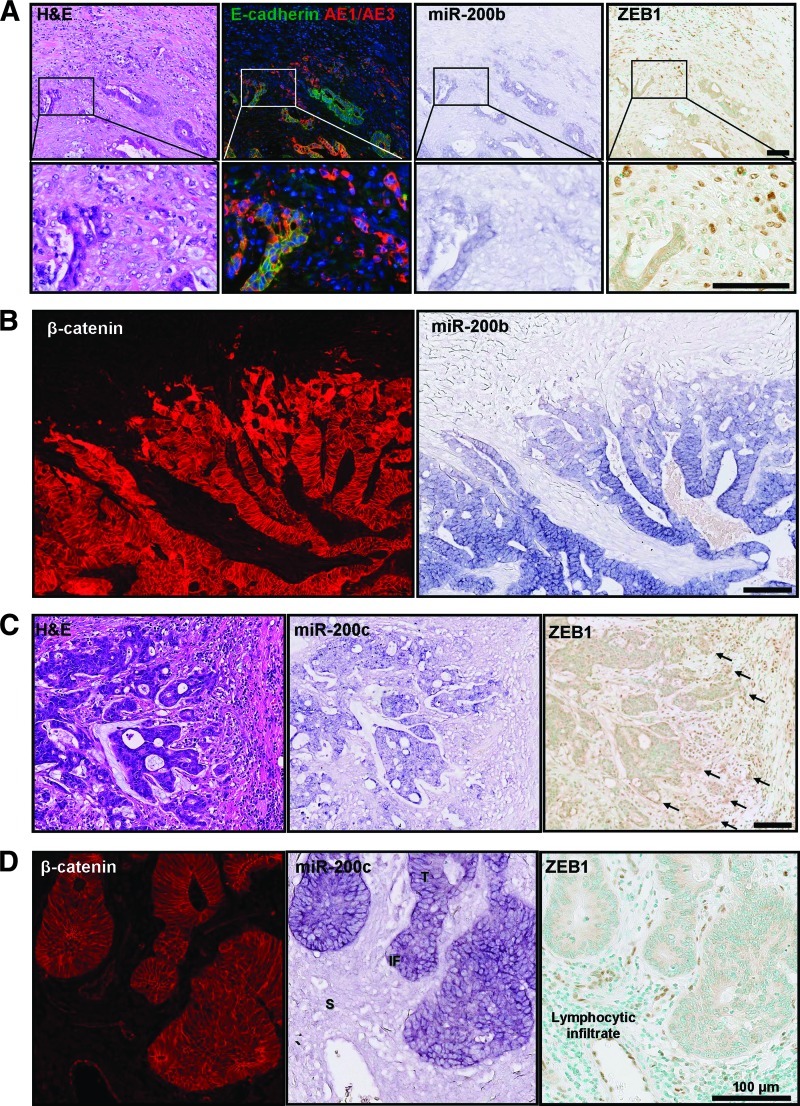
Histologic assessment of the miR-200b and miR-200c stains revealed strong expression in the tumor core (defined here as the bulk of the tumor excluding the invasive front). Both miR-200b and miR-200c were downregulated at the invasive front of adenocarcinomas in regions showing nuclear β-catenin and loss of E-cadherin (Figure 2) and degradation of the basement membrane (Figure 3). This reduction of miR-200 intensity at the invasive front was approximately 20 cells deep and similar in extent to the cytosolic and nuclear expression pattern of β-catenin (Figure 2B). In addition, ZEB1, which is a posttranscriptional target of the miR-200 family, was detected at the invasive front of some cancers that showed loss of miR-200 expression and degraded basement membranes (Figures 2, 3, W1, and W2). To verify that the reduced ISH staining at the invasive front was specific for miR-200, we also stained adjacent sections for the ubiquitous small nuclear RNA, snRNA U6. There was no decrease in snRNA U6 staining at the leading edge (Figure W3). A non-targeting scrambled probe showed minimal background (Figure W3). As an internal control for miR-200 staining, we measured the miR-200 stain intensity across normal epithelial colonic crypts matched to each adenocarcinoma specimen. The intensity of miR-200 in normal crypts was maintained from the base of the crypt to the lumen (Figure W3).
Figure 2.
MicroRNA-200 expression is reduced at the invasive front of colorectal cancer where EMT is evident. ISH of miR-200b and miR-200c was performed on FFPE human colorectal adenocarcinomas. Adjacent sections were immunohistochemically stained for cytokeratins (AE1/AE3) and EMT markers, including E-cadherin, β-catenin, and ZEB1. (A) Panel of consecutive serial sections stained with H&E; E-cadherin (green), AE1/AE3 (red), and DAPI (blue); miR-200b; and ZEB1 counterstained with methyl green. (B) Adjacent sections of an adenocarcinoma with a degraded basement membrane stained for β-catenin and miR-200b. (C) H&E, miR-200c, and ZEB1 at the invasive front of an adenocarcinoma with a degraded basement membrane. Arrows indicate ZEB1 expression. (D) β-catenin, miR-200c, and ZEB1 in an adenocarcinoma with an intact basement membrane. Scale bars represent 100 µm. This staining pattern was representative of 10 tumors examined in this way.
