Abstract
Our previous studies have shown the role of radiation-induced urokinase plasminogen activator (uPA) expression in the progression of meningioma. In the present study, we investigated whether modulation of DNA methylation profiles could regulate uPA expression. Initially, radiation treatment was found to induce hypomethylation in meningioma cells with a decrease in DNA (cytosine-5)-methyltransferase 1 (DNMT1) and methyl-CpG binding domain protein (MBD) expression. However, oxidative damage by H2O2 or pretreatment of irradiated cells with N-acetyl cysteine (NAC) did not show any influence on these proteins, thereby indicating a radiation-specific change in the methylation patterns among meningioma cells. Further, we identified that hypomethylation is coupled to an increase in uPA expression in these cells. Azacytidine treatment induced a dose-dependent surge of uPA expression, whereas pre-treatment with sodium butyrate inhibited radiation-induced uPA expression, which complemented our prior results. Methylation-specific polymerase chain reaction on bisulfite-treated genomic DNA revealed a diminished methylation of uPA promoter in irradiated cells. Transfection with small hairpin RNA (shRNA)-expressing plasmids targeting CpG islands of the uPA promoter showed a marked decline in uPA expression with subsequent decrease in invasion and proliferation of meningioma cells. Further, radiation treatment was found to recruit SP1 transcription factor, which was abrogated by shRNA treatment. Analysis on signaling events demonstrated the activation of MAP kinase kinase (MEK)-extracellular signal-regulated kinase (ERK) in radiation-treated cells, while U0126 (MEK/ERK inhibitor) blocked hypomethylation, recruitment of SP1, and uPA expression. In agreement with our in vitro data, low DNMT1 levels and high uPA were found in intracranial tumors treated with radiation compared to untreated tumors. In conclusion, our data suggest that radiation-mediated hypomethylation triggers uPA expression in meningioma cells.
Introduction
DNA methylation is essential for growth, development, and environmental responsiveness of mammalian cells. Cellular phenomena such as changes in gene expression, chromatin structure alterations, activation of transposable elements, genomic imprinting, and carcinogenesis have been shown to occur along with DNA methylation [1]. Both hypomethylation and hypermethylation of genomic DNA induce significant epigenetic and genetic changes in the cell [2]. It is increasingly apparent that cancer development depends not only on genetic alterations but also on a heritable cellular memory or epigenetic changes that are critical for tumor initiation and progression [3]. From an epigenetics perspective, during carcinogenesis, DNA undergoes genome-wide hypomethylation and regional hypermethylation of CpG islands in tandem, offering perspective advantage for the preliminary tumor cell. Localized hypermethylation, which represses transcription of the promoter regions of tumor suppressor genes, and global hypomethylation have been recognized as strategic events that typify many cancers. There are several protective mechanisms that prevent the hypermethylation of the CpG islands including active transcription, active demethylation, replication timing, and local chromatin structure, thereby preventing access to the DNA methyltransferase. However, the mechanisms by which hypomethylation contributes to malignancy are oncogene activation, loss of imprinting, and promotion of genomic instability through unmasking of repetitive elements. Hypomethylation is common in solid tumors such as metastatic hepatocellular cancer, cervical cancer, and prostate tumors, as well as hematologic malignancies such as B cell chronic lymphocytic leukemia [4]. A number of cancers, such as breast, cervical, and brain, usually show a progressive increase of hypomethylation corresponding with the grade of malignancy. New information about the mechanism of methylation and its control has led to the discovery of many regulatory proteins and enzymes. All evidence indicates that the DNA (cytosine-5)-methyltransferase 1 (DNMT1) enzyme acts as a maintenance methyltransferase to prevent binding of transcription factors, whereas methyl-CpG binding domain protein 1 (MBD), MBD2, methyl CpG-binding protein 2 (MeCP2), and Kaiso have been shown to repress transcription of target genes.
It has been acknowledged for many years that radiation exposure induces delayed nontargeted effects in the progeny of the irradiated cell. Evidence is beginning to demonstrate that among these delayed effects of radiation are epigenetic aberrations including altered DNA methylation [5]. Although somewhat preliminary, multiple studies have shown how signaling events are involved in abnormal DNA methylation in cancer. Many signal transduction pathways that drive cell transformation and tumor progression lead to the up-regulation of CpG and/or components of the DNA methylation machinery [6]. Specifically, increased methylation of the urokinase plasminogen activator (uPA) promoter was found to associate significantly with lower levels of uPA and the transcription pattern of uPA in meningiomas; this might, in part, be controlled by promoter methylation [7]. Recent studies provide evidence that RNA interference can also direct DNA methylation and transcriptional gene silencing (TGS) in human cells [8–10], thereby suggesting a potential and additional mechanism for transcriptional regulation in mammals. Data are also accruing for a central role of transcription factors in epigenetically regulated processes. These processes include control of organization and placement of proteins that determine accessibility and transcriptional competency of genomic sequences for expression. As such, these processes support proliferation, growth, phenotype, and homeostatic regulation at both the transcriptional and the post-transcriptional levels [11]. Further, DNA methylation has been shown to determine access of transcription factors to gene regulatory sequences [11].
We have reported a radiation-induced overexpression of uPA in meningioma cells, which could only partly be attributed to mitogen-activated protein kinase (MAPK) signaling, suggesting additional level of regulation [15,16]. However, methylated promoter was shown to have significant negative correlation with uPA expression in meningioma [7] and radiation treatment was shown to induce epigenetic aberrations in human cells [5]. Therefore, we hypothesized that radiation-induced uPA expression in meningioma could be a result of hypomethylation. We performed this study to demonstrate that changes in the promotermethylation patterns do indeed contribute to meningioma progression in vitro and in vivo.
Materials and Methods
Cell Culture
We used the human meningioma IOMM-Lee and SF3061 cell lines (kindly provided by Dr Ian E. McCutcheon, University of Texas M.D. Anderson Cancer Center, Houston, TX, and Dr Anitha Lal, University of California, San Francisco, CA, respectively). Both cell lines were maintained in Dulbecco's modified Eagle's medium (Thermo Scientific, San Jose, CA) supplemented with 10% FBS, 100 U/ml streptomycin, and 100 U/ml penicillin (Invitrogen, Carlsbad, CA). Cells were maintained in a humidified atmosphere containing 5% CO2 at 37°C. Cells were treated with zebularine (VWR Scientific, Chicago, IL), azacytidine, sodium butyrate, and U0126 and incubated for the indicated period of time in complete medium. DNMT1, phospho extracellular signal-regulated kinase (pERK), ERK, MEK, phospho MAP kinase kinase (pMEK), cRaf, pcRAF, uPA, and glyceraldehyde 3-phosphate dehydrogenase (GAPDH) antibodies and HRP-conjugated secondary antibodies were obtained from Santa Cruz Biotechnology (Santa Cruz, CA).
Radiation Treatment
The RS 2000 Biological Irradiator (Rad Source Technologies, Inc, Boca Raton, FL) X-ray unit operated at 150 kV/50 mA and delivering 0.71 Gy/min was used for radiation treatments. Cells were given a single dose of 7-Gy radiation, and for experiments involving the inhibitors, radiation was given after 1 hour of inhibitor treatment.
Construction of Small Hairpin RNA Plasmids
The uPA promoter was identified on the 10th chromosome. About 10,000 bases upstream of the transcription site were annotated for the presence of promoter-specific elements. The promoter sequence was predicted using The Markov Chain Promoter Prediction Server, which is freely available for the public's use from Duke University (Durham, NC). The predicted sequences were screened with a cutoff of 0.90 and 1.0 being the highest. The presence of CpG islands were defined as sequence ranges that give a y value of greater than 0.6 and a guanine-cytosine (GC) content of greater than 50% using sequence manipulation suite (Stothard 2000). The plasmid vector, pSilencer 4.1-CMV (Ambion, Austin, TX), was used in the construction of the shRNA-expressing vector, and the human uPA promoter sequence rich in CpG islands AGGAAGCACGGAGAATTTACAAG was the shRNA target sequence. Inverted repeat sequences were synthesized. The inverted repeats were laterally symmetrical, making them self-complimentary with a 9-bp mismatch in the loop region that would aid in the loop formation of the shRNA. Oligonucleotides were heated in a boiling water bath in 6x SSC for 5 minutes and self-annealed by slow cooling to room temperature. The annealed oligonucleotides were ligated to pSilencer vector at BamHI and HindIII sites. Cells at 60% to 70% confluence in 100-mm tissue culture plates were transfected with 7 µg of shRNA-expressing plasmid constructs using FuGENE HD following the manufacturer's instructions (Roche Applied Science, Indianapolis, IN).
