Abstract
In epithelial tumors, the platelet-derived growth factor receptor B (PDGFRB) is mainly expressed by stromal cells of mesenchymal origin. Tumor cells may also acquire PDGFRB expression following epithelial-to-mesenchymal transition (EMT), which occurs during metastasis formation. Little is known about PDGFRB signaling in colorectal tumor cells. We studied the relationship between PDGFRB expression, EMT, and metastasis in human colorectal cancer (CRC) cohorts by analysis of gene expression profiles. PDGFRB expression in primary CRC was correlated with short disease-free and overall survival. PDGFRB was co-expressed with genes involved in platelet activation, transforming growth factor beta (TGFB) signaling, and EMT in three CRC cohorts. PDGFRB was expressed in mesenchymal-like tumor cell lines in vitro and stimulated invasion and liver metastasis formation in mice. Platelets, a major source of PDGF, preferentially bound to tumor cells in a non-activated state. Platelet activation caused robust PDGFRB tyrosine phosphorylation on tumor cells in vitro and in liver sinusoids in vivo. Platelets also release TGFB, which is a potent inducer of EMT. Inhibition of TGFB signaling in tumor cells caused partial reversion of the mesenchymal phenotype and strongly reduced PDGFRB expression and PDGF-stimulated tumor cell invasion. These results suggest that PDGFRB may contribute to the aggressive phenotype of colorectal tumors with mesenchymal properties, most likely downstream of platelet activation and TGFB signaling.
Introduction
Multiple receptor tyrosine kinases and their growth factor ligands have been implicated in cancer progression and metastasis. Among these are the platelet-derived growth factor receptors (PDGFRs) [1]. Stimulation of the PDGFR leads to activation of intracellular signaling pathways that can promote cell migration, invasion, survival, and proliferation [2,3].
Expression of PDGFRs is mainly restricted to mesenchymal cell types [2]. Activating mutations in PDGFRs are found in gastrointestinal stromal tumors [1]. In colorectal carcinomas, PDGFR expression appears to be mainly expressed by stromal cells and pericytes [4,5]. However, PDGFRB can also be expressed by colorectal tumor cell lines [3,6], similar to the mesenchymal marker vimentin [7]. We have recently shown that PDGFRB primarily signals invasion in colorectal tumor cells [3]. In line with this, PDGFR signaling contributes to the aggressive behavior of other epithelial tumor types such as breast, liver, and pancreas carcinomas [8–10]. High PDGFR expression correlates with advanced stage disease and poor prognosis in all these tumor types [6,8–11].
While most epithelial tumor cells do not express PDGFRs, they may acquire PDGFR expression following epithelial-to-mesenchymal transition (EMT) [12–19]. EMT is thought to contribute to metastasis formation in multiple tumor types by generating tumor cells with decreased cell-cell adhesion and enhanced invasive and clonogenic properties [20]. In breast cancer, circulating tumor cells with a mesenchymal-like phenotype are associated with poor survival [21–23]. Breast and lung cancer cells acquire PDGFR expression following EMT, which is essential for their metastatic potential [18,24]. A prominent inducer of EMT is transforming growth factor beta (TGFB) [20]. Recently, it was shown that TGFB stimulation of colorectal cancer (CRC) cells promotes invasive mesenchymal-like growth of murine colorectal tumor cells and increased metastatic capacity [25]. A potential role for the PDGFR was not investigated in that study. Taken together, the above studies suggest that PDGFR expression, like EMT, could be a transient phenomenon that may facilitate the metastatic process. This could play a role at the primary tumor site but also in the circulation and/or at the secondary organ site.
Disseminated tumor cells are surrounded by platelets, which are a major source of PDGF [2]. Tumor cell-associated platelet aggregation and microthrombus formation occurs when tumor cells get trapped in the microvasculature of the distant organ [26–30]. This can be mediated by cancer-specific mucins that contain multiple binding sites for platelets, leukocytes, and endothelial cells [31]. Once activated, platelets promote metastasis formation by releasing specific growth factors, including lysophosphatidic acid and TGFB [25,32–34]. In line with their prominent pro-metastatic activity, high platelet counts are associated with poor prognosis in many cancer types, including CRC [32,35,36].
In the present report, we show that PDGFRB expression in human CRC is strongly correlated with platelet activation, TGFB signaling, and EMT. We also show that PDGFRB signaling in mesenchymal-like tumor cells contributes to invasion and liver metastasis formation.
Materials and Methods
Bioinformatic Analyses
Most analyses were performed using the R2 microarray analysis and visualization platform (http://r2.amc.nl). Expression of PDGFR and epithelial growth factor receptor (EGFR) across data sets was done by choosing the “Megasampler“ option in R2 and selecting all nine colorectal tumor data sets.
Disease-free survival (DFS) data are available for three of the data sets (Jorissen et al. [37], Smith et al. [38], and Snoeren et al. [39]). In addition, two data sets contain overall survival (OS) data (Smith et al. and Snoeren et al.). The association of PDGFRB expression with DFS and OS was determined by using the Kaplan-Meier option in R2. Median PDGFR expression levels were used as cutoff values. P values were determined by log-rank test as described in Bewick et al. [40].
Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis was performed by choosing the “KEGG Pathway Finder“ option and setting the single gene association (Pearson correlation) P values to <E-10 (P value determined by t = r/sqrt((1 - r2)/(n - 2)), distributed approximately as t with n - 2 degrees of freedom) for each of the three data sets. The KEGG pathways that were significantly (P < .01 chi-square goodness-of-fit test) enriched in at least two of three lists of PDGFR-associated genes were then identified and ordered according to significance, based on the combined P values (Stouffer z-trend) that were calculated with theWeb-basedMetaP application (http://compute1.lsrc.duke.edu/softwares/MetaP/metap.php). A similar approach was used to identify Gene Ontology (GO) terms significantly enriched in the sets of PDGFR-associated genes. Here, the P values for single gene associations was set to <E-7 for each data set.
Genes that were significantly associated with PDGFRB within each of the identified KEGG pathways (P < .01; chi square goodness of fit test) were identified by making use of the KEGG pathway gene filter option in R2. GeneVenn was then used to identify subsets of genes showing significant association with PDGFR expression in at least two of three data sets. The functional interconnectivity between the genes in these lists was visualized using the STRING tool for known and predicted protein-protein interactions (www.string-db.org). The lists were reimported into R2 to generate heat maps in which the tumors were ordered according to PDGFR expression levels from low to high using the largest data set available (Jorissen et al.; 290 tumors).
All human experiments were carried out with informed consent of the volunteers and under approval and accordance with the guidelines of the Medical Ethical Committee of the University Medical Center Utrecht (Utrecht, The Netherlands).
Cell Culture
C26 and MC38 murine colorectal tumor cells were cultured in Dulbecco's modified Eagle's medium (Dulbecco, ICM Pharmaceuticals, Costa Mesa, CA) supplemented with 5% (vol/vol) fetal calf serum (FCS), 2 mM ultraglutamine, 0.3 mg/ml streptomycin, and 100 U/ml penicillin. Cells were kept at 37°C in a humidified atmosphere containing 5% CO2.
C26 luciferase were described before [41]. C26GFP and MC38GFP cell lines were generated by lentiviral transduction using pWPT-green fluorescent protein (GFP; a kind gift from D. Trono).
Antibodies and Reagents
The following antibodies were obtained fromCell Signaling Technology, Inc (Beverly, MA): rabbit pY1021-PDGFRB (#2227) and rabbit PDGFRB (#4564); from BD Pharmingen (Breda, The Netherlands): PE-Cy5 mouse CD42b (551141), PE mouse P-selectin (Cd62P) (#555524), rat CD41 (#553847), mouse N-cadherin (#610920), mouse Fibronectin (#610077), rabbit active caspase-3 (#559565); from Santa Cruz Biotechnology, Inc (Santa Cruz, CA): rabbit pY1021-PDGFRB (sc-12909-R); from Thermo Fisher Scientific (Amsterdam, The Netherlands): rabbit KI67 (rm-9106-s); from Sigma-Aldrich (Zwijndrecht, The Netherlands): PDGFRB (HPA028499); from Emfret Analytics (Eibelstadt, Germany): rat anti-mouse P-selectin (#M130-2). Secondary peroxidase-conjugated antibodies were from Dako (Heverlee, Belgium). The following reagents were used in this study:PDGF-BB (PGM0044; Invitrogen, Bleiswijk, TheNetherlands), TGFB1 (H8541; Sigma-Aldrich), thrombin receptor-activating peptide (TRAP, SFLLRN; Bachem AG, Bubendorf, Switzerland), prostaglandin I2 (PGI2) analog iloprost (Ilomedine; Bayer Schering Pharma AG, Berlin, Germany), collagen-related peptide (CRP) generated as described earlier [42], and SB431524 (#S4317; Sigma-Aldrich). Short interfering RNA (siRNA) OTP SMARTpools from Dharmacon (Amsterdam, The Netherlands) were transfected using reverse transfection with Hiperfect (Qiagen, Venlo, The Netherlands) according to the manufacturers' guidelines. Gene names and siRNA sequences of the siRNA library are listed in Figure W4C.
Platelet Isolation
Human platelets were isolated as previously described [43]. In short, fresh whole blood was drawn from healthy volunteers into 3.2% trisodium citrate tubes (Greiner Bio-One GmbH, Frickenhausen, Germany). Platelet-rich plasma was prepared within 1 hour after collection by centrifugation at 160g for 15 minutes at 20°C. Subsequently, 0.1 volume of acid citrate dextrose (containing 2.5% trisodium citrate, 1.5% citric acid, and 2% d-glucose) was added to lower the pH to 6.5. Platelet-rich plasma was spun down by centrifugation at 340g for 15 minutes at 20°C. The platelet pellet was resuspended in Hepes-Tyrode buffer (containing 145 mM NaCl, 5 mM KCl, 0.5 mM Na2HPO4, 1 mM MgSO4, 10 mM Hepes, and 5 mM d-glucose, pH 6.5). Prostacyclin (PGI2; Cayman Chemical Company, Ann Arbor, MI) was added to a final concentration of 10 ng/ml and platelets were spun down at 340g for 15 minutes at 20°C. The pellet was resuspended in Hepes-Tyrode buffer (pH 7.2) at a platelet count of 2.0 x 1011 platelets/l. Platelets were allowed to return to a resting state for at least 30 minutes before usage.