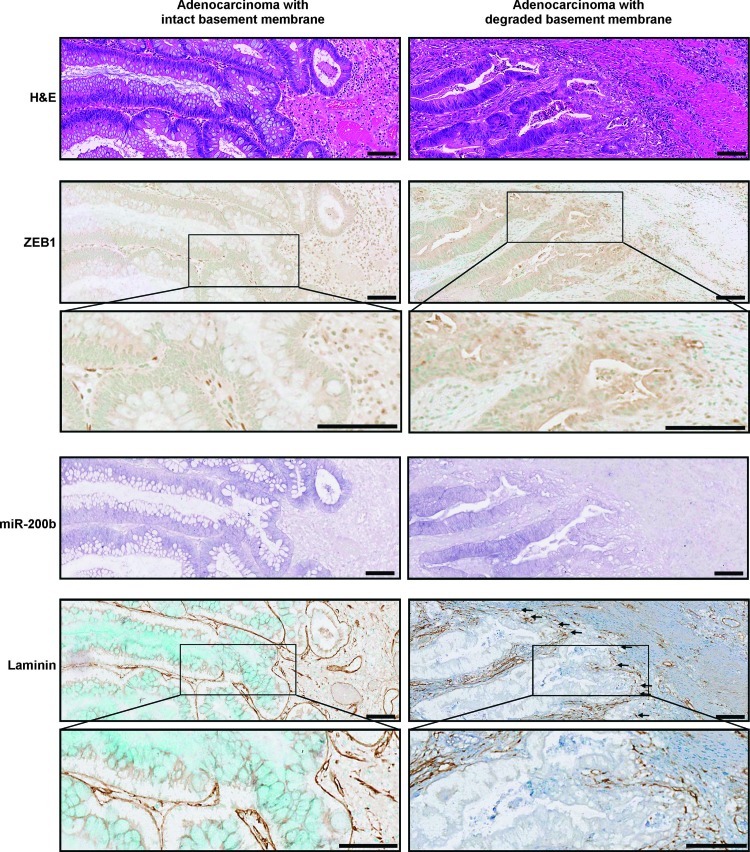
Figure 3.
Loss of miR-200 is associated with up-regulation of ZEB1 and degradation of the basement membrane. Comparison of ZEB1, miR-200b, and basement membrane status in two primary adenocarcinomas. Serial sections show H&E, ZEB1 and methyl green, miR-200b, and laminin and methyl green in an adenocarcinoma with an intact basement membrane (left panels) and in an adenocarcinoma with a degraded basement membrane (right panels). Arrows indicate regions of basement membrane degradation. Scale bars represent 100 µm. These images are representative of seven tumors examined.
In contrast to the pattern of staining seen across the invasive front of adenocarcinomas with a degraded basement membrane, adenomas and adenocarcinomas with intact basement membranes showed no loss of miR-200 staining intensity at the tumor-host interface, no nuclear β-catenin, and no evidence of ZEB1 expression in tumor cells (Figures 2D and 3).
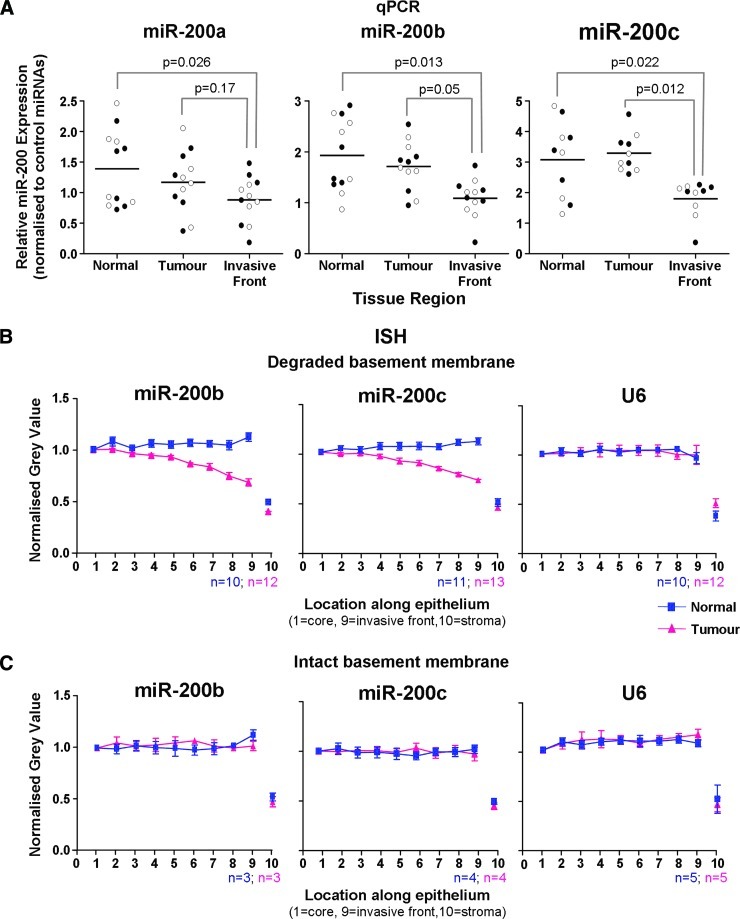
To quantitate the miR-200 ISH staining, we performed image analysis to measure staining intensity from the tumor core to the tumor margin and comparing this to changes in the miR-200 intensity across normal epithelium (Figure 4). In adenocarcinomas displaying extensive degradation of the basement membrane, there was significant reduction in miR-200 staining intensity at the invasive edge (example shown in Figure 4, A–C). In contrast, adenomas and adenocarcinomas with intact basement membranes showed uniform expression of miR-200 from the tumor core to the tumor-host interface (Figure 4, D–F). In summary, histologic analysis of miR-200 expression in colorectal tumors revealed a decreasing gradient of expression toward the invasive front of adenocarcinomas that showed evidence of EMT and degradation of the basement membrane. In contrast, miR-200 expression was maintained in adenomas and adenocarcinomas that showed no signs of EMT and had not invaded beyond the basement membrane.