Transfection Studies
All transfection experiments were performed with FuGENE HD transfection reagent as per the manufacturer's protocol (Roche Applied Science). IOMM-Lee and SF3061 cells were transfected with plasmid constructs containing scrambled sequence [scrambled vector (SV)] or shRNA for uPA promoter (puPA)-expressing sequences. After 6 hours of transfection, complete medium was added, and cells were incubated for 36 hours. Later, the cells were irradiated (7 Gy) and incubated overnight before being subjected to further analysis.
Fibrin Zymography
Conditioned media containing 0.1 µg (IOMM-Lee) and 5.0 µg (SF3061 cells) of protein from different experiments were subjected to zymography. Samples were resolved on 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis containing plasminogen and fibrinogen and stained as described previously [12].
Matrigel Invasion Assay
Invasion assays were performed in Boyden chambers, using 8-µm pore size polyvinyl pyrrolidone-free (PVDF) polycarbonate matrigel-coated filters. IOMM-Lee and SF3061 cells from different treatment groups were added in the upper chamber in serum-free medium, and complete medium was added in the lower chamber. Cells were allowed to migrate overnight at 37°C and 5% CO2. Cells on the lower surface of the filter were then fixed in ethanol, stained with hematoxylin, and observed under a light microscope [12].
Western Blot Analysis
Cells were collected from treated and untreated experimental groups. Cell lysates were prepared in RIPA buffer and nuclear extracts were prepared using nuclear extraction buffer. Equal amounts of protein were then subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis using gels with appropriate percentage of acrylamide followed by transfer of protein to nitrocellulose membranes (Bio-Rad, Hercules, CA). Membranes were processed with primary antibodies at a 1:1000 dilution and HRP-conjugated secondary antibodies at a 1:2000 dilution as described previously [13]. The membranes were developed following an enhanced chemiluminescence protocol (Amersham Biosciences, Piscataway, NJ) and were further probed for GAPDH, which was used as a loading control.
Bisulfite Conversion
The genomic DNA from IOMM-Lee and SF3061 cells was extracted using DNeasy Blood And Tissue Kit (Qiagen, Valencia, CA) following the manufacturer's recommendations. Equal quantities of DNA were subjected to bisulfite conversion using EpiTect Bisulfite Kit (Qiagen).
3-(4,5-Dimethylthiazol-2-yl)-2,5-Diphenyl Tetrazolium Bromide (MTT) Proliferation Assay
IOMM-Lee and SF3061 cells seeded in six-well plates were treated with different agents and irradiated as described earlier. Six hours later, cells were trypsinized, counted, and seeded at 1 x 104 cells per well in 96-well plates (three wells per treatment group). After the indicated hours of incubation in conditioned medium, 20 µl of MTT reagent was added to the cells, followed by another 4 hours of incubation at 37°C. Acid-isopropanol (0.04 MHCl/isopropanol) was added to all wells and mixed vigorously so that the formazan crystals dissolved effectively. Absorbance was measured on a microtiter plate reader (Model 680; Bio-Rad) with a test wavelength of 550 nm and a reference wavelength of 655 nm.
Reverse Transcription-Polymerase Chain Reaction
Total RNA extracted from frozen brain tissues and meningioma cells was subjected to cDNA synthesis using Transcriptor First Strand cDNA Synthesis Kit (Roche Applied Science). Polymerase chain reaction (PCR) was performed using primers specific for human uPA, DNMT1, MeCP, and MBD2, and products were analyzed on 1.8% agarose gels.
Methylation-Specific PCR
DNA was extracted using standard methods (Qiagen DNA Kit) from meningioma cells or snap-frozen tumor tissues and subjected to bisulfite treatment with the EpiTect Bisulfite Kit (Qiagen) in accordance with the manufacturer's recommended procedure. A 365-bp (-430 to -66) or a 131-bp fragment (-145 to -14) of the uPA promoter (NCBI Reference Sequence: NG_011904.1: 4596–4960) was PCR-amplified using methylation-specific (MS) primers or unmethylated specific (UM) primers.
Restriction Digestion
The selected 131-bp uPA promoter contains three potential Sp1 binding sites that are part of AciI recognition sites; these AciI sites are destroyed by bisulfite treatment unless protected by CpG methylation. Hence, the band pattern after AciI restriction will reflect the degree of methylation of this region. Increasing levels of DNA methylation will result in AciI digest and smaller DNA fragments. The amplicons obtained from pSV-transfected cells using MS primers and those from irradiation (IR)-treated cells using UM primers were purified (Qiagen Gel Extraction Kit). A 25-µl reaction containing 250 ng of DNA and 10 units of AciI was incubated for 3 hours at 37°C and the enzyme was inactivated as per the manufacturer's recommendation. The resultant mixtures were analyzed on 2% agarose gels, and the patterns of digestion were documented.
Immunocytochemistry
IOMM-Lee cells and SF3061 cells (5 x 103) were seeded onto eight-well chamber slides and transfected with puPA or pSV. Twenty-four hours after transfection, cells were irradiated and incubated overnight. The cells were washed, fixed with 4% buffered paraformaldehyde, and permeabilized with freshly prepared 0.1% Triton X-100 containing 0.1% sodium citrate. Later, the cells were blocked with 2% BSA for 1 hour followed by overnight incubation with uPA antibody (1:100 dilution) at 4°C. Next, the chamber slides were treated with HRP-conjugated secondary antibodies (1:200) for 45 minutes at room temperature. Immunolocalization was accomplished by exposing sections to 0.05% 3,3-diaminobezidine tetrahydrochloride as the chromogen. All microscopy studies were performed using a microscope attached to a closed circuit (CC) camera.
In Vivo Studies
The Institutional Animal Care and Use Committee at the University of Illinois College of Medicine in Peoria approved all experimental procedures involving the use of animals. Intracranial implantation of meningioma cells was accomplished as described previously [14]. Irradiated cells were allowed to recover for 72 hours with a regular replenishment of fresh medium every 24 hours and infused into one group of animals (n = 9). Nude mice infused with nonirradiated cells served as controls for the respective groups. The animals were observed for changes in morphologic characteristics for 4 weeks. After 4 weeks, the brains were harvested and either snap frozen or fixed in formalin for further analysis.
Immunohistochemistry
Sections of formalin-fixed and paraffin-embedded brain tissue were subjected to hematoxylin and eosin (H&E) staining to verify tumor formation and immunostaining for cyclin B1 (1:100 dilution) and HRP-conjugated secondary antibody (1:200 dilution) as described elsewhere [15].
Statistical Analysis
All data are presented as means ± SEs of at least three independent experiments (each performed at least in triplicate). One-way analysis of variance combined with the Tukey post-hoc test of means were used for multiple comparisons of cell culture experiments. Statistical differences are presented at probability levels of P < .05.