Tumor Cell Platelet Binding
GFP-expressing tumor cells were harvested by brief trypsinization, washed, and suspended in Hepes-Tyrode buffer to a final concentration of 1 x 105 cells/ml. To exclude clumps of cells, the mixture was passed through a 40-µm cell strainer. Tumor cells and platelets were mixed in a 1:10 ratio and incubated for 30 minutes at room temperature. Hereafter, blocking was performed with FCS after which cells were washed with Hepes-Tyrode buffer by centrifugation at 400g for 10 minutes. Antibody binding (1:50) was allowed in Hepes-Tyrode buffer. After 30 minutes at room temperature, the cell platelet suspension was washed and resuspended in Hepes-Tyrode buffer containing 2% formaldehyde. Cells were analyzed using flow cytometry.
Flow Cytometry
The expression of a panel of cell surface markers was analyzed using a FACSCalibur (BD, Franklin Lakes, NJ). All antibody incubation steps were carried out at room temperature. GFP-expressing tumor cells were used and selected on fluorescent intensity. Doublets and clumps of tumor cells were excluded by size using doublet discrimination gating. For analysis of inactive platelets, PE-Cy5 anti-CD42b was used. For analysis of active platelets, PE anti-P-selectin was used as a marker. All samples were analyzed by bivariate flow cytometry using Cell Quest software (BD).
Western Blot Analysis
Western blot analysis was performed exactly as described before [41,44].
Invasion Assay
For in vitro invasion assays, 24-well BioCoat Matrigel invasion chambers (#354480; BD), with an 8-µm pore PET membrane coated with Matrigel basement membrane matrix, were used according to the manufacturer's protocol. Cells were kept under serum-free conditions overnight. In the upper compartment, 5 x 104 cells/well were plated onto 0.5 ml of serum-free medium. The lower compartment contained 0.75 ml of medium with 0% FCS. PDGF (10 ng/ml) was added to the upper and/or lower compartment. For inhibitor experiments, cells were pretreated with 10 µg/ml SB431524 overnight before plating and during the experiment in both the upper and lower compartments. Invasion chambers were incubated for 8 hours at 37°C in a humidified incubator with 5% CO2. Remaining cells in the upper compartment were removed with a cotton swap. The transmigrated cells were fixed in 3.7% formaldehyde, stained with 4′,6-diamidino-2-phenylindole (DAPI), and counted by analyzing microscopic images (five to six fields per transwell membrane; magnification, x10). Data are expressed relative to control. All assays were performed in duplicate and were repeated twice.
In Vitro Proliferation Assay
Cells (5000 cells/96-well plate) were plated and incubated at 37°C in a humidified atmosphere containing 5% CO2. Proliferation was analyzed every 24 hours for 4 days by 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide assays (Roche Diagnostics, Basel, Switzerland) according to the manufacturer's instructions.
Immunohistochemistry
After harvesting, organs were either snap frozen in Tissue-Tek OCT using liquid nitrogen or fixed in 4% paraformaldehyde and paraffin embedded. Frozen tissue was sectioned using a Leica cryotome CM3050 (Mannheim, Germany), fixed with acetone, and an immunostaining was performed. Paraffin-embedded tissues were sectioned by a Leica microtome RM 2235 and stained according to standard histology protocols. For immunofluorescence, image acquisition and analysis was performed using a Zeiss Axiovert 200M and Zeiss LSM 510 Software.
Animals and Surgery
Male Balb/c mice (10–12 weeks) were purchased from Charles River (Wilmington, MA). Mice were housed under standard laboratory conditions and received food and water ad libitum. All surgical procedures were performed under isoflurane inhalation anesthesia. Before surgery, buprenorfine was administered intramuscularly to provide sufficient perioperative analgesia. All animal experiments were carried out in accordance with the guidelines of the Animal Welfare Committee of the University Medical Center Utrecht.
Liver Metastasis Mouse Model
C26 cells expressing GFP or luciferase were harvested by brief trypsinization. Colorectal liver metastases were induced as previously described [45,46]. In brief, single cell suspensions were prepared in phosphate-buffered saline to a final concentration of 7.5 x 104 cells/100 µl. Cells were injected into the parenchyma of the spleen. Ten minutes after injection, the spleen was removed. Induction and bioluminescence imaging of colorectal liver metastases was performed as described [41].
Hepatic Replacement Area
Tumor load in the liver was assessed in all liver lobes. Tumor load was scored as hepatic replacement area (HRA), that is, the percentage of liver tissue that had been replaced by tumor tissue, exactly as described before [47].
In brief, on hematoxylin and eosin-stained sections, at least 100 fields were selected using an interactive video overlay system, including an automated microscope (Q-Prodit; Leica Microsystems) at a x40 magnification. Using a four-point grid overlay, the ratio of tumor cells versus normal hepatocytes was determined for each field. Tumor load (HRA) was expressed as the average area ratio of all fields.
Statistical Analysis
Statistical differences between groups were analyzed by an unpaired two-sided t test. Data are expressed as means ± SEM. A P value of <.05 was considered statistically significant (indicated by an asterisk).
Results
PDGFRB Expression in Primary CRC Is Associated with Poor Prognosis
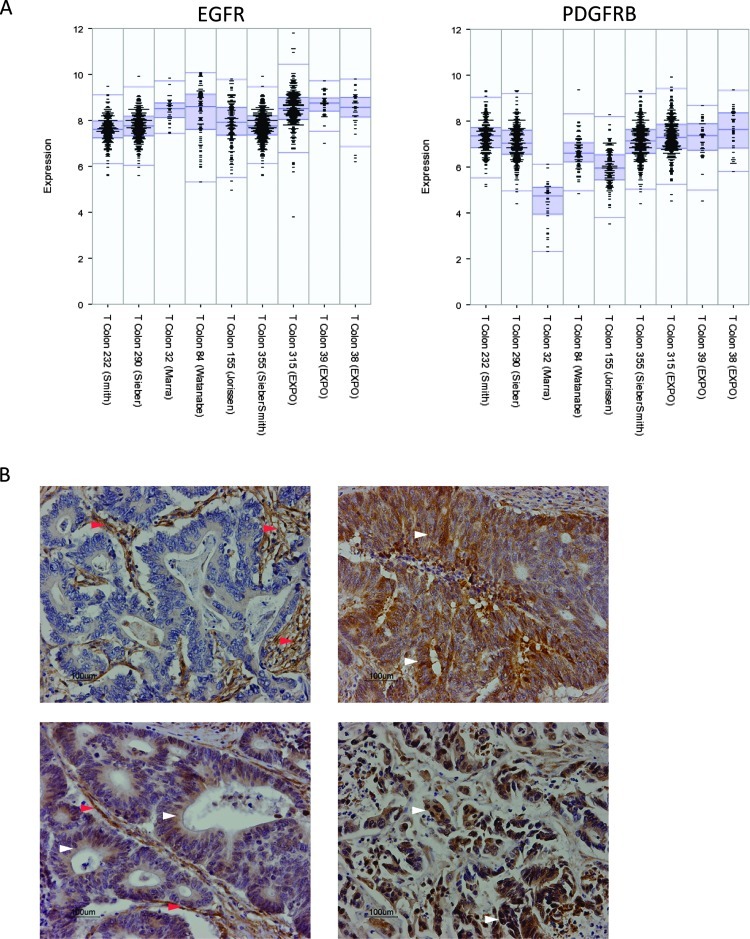
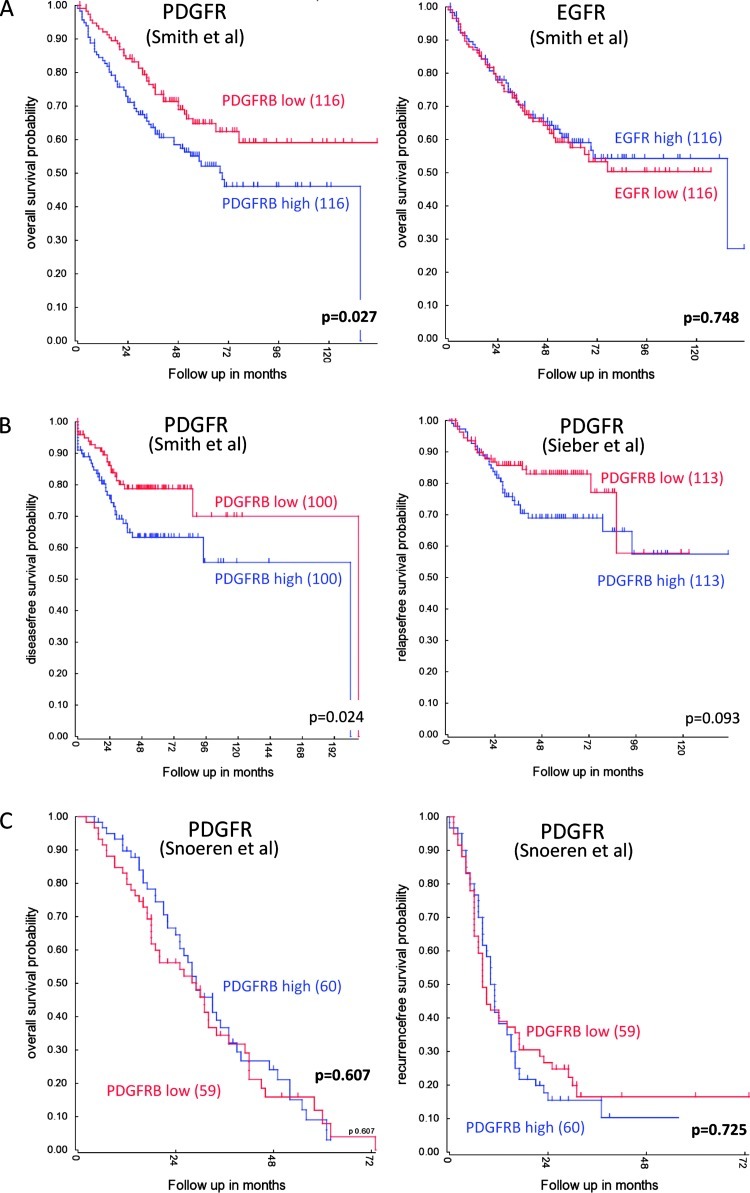
Our previous results have implicated PDGFR signaling in the invasion of colorectal tumor cells in vitro [3]. Very little is known about the impact of PDGFR signaling on human colorectal tumor behavior. We first analyzed gene expression profiles of nine different tumor cohorts to assess whether PDGFRB expression would be associated with clinical outcome. Figure 1A shows that PDGFRB and EGFR are expressed at considerable levels in colorectal tumors, although a direct comparison of the expression levels of both receptor tyrosine kinases is not possible based on microarray data. One cohort of colorectal adenomas (T Colon 32 (Marra)) shows considerable less expression of PDGFRB. Immunohistochemistry staining for PDGFRB in CRC specimens demonstrated the expected stromal staining (Figure 1B). In addition, PDGFRB expression was also clearly observed in the tumor cells of 8 of 10 human CRC tumors (Figures 1B and W1). We next assessed whether PDGFRB expression was correlated with survival in two cohorts of 232 and 290 primary CRC tumors [37,38] and a cohort of 119 liver metastases [39]. Using mean PDGFR levels as the cutoff value, we found that high PDGFRB expression in primary CRC tumors was correlated with shorter DFS and OS (Figure 2, A and B). This was not observed in liver metastases (Figure 2C).