qPCR of miR-200 from Laser Capture Microdissected Adenocarcinomas
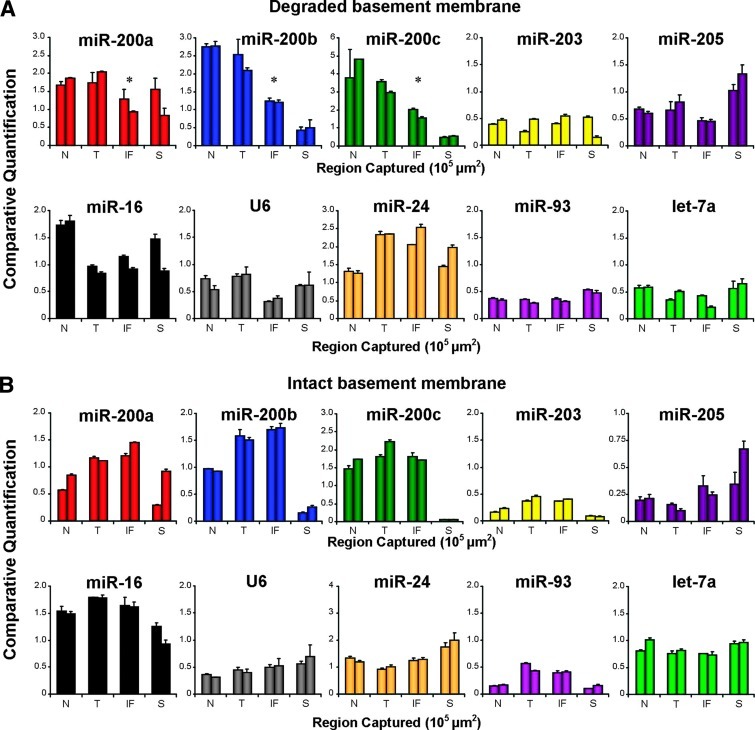
To further verify that the reduced miR-200 staining at the invasive front is due to lower levels of the miRNAs, we microdissected a series of adenocarcinomas by laser capture microscopy (Figure W4) and measured the miRNA levels by qPCR with multiplex preamplification. We quantified miRNA levels in normal colonic crypts, the tumor core, and the invasive front, as well as nearby stromal tissue. The expression levels of nine microRNAs (miR-200a, miR-200b, miR-200c, miR-203, miR-205, miR-24, miR-16, miR-93, and let-7a) and snRNA U6 were measured and normalized to a pool of controls (miR-16, miR-24, miR-93, and let-7a). Figure 5 shows the qPCR data from the same representative tumors shown in Figure 4. Concordant with the ISH data, qPCR revealed that miR-200 expression was down-regulated in malignant cells at the invasive front of the adenocarcinoma with a degraded basement membrane (Figure 5A), whereas the adenocarcinoma with an intact basement membrane showed no loss of miR-200 at the tumor-host interface by qPCR (Figure 5B). We also measured miR-205 and miR-203, which are downregulated during EMT in vitro [22,35]. However, expression of miR-205 and miR-203 in laser microdissected colorectal tissue was at the lower limit of qPCR detection and no significant difference in expression between the four tissue areas was found. Pooling the qPCR data from six adenocarcinomas with degraded basement membranes revealed a significant reduction in miR-200b and miR-200c expression at the invasive front compared to the tumor core and normal epithelium (Figure 6A). These data were consistent with the pooled ISH data showing loss of miR-200b and miR-200c expression toward the invasive front (Figure 6, B and C). Additionally, miR-200a showed a trend toward reduction at the invasive front by qPCR but did not reach statistical significance (Figure 6A). Interestingly, qPCR revealed that some tumors that did not display differential expression of miR-200a and/or miR-200b between the tumor core and the invasive front already had reduced miR-200a and/or miR-200b in the tumor compared to normal epithelium (Table W1).