Results
Radiation Induces Hypomethylation of the uPA Promoter in Meningioma
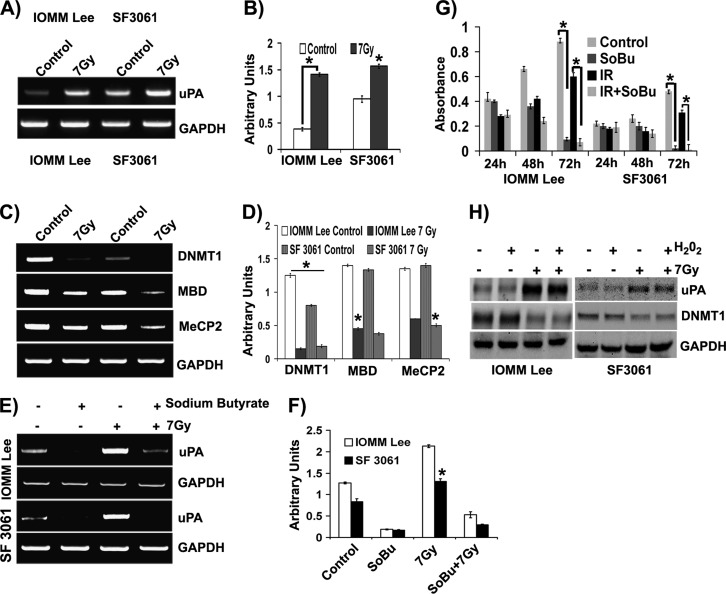
Our previous studies have demonstrated that radiation induces uPA expression at both the protein and transcript levels in meningioma cells [16]. In the present study, we questioned whether hypomethylation has any role in this radiation-induced expression of uPA. To this end, we analyzed the methylation pattern of the uPA promoter by MS PCR (MSP) using primers designed specifically to amplify the converted sequence within the uPA promoter upstream to the TATA box. Genomic DNA extracted from IOMM-Lee and SF3061 cells was subjected to bisulfite conversion, and then MSP was performed. A significant decrease (more than two-fold) in the methylation within CG dinucleotides (indicated by higher band intensity) of uPA promoter was observed in both cell lines, suggesting that X-rays can induce hypomethylation in vitro (Figure 1, A and B). We continued our investigation to determine the enzymes and proteins that support methylation of DNA. Reverse transcription (RT)-PCR analysis for DNMT1, MBD, and MeCP2 revealed that these factors were adversely affected by radiation treatment (Figure 1C). The decrease in the levels was drastic, with DNMT1 showing maximum response compared to MBD and MeCP2 (Figure 1, C and D). Since the methylation of CpG sites within the uPA promoter was shown to reversibly inactivate transcription of uPA gene, we next used sodium butyrate to methylate the uPA promoter in these cells. Cells pretreated with sodium butyrate showed an apparent increase in the methylation of the uPA promoter (indicated by decrease in the band intensity), suggesting the reversible nature of promoter methylation and substantiating hypomethylation as a key event in radiation treatment (Figure 1, E and F). Next, we performed cell proliferation assays on sodium butyrate-treated cells to determine the effect of hypermethylation on cell survival. After 72 hours of treatment, we noticed a drastic decrease in the number of cells, while untreated and irradiated cells continue to divide at markedly different rates (Figure 1G). We then extended our studies to confirm whether oxidative damage, which is often associated with radiation treatment, had any role in DNA demethylation and uPA expression. Treatment with hydrogen peroxide did not result in any difference in uPA or DNMT1 expression patterns, as confirmed by immunoblot analysis (Figure 1H). Similarly, pretreatment of cells with N-acetyl-l-cysteine also did not result in any significant change in expression profiles (data not shown).
Figure 1.
Radiation induces hypomethylation of uPA promoter in meningioma. (A) Genomic DNA extracted from control and irradiated meningioma cells (1.5 x 105) was subjected to bisulfite conversion followed by MSP targeting the uPA promoter and agarose gel analysis. (B) The band intensities were quantified with ImageJ software and represented in arbitrary units. (C) Total RNA extracted from control and irradiated meningioma cells (1.5 x 105) was subjected to cDNA conversion followed by RT-PCR and agarose gel analysis. (D) The band intensities were quantified with ImageJ software and represented as arbitrary units. (E) IOMM-Lee and SF3061 (1 x 105) cells were treated with sodium butyrate (2 mM) for 24 hours before irradiation. Bisulfite-treated genomic DNA was subjected to MSP and followed by agarose gel analysis. (F) Band intensities were quantified by ImageJ software as arbitrary units. (G) IOMM-Lee and SF3061 (1 x 105) cells were treated with sodium butyrate (2 mM) for 24 hours before irradiation, and then MTT assays were performed for 72 hours at 24-hour intervals. Mean absorbance values were plotted against time. (H) IOMM-Lee and SF3061 (1 x 105) cells were treated with H2O2 (150 mM) and 7-Gy radiation as indicated. After overnight incubation, the cell lysates were prepared and analyzed for uPA and DNMT1 by Western blot analysis. GAPDH served as a loading control. Each blot/image is representative of three independent experiments, and each column represents a mean of three values (asterisk represents significant value of P < .05).
Hypomethylation Is Linked to uPA Expression in Meningioma
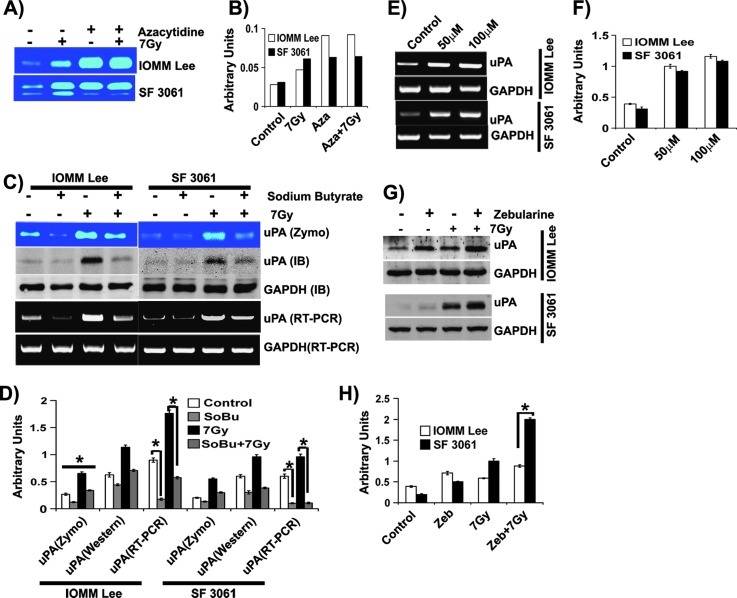
To determine whether demethylation of the uPA promoter is responsible for induction of uPA expression in these tumors, we tested the culture supernatants of IOMM-Lee and SF3061 cells treated with azacytidine and radiation for uPA activity. Treatment with azacytidine increased uPA activity in both cell lines compared to the untreated controls (Figure 2A). The increase of uPA activity in azacytidine-treated cells was significant and greater than five-fold in these cells (Figure 2, A and B). Radiation treatment enhanced uPA levels in untreated cells, whereas azacytidine and radiation did not further increase uPA activity (Figure 2, A and B). These results suggest that radiation-augmented uPA expression is a result of hypomethylation.
Figure 2.
Hypomethylation is linked to uPA expression in meningioma. (A) IOMM-Lee and SF3061 (1 x 105) cells were treated with azacytidine (10 µM) for 24 hours before irradiation followed by incubation in serum-free medium overnight. Conditioned media were collected and 0.1 µg of protein was analyzed for uPA activity using fibrin zymography. (B) The band intensities were quantified and represented as arbitrary units. (C) IOMM-Lee and SF3061 (1 x 105) cells were treated with sodium butyrate (2 mM) for 24 hours before irradiation. Conditioned media were subjected to fibrin zymography, total RNA was used to perform RT-PCR, and cell lysates were used for immunoblot analysis. (D) The band intensities were quantified and represented as arbitrary units. (E) IOMM-Lee and SF3061 (1 x 105) cells were treated with zebularine for 24 hours. Total RNA was used to perform RT-PCR, and cell lysates were used for immunoblot analysis for uPA. (F) The band intensities were quantified and represented as arbitrary units. (G) Meningioma cells were pretreated with zebularine (50 µM) for 24 hours, and the cell lysates were used to perform immunoblot analysis for uPA and GAPDH. (H) The band intensities were quantified and represented as arbitrary units. Each blot/image is representative of three independent experiments, and each column represents a mean of three values (asterisk represents significant value of P < .05).
We next sought to analyze the effect of methylating agents on uPA expression by treating cells with sodium butyrate. This treatment downregulated uPA activity noticeably in both control and irradiated cells (Figure 2, C and D). The reduction in uPA activity was observed to be a result of declines in protein and transcript levels as shown by immunoblot analysis and RT-PCR, respectively (Figure 2, C and D). In the subsequent experiments, we used different concentrations of methyltransferase inhibitor (zebularine) to determine its influence on uPA expression. We noticed a dose-dependent increase at the indicated concentrations in both cell lines (Figure 2, E and F). However, at higher concentrations, cell proliferation was adversely affected (data not shown). In addition, we performed experiments with zebularine in combination with radiation and found that the combination treatment further increased uPA expression in irradiated groups of both cell lines (Figure 2, G and H). Data from all of these experiments suggest that hypomethylation is the underlying event in radiation-induced uPA expression in meningioma cells.