Figure 1.
Expression of PDGFRB in human CRC. (A) mRNA levels of PDGFRB and EGFR of all tumors from nine different cohorts were plotted. With the exception of one adenoma cohort, PDGFRB is expressed at considerable levels in multiple CRC cohorts. (B) Immunohistochemistry analysis of the expression of PDGFRB in human colorectal tumors. Examples of tumors are shown in which PDGFRB is predominantly expressed in stromal cells (left upper panel) in both tumor cell and stromal compartments (left lower panel) and in tumor cells only (right upper and lower panels). Red arrowheads indicate expression in stromal tissue. White arrowheads indicate expression in tumor tissue.
Figure 2.
PDGFRB expression in primary CRC is associated with shorter DFS and OS. (A) Expression levels of PDGFRB and EGFR were correlated with OS in a cohort of 232 primary CRCs [38]. Median expression was used as a cutoff. Kaplan-Meier curves show that expression of PDGFRB, but not EGFR, is associated with a significantly shorter OS. (B) Expression levels of PDGFRB were correlated with DFS in a cohort of 232 primary CRCs [38] and a cohort of 290 primary CRCs [37]. Median expression was used as a cutoff. Kaplan-Meier curves show that expression of PDGFRB is associated with shorter DFS in both cohorts. (C) A possible correlation between PDGFRB levels with OS and DFS was analyzed in a cohort of 119 colorectal liver metastases [39]. Median expression was used as a cutoff. Kaplan-Meier curve shows that expression of PDGFRB is not significantly associated with shorter survival.
PDGFRB Expression Is Associated with Extracellular Matrix-Receptor Signaling, Platelet Activation, TGFB Signaling, and EMT
We next used bioinformatics tools of the R2 Web application (http://r2.amc.nl) to search for processes, pathways, and single genes associated with expression of PDGFRB in CRC. Searching the KEGG pathway database revealed seven pathways that were significantly associated with PDGFRB expression in the three separate tumor cohorts (Table 1). These include “Extracellular Matrix-Receptor Interaction,“ “Coagulation,“ and “TGFB signaling.“ Within the “ECM-Receptor“ pathway, 46 genes were significantly correlated with PDGFRB in at least two of three tumor cohorts (Figure W2A; P = 6.7E–76). This gene set contains 15 collagens, 6 laminins, 8 integrins, and 4 thrombospondins (Table W1 and Figure W2A). All of the genes in this category were positively (rather than negatively) correlated with PDGFRB expression (Figure 3).
Table 1.
Pathway Analysis of Genes Co-expressed with PDGFRB in CRC.
| P Value, Snoeren et al. | P Value, Smith et al. | P Value, Jorissen et al. | P Value, Combined | |
| KEGG pathway | ||||
| ECM-receptor interaction | 4.20E-12 | 3.90E-32 | 8.10E-15 | 3.33E-50 |
| Focal adhesion | 2.20E-10 | 5.70E-24 | 5.40E-11 | 1.42E-37 |
| Malaria | 7.90E-05 | 3.30E-15 | 5.40E-08 | 3.07E-23 |
| Complement and coagulation cascades | 1.30E-04 | 1.50E-08 | 7.10E-06 | 3.56E-15 |
| Glycosaminoglycan biosynthesis | 2.00E-02 | 2.80E-09 | 6.40E-06 | 2.97E-14 |
| Amebiasis | 7.50E-05 | 1.10E-10 | 1.30E-03 | 1.08E-13 |
| Phagosome | 1.00E-02 | 1.00E-08 | 1.20E-04 | 1.76E-12 |
| Leukocyte transendothelial migration | 2.00E-02 | 1.50E-06 | 1.90E-05 | 1.47E-11 |
| Cell adhesion molecules | 4.00E-02 | 1.10E-04 | 1.50E-03 | 1.25E-07 |
| Regulation of actin cytoskeleton | 3.00E-02 | 1.10E-04 | 7.30E-03 | 7.23E-07 |
| Vascular smooth muscle contraction | 2.00E-03 | 5.40E-03 | 2.00E-02 | 1.54E-05 |
| TGFB signaling pathway | 4.40E-05 | 4.60E-03 | 7.00E-02 | 1.95E-05 |
| GO pathway | ||||
| ECM | 8.9E-44 | 1.4E-98 | 6.8E-27 | 5.2E-135 |
| PDGF binding | 2.7E-22 | 2.3E-18 | 4.5E-8 | 2.7E-34 |
| Platelet activation | 7.2E-9 | 5.1E-10 | 1.2E-9 | 1.9E-23 |
| Platelet alpha-granule | 2.2E-6 | 3.7E-13 | 1.2E-6 | 7.2E-21 |
| Platelet degranulation | 2.9E-3 | 3.8E-10 | 2.4E-9 | 1.2E-19 |
| TGFB signaling pathway | 4.4E-6 | 4.1E-4 | 1.7E-7 |
PDGFRB-associated genes in each separate tumor cohortwere analyzed for overrepresentation ofKEGGpathways andGOterms by using R2. All KEGGpathways significantly associatedwith PDGFRB expression in at least two of three data sets are shown. A limited set of GO terms that are related to those KEGG pathways is shown in addition. Combined P values were calculated with the Web-based MetaP software.
Figure 3.
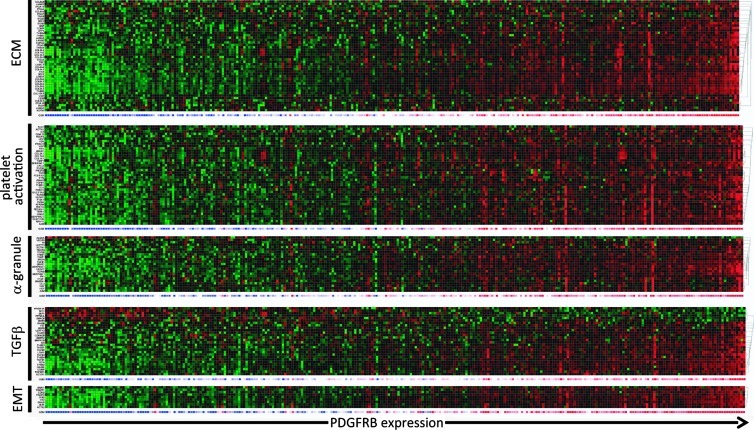
Co-expression of PDGFRB with genes governing ECM-receptor interaction, platelet activation, TGFB pathway, and EMT in colorectal tumors. Heat map of the expression of genes co-expressed with the PDGFR in at least two of three tumor cohorts in the categories “ECM-Receptor Interaction,“ “Platelet Activation,“ “Platelet alpha-Granule,“ “TGFB pathway,“ and “EMT.“ The overlapping genes were identified by GeneVenn (Figure W1). An overview of the individual genes in these categories and their correlation with PDGFRB is presented in Table W1.
We noted that many of the genes within the extracellular matrix (ECM)-Receptor class promote platelet activation, including collagens, laminins, thrombospondins, von Willebrand factor, and fibronectin (Table W1). GO analysis of the genes co-expressed with PDGFRB in the separate tumor cohorts revealed that “Platelet Activation,“ “Platelet alpha-granule,“ and “Platelet Degranulation“ were indeed significantly overrepresented in all three cohorts (Table 1; P = 1.9E–23). We identified a set of 41 “Platelet activation“ genes that were significantly correlated with PDGFR expression in at least two of three tumor cohorts. All of these correlations were positive (Table W1 and Figure W2B). In addition, all except one (ALDOA) “Platelet alpha-granule“ genes were positively correlated with PDGFRB expression (Figure 3).
Platelet activation promotes the metastatic capacity of tumor cells [32]. TGFB signaling has recently been identified as a major plateletreleased pro-metastatic growth factor in a model of colorectal metastasis formation [25]. TGFB signaling was also one of the seven KEGG pathways that were significantly associated with PDGFRB expression in all three tumor cohorts (Tables 1 and W1 and Figure W2C; P = 1.2E–8). A set of 28 overlapping “TGFB pathway genes“ was identified of which 22 were positively correlated with PDGFRB, including TGFB1, TGFB2, TGFB3, and TGFB receptor 1 (TGFBR1). Mapping the PDGFRB-co-expressed genes on a TGFB pathway image shows enrichment of ligands, receptors, and signaling intermediates (Figure W3).