Figure 5.
qPCR confirms that expression of miR-200 is downregulated at the invasive front of adenocarcinomas with degraded basement membranes and maintained in tumors with intact borders. Multiplex qPCR of miRNAs from duplicate regions of normal (N), tumor (T), invasive front (IF), and stroma (S). (A) Data correspond to the specimen shown in Figure 4A, which had a degraded basement membrane and loss of miR-200 at the invasive front indicated by ISH. Asterisks represent significant loss (P < .05) of miR-200 at the invasive front compared to both normal and tumor core epithelial tissues. (B) Data correspond to the specimen shown in Figure 4B, which had an intact basement membrane and uniform expression of miR-200 from the tumor core to the invasive front by ISH. All miRNAs were normalized to a pool of control miRNAs that are unchanged during EMT (miR-16, 24, 93, and let-7a).
Figure 6.
Pooled data show a statistically significant reduction in miR-200 expression at the invasive front of adenocarcinomas with degraded basement membranes. (A) qPCR analysis of laser microdissected epithelium from normal crypts, the tumor core, and the invasive front of six adenocarcinomas with degraded basement membranes. For each specimen and region, two separate laser micro-dissection samples were captured and separately analyzed. These are represented separately by black and white dots. A linear mixed effects model was used to compare expression of miR-200a, miR-200b, and miR-200c. (B) ISH data pooled from 10 to 13 adenocarcinomas with degraded basement membranes. (C) Pooled ISH data from three to five adenomas and adenocarcinomas with intact basement membranes. Error bars in B and C represent SEM.
miR-200 Is Expressed in Regional Lymph Node Metastases and Vascular Lesions
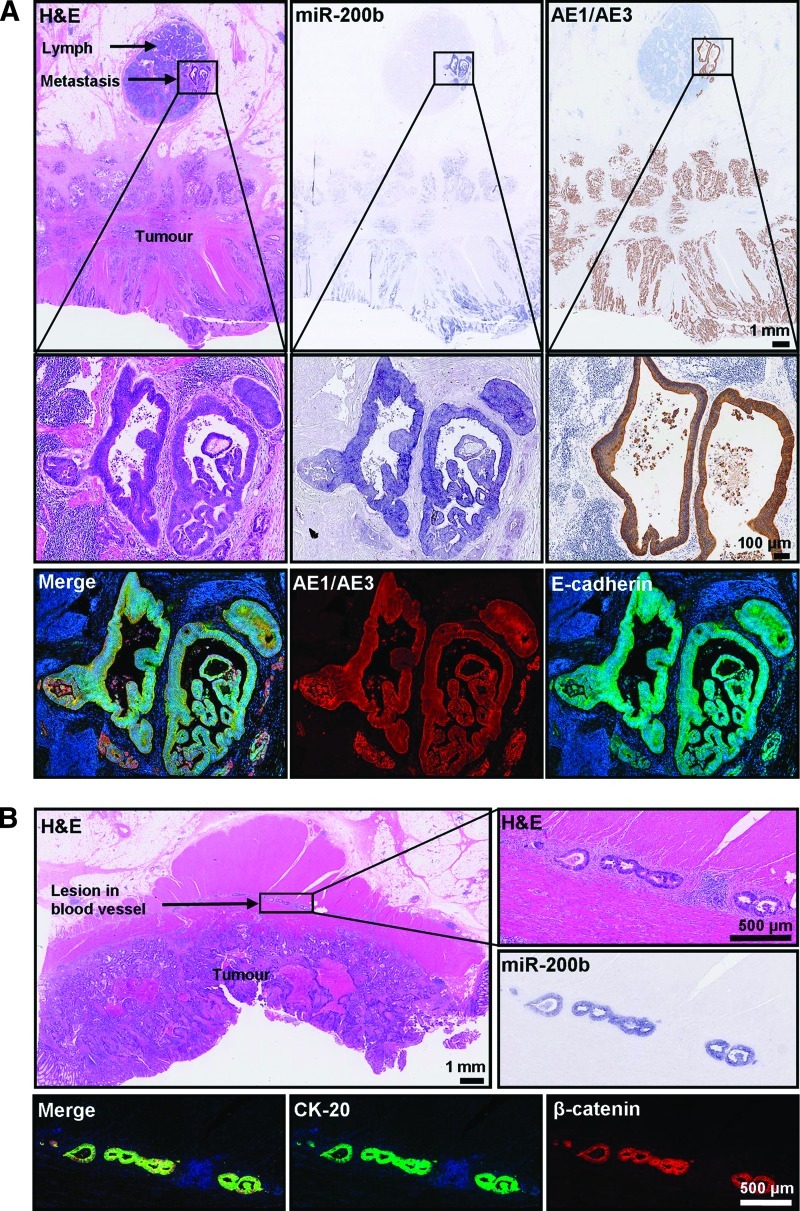
Among the 28 specimens analyzed by ISH in this study, five displayed regional lymph node metastases or vascular deposits proximal enough to be present on the primary adenocarcinoma section. One example each of lymph node and vascular lesions is shown in Figure 7. Histologic assessment revealed intense miR-200 staining in these malignant lesions, despite a decline of miR-200 toward the invasive front of the primary adenocarcinoma (Figure 7). This is consistent with the notion that metastasis occurs through a cascade of events involving dedifferentiation (EMT) at the primary tumor site and redifferentiation (MET) at the secondary site [6,21]. Consistent with a reversion to the epithelial phenotype at the secondary site, the lymph node metastases and vascular lesions arising from primary adenocarcinomas expressed E-cadherin and β-catenin at the cell membrane and stained positively for the pan-cytokeratin antibody AE1/AE3, as well as cytokeratin-20 (CK-20), and miR-200 (Figure 7). These results show that the miR-200 family is expressed in metastases and collectively, with the ISH and qPCR data showing loss of miR-200 at the invasive front of primary adenocarcinomas, add further support to the concept that EMT involving downregulation of miR-200 promotes local invasion, but up-regulation of miR-200 and MET occur at distant sites of metastatic growth.