Radiation-Induced Hypomethylation Is Accompanied by MEK/ERK Activation
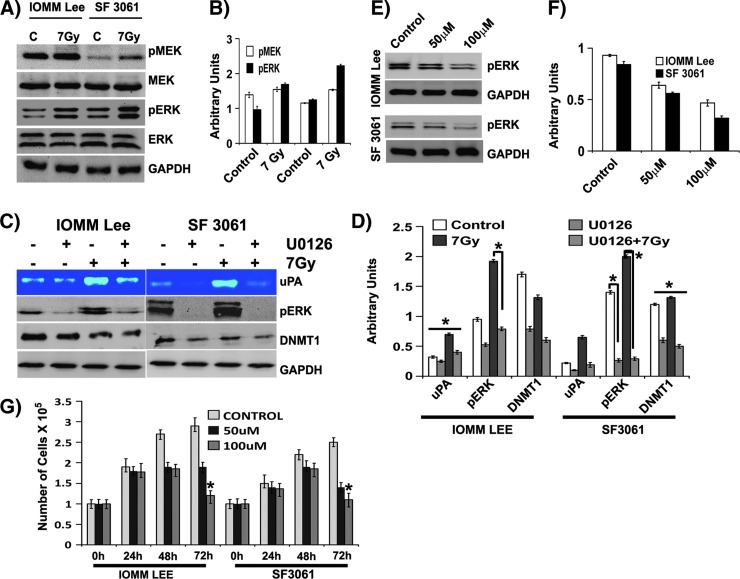
On the basis of studies of human and mouse embryonic stem cells, several signaling pathways have emerged as major candidate regulators of epigenetic remodeling of gene expression [6]. In earlier studies, we observed modulation of uPA expression in an ERK-dependent manner [16,17]. We questioned whether this ERK dependence has any role in radiation-induced hypomethylation. Radiation treatment stimulated the phosphorylation of ERK 1 and 2 in both cell lines, while the total forms remain unaffected (Figure 3A). The upstream molecule MEK also responded similarly to ERK, showing increase in activated MEK levels while the total forms remain unchanged (Figure 3A). The increase in phosphorylation was found to be significant and more than two-fold in both cases (Figure 3, A and B). To establish the upstream effects of these molecules, we also probed for variations in epidermal growth factor receptor (EGFR) and Ras levels. However, the results were not consistent (data not shown). Next, we examined the effect of the ERK phosphorylation inhibitor (U0126). The inhibitor-treated cells showed a significant reduction in the expression of uPA irrespective of radiation treatment (Figure 3, C and D). An obvious decrease in the phosphorylation of MEK and ERK coupled with a decline in the DNMT1 levels indicated the plausible involvement of ERK in regulating the methylation status of DNA in meningioma cells (Figure 3, C and D). Next, we analyzed the influence of zebularine on ERK phosphorylation and found a gradual decrease of pERK levels in a dose-dependent manner, suggesting a mutual dependence of ERK and DNMT1 on one another (Figure 3, E and F). We extended our investigation to determine the effect of zebularine on cellular proliferation. A decrement in the cellular proliferation was apparent in both cell lines after 48 hours of treatment (Figure 3G), while increasing the dosage of zebularine killed most of the cells after 72 hours of treatment (Figure 3G).
Figure 3.
Radiation-induced hypomethylation is accompanied by MEK/ERK activation. (A) Meningioma cells were irradiated at 7 Gy and the cell lysates were immunoblotted for different molecules as indicated. (B) The band intensities were quantified and represented as arbitrary units. (C) Meningioma cells were pretreated with U0126 (10 µM) for 30 minutes before radiation treatment and incubated overnight in serum-free medium. Conditioned media were probed for uPA activity, and cell lysates were immunoblotted for different molecules. (D) The band intensities were quantified and represented as arbitrary units. (E) Meningioma cells were pretreated with zebularine (50 µM) for 24 hours, and the cell lysates were immunoblotted for pERK and GAPDH. (F) The band intensities were quantified and represented as arbitrary units. (G) Meningioma cells were pretreated with zebularine (50 µM) for 24 hours, and MTT assays were performed. Each blot/image is representative of three independent experiments, and each column represents a mean of three values (asterisk represents significant value of P < .05).
Small Interfering RNA (siRNA) Directed toward uPA Promoter Prevents Radiation-Induced Hypomethylation
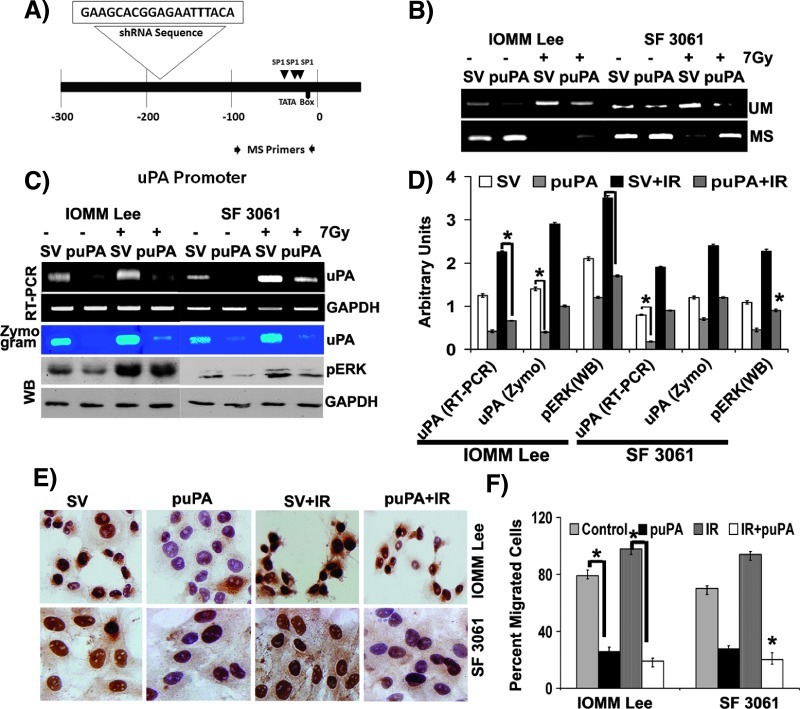
To identify the role of methylation in the TGS of uPA in meningioma cells, we constructed a plasmid encoding shRNA against the upstream sequences of the TATA box (Figure 4A). We also constructed and examined the effect of a vector containing scrambled shRNA sequences on the methylation of uPA promoter. The constructs were sequence verified and used for further transfection studies. We first analyzed methylation status of the targeted regions. Genomic DNA from transfected cells was chemically modified with bisulfite treatment and subjected to MSP. Primers designed to amplify the methylated regions yielded prominently high levels of amplicons in puPA-transfected cells compared to respective untreated cells (Figure 4B, MS primers). Cells treated with a combination of radiation and pSV showed a remarkable decrease in the amplification, illustrating the demethylation of target region in both cell lines (Figure 4B, MS primers). In contrast, primers targeting unmethylated regions displayed significantly greater band intensities in cells transfected with pSV and treated with radiation (Figure 4B, UM primers). Accordingly, we observed very few or no amplicons in puPA-transfected cells with these primers, which signifies siRNA-mediated methylation in these cells (Figure 4B, UM primers). In addition, transfection with puPA failed to induce DNA methylation in a nontargeted 14-3-3 σ promoter, which is normally hypomethylated in cells, indicating specific methylation of target regions (data not shown).
Figure 4.
siRNA directed toward the uPA promoter prevents radiation-induced hypomethylation. (A) Schematic depiction of the uPA promoter showing shRNA target region, Sp1 binding sites, and the region selected for MSP. (B) IOMM-Lee and SF3061 (1 x 105) cells were transfected with pSV or puPA for 36 hours before irradiation. After overnight incubation, bisulfite-treated genomic DNA was subjected to MSP and followed by agarose gel analysis. (C) IOMM-Lee and SF3061 (1 x 105) cells were transfected with pSV or puPA for 36 hours before irradiation. After overnight incubation, conditioned media were subjected to fibrin zymography, total RNA was used to perform RT-PCR, and cell lysates were used for immunoblot analysis as indicated. (D) The band intensities were quantified and represented as arbitrary units. (E) IOMM-Lee and SF3061 (1 x 105) cells were transfected with pSV or puPA for 36 hours before irradiation. After overnight incubation, cells were fixed and subjected to immunocytochemistry with anti-uPA antibody (1:200) followed by HRP-conjugated secondary antibody (1:500). The slides were treated with DAB, counterstained with hematoxylin, and observed under x25 objective. Representative images of each treatment group are shown. (F) IOMM-Lee and SF3061 (1 x 105) cells were transfected with pSV or puPA for 36 hours before irradiation. After overnight incubation, cells were transferred to transwell chambers coated with matrigel (1 mg/ml) and allowed to invade for 24 hours. Percentage of invaded cells in each treatment group was calculated and plotted. Each blot/image is representative of three independent experiments, and each column represents a mean of three values (asterisk represents significant value of P < .05).