TGFB stimulation of epithelial tumor cells may lead to the acquisition of a more mesenchymal phenotype, which is associated with increased invasion and metastatic potential. Moreover, activated platelets promote EMT in tumor cells by secreting TGFB [25] that stimulates expression of the core transcription factors that mediate EMT (SNAI1, SNAI2, ZEB1, ZEB2, TWIST1, and TWIST2; [48]). Therefore, we next analyzed whether PDGFRB expression would be associated with these EMT-driving transcription factors. Indeed, PDGFRB was strongly associated with all six EMT-inducing transcription factors and with mesenchymal genes, such as vimentin and N-cadherin (CDH2; Table W1 and Figure 3). Conversely, PDGFRB was negatively correlated with epithelial genes, including E-cadherin (CDH1), plakophilin-2, and occludin (Table W1).
PDGFRB in Colorectal Tumor Cells Signals Invasion and Metastasis Formation
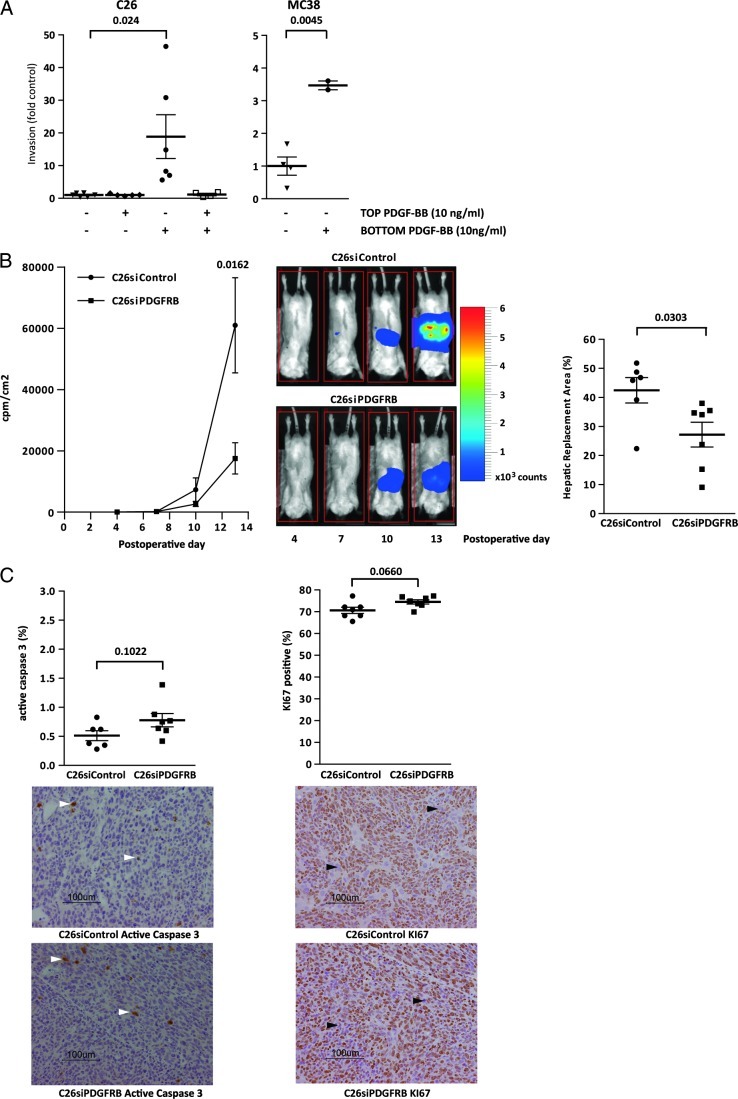
The above results link PDGFRB expression to platelet activation, TGFB signaling, EMT, and poor survival in human CRC. To test the function of PDGFRB in CRC cells, we performed Matrigel Transwell assays using PDGF-responsive C26 and MC38 CRC cells. Checker-board analysis revealed that PDGF strongly promotes directed tumor cell migration (chemotaxis), whereas non-directed migration (chemokinesis) was not affected (Figure 4A). PDGF stimulation did not affect the growth rate of MC38 or C26 cells (Figure W4A).
Figure 4.
PDGFRB stimulates invasion and liver metastasis formation in CRC cell lines. (A) C26 and MC38 cells were cultured in transwell chambers, and invasion through Matrigel was assessed following addition of PDGF-BB (10 ng/ml; 8 hours) to the insert (top) or to the well (bottom) of the invasion chamber. All conditions were tested in duplicate in two independent experiments. Numbers represent fold change of the number of invaded cells relative to control (no PDGF-BB in either compartment). (B) C26 cells expressing luciferase and either siRNAs targeting PDGFRB or control siRNAs were injected into the splenic parenchyma followed by splenectomy. The formation of liver metastases was then followed over time using bioluminescence imaging. Representative bioluminescence images of liver metastasis formation over time are depicted (scale bar represents bioluminescence counts). Two weeks after tumor cell injection, the livers were harvested and the HRA (liver area occupied/replaced by tumor tissue) was determined morphometrically (n = 7 mice per group). (C) Sections of the livers harvested in B were analyzed for the presence of apoptotic caspase-3-positive cells (left panel; white arrowhead indicates active caspase-3-positive cell) and proliferating Ki67-positive cells (right panel; black arrowhead indicates Ki67-negative cell) by immunohistochemistry. The bar graphs show the percentage positive cells measured in five high-power fields per mouse in six mice per condition. All error bars represent SEM. Significance was tested using Student's t test (unpaired; double sided).
Next, we assessed the contribution of PDGFRB to livermetastasis formation. To this end, PDGFRB expression was suppressed by transfecting siRNAs into C26 cells expressing firefly luciferase. This resulted in efficient suppression of PDGFRB expression over a period of at least 4 days (Figure W5A). Expression levels returned to normal 6 days after transfection (Figure W5B). C26-siPDGFRB and control cells expressing scrambled siRNAs were injected into the spleens of syngeneic Balb/c mice 2 days after transfection. Bioluminescence imaging over time showed that PDGFRB knockdown significantly reduced the outgrowth of liver metastases (Figure 4B). After 13 days, the livers were removed and the HRA (percentage of liver tissue occupied by tumor) was analyzed. PDGFRB knockdown had caused a significant drop in metastatic load (Figure 4B). Immunohistochemistry for active caspase-3 and KI67 on tumor tissue sections showed that suppression of PDGFRB expression had no effect on apoptosis or proliferation in liver metastases (Figure 4C). Likewise, PDGFRB knockdown had no effect on the in vitro growth rate of C26 or MC38 cells (Figure W4B).
ALK5 Inhibition Reduces PDGFRB Expression and PDGF-Stimulated Invasion
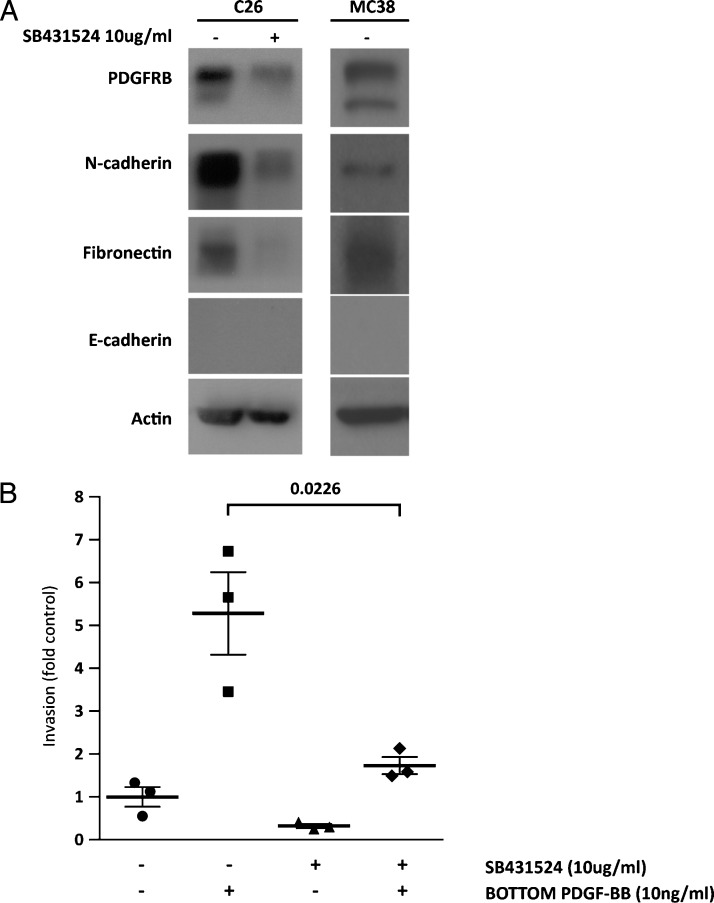
The above results show that PDGFRB promotes invasion and liver metastasis formation. The C26 and MC38 cells that were used in this study display mesenchymal features including low expression of E-cadherin and high expression of N-cadherin and fibronectin (Figure 5A). Inhibition of constitutive TGFB signaling in C26 cells has previously been shown to lead to loss of the mesenchymal phenotype and to a concomitant loss ofmetastatic potential [49]. Indeed, treatment with the ALK5 inhibitor SB431524 blocked TGFB signaling in these cells (Figure W6) and strongly reduced basal fibronectin and N-cadherin expression, although this was not accompanied by reexpression of E-cadherin (Figure 5A). Importantly, ALK5 inhibition reduced PDGFRB expression and PDGF-stimulated tumor cell invasion (Figure 5, A and B), indicating that PDGFRB expression and signaling in these cells requires TGFB signaling.
Figure 5.