Figure 7.
miR-200 expression in lymph node metastases and vascular lesions. (A) A diffuse adenocarcinoma with a lymph node metastasis. Adjacent sections immunostained for cytokeratins (AE1/AE3) and EMT markers, including E-cadherin miR-200. (Top) Panel of consecutive serial sections stained with H&E, miR-200b, and for the pan-cytokeratin antibody AE1/AE3 (counterstained with hematoxylin; scale bars represent 1 mm). (Middle) Magnified images of the lymph node metastasis (scale bars represent 100 µm). (Lower) Merged and separate images of the lymph node immunostained with AE1/AE3 antibody (red) and E-cadherin (green). (B) Adenocarcinoma with malignant lesions in a proximal blood vessel. Adjacent sections stained with H&E (top), miR-200b (center, right), and CK-20 (green) and β-catenin (red).
Discussion
In this study, we explored the role of miR-200 in the progression of colorectal cancer. On the basis of the notion that invasion of leading edge cells through the basement membrane occurs as a direct result of EMT and that down-regulation of miR-200 is an essential component of EMT in vitro, we predicted that miR-200 would be down-regulated in malignant cells undergoing EMT at the tumor-host interface. This was observed in 18 of 18 adenocarcinomas with degraded basement membranes that had a sufficient in situ signal to quantify (Table W1). Given miR-200 is a potent regulator of EMT, its loss at the invasive front of carcinomas further supports the concept that EMT underlies tumor invasion.
Our ISH and qPCR analyses showed that expression of miR-200b and miR-200c declines toward the invasive front of adenocarcinomas displaying budding cells. Most importantly, the invasive margins of these tumors lacked the strong boundaries provided by an intact basement membrane. This finding complements the work of Spaderna et al. [20], who demonstrated that loss of the basement membrane in colorectal adenocarcinomas correlates with metastasis and poor survival. In addition, they showed that ZEB1 is upregulated in invasive front regions deficient of a basement membrane and that ZEB1 directly represses transcription of LAMA3, which encodes an essential basement membrane protein. Here, we demonstrate that down-regulation of miR-200 is also associated with expression of ZEB1 and degradation of the basement membrane at the invasive front of colorectal adenocarcinomas. Furthermore, these findings are consistent with the miR200 family participating in a double-negative feedback loop with ZEB1 [23,24].
Our results indicate that the altered expression patterns of miR-200 and β-catenin occur to a similar degree in cells at the invasive front (Figure 2B). Specifically, the depth of the declining gradient of miR200 toward the leading edge was similar in extent to the expression pattern of β-catenin, which shifted from membrane-associated in the tumor core to cytoplasmic and nuclear localization at the invasive front. Noteworthy, Xia et al. [30] reported that β-catenin mRNA is a target of miR-200a; therefore, loss of miR-200a expression in leading edge cells could directly contribute to the accumulation of cytosolic and nuclear β-catenin. Interestingly, the neural cell adhesion marker L1-CAM, whose expression is generally confined to nerve cells, has been found to be upregulated at the invasive front of colorectal cancer [31]. L1-CAM is a β-catenin target gene, a promoter of cell motility, and is positively correlated with metastasis [32]. It is not expressed in the tumor core or normal epithelium, nor is it a predicted target for the miR-200 family; however, it does show a remarkable inverse expression pattern to miR200 in colorectal cancer [31–33].