We next tested if these changes in methylation patterns were transformed to transcriptional gene regulation of uPA. Induction of methylation by puPA significantly reduced uPA expression at both the mRNA and protein levels in these cells. Both uPA mRNA and protein were undetectable within 2 days of puPA transfection when compared with pSV-transfected cells (Figure 4C). We noticed a significant decrease in the expression patterns of uPA expression in the cells treated with a combination of puPA and radiation compared to radiation treatment alone (Figure 4, C and D). Moreover, we observed a marked reduction in the phosphorylation of ERK in puPA-transfected cells (Figure 4C). Next, we analyzed the in situ cellular expression of uPA in cells transfected with puPA and pSV and treated with radiation. As anticipated, we saw a remarkable decrease in the overall expression of uPA in puPA-transfected cells of both cell lines (Figure 4E).
We further investigated whether the decrease in uPA expression translated to the functional aspects of cells using invasion assays. Cells transfected with puPA showed a more than 50% decrease in the number of invaded cells. Not surprisingly, irradiated cells transfected with puPA showed a more than 70% decline in the invaded cells compared to the cells treated with radiation alone (Figure 4F). These data indicate that induction of DNA demethylation of the uPA gene promoter during radiation treatment could be blocked by methylation induced by puPA transfection.
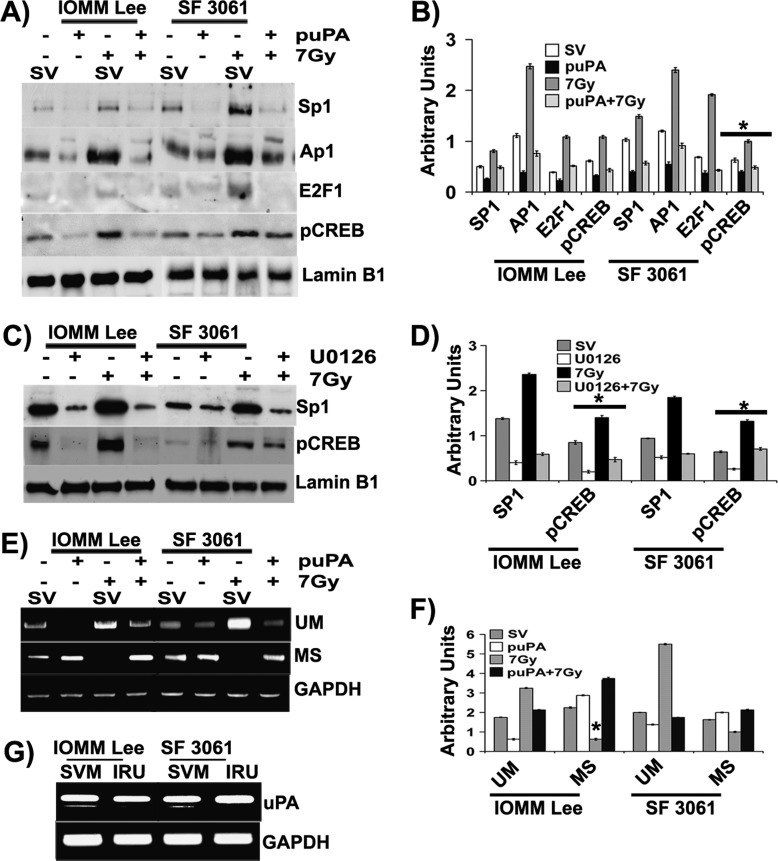
MS Transcription Factors Drive uPA Expression in Meningioma
Previous studies using cell-free and in situ assays showed that multiple transcription factors can bind to the uPA promoter and thus regulate transcription of uPA gene expression [18]. Thus, we first analyzed the modulation of Sp1, Ap1, phospho-cAMP response element-binding protein (pCREB), and E2F1 in the nuclear extracts of IOMM-Lee and SF3061 cell lines. Nuclear fractions of irradiated cells showed the highest levels of Sp1, Ap1, and pCREB compared to the other treatment groups (Figure 5, A and B). In contrast, nuclear localization was apparently less in puPA-transfected cells treated with or without radiation (Figure 5, A and B). Later, we tested the influence of ERK activation on the recruitment of Ap1 and pCREB into the nucleus by treating the cells with U0126. Similar to puPA-treated cells, we saw a drastic decrease in Sp1 and pCREB among U0126-treated cells in both cell lines, which demonstrates the role of ERK in Sp1 recruitment (Figure 5, C and D). We extended our studies to determine the methylation status of Sp1 binding sites upstream to TATA box of the uPA promoter (Figure 4A). We designed two sets of primers to amplify a fragment of 120 bp including three binding sites of Sp1 for MSP on bisulfite-treated genomic DNA. We obtained amplification patterns similar to those described in Figure 4B (Figure 5E). Nevertheless, amplicons obtained in both reactions (with MS primers and UM primers) were subjected to restriction digestion with AciI after gel purification. Agarose gel analysis of the digested molecules showed that the amplicons obtained by using MS primers were digested to a smaller fragment compared to undigested controls (Figure 5F). In contrast, the amplicons obtained by primers specific for unmethylated DNA were not digested during the AciI treatment, thereby indicating the lack of target site (Figure 5E). These results suggest that radiation treatment induces hypomethylation, which, in turn, facilitates Sp1 binding to the consensus site and drives uPA transcription.
Figure 5.
MS transcription factors drive uPA expression in meningioma. (A) IOMM-Lee and SF3061 (1 x 105) cells were transfected with pSV or puPA for 36 hours before irradiation. After overnight incubation, nuclear extracts were prepared and immunoblotted for the indicated molecules. (B) The band intensities were quantified and represented as arbitrary units. (C) Meningioma cells were pretreated with U0126 (10 µM) for 30 minutes before radiation treatment, and nuclear extracts were immunoblotted for different transcription factors. (D) The band intensities were quantified and represented as arbitrary units. (E) IOMM-Lee and SF3061 (1 x 105) cells were transfected with SV or puPA for 36 hours before irradiation. After overnight incubation, bisulfite-treated genomic DNA was subjected to MSP and unmethylated PCR. (F) The band intensities were quantified and represented as arbitrary units. (G) The gel-purified amplicons from previous experiments were digested with AciI and analyzed on 2% agarose gels. Each blot/image is representative of three independent experiments, and each column represents a mean of three values (asterisk represents significant value of P < .05). Lamin B1 was used as a loading control for nuclear extracts. SVM, scrambled vector methylation PCR amplicons; IRU, irradiation unmethylated PCR amplicons.
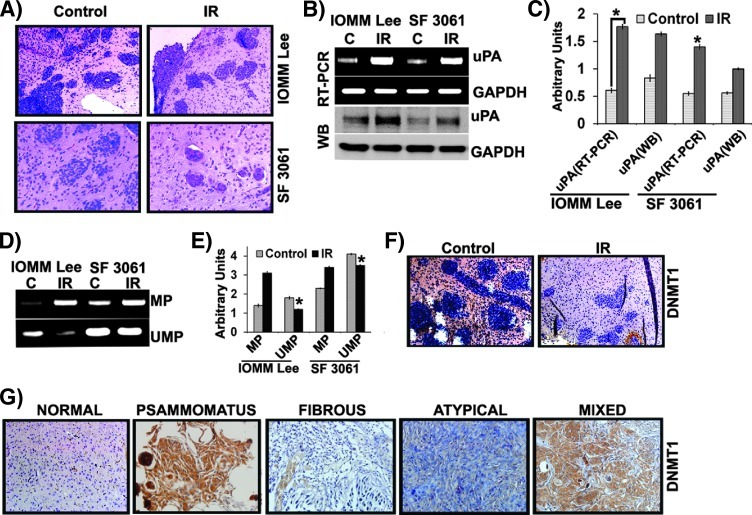
Radiation Treatment Induces Hypomethylation In Vivo
We extended our studies on animal models to test whether the in vitro observations held true in orthotopic tumors. Initially, the formalin-fixed brains were sectioned, and H&E staining was performed to confirm tumor formation. Apparent tumor formation was evident in both control and irradiated groups of animals (Figure 6A). Subsequently, snap-frozen brains were analyzed for uPA expression at the RNA and protein levels; we found a marked increase of uPA expression in the irradiated tumors compared to the control (Figure 6B). The increase was found to be greater than five-fold among irradiated tumors (Figure 6, B and C). Further, we extracted the genomic DNA from these tumors and subjected it to bisulfite conversion followed by MSP to assess the methylation status. As anticipated, hypomethylation was pronounced in irradiated cells compared to the controls (Figure 6D). There was a two-fold change in hypomethylation among the irradiated cells (Figure 6, D and E). Later, we performed immunohistochemical studies to visualize the levels of DNMT1 in formalin-fixed and paraffin-embedded brain sections and found that the expression was very low in irradiated brain sections compared to controls (Figure 6F). Concomitantly, we performed immunohistochemical analysis for DNMT1 in (malignant) meningioma tissue array containing 80 clinical samples as well as 20 tumor sections to test whether global hypomethylation in meningioma contributes to tumor progression. We assessed the expression of DNMT1 in 60 samples. Among the positive sections, we observed varied expression of DNMT1 in meningioma tissues with psammomatous tumors showing intense reactivity and fibrous tumors with the least expression (Figure 6G). Mixed and atypical tumors showed moderate expression, whereas normal brain sections showed little expression (Figure 6G).