The ALK5 inhibitor SB431524 suppresses PDGFRB expression and PDGF-stimulated invasion. (A) C26 cells were incubated with SB431524 (10 ng/ml) or vehicle for 1 week. Cells were lysed and expression of PDGFRB, N-cadherin, fibronectin, E-cadherin, and actin was assessed by Western blot analysis. (B) C26 cells were cultured in transwell chambers and invasion throughMatrigel was assessed following addition of PDGF-BB (10 ng/ml; 8 hours) to the bottom well of the invasion chamber, in the presence or absence of SB431524 (10 ng/ml). All conditions were tested in triplicate in two independent experiments. Numbers represent fold change of the number of invaded cells relative to control (no PDGF-BB added). All error bars represent SEM. Significance was tested using Student's t test (unpaired; double sided).
PDGF from Platelets Activates the PDGFRB on Tumor Cells
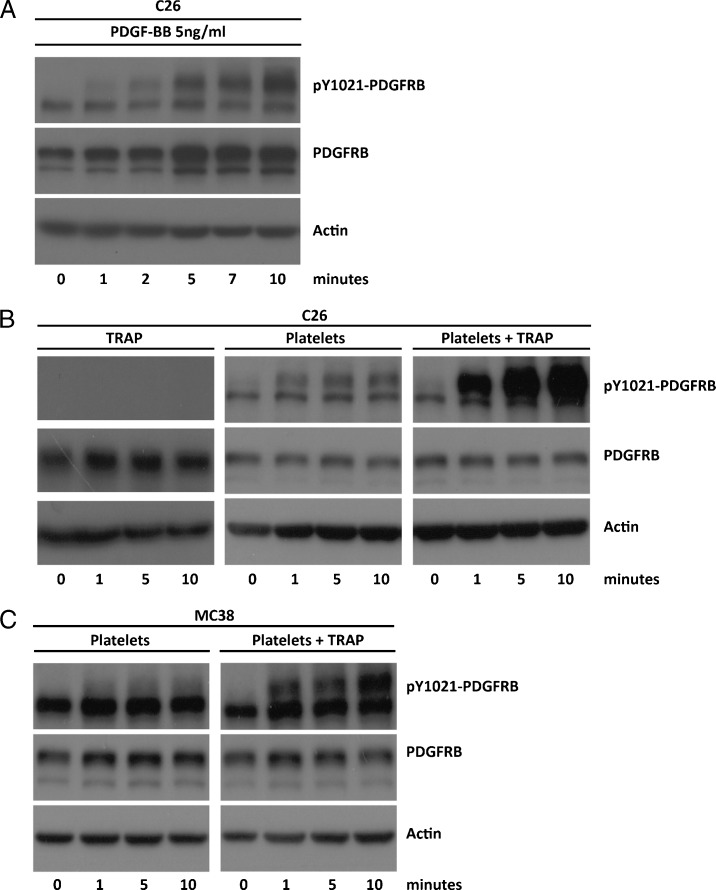
Circulating platelets are a major source of PDGF and have strong pro-metastatic activity [2,32]. Therefore, we assessed whether PDGF from platelets could stimulate the PDGFRB expressed on tumor cells. To this end, C26 and MC38 cells were exposed to PDGF or to platelets. Platelets were isolated from blood and were either left unstimulated or pre-stimulated with TRAP. As expected, PDGF caused rapid tyrosine phosphorylation of the PDGFRB on C26 cells (Figure 6A). Non-stimulated platelets also induced some PDGFRB phosphorylation on C26 and MC38 cells, but this was drastically increased when platelets were pre-activated (Figure 6, B and C). The time course of PDGFRB activation with (activated) platelets was similar to that induced by purified PDGF.
Figure 6.
Activation of PDGFRB on CRC cell lines by activated platelets. C26 cells were cultured in serum-free medium overnight and were subsequently stimulated with purified 5 ng/ml PDGF-BB for different lengths of time (A), with 625 µM TRAP alone, or with platelets (25 x 106/ml) that had been pre-activated or not (B). Platelet activation was performed by exposing them to 625 µM TRAP. The same experiment was performed using MC38 cells (C). Stimulus-induced changes in expression and tyrosine 1021 phosphorylation of PDGFRB (pY1021-PDGFRB) was analyzed over time (0.10 minutes) by Western blot analysis.
Tumor Cells Preferentially Bind Inactive Platelets
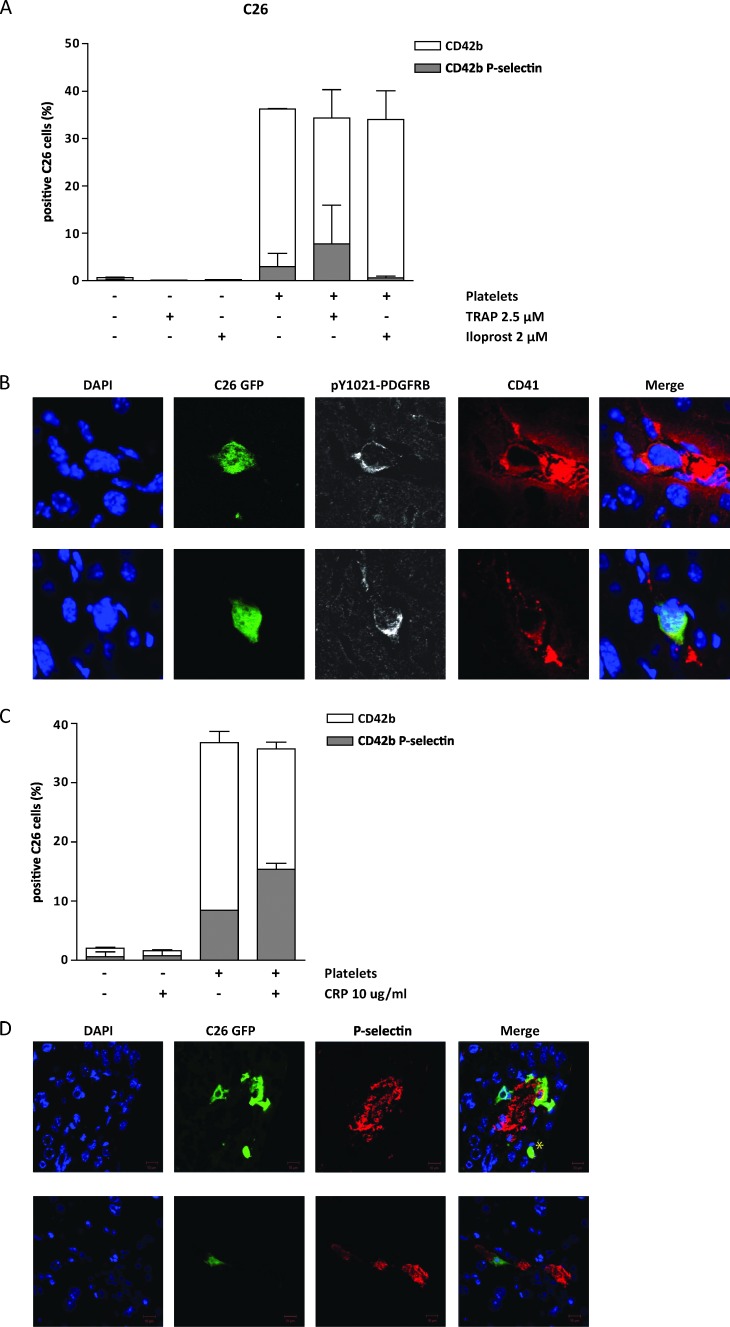
Next, we used flow cytometry to study a potential interaction between tumor cells and platelets. Inactive platelets were isolated from donor blood andmixed with GFP-expressing tumor cells in a 10:1 ratio. Of all GFP-positive C26 and MC38 cells, 30% to 40% were also positive for the platelet marker CD42b (Figures 7A and W7B). Additionally, 5% to 10%of tumor cells were also positive for P-selectin, a marker for activated platelets (Figures 7A and W7B). Activation of platelets with TRAP beforemixing with tumor cells did not change the total percentage of platelet-tumor cell events. Although 95% of the platelets were activated (Figure W7A), the majority of tumor cell-bound platelets were inactive (Figures 7A and W7B). Prevention of platelet activation by treatment with iloprost did not change the total percentage of platelet-bound tumor cells but reduced the number of activated platelet-tumor cell events to undetectable levels (Figures 7A and W7B). Together, these results suggest that tumor cells preferentially bind inactive platelets.
Figure 7.
Platelets bind tumor cells and cause PDGFRB activation on tumor cells lodged in the liver sinusoids. (A) GFP-expressing tumor cells and platelets were mixed in a 1:10 ratio. Before mixing, platelets were non-treated, inactivated by iloprost (2 µM), or activated by TRAP (2.5 µM) treatment as indicated. Two independent experiments were performed in duplicate. The fraction of tumor cells that were bound to non-activated and activated platelets was then analyzed by flow cytometry for GFP, CD42b, and P-selectin. The white bars represent all tumor cell (GFP)-platelet (CD42b+ve) events, whereas the gray bars represent tumor cell (GFP)-activated platelet (CD42b+ve; P-selectin+ve) events. (B) GFP-expressing C26 tumor cells were injected into the spleen. After 2 hours, the liver was perfused with periodate-lysineparaformaldehyde (PLP), harvested, and processed for cryosectioning and immunofluorescence using anti-pY1021-PDGFRB (white) and CD41 (red) antibodies and DAPI (blue). CD41-positive platelet clusterswere observed around ∼52% of all sinusoid-arrested single tumor cells. These tumor cells were also positive for pPDGFRB. Representative images are shown. (C) Tumor cells were mixed with platelets exactly as in A. Hereafter, the mixture was treated with CRP (10 µm/ml) as indicated. Two independent experiments were performed in duplicate. The mixture was subsequently analyzed by flow cytometry exactly as described in A. (D) As in B, cryosections were processed for immunofluorescence using anti-P-selectin (red) and DAPI (blue). P-selectin-positive tumor cells were identified surrounding ∼50%of all GFP-positive tumor cells. Representative images are shown; a tumor cell negative for P-selectin is indicated by asterisk.