While in vitro EMT models in carcinoma cell lines demonstrate that reduced miR-200 expression promotes invasion and migration [22,30], investigations in vivo have shown the miR-200 family can encourage or minimize metastasis in mouse models of breast, lung, and pancreatic cancers [34–36]. Collectively, these studies indicate that constitutive overexpression of miR-200 is protective against invasion and metastasis (e.g., [34]), yet that miR-200 is required for metastasis [36]. On the basis of our data 1) showing loss of miR-200 by ISH and qPCR at the invasive front of advanced primary colorectal adenocarcinomas lacking intact basement membranes and 2) revealing strong expression of miR-200 in local metastases, we postulate that high expression of miR-200 in the primary tumor favors metastasis formation, provided the cells at the invasive front can undergo EMT, and that these budding mesenchymal cells retain the ability to switch from the mesenchymal state back to the epithelial state to favor colonization in a foreign microenvironment. Our data are consistent with the work of Korpal et al. [37], who demonstrated a pro-metastatic role for miR-200. They found that increased miR-200 in human breast cancers correlated with poor distant disease-free survival and, most remarkably, showed that in a mouse model of breast cancer metastasis, elevated miR-200 in primary mammary tissues favored metastasis, despite inhibiting invasion and intravasation into the blood at the primary carcinoma site. They postulated that the miR-200 family is able to minimize invasion by inhibiting EMT, but if these malignant cells do intravasate into the blood, they have an increased propensity to colonize at distant sites due to their highly epithelial origin. More recently, Hur et al. [38] showed that miR-200c is downregulated at the invasive front of colorectal cancer and upregulated in liver metastasis. Here, we showed that miR-200b is also downregulated at the invasive front. Taken together, our findings and those of Korpal et al. [37] and Hur et al. [38] indicate that altered expression of the miR-200 family contributes to poor outcome.
In summary, this study complements a large volume of work on expression of EMT markers at the invasive front of colorectal adenocarcinomas and further implicates the miR-200 family, a strong regulator of EMT and MET, in invasion and metastasis. With an aging population, rising levels of obesity, and poor dietary and lifestyle habits, the incidence of colorectal cancer is likely to increase; therefore, we must look to novel targets to treat this disease. The metastasis cascade consists of a broad range of molecules that anticancer therapies could potentially target to prevent invasion (EMT) and metastasis (MET). Moreover, the ZEB-miR-200 axis holds much potential as an anticancer target because of its ability to regulate migratory cancer stem cells.
Supplementary Material
Acknowledgments
We thank Cameron Bracken and Philip Gregory for useful discussions.
Footnotes
This work was supported by grants from Cancer Council SA and the National Health & Medical Research Council to G.J.G. and Y.K.-G. E.L.P. was the recipient of a Dawes Scholarship from the Royal Adelaide Hospital Research Fund. The authors declare no conflict of interest.
This article refers to supplementary materials, which are designated by Table W1 and Figures W1 to W4 and are available online at www.neoplasia.com.
References
- 1.Huber MA, Kraut N, Beug H. Molecular requirements for epithelialmesenchymal transition during tumor progression. Curr Opin Cell Biol. 2005;17:548–558. doi: 10.1016/j.ceb.2005.08.001. [DOI] [PubMed] [Google Scholar]
- 2.Cannito S, Novo E, di Bonzo LV, Busletta C, Colombatto S, Parola M. Epithelial-mesenchymal transition: from molecular mechanisms, redox regulation to implications in human health and disease. Antioxid Redox Signal. 2010;12:1383–1430. doi: 10.1089/ars.2009.2737. [DOI] [PubMed] [Google Scholar]
- 3.Bhowmick NA, Ghiassi M, Bakin A, Aakre M, Lundquist CA, Engel ME, Arteaga CL, Moses HL. Transforming growth factor-β1 mediates epithelial to mesenchymal transdifferentiation through a RhoA-dependent mechanism. Mol Biol Cell. 2001;12:27–36. doi: 10.1091/mbc.12.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Morita T, Mayanagi T, Sobue K. Dual roles of myocardin-related transcription factors in epithelial-mesenchymal transition via slug induction and actin remodeling. J Cell Biol. 2007;179:1027–1042. doi: 10.1083/jcb.200708174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kalluri R, Weinberg RA. The basics of epithelial-mesenchymal transition. J Clin Invest. 