Figure 6.
Radiation treatment induces hypomethylation in vivo. Nude mice were intracranially implanted with IOMM-Lee cells as described in the Materials and Methods section. (A) Formalin-fixed brain sections were subjected to H&E staining and observed under x20 objective. Each image represents a brain section of five animals. (B) Total RNA was extracted from frozen brain tissues, and RT-PCR was performed for uPA and GAPDH. Tissue lysates were analyzed for uPA expression by Western blot analysis. (C) The band intensities of RT-PCR gels and Western blots were quantified and represented as arbitrary units. (D) Genomic DNA from frozen brain tissues was extracted, treated with bisulfite, and subjected to MSP targeting the uPA promoter (MP, methylated primer; UMP, unmethylated primer). (E) The band intensities were quantified and represented as arbitrary units. (F) Immunostaining with anti-DNMT1 antibody and non-specific IgG (NsIgG) was performed on formalin-fixed brain sections from animals implanted with IOMM-Lee cells followed by treatment with HRP-conjugated secondary antibody and DAB staining. Representative photomicrographs (x200) are shown in comparison to normal mouse brain sections. (G) Immunohistochemical analysis for DNMT1 was performed on tissue microarrays and clinical meningioma samples. Representative images (x400) are shown. Each column represents a mean value of brain tissues from three different animals (*P < .05, significant difference from respective normal tissues).
Discussion
Epigenetic mechanisms are known to contribute significantly to the progression of meningioma with grade I and II tumors having more aberrantly methylated loci [19]. Although not all aberrantly methylated CpG islands are expected to have an effect on gene expression, it is evident that epigenetic mechanisms have a contributory role in meningioma. It is well known that radiation treatment induces genomic instability and manifests itself in induction of chromosomal aberrations, aneuploidy, gene mutations and amplifications, microsatellite instability, and cell death [20,21]. Further, prominent hypomethylation during X-ray exposure, subsequent genome instability, and resultant recruitment of repair machinery contributing significantly to carcinogenesis have been reported in different tissues [22,23]. It is also evident that most of the unmethylated CpGs are located in the promoters of transcribed genes [24], and uPA promoter hypomethylation has been recognized to be a causal, necessary, and sufficient mechanism to drive transcription in prostate and breast cancer cells [25–27]. In the present study, we hypothesized that radiation-induced uPA expression in meningioma cells could be because of promoter hypomethylation. Our initial studies on methylation status indicated that radiation did induce hypomethylation in meningioma cells as evidenced by depleted expression of DNMT1, MBP, and Mecp2 transcript levels. Similar to our observations, an earlier study on hypomethylation induced by radiation treatment indicated a decrease in DNMTs and Mecp2 in mammary tissues [22]. Further, pretreatment of cells with sodium butyrate quenched radiation-induced expression of uPA, thereby providing evidence that methylation has a role in uPA expression. Since oxidative damage has been shown to interfere with MBPs resulting in heritable epigenetic alterations [28], we treated cells with H2O2. Our results demonstrate that uPA expression is not influenced by oxidative damage. This observation suggests that radiation treatment induces changes in the methylation patterns in an oxidative damage-independent fashion among meningioma cells. In contrast to our findings, DNA methylation was found to be one of the determinants of γ radiation-induced gene expression [29] and ultra violet B (UVB) radiation induced DNMT activity to silence tumor suppressor genes, thereby supporting tumor growth [30], which perhaps suggests a complex network of epigenetic events that can be initiated by ionizing radiation.
Genome-wide reduction in 5-methylcytosine is an epigenetic hallmark of human tumorigenesis and experimentally induced hypomethylation in mice was shown to be sufficient to initiate tumorigenesis [4]. Our experiments using MSP demonstrated that the uPA promoter region underwent extensive hypomethylation after radiation treatment. Similarly, the direct association between hypomethylation and uPA expression was also confirmed by azacytidine treatment studies wherein pretreatment of cells with azacytidine resulted in induction of uPA, which could not be further increased by radiation treatment. These results indicate that the azacytidine treatment induced total demethylation. Further, we have demonstrated that demethylation within the uPA promoter can be reversibly blocked by sodium butyrate (methylation agent) as seen by a decrease in uPA expression, therefore suggesting that radiation has a direct influence on the methylation patterns of the uPA gene promoter. Apart from our present study, increase in methylation of uPA promoter was found to correlate with lower levels of uPA and transcription pattern of uPA in meningiomas [7], suggesting that uPA expression might at least, in part, be controlled by promoter methylation.
Maintenance of the methylation patterns is sensitive to exogenous and endogenous influences. However, signaling pathways involved in DNA methylation/demethylation are not well understood. Experimental overactivation of the Ras pathway [31], overexpression of c-Myc [32], and overactivation of Shh [33], Wnt [34], and Notch pathways [35] have been shown to perform a dynamic role in methylation among cancer cells. In light of these reports, and our prior studies on ERK activation in meningioma cells [16], we assessed the role of the ERK pathway in hypomethylation of the uPA promoter. MEK kinase inhibitor treatment consistently blocked the expression of uPA with concomitant decrease in DNMT1 levels signifying the role of the ERK pathway in uPA expression. Further, a dose-dependent decrement in the activation of ERK upon zebularine treatment also verified the association of ERK pathway with hypomethylation in meningioma cells. In contradiction to our results, sodium arsenite was shown to induce death-associated protein kinase (DAPK) promoter hypermethylation through ERK1/2 phosphorylation in human uroepithelial cells [36,37]. Likewise, down-regulation of Ras activity reverted transformed morphology of Y1 cells with a concomitant reduction in DNA methylation, DNA methyltransferase mRNA, and enzymatic activity [38]. These conflicting findings illustrate that the cross talk between cell signaling and epigenetic control is intricate and may often be tangled.
The development of promoter-targeted siRNA to achieve TGS allows stable, long-lasting, and heritable epigenetic silencing of tumor-promoting genes. In the present study, we designed siRNA targeting CpG islands of the uPA promoter to inhibit uPA expression. Kawasaki and Taira [9] reported that siRNA targeted to E-cadherin promoter induced significant DNA methylation, which was dependent on the DNA methyltransferases DNMT1 and DNMT3b, suggesting that it occurred at the transcriptional level. Likewise, Morris et al. [39] showed TGS of eukaryotic translation elongation factor-1 alpha (EF-1α) in human embryonic kidney 293 (HEK293) cells using promoter-specific siRNA. The studies of Castanotto et al. [8] demonstrated that shRNA to the RASSF1A promoter induced partial gene silencing. Murayama et al. [40] showed that shRNA against the hIL2 promoter dramatically reduced endogenous hIL2 expression. Taken together, these studies suggest that TGS is an effective mechanism for regulating gene expression in human cells. A marked reduction of uPA transcription in siRNA-treated cells compared to respective controls ensured that these meningioma cells already possess hemimethylated or partially methylated CpG islands in the uPA promoter. Upon siRNA treatment, these CpG islands were completely methylated, thereby abolishing uPA transcription. Additionally, our experiment involving the pretreatment of cells with siRNA before irradiation demonstrated the dominant effect of TGS over radiation treatment. These results confirm that radiation-induced hypomethylation can be surpassed by DNA methylation and quench uPA transcription.