PDGFRB Phosphorylation on Platelet-Covered Tumor Cells in the Liver Sinusoids
To study the platelet-tumor cell interaction in vivo, C26-GFP cells were injected into the spleen and the livers were harvested 2 hours after injection. Immunofluorescence microscopy confirmed intrasinusoidal localization of GFP-positive tumor cells (Figure 7B). Costaining with CD41 showed that approximately 52%of all green tumor cells were surrounded by platelets. These cells also showed tyrosine phosphorylation of the PDGFRB (Figure 7B). By contrast, tumor cells that were not surrounded by CD41-positive platelets were negative for PDGFRB phosphorylation.
Activation of Platelets in Complex with Tumor Cells
To investigate if tumor cell-associated platelets are still responsive to activation by ECM components, tumor cells were mixed with isolated platelets. To mimic ECM exposure, CRP was added to the mixture and flow cytometry was used to study activation of platelets on platelet-tumor cell complexes. Approximately 40% of all tumor cells were covered by platelets, of which 8% was also positive for p-Sel. Thus, ∼20% of tumor cell-bound platelets was activated (Figure 7C). After exposure to CRP, the percentage of platelet-tumor cell complexes positive for P-selectin increased to ±45% (Figure 7C). This indicates that platelets associated to tumor cells are still responsive to activation by ECM components such as collagen.
Next, we analyzed C26-GFP tumor cells arrested in mouse liver sinusoids 2 hours after intrasplenic injection for P-selectin positivity by immunofluorescence (Figure 7D). Of 208 arrested tumor cells, 105 (∼50%) were also positive for P-selectin (Figure 7D), similar to the total percentage of platelet-covered tumor cells (52%, Figure 7B). Together, these results indicate that platelets in complex with tumor cells are activated following tumor cell lodging in the sinusoids and that this may be mediated by exposure to ECM components.
Discussion
In the present report, we provide evidence that mesenchymal-like colorectal tumor cells express PDGFRB that stimulates invasion and contributes to the metastatic capacity of such cells. Mounting evidence suggests that epithelial cells have to undergo EMT to efficiently seed metastases [18,20,50,51]. In a mouse model for spontaneous metastatic pancreas carcinoma, the metastasizing tumor cells undergo EMT transiently and revert back to an epithelial phenotype in the distant organ [50]. Furthermore, recent studies in breast cancer patients have shown that a subpopulation of circulating tumor cells indeed has mesenchymal features and that high numbers of such cells predict poor prognosis [50,52–55]. The transient nature of EMTmay explain why this phenomenon is hard to detect by immunohistochemistry on human tumor tissue sections. In addition, epithelial tumor cells with mesenchymal properties are distinct from true mesenchymal cells and may be hard to discriminate morphologically from their epithelial neighbors on tissue sections. Indeed, a considerable proportion of human colorectal tumors express moderate to high levels of the mesenchymal marker vimentin in the tumor cells without appearing mesenchymal. This was associated with the presence of nuclear B-catenin, which helps drive the EMT-like process, but not with clear changes in epithelial morphology [7]. Likewise, PDGFRB expression in colorectal tumor cells has previously been documented [6]. In the current study, we show that vimentin expression is strongly associated with PDGFRB expression and that this identifies an aggressive subset of CRC tumors (Table W1). We found that high PDGFRB expression in primary tumors correlates with tumor recurrence (metastasis formation) but also that this association was not found in already established metastases. Therefore, PDGFRB may primarily play a role in establishing distant metastases rather than in promoting the growth of established lesions.
Although the contribution of stromal cells to gene expression profiles of colorectal tumors is relatively small [56], we cannot exclude that stromal cells have contributed to PDGFRB expression in our analyses of the human tumor cohorts. Whatever that contribution may be, there is no doubt that EMT of tumor cells from diverse epithelial origins generates PDGFRB-expressing tumor cells with mesenchymal properties [12–14,16,18,24] (this study). Importantly, such cells become dependent on PDGFR signaling for efficient metastasis formation [18,24]. Expression of the PDGFRB in epithelial tumor cells is stimulated by TGFB [18,24] and by the EMT-driving transcription factor SNAIL [57]. Indeed, PDGFRB expression in CRC was strongly correlated with both TGFB signaling and with key EMT-driving transcription factors. Furthermore, inhibition of TGFB signaling reduced PDGFRB signaling in mesenchymal-like CRC cells (this study) and suppressed metastasis formation [49]. Vice versa, restoration of TGFB receptor signaling in human epithelial colorectal tumor cells with a mutation in TGFBR2 greatly enhanced tumor cell invasion [49].
EMT induced by TGFB or SNAIL results in enhanced expression of PDGFRB and ECM genes including collagens and fibronectin [57–59]. ECM deposition promotes platelet activation, which stimulates the metastatic process [32–34]. However, it is less clear during which steps in the metastatic cascade platelets may play a role. Most of the available evidence supports a role for platelets during initial metastasis establishment at distant sites. Microthrombi containing activated platelets are formed around tumor cells that are trapped in the microvasculature of the liver or the lungs [26–30] (this study). This may help tumor cell extravasation and/or early establishment of novel metastatic lesions. Interestingly, at least two key components of the metastatic niche, fibronectin and tenascin-C, are also potent inducers of platelet activation [60–63]. Furthermore, both ECM components are co-expressed with PDGFRB in CRC (Table W1). Platelet activation and subsequent EMT may also help tumor cells to detach from the primary tumor to disseminate.
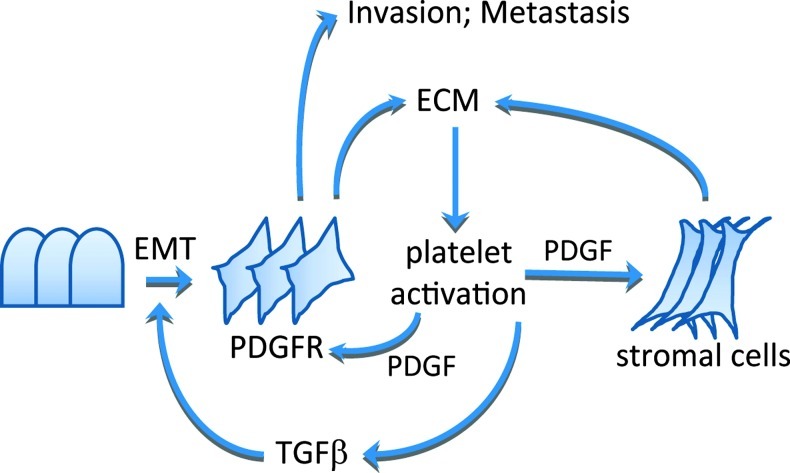
We propose that aggressive colorectal tumors with high expression of PDGFRB and EMT genes may sustain their phenotype by high-level matrix deposition resulting in an increased propensity for platelet activation. Activated platelets release TGFB that subsequently promotes EMT and PDGFR signaling in tumor and stromal cells (Figure 8). This self-sustaining series of events may operate both within the primary tumor and at distant sites.
Figure 8.
Working model for PDGFR signaling in metastatic CRC. Tumors with high ECM content, including collagens, fibronectin, and tenascin-C, are prone to platelet activation. Activated platelets produce TGFB that stimulates EMT and PDGFR expression in tumor cells. Platelets also produce PDGF that can then stimulate tumor cells and stromal cells in a paracrine fashion. Mesenchymal-like tumor cells are invasion-prone and have a relatively high capacity to seed distant metastases.
Supplementary Material
Abbreviations
- CRC
colorectal cancer
- CRP
collagen-related peptide
- DAPI
4′,6-diamidino-2-phenylindole
- DFS
disease-free survival
- ECM
extracellular matrix
- EGFR
epithelial growth factor receptor
- EMT
epithelial-to-mesenchymal transition
- FCS
fetal calf serum
- GFP
green fluorescent protein
- HRA
hepatic replacement area
- OS
overall survival
- PDGF
platelet-derived growth factor
- PDGFR
platelet-derived growth factor receptor
- TGFBR
transforming growth factor beta receptor
- TRAP
thrombin receptor-activating peptide
Footnotes
This work was supported by grants from the Dutch Cancer Society (E.J.A.S.; 2009-4379/D.A.R.; 2009-4417/B.L.E.; 2009-4367/K.M.G.; 2010-4608/N.S.; 2007-3923), the PON Foundation, and the Netherlands Organization for Scientific Research (C.M.; 016-126-159). Conflict of interest: None.
This article refers to supplementary materials, which are designated by Table W1 and Figures W1 to W7 and are available online at www.neoplasia.com.