2009;119:1420–1428. doi: 10.1172/JCI39104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brabletz T, Jung A, Reu S, Porzner M, Hlubek F, Kunz-Schughart LA, Knuechel R, Kirchner T. Variable β-catenin expression in colorectal cancers indicates tumor progression driven by the tumor environment. Proc Natl Acad Sci USA. 2001;98:10356–10361. doi: 10.1073/pnas.171610498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aigner K, Dampier B, Descovich L, Mikula M, Sultan A, Schreiber M, Mikulits W, Brabletz T, Strand D, Obrist P, et al. The transcription factor ZEB1 (δEF1) promotes tumour cell dedifferentiation by repressing master regulators of epithelial polarity. Oncogene. 2007;26:6979–6988. doi: 10.1038/sj.onc.1210508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Spaderna S, Schmalhofer O, Wahlbuhl M, Dimmler A, Bauer K, Sultan A, Hlubek F, Jung A, Strand D, Eger A, et al. The transcriptional repressor ZEB1 promotes metastasis and loss of cell polarity in cancer. Cancer Res. 2008;68:537–544. doi: 10.1158/0008-5472.CAN-07-5682. [DOI] [PubMed] [Google Scholar]
- 9.Hlubek F, Brabletz T, Budczies J, Pfeiffer S, Jung A, Kirchner T. Heterogeneous expression of Wnt/β-catenin target genes within colorectal cancer. Int J Cancer. 2007;121:1941–1948. doi: 10.1002/ijc.22916. [DOI] [PubMed] [Google Scholar]
- 10.Nakamura T, Mitomi H, Kikuchi S, Ohtani Y, Sato K. Evaluation of the usefulness of tumor budding on the prediction of metastasis to the lung and liver after curative excision of colorectal cancer. Hepatogastroenterology. 2005;52:1432–1435. [PubMed] [Google Scholar]
- 11.Suzuki A, Togashi K, Nokubi M, Koinuma K, Miyakura Y, Horie H, Lefor AT, Yasuda Y. Evaluation of venous invasion by Elastica van Gieson stain and tumor budding predicts local and distant metastases in patients with T1 stage colorectal cancer. Am J Surg Pathol. 2009;33:1601–1607. doi: 10.1097/PAS.0b013e3181ae29d6. [DOI] [PubMed] [Google Scholar]
- 12.Ogawa T, Yoshida T, Tsuruta T, Tokuyama W, Adachi S, Kikuchi M, Mikami T, Saigenji K, Okayasu I. Tumor budding is predictive of lymphatic involvement and lymph node metastases in submucosal invasive colorectal adenocarcinomas and in non-polypoid compared with polypoid growths. Scand J Gastroenterol. 2009;44:605–614. doi: 10.1080/00365520902718911. [DOI] [PubMed] [Google Scholar]
- 13.Okuyama T, Oya M, Ishikawa I. Budding as a risk factor for lymph node metastasis in pT1 or pT2 well-differentiated colorectal adenocarcinoma. Dis Colon Rectum. 2002;45:628–634. doi: 10.1007/s10350-004-6259-0. [DOI] [PubMed] [Google Scholar]
- 14.Uchida H, Hasegawa H, Nishibori H, Ishii Y, Endo T, Mukai M, Kitajima M. Area-specific tumor budding in T2 colorectal cancer: an independent predictive factor for lymph node metastasis. Dis Colon Rectum. 2007;50:768. [Google Scholar]
- 15.Prall F, Nizze H, Barten M. Tumour budding as prognostic factor in stage I/II colorectal carcinoma. Histopathology. 2005;47:17–24. doi: 10.1111/j.1365-2559.2005.02161.x. [DOI] [PubMed] [Google Scholar]
- 16.Prall F, Ostwald C, Linnebacher M. Tubular invasion and the morphogenesis of tumor budding in colorectal carcinoma. Hum Pathol. 2009;40:1510–1512. doi: 10.1016/j.humpath.2009.06.015. [DOI] [PubMed] [Google Scholar]
- 17.Masaki T, Matsuoka H, Kobayashi T, Sugiyama M, Atomi Y. Actual number of tumor budding; a novel indicator of lymph node metastasis in T1 colorectal carcinoma. J Gastroenterol Hepatol. 2009;24:A66. doi: 10.1111/j.1440-1746.2006.04073.x. [DOI] [PubMed] [Google Scholar]
- 18.Lugli A, Zlobec I, Baker K, Roth S, Minoo P, Hayashi S, Jass JR, Terracciano L. Tumor budding and lymphocytic infiltration in colorectal cancer: pathobiological interactions at the tumor margin and clinical impact. Pathol Res Pract. 2007;203:85. [Google Scholar]
- 19.Kazama S, Watanabe T, Ajioka Y, Kanazawa T, Nagawa H. Tumour budding at the deepest invasive margin correlates with lymph node metastasis in submucosal colorectal cancer detected by anticytokeratin antibody CAM5.2. Br J Cancer. 2006;94:293–298. doi: 10.1038/sj.bjc.6602927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Spaderna S, Schmalhofer O, Hlubek F, Berx G, Eger A, Merkel S, Jung A, Kirchner T, Brabletz T. A transient, EMT-linked loss of basement membranes indicates metastasis and poor survival in colorectal cancer. Gastroenterology. 2006;131:830–840. doi: 10.1053/j.gastro.2006.06.016. [DOI] [PubMed] [Google Scholar]
- 21.Brabletz T, Jung A, Kirchner T. β-Catenin and the morphogenesis of colorectal cancer. Virchows Arch. 2002;441:1–11. doi: 10.1007/s00428-002-0642-9. [DOI] [PubMed] [Google Scholar]
- 22.Gregory PA, Bert AG, Paterson EL, Barry SC, Tsykin A, Farshid G, Vadas MA, Khew-Goodall Y, Goodall GJ. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol. 2008;10:593–601. doi: 10.1038/ncb1722. [DOI] [PubMed] [Google Scholar]