It is well known that certain transcription factors, such as c-Myc, do not bind to their recognition sequences in the methylated condition, suggesting that CpG methylation may affect the ability of Myc to bind to multiple sites within the genome [41]. Given that Myc can influence chromatin structure [42], it is certainly plausible that inappropriate methylation of its recognition sites could have profound implications on the cancer epigenome. Likewise, methylation of cytosine in the flanking region adjacent to Sp1-binding consensus directly reduces Sp1 binding [43]. This reduction in binding was revealed to diminish p21Cip1 expression in response to depsipeptide treatment [44]. Our results on nuclear extracts showing a rise of Sp1 levels during radiation treatment and occurrence of three Sp1 binding sites upstream to the transcription initiation site of uPA gene led us to investigate the alterations in the methylation patterns on SP1 consensus. Further, in addition to direct activation of uPA gene expression, Sp1 also interacts with and recruits other transcription factors to the uPA promoter [45]. AciI digestion patterns of amplicons suggested that radiation induces hypomethylation in Sp1 binding sites. However, puPA eradicated the recruitment of Sp1 into nucleus. These results indicate the modulation of uPA promoter by Sp1, which, in turn, is dependent on the methylation status of its binding sites. Although the tumors obtained from animal models revealed methylation patterns similar to our observations in cell lines, the patterns of DNMT1 expression in clinical samples seem to fluctuate, suggesting complex and context-dependent epigenetic regulation in these tumors. Recently, hypermethylation of HOXA6, HOXA9, PENK, UPK3A, and IGF2BP1 genes were shown to correlate with the recurrence of World Health Organization grade I and II meningioma tumors [46]. Nevertheless, our investigation reveals that hypomethylation of uPA promoter is a causal and necessary event for the irradiation-induced aggressive behavior of meningioma.
Although DNA repair proteins were shown to contribute indirectly to radiation-induced tumorigenesis, this study shows that the irradiated cells acquire epigenetic changes, which suggests that epigenetic aberrations may arise in meningioma cells without initiating chromosomal instability. Further, preferential epigenetic changes seem to be more frequent than previously thought and suggest additional levels of uPA gene regulation.
Acknowledgments
We thank Debbie McCollum and Susan Renner for manuscript preparation and Diana Meister and Sushma Jasti for manuscript review.
Footnotes
This research was funded by National Institute of Neurological Disorders and Stroke (NS061835) to Jasti S. Rao.
References
- 1.Goll MG, Bestor TH. Eukaryotic cytosine methyltransferases. Annu Rev Biochem. 2005;74:481–514. doi: 10.1146/annurev.biochem.74.010904.153721. [DOI] [PubMed] [Google Scholar]
- 2.Jaenisch R, Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat Genet. 2003;33 S:245–254. doi: 10.1038/ng1089. [DOI] [PubMed] [Google Scholar]
- 3.Iacobuzio-Donahue CA. Epigenetic changes in cancer. Annu Rev Pathol. 2009;4:229–249. doi: 10.1146/annurev.pathol.3.121806.151442. [DOI] [PubMed] [Google Scholar]
- 4.Das PM, Singal R. DNA methylation and cancer. J Clin Oncol. 2004;22:4632–4642. doi: 10.1200/JCO.2004.07.151. [DOI] [PubMed] [Google Scholar]
- 5.Goetz W, Morgan MN, Baulch JE. The effect of radiation quality on genomic DNA methylation profiles in irradiated human cell lines. Radiat Res. 2011;175:575–587. doi: 10.1667/RR2390.1. [DOI] [PubMed] [Google Scholar]
- 6.Mohammad HP, Baylin SB. Linking cell signaling and the epigenetic machinery. Nat Biotechnol. 2010;28:1033–1038. doi: 10.1038/nbt1010-1033. [DOI] [PubMed] [Google Scholar]
- 7.Kandenwein JA, Park-Simon TW, Schramm J, Simon M. uPA/PAI-1 expression and uPA promoter methylation in meningiomas. J Neurooncol. 2011;103:533–539. doi: 10.1007/s11060-010-0411-6. [DOI] [PubMed] [Google Scholar]
- 8.Castanotto D, Tommasi S, Li M, Li H, Yanow S, Pfeifer GP, Rossi JJ. Short hairpin RNA-directed cytosine (CpG) methylation of the RASSF1A gene promoter in HeLa cells. Mol Ther. 2005;12:179–183. doi: 10.1016/j.ymthe.2005.03.003. [DOI] [PubMed] [Google Scholar]
- 9.Kawasaki H, Taira K. Induction of DNA methylation and gene silencing by short interfering RNAs in human cells. Nature. 2004;431:211–217. doi: 10.1038/nature02889. [DOI] [PubMed] [Google Scholar]
- 10.Pulukuri SM, Rao JS. Small interfering RNA directed reversal of urokinase plasminogen activator demethylation inhibits prostate tumor growth and metastasis. Cancer Res. 2007;67:6637–6646. doi: 10.1158/0008-5472.CAN-07-0751. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 11.Stein GS, Stein JL, Van Wijnen AJ, Lian JB, Montecino M, Croce CM, Choi JY, Ali SA, Pande S, Hassan MQ, et al. Transcription factor-mediated epigenetic regulation of cell growth and phenotype for biological control and cancer. Adv Enzyme Regul. 2010;50:160–167. doi: 10.1016/j.advenzreg.2009.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gondi CS, Lakka SS, Yanamandra N, Siddique K, Dinh DH, Olivero WC, Gujrati M, Rao JS. Expression of antisense uPAR and antisense uPA from a bicistronic adenoviral construct inhibits glioma cell invasion, tumor growth, and angiogenesis. Oncogene. 2003;22:5967–5975. doi: 10.1038/sj.onc.1206535. [DOI] [PubMed] [Google Scholar]
- 13.Velpula KK, Dasari VR, Tsung AJ, Dinh DH, Rao JS. Transcriptional repression of MAD-MAX complex by human umbilical cord blood stem cells downregulates extracellular signal-regulated kinase in glioblastoma. Stem Cells Dev. 2012;21:1779–1793. doi: 10.1089/scd.2011.0424. doi:10.10891scd.2011.0424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McCutcheon IE, Friend KE, Gerdes TM, Zhang BM, Wildrick DM, Fuller GN. Intracranial injection of human meningioma cells in athymic mice: an orthotopic model for meningioma growth. J Neurosurg. 2000;92:306–314. doi: 10.3171/jns.2000.92.2.0306. [DOI] [PubMed] [Google Scholar]
- 15.Gogineni VR, Nalla AK, Gupta R, Gorantla B, Gujrati M, Dinh DH, Rao JS. Radiation-inducible silencing of uPA and uPAR in vitro and in vivo in meningioma. Int J Oncol. 2010;36:809–816. doi: 10.3892/ijo_00000557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kargiotis O, Chetty C, Gogineni V, Gondi CS, Pulukuri SM, Kyritsis AP, Gujrati M, Klopfenstein JD, Dinh DH, Rao JS. uPA/uPAR downregulation inhibits radiation-induced migration, invasion and angiogenesis in IOMM-Lee meningioma cells and decreases tumor growth in vivo. Int J Oncol. 2008;33:937–947. [PMC free article] [PubMed] [Google Scholar]
- 17.Nalla AK, Gogineni VR, Gupta R, Dinh DH, Rao JS. Suppression of uPA and uPAR blocks radiation-induced MCP-1 mediated recruitment of endothelial cells in meningioma. Cell Signal. 2011;23:1299–1310. doi: 10.1016/j.cellsig.2011.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Irigoyen JP, Munoz-Canoves P, Montero L, Koziczak M, Nagamine Y. The plasminogen activator system: biology and regulation. Cell Mol Life Sci. 1999;56:104–132. doi: 10.1007/PL00000615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jun P, Hong C, Lal A, Wong JM, McDermott MW, Bollen AW, Plass C, Held WA, Smiraglia DJ, Costello JF. Epigenetic silencing of the kinase tumor suppressor WNK2 is tumor-type and tumor-grade specific. Neuro Oncol. 2009;11:414–422. doi: 10.1215/15228517-2008-096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang L, Snyder AR, Morgan WF. Radiation-induced genomic instability and its implications for radiation carcinogenesis. Oncogene. 2003;22:5848–5854. doi: 10.1038/sj.onc.1206697. [DOI] [PubMed] [Google Scholar]