References
- 1.Ostman A, Heldin CH. PDGF receptors as targets in tumor treatment. Adv Cancer Res. 2007;97:247–274. doi: 10.1016/S0065-230X(06)97011-0. [DOI] [PubMed] [Google Scholar]
- 2.Heldin CH, Westermark B. Mechanism of action and in vivo role of platelet-derived growth factor. Physiol Rev. 1999;79:1283–1316. doi: 10.1152/physrev.1999.79.4.1283. [DOI] [PubMed] [Google Scholar]
- 3.Steller EJ, Ritsma L, Raats DA, Hoogwater FJ, Emmink BL, Govaert KM, Laoukili J, Borel Rinkes IH, van Rheenen J, Kranenburg O. The death receptor CD95 activates the cofilin pathway to stimulate tumour cell invasion. EMBO Rep. 2011;12:931–937. doi: 10.1038/embor.2011.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lindmark G, Sundberg C, Glimelius B, Pahlman L, Rubin K, Gerdin B. Stromal expression of platelet-derived growth factor beta-receptor and platelet-derived growth factor B-chain in colorectal cancer. Lab Invest. 1993;69:682–689. [PubMed] [Google Scholar]
- 5.Sundberg C, Ljungstrom M, Lindmark G, Gerdin B, Rubin K. Microvascular pericytes express platelet-derived growth factor-β receptors in human healing wounds and colorectal adenocarcinoma. Am J Pathol. 1993;143:1377–1388. [PMC free article] [PubMed] [Google Scholar]
- 6.Wehler TC, Frerichs K, Graf C, Drescher D, Schimanski K, Biesterfeld S, Berger MR, Kanzler S, Junginger T, Galle PR, et al. PDGFRα/β expression correlates with the metastatic behavior of human colorectal cancer: a possible rationale for a molecular targeting strategy. Oncol Rep. 2008;19:697–704. [PubMed] [Google Scholar]
- 7.Veenendaal LM, Kranenburg O, Smakman N, Klomp A, Borel Rinkes IH, van Diest PJ. Differential Notch and TGFbeta signaling in primary colorectal tumors and their corresponding metastases. Cell Oncol. 2008;30:1–11. doi: 10.1155/2008/839076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hwang RF, Yokoi K, Bucana CD, Tsan R, Killion JJ, Evans DB, Fidler IJ. Inhibition of platelet-derived growth factor receptor phosphorylation by STI571 (Gleevec) reduces growth and metastasis of human pancreatic carcinoma in an orthotopic nude mouse model. Clin Cancer Res. 2003;9:6534–6544. [PubMed] [Google Scholar]
- 9.Maass T, Thieringer FR, Mann A, Longerich T, Schirmacher P, Strand D, Hansen T, Galle PR, Teufel A, Kanzler S. Liver specific overexpression of platelet-derived growth factor-B accelerates liver cancer development in chemically induced liver carcinogenesis. Int J Cancer. 2011;128:1259–1268. doi: 10.1002/ijc.25469. [DOI] [PubMed] [Google Scholar]
- 10.Seymour L, Bezwoda WR. Positive immunostaining for platelet derived growth factor (PDGF) is an adverse prognostic factor in patients with advanced breast cancer. Breast Cancer Res Treat. 1994;32:229–233. doi: 10.1007/BF00665774. [DOI] [PubMed] [Google Scholar]
- 11.Kitadai Y, Sasaki T, Kuwai T, Nakamura T, Bucana CD, Hamilton SR, Fidler IJ. Expression of activated platelet-derived growth factor receptor in stromal cells of human colon carcinomas is associated with metastatic potential. Int J Cancer. 2006;119:2567–2574. doi: 10.1002/ijc.22229. [DOI] [PubMed] [Google Scholar]
- 12.Campbell CI, Moorehead RA. Mammary tumors that become independent of the type I insulin-like growth factor receptor express elevated levels of platelet-derived growth factor receptors. BMC Cancer. 2011;11:480. doi: 10.1186/1471-2407-11-480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eckert MA, Lwin TM, Chang AT, Kim J, Danis E, Ohno-Machado L, Yang J. Twist1-induced invadopodia formation promotes tumor metastasis. Cancer Cell. 2011;19:372–386. doi: 10.1016/j.ccr.2011.01.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fischer AN, Fuchs E, Mikula M, Huber H, Beug H, Mikulits W. PDGF essentially links TGF-β signaling to nuclear β-catenin accumulation in hepatocellular carcinoma progression. Oncogene. 2007;26:3395–3405. doi: 10.1038/sj.onc.1210121. [DOI] [PubMed] [Google Scholar]
- 15.Jechlinger M, Grunert S, Tamir IH, Janda E, Ludemann S, Waerner T, Seither P, Weith A, Beug H, Kraut N. Expression profiling of epithelial plasticity in tumor progression. Oncogene. 2003;22:7155–7169. doi: 10.1038/sj.onc.1206887. [DOI] [PubMed] [Google Scholar]
- 16.Lahsnig C, Mikula M, Petz M, Zulehner G, Schneller D, van Zijl F, Huber H, Csiszar A, Beug H, Mikulits W. ILEI requires oncogenic Ras for the epithelial to mesenchymal transition of hepatocytes and liver carcinoma progression. Oncogene. 2009;28:638–650. doi: 10.1038/onc.2008.418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Smith CL, Baek ST, Sung CY, Tallquist MD. Epicardial-derived cell epithelial-to-mesenchymal transition and fate specification require PDGF receptor signaling. Circ Res. 2011;108:e15–e26. doi: 10.1161/CIRCRESAHA.110.235531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thomson S, Petti F, Sujka-Kwok I, Epstein D, Haley JD. Kinase switching in mesenchymal-like non-small cell lung cancer lines contributes to EGFR inhibitor resistance through pathway redundancy. Clin Exp Metastasis. 2008;25:843–854. doi: 10.1007/s10585-008-9200-4. [DOI] [PubMed] [Google Scholar]
- 19.Thomson S, Petti F, Sujka-Kwok I, Mercado P, Bean J, Monaghan M, Seymour SL, Argast GM, Epstein DM, Haley JD. A systems view of epithelialmesenchymal transition signaling states. Clin Exp Metastasis. 2011;28:137–155. doi: 10.1007/s10585-010-9367-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kalluri R, Weinberg RA. The basics of epithelial-mesenchymal transition. J Clin Invest. 2009;119:1420–1428. doi: 10.1172/JCI39104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cristofanilli M, Hayes DF, Budd GT, Ellis MJ, Stopeck A, Reuben JM, Doyle GV, Matera J, Allard WJ, Miller MC, et al. Circulating tumor cells: a novel prognostic factor for newly diagnosed metastatic breast cancer. J Clin Oncol. 2005;23:1420–1430. doi: 10.1200/JCO.2005.08.140. [DOI] [PubMed] [Google Scholar]
- 22.Gradilone A, Raimondi C, Nicolazzo C, Petracca A, Gandini O, Vincenzi B, Naso G, Agliano AM, Cortesi E, Gazzaniga P. Circulating tumour cells lacking cytokeratin in breast cancer: the importance of being mesenchymal. J Cell Mol Med. 2011;15:1066–1070. doi: 10.1111/j.1582-4934.2011.01285.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Raimondi C, Gradilone A, Naso G, Vincenzi B, Petracca A, Nicolazzo C, Palazzo A, Saltarelli R, Spremberg F, Cortesi E, et al. Epithelial-mesenchymal transition and stemness features in circulating tumor cells from breast cancer patients. Breast Cancer Res Treat. 2011;130:449–455. doi: 10.1007/s10549-011-1373-x. [DOI] [PubMed] [Google Scholar]
- 24.Jechlinger M, Sommer A, Moriggl R, Seither P, Kraut N, Capodiecci P, Donovan M, Cordon-Cardo C, Beug H, Grunert S. Autocrine PDGFR signaling promotes mammary cancer metastasis. J Clin Invest. 2006;116:1561–1570. doi: 10.1172/JCI24652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Labelle M, Begum S, Hynes RO. Direct signaling between platelets and cancer cells induces an epithelial-mesenchymal-like transition and promotes metastasis. Cancer Cell. 2011;20:576–590. doi: 10.1016/j.ccr.2011.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Erpenbeck L, Nieswandt B, Schon M, Pozgajova M, Schon MP. Inhibition of platelet GPIbα and promotion of melanoma metastasis. J Invest Dermatol. 2010;130:576–586. doi: 10.1038/jid.2009.278. [DOI] [PubMed] [Google Scholar]
- 27.Im JH, Fu W, Wang H, Bhatia SK, Hammer DA, Kowalska MA, Muschel RJ. Coagulation facilitates tumor cell spreading in the pulmonary vasculature during early metastatic colony formation. Cancer Res. 2004;64:8613–8619. doi: 10.1158/0008-5472.CAN-04-2078. [DOI] [PubMed] [Google Scholar]
- 28.Kawaguchi T, Nakamura K. Analysis of the lodgement and extravasation of tumor cells in experimental models of hematogenous metastasis. Cancer Metastasis Rev. 1986;5:77–94. doi: 10.1007/BF00046424. [DOI] [PubMed] [Google Scholar]
- 29.Kim YJ, Borsig L, Varki NM, Varki A. P-selectin deficiency attenuates tumor growth and metastasis. Proc Natl Acad Sci USA. 1998;95:9325–9330. doi: 10.1073/pnas.95.16.9325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tanaka K, Morimoto Y, Toiyama Y, Okugawa Y, Inoue Y, Uchida K, Kimura K, Mizoguchi A, Kusunoki M. Intravital dual-colored visualization of colorectal liver metastasis in living mice using two photon laser scanning microscopy. Microsc Res Tech. 2012;75:307–315. doi: 10.1002/jemt.21059. [DOI] [PubMed] [Google Scholar]
- 31.Kim YJ, Borsig L, Han HL, Varki NM, Varki A. Distinct selectin ligands on colon carcinoma mucins can mediate pathological interactions among platelets, leukocytes, and endothelium. Am J Pathol. 1999;155:461–472. doi: 10.1016/S0002-9440(10)65142-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gay LJ, Felding-Habermann B. Contribution of platelets to tumour metastasis. Nat Rev Cancer. 2011;11:123–134. doi: 10.1038/nrc3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Karpatkin S, Pearlstein E, Ambrogio C, Coller BS. Role of adhesive proteins in platelet tumor interaction in vitro and metastasis formation in vivo. J Clin Invest. 1988;81:1012–1019. doi: 10.1172/JCI113411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pearlstein E, Ambrogio C, Karpatkin S. Effect of antiplatelet antibody on the development of pulmonary metastases following injection of CT26 colon adenocarcinoma, Lewis lung carcinoma, and B16 amelanotic melanoma tumor cells into mice. Cancer Res. 1984;44:3884–3887. [PubMed] [Google Scholar]