- 23.Bracken CP, Gregory PA, Kolesnikoff N, Bert AG, Wang J, Shannon MF, Goodall GJ. A double-negative feedback loop between ZEB1-SIP1 and the microRNA-200 family regulates epithelial-mesenchymal transition. Cancer Res. 2008;68:7846–7854. doi: 10.1158/0008-5472.CAN-08-1942. [DOI] [PubMed] [Google Scholar]
- 24.Burk U, Schubert J, Wellner U, Schmalhofer O, Vincan E, Spaderna S, Brabletz T. A reciprocal repression between ZEB1 and members of the miR-200 family promotes EMT and invasion in cancer cells. EMBO Rep. 2008;9:582–589. doi: 10.1038/embor.2008.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aigner K, Descovich L, Mikula M, Sultan A, Dampier B, Bonne S, Van Roy F, Mikulits W, Schreiber M, Brabletz T, et al. The transcription factor ZEB1 (δEF1) represses Plakophilin 3 during human cancer progression. FEBS Lett. 2007;581:1617–1624. doi: 10.1016/j.febslet.2007.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Darling DS, Stearman RP, Qi YC, Qiu MS, Feller JP. Expression of Zfhep/δEF1 protein in palate, neural progenitors, and differentiated neurons. Gene Expr Patterns. 2003;3:709–717. doi: 10.1016/s1567-133x(03)00147-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tomita N, Wei J, Hibshoosh H, Warburton D, Kahn SM, Weinstein IB. Isolation and characterization of a highly malignant variant of the SW480 human colon cancer cell line. Cancer Res. 1992;52:6840–6847. [PubMed] [Google Scholar]
- 28.Biazik JM, Jahn KA, Su YY, Wu YN, Braet F. Unlocking the ultrastructure of colorectal cancer cells in vitro using selective staining. World J Gastroenterol. 2010;16:2743–2753. doi: 10.3748/wjg.v16.i22.2743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Palmer HG, Gonzalez-Sancho JM, Espada J, Berciano MT, Puig I, Baulida J, Quintanilla M, Cano A, de Herreros AG, Lafarga M, et al. Vitamin D3 promotes the differentiation of colon carcinoma cells by the induction of E-cadherin and the inhibition of β-catenin signaling. J Cell Biol. 2001;154:369–387. doi: 10.1083/jcb.200102028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xia HP, Ng SS, Jiang SS, Cheung WKC, Sze J, Bian XW, Kung HF, Lin MC. miR-200a-mediated downregulation of ZEB2 and CTNNB1 differentially inhibits nasopharyngeal carcinoma cell growth, migration and invasion. Biochem Biophys Res Commun. 2010;391:535–541. doi: 10.1016/j.bbrc.2009.11.093. [DOI] [PubMed] [Google Scholar]
- 31.Gavert N, Conacci-Sorrell M, Gast D, Schneider A, Altevogt P, Brabletz T, Ben-Ze'ev A. L1, a novel target of β-catenin signaling, transforms cells and is expressed at the invasive front of colon cancers. J Cell Biol. 2005;168:633–642. doi: 10.1083/jcb.200408051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gavert N, Sheffer M, Raveh S, Spaderna S, Shtutman M, Brabletz T, Barany F, Paty P, Notterman D, Domany E, et al. Expression of L1-CAM and ADAM10 in human colon cancer cells induces metastasis. Cancer Res. 2007;67:7703–7712. doi: 10.1158/0008-5472.CAN-07-0991. [DOI] [PubMed] [Google Scholar]
- 33.Gavert N, Ben-Shmuel A, Raveh S, Ben-Ze'ev A. L1-CAM in cancerous tissues. Expert Opin Biol Ther. 2008;8:1749–1757. doi: 10.1517/14712598.8.11.1749. [DOI] [PubMed] [Google Scholar]
- 34.Gibbons DL, Lin W, Creighton CJ, Rizvi ZH, Gregory PA, Goodall GJ, Thilaganathan N, Du LQ, Zhang YQ, Pertsemlidis A, et al. Contextual extracellular cues promote tumor cell EMT and metastasis by regulating miR200 family expression. Genes Dev. 2009;23:2140–2151. doi: 10.1101/gad.1820209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wellner U, Schubert J, Burk UC, Schmalhofer O, Zhu F, Sonntag A, Waldvogel B, Vannier C, Darling D, zur Hausen A, et al. The EMT-activator ZEB1 promotes tumorigenicity by repressing stemness-inhibiting microRNAs. Nat Cell Biol. 2009;11:1487–1495. doi: 10.1038/ncb1998. [DOI] [PubMed] [Google Scholar]
- 36.Dykxhoorn DM, Wu YC, Xie HM, Yu FY, Lal A, Petrocca F, Martinvalet D, Song E, Lim B, Lieberman J. miR-200 enhances mouse breast cancer cell colonization to form distant metastases. Plos One. 2009;4:e7181. doi: 10.1371/journal.pone.0007181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Korpal M, Ell BJ, Buffa FM, Ibrahim T, Blanco MA, Celia-Terrassa T, Mercatali L, Khan Z, Goodarzi H, Hua Y, et al. Direct targeting of Sec23a by miR-200s influences cancer cell secretome and promotes metastatic colonization. Nat Med. 2011;17:1101–1108. doi: 10.1038/nm.2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hur K, Toiyama Y, Takahashi M, Balaguer F, Nagasaka T, Koike J, Hemmi H, Koi M, Boland CR, Goel A. MicroRNA-200c modulates epithelial-to-mesenchymal transition (EMT) in human colorectal cancer metastasis. Gut. 2012 doi: 10.1136/gutjnl-2011-301846. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.