- 21.Morgan WF. Non-targeted and delayed effects of exposure to ionizing radiation: II. Radiation-induced genomic instability and bystander effects in vivo, clastogenic factors and transgenerational effects. Radiat Res. 2003;159:581–596. doi: 10.1667/0033-7587(2003)159[0581:nadeoe]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 22.Loree J, Koturbash I, Kutanzi K, Baker M, Pogribny I, Kovalchuk O. Radiation-induced molecular changes in rat mammary tissue: possible implications for radiation-induced carcinogenesis. Int J Radiat Biol. 2006;82:805–815. doi: 10.1080/09553000600960027. [DOI] [PubMed] [Google Scholar]
- 23.Pogribny I, Koturbash I, Tryndyak V, Hudson D, Stevenson SM, Sedelnikova O, Bonner W, Kovalchuk O. Fractionated low-dose radiation exposure leads to accumulation of DNA damage and profound alterations in DNA and histone methylation in the murine thymus. Mol Cancer Res. 2005;3:553–561. doi: 10.1158/1541-7786.MCR-05-0074. [DOI] [PubMed] [Google Scholar]
- 24.Saxonov S, Berg P, Brutlag DL. A genome-wide analysis of CpG dinucleotides in the human genome distinguishes two distinct classes of promoters. Proc Natl Acad Sci USA. 2006;103:1412–1417. doi: 10.1073/pnas.0510310103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pakneshan P, Szyf M, Rabbani SA. Hypomethylation of urokinase (uPA) promoter in breast and prostate cancer: prognostic and therapeutic implications. Curr Cancer Drug Targets. 2005;5:471–488. doi: 10.2174/156800905774574011. [DOI] [PubMed] [Google Scholar]
- 26.Pakneshan P, Szyf M, Farias-Eisner R, Rabbani SA. Reversal of the hypomethylation status of urokinase (uPA) promoter blocks breast cancer growth and metastasis. J Biol Chem. 2004;279:31735–31744. doi: 10.1074/jbc.M401669200. [DOI] [PubMed] [Google Scholar]
- 27.Pulukuri SM, Estes N, Patel J, Rao JS. Demethylation-linked activation of urokinase plasminogen activator is involved in progression of prostate cancer. Cancer Res. 2007;67:930–939. doi: 10.1158/0008-5472.CAN-06-2892. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 28.Valinluck V, Tsai HH, Rogstad DK, Burdzy A, Bird A, Sowers LC. Oxidative damage to methyl-CpG sequences inhibits the binding of the methyl-CpG binding domain (MBD) of methyl-CpG binding protein 2 (MeCP2) Nucleic Acids Res. 2004;32:4100–4108. doi: 10.1093/nar/gkh739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kumar A, Rai PS, Upadhya R, Vishwanatha, Prasada KS, Rao BS, Satyamoorthy K. γ-Radiation induces cellular sensitivity and aberrant methylation in human tumor cell lines. Int J Radiat Biol. 2011;87:1086–1096. doi: 10.3109/09553002.2011.605417. [DOI] [PubMed] [Google Scholar]
- 30.Nandakumar V, Vaid M, Tollefsbol TO, Katiyar SK. Aberrant DNA hypermethylation patterns lead to transcriptional silencing of tumor suppressor genes in UVB-exposed skin and UVB-induced skin tumors of mice. Carcinogenesis. 2011;32:597–604. doi: 10.1093/carcin/bgq282. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 31.Gazin C, Wajapeyee N, Gobeil S, Virbasius CM, Green MR. An elaborate pathway required for Ras-mediated epigenetic silencing. Nature. 2007;449:1073–1077. doi: 10.1038/nature06251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Opavsky R, Wang SH, Trikha P, Raval A, Huang Y, Wu YZ, Rodriguez B, Keller B, Liyanarachchi S, Wei G, et al. CpG island methylation in a mouse model of lymphoma is driven by the genetic configuration of tumor cells. PLoS Genet. 2007;3:1757–1769. doi: 10.1371/journal.pgen.0030167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu S, Dontu G, Mantle ID, Patel S, Ahn NS, Jackson KW, Suri P, Wicha MS. Hedgehog signaling and Bmi-1 regulate self-renewal of normal and malignant human mammary stem cells. Cancer Res. 2006;66:6063–6071. doi: 10.1158/0008-5472.CAN-06-0054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fre S, Huyghe M, Mourikis P, Robine S, Louvard D, Artavanis-Tsakonas S. Notch signals control the fate of immature progenitor cells in the intestine. Nature. 2005;435:964–968. doi: 10.1038/nature03589. [DOI] [PubMed] [Google Scholar]
- 35.Reya T, Clevers H. Wnt signalling in stem cells and cancer. Nature. 2005;434:843–850. doi: 10.1038/nature03319. [DOI] [PubMed] [Google Scholar]
- 36.Huang YC, Hung WC, Chen WT, Yu HS, Chai CY. Effects of DNMT and MEK inhibitors on the expression of RECK, MMP-9, -2, uPA and VEGF in response to arsenite stimulation in human uroepithelial cells. Toxicol Lett. 2011;201:62–71. doi: 10.1016/j.toxlet.2010.12.006. [DOI] [PubMed] [Google Scholar]
- 37.Huang YC, Hung WC, Chen WT, Yu HS, Chai CY. Sodium arsenite-induced DAPK promoter hypermethylation and autophagy via 1ERK1/2 phosphorylation in human uroepithelial cells. Chem Biol Interact. 2009;181:254–262. doi: 10.1016/j.cbi.2009.06.020. [DOI] [PubMed] [Google Scholar]
- 38.Deng C, Yang J, Scott J, Hanash S, Richardson BC. Role of the ras-MAPK signaling pathway in the DNA methyltransferase response to DNA hypomethylation. Biol Chem. 1998;379:1113–1120. doi: 10.1515/bchm.1998.379.8-9.1113. [DOI] [PubMed] [Google Scholar]
- 39.Morris KV, Chan SW, Jacobsen SE, Looney DJ. Small interfering RNA-induced transcriptional gene silencing in human cells. Science. 2004;305:1289–1292. doi: 10.1126/science.1101372. [DOI] [PubMed] [Google Scholar]
- 40.Murayama A, Sakura K, Nakama M, Yasuzawa-Tanaka K, Fujita E, Tateishi Y, Wang Y, Ushijima T, Baba T, Shibuya K, et al. A specific CpG site demethylation in the human interleukin 2 gene promoter is an epigenetic memory. EMBO J. 2006;25:1081–1092. doi: 10.1038/sj.emboj.7601012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jones PA, Baylin SB. The epigenomics of cancer. Cell. 2007;128:683–692. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Knoepfler PS, Zhang XY, Cheng PF, Gafken PR, McMahon SB, Eisenman RN. Myc influences global chromatin structure. EMBO J. 2006;25:2723–2734. doi: 10.1038/sj.emboj.7601152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Clark SJ, Harrison J, Molloy PL. Sp1 binding is inhibited by mCpmCpG methylation. Gene. 1997;195:67–71. doi: 10.1016/s0378-1119(97)00164-9. [DOI] [PubMed] [Google Scholar]
- 44.Zhu WG, Srinivasan K, Dai Z, Duan W, Druhan LJ, Ding H, Yee L, Villalona-Calero MA, Plass C, Otterson GA. Methylation of adjacent CpG sites affects Sp1/Sp3 binding and activity in the p21Cip1 promoter. Mol Cell Biol. 2003;23:4056–4065. doi: 10.1128/MCB.23.12.4056-4065.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Belaguli NS, Aftab M, Rigi M, Zhang M, Albo D, Berger DH. GATA6 promotes colon cancer cell invasion by regulating urokinase plasminogen activator gene expression. Neoplasia. 2010;12:856–865. doi: 10.1593/neo.10224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kishida Y, Natsume A, Kondo Y, Takeuchi I, An B, Okamoto Y, Shinjo K, Saito K, Ando H, Ohka F, et al. Epigenetic subclassification of meningiomas based on genome-wide DNA methylation analyses. Carcinogenesis. 2012;33:436–441. doi: 10.1093/carcin/bgr260. [DOI] [PubMed] [Google Scholar]