- 35.Lin MS, Huang JX, Zhu J, Shen HZ. Elevation of platelet count in patients with colorectal cancer predicts tendency to metastases and poor prognosis. Hepatogastroenterology. 2012;59:1687–1690. doi: 10.5754/hge12277. [DOI] [PubMed] [Google Scholar]
- 36.Sasaki K, Kawai K, Tsuno NH, Sunami E, Kitayama J. Impact of preoperative thrombocytosis on the survival of patients with primary colorectal cancer. World J Surg. 2012;36:192–200. doi: 10.1007/s00268-011-1329-7. [DOI] [PubMed] [Google Scholar]
- 37.Jorissen RN, Gibbs P, Christie M, Prakash S, Lipton L, Desai J, Kerr D, Aaltonen LA, Arango D, Kruhoffer M, et al. Metastasis-associated gene expression changes predict poor outcomes in patients with Dukes stage B and C colorectal cancer. Clin Cancer Res. 2009;15:7642–7651. doi: 10.1158/1078-0432.CCR-09-1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Smith JJ, Deane NG, Wu F, Merchant NB, Zhang B, Jiang A, Lu P, Johnson JC, Schmidt C, Bailey CE, et al. Experimentally derived metastasis gene expression profile predicts recurrence and death in patients with colon cancer. Gastroenterology. 2010;138:958–968. doi: 10.1053/j.gastro.2009.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Snoeren N, van Hooff SR, Adam R, van Hillegersberg R, Voest EE, Guettier C, van Diest PJ, Nijkamp MW, Brok MO, van Leenen D, et al. Exploring gene expression signatures for predicting disease free survival after resection of colorectal cancer liver metastases. PLoS One. 2012;7:e49442. doi: 10.1371/journal.pone.0049442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bewick V, Cheek L, Ball J. Statistics review 12: survival analysis. Crit Care. 2004;8:389–394. doi: 10.1186/cc2955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Smakman N, Martens A, Kranenburg O, Borel Rinkes IH. Validation of bioluminescence imaging of colorectal liver metastases in the mouse. J Surg Res. 2004;122:225–230. doi: 10.1016/j.jss.2004.05.021. [DOI] [PubMed] [Google Scholar]
- 42.Knight CG, Morton LF, Onley DJ, Peachey AR, Ichinohe T, Okuma M, Farndale RW, Barnes MJ. Collagen-platelet interaction: Gly-Pro-Hyp is uniquely specific for platelet Gp VI and mediates platelet activation by collagen. Cardiovasc Res. 1999;41:450–457. doi: 10.1016/s0008-6363(98)00306-x. [DOI] [PubMed] [Google Scholar]
- 43.Korporaal SJ, Van Eck M, Adelmeijer J, Ijsseldijk M, Out R, Lisman T, Lenting PJ, Van Berkel TJ, Akkerman JW. Platelet activation by oxidized low density lipoprotein is mediated by CD36 and scavenger receptor-A. Arterioscler Thromb Vasc Biol. 2007;27:2476–2483. doi: 10.1161/ATVBAHA.107.150698. [DOI] [PubMed] [Google Scholar]
- 44.Kranenburg O, Verlaan I, Moolenaar WH. Regulating c-Ras function: cholesterol depletion affects caveolin association, GTP loading, and signaling. Curr Biol. 2001;11:1880–1884. doi: 10.1016/s0960-9822(01)00582-6. [DOI] [PubMed] [Google Scholar]
- 45.Drixler TA, Borel Rinkes IH, Ritchie ED, van Vroonhoven TJ, Gebbink MF, Voest EE. Continuous administration of angiostatin inhibits accelerated growth of colorectal liver metastases after partial hepatectomy. Cancer Res. 2000;60:1761–1765. [PubMed] [Google Scholar]
- 46.te Velde EA, Vogten JM, Gebbink MF, van Gorp JM, Voest EE, Borel Rinkes. Enhanced antitumour efficacy by combining conventional chemotherapy with angiostatin or endostatin in a liver metastasis model. Br J Surg IH. 2002;89:1302–1309. doi: 10.1046/j.1365-2168.2002.02183.x. [DOI] [PubMed] [Google Scholar]
- 47.van der Bilt JD, Kranenburg O, Nijkamp MW, Smakman N, Veenendaal LM, te Velde EA, Voest EE, van Diest PJ, Borel Rinkes IH. Ischemia/reperfusion accelerates the outgrowth of hepatic micrometastases in a highly standardized murine model. Hepatology. 2005;42:165–175. doi: 10.1002/hep.20739. [DOI] [PubMed] [Google Scholar]
- 48.Peinado H, Olmeda D, Cano A. Snail, Zeb and bHLH factors in tumour progression: an alliance against the epithelial phenotype? Nat Rev Cancer. 2007;7:415–428. doi: 10.1038/nrc2131. [DOI] [PubMed] [Google Scholar]
- 49.Oft M, Heider KH, Beug H. TGFβ signaling is necessary for carcinoma cell invasiveness and metastasis. Curr Biol. 1998;8:1243–1252. doi: 10.1016/s0960-9822(07)00533-7. [DOI] [PubMed] [Google Scholar]
- 50.Rhim AD, Mirek ET, Aiello NM, Maitra A, Bailey JM, McAllister F, Reichert M, Beatty GL, Rustgi AK, Vonderheide RH, et al. EMT and dissemination precede pancreatic tumor formation. Cell. 2012;148:349–361. doi: 10.1016/j.cell.2011.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Siegel PM, Massague J. Cytostatic and apoptotic actions of TGF-β in homeostasis and cancer. Nat Rev Cancer. 2003;3:807–821. doi: 10.1038/nrc1208. [DOI] [PubMed] [Google Scholar]
- 52.Aktas B, Tewes M, Fehm T, Hauch S, Kimmig R, Kasimir-Bauer S. Stem cell and epithelial-mesenchymal transition markers are frequently overexpressed in circulating tumor cells of metastatic breast cancer patients. Breast Cancer Res. 2009;11:R46. doi: 10.1186/bcr2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hou JM, Krebs MG, Lancashire L, Sloane R, Backen A, Swain RK, Priest LJ, Greystoke A, Zhou C, Morris K, et al. Clinical significance and molecular characteristics of circulating tumor cells and circulating tumor microemboli in patients with small-cell lung cancer. J Clin Oncol. 2012;30:525–532. doi: 10.1200/JCO.2010.33.3716. [DOI] [PubMed] [Google Scholar]
- 54.Kallergi G, Papadaki MA, Politaki E, Mavroudis D, Georgoulias V, Agelaki S. Epithelial to mesenchymal transition markers expressed in circulating tumour cells of early andmetastatic breast cancer patients. Breast Cancer Res. 2011;13:R59. doi: 10.1186/bcr2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bonnomet A, Syne L, Brysse A, Feyereisen E, Thompson EW, Noel A, Foidart JM, Birembaut P, Polette M, Gilles C. A dynamic in vivo model of epithelial-to-mesenchymal transitions in circulating tumor cells and metastases of breast cancer. Oncogene. 2011;31:3741–3753. doi: 10.1038/onc.2011.540. [DOI] [PubMed] [Google Scholar]
- 56.de Bruin EC, van de Pas S, Lips EH, van Eijk R, van der Zee MM, Lombaerts M, van Wezel T, Marijnen CA, van Krieken JH, Medema JP, et al. Macrodissection versus microdissection of rectal carcinoma: minor influence of stroma cells to tumor cell gene expression profiles. BMC Genomics. 2005;6:142. doi: 10.1186/1471-2164-6-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Argast GM, Krueger JS, Thomson S, Sujka-Kwok I, Carey K, Silva S, O'Connor M, Mercado P, Mulford IJ, Young GD, et al. Inducible expression of TGFβ, snail and Zeb1 recapitulates EMT in vitro and in vivo in a NSCLC model. Clin Exp Metastasis. 2011;28:593–614. doi: 10.1007/s10585-011-9394-8. [DOI] [PubMed] [Google Scholar]
- 58.Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139:871–890. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 59.Zeisberg M, Yang C, Martino M, Duncan MB, Rieder F, Tanjore H, Kalluri R. Fibroblasts derive from hepatocytes in liver fibrosis via epithelial to mesenchymal transition. J Biol Chem. 2007;282:23337–23347. doi: 10.1074/jbc.M700194200. [DOI] [PubMed] [Google Scholar]
- 60.Kaplan RN, Riba RD, Zacharoulis S, Bramley AH, Vincent L, Costa C, MacDonald DD, Jin DK, Shido K, Kerns SA, et al. VEGFR1-positive haematopoietic bone marrow progenitors initiate the pre-metastatic niche. Nature. 2005;438:820–827. doi: 10.1038/nature04186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Oskarsson T, Acharyya S, Zhang XH, Vanharanta S, Tavazoie SF, Morris PG, Downey RJ, Manova-Todorova K, Brogi E, Massague J. Breast cancer cells produce tenascin C as a metastatic niche component to colonize the lungs. Nat Med. 2011;17:867–874. doi: 10.1038/nm.2379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schaff M, Receveur N, Bourdon C, Wurtz V, Denis CV, Orend G, Gachet C, Lanza F, Mangin PH. Novel function of tenascin-C, a matrix protein relevant to atherosclerosis, in platelet recruitment and activation under flow. Arterioscler Thromb Vasc Biol. 2011;31:117–124. doi: 10.1161/ATVBAHA.110.206375. [DOI] [PubMed] [Google Scholar]
- 63.Sleeman JP. The metastatic niche and stromal progression. Cancer Metastasis Rev. 2012;31:429–440. doi: 10.1007/s10555-012-9373-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Supplemental Reference List
- 1.Pirooznia M, Nagarajan V, Deng Y. GeneVenn—a web application for comparing gene lists using Venn diagrams. Bioinformation. 2007;1:420–422. doi: 10.6026/97320630001420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jensen LJ, Kuhn M, Stark M, Chaffron S, Creevey C, Muller J, Doerks T, Julien P, Roth A, Simonovic M, et al. STRING 8—a global view on proteins and their functional interactions in 630 organisms. Nucleic Acids Res. 2009;37:D412–D416. doi: 10.1093/nar/gkn760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jorissen RN, Gibbs P, Christie M, Prakash S, Lipton L, Desai J, Kerr D, Aaltonen LA, Arango D, Kruhoffer M, et al. Metastasis-associated gene expression changes predict poor outcomes in patients with Dukes stage B and C colorectal cancer. Clin Cancer Res. 2009;15:7642–7651. doi: 10.1158/1078-0432.CCR-09-1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.