Abstract
Experimental investigations into the effects of traumatic brain injury (TBI) have demonstrated significant alterations in dopaminergic systems. Dopaminergic fibers originating within the substantia nigra and ventral tegmental area (VTA) are important for reward learning, addiction, movement, and behavior. However, little is known about the effect of TBI on substantia nigra and VTA function. Environmental enrichment (EE) has been shown to improve functional outcome after TBI, and a number of studies suggest that it may exert some benefits via dopaminergic signaling. To better understand the role of dopamine in chronic TBI pathophysiology and the effect of EE, we examined the mRNA expression profile within the substantia nigra and VTA at 4 weeks post-injury. Specifically, three comparisons were made: 1) TBI versus sham, 2) sham+EE versus sham+standard (STD) housing, and 3) TBI+EE versus TBI+STD. There were differential expressions of 25, 4, and 40 genes in these comparisons, respectively. Chronic alterations in genes post-injury within the substantia nigra and VTA included genes important for cellular membrane homeostasis and transcription. EE-induced gene alterations after TBI included genes important for signal transduction, in particular calcium signaling pathways, membrane homeostasis, and metabolism. Elucidation of these alterations in gene expression within the substantia nigra and VTA provides new insights into chronic changes in dopamine signaling post-TBI, and the potential role of EE in TBI rehabilitation.
Key words: dopamine, EE, microarray, TBI
Introduction
Cognitive dysfunction following traumatic brain injury (TBI) has been well described in clinical settings and in animal models.1,2 Persistent deficits in learning, memory, and executive function are leading factors in continued disability among TBI survivors.3–7 Alterations in dopamine (DA) signaling may be a potential mechanism for the persistent cognitive dysfunction seen after TBI.8 Prior research has demonstrated that TBI can lead to changes in evoked DA release,9,10 altered DA transporter activity,11 and changes in DA-targeted phosphorylation systems within striatal neurons.12 Strategies to target dopaminergic (DAergic) signaling post-TBI have shown promise in animal models13–15 and clinical studies.16–21 In addition to pharmacologic manipulation, cognitive rehabilitation strategies have been employed with modest success in TBI patient populations.22,23 However, there are relatively few studies examining the effects of TBI on the substantia nigra and ventral tegmental area (VTA), the regions within the brain that have a significant number of DAergic projections to other brain structures. Alterations in either intrinsic function or afferent signaling to the substantia nigra and VTA could have widespread implications for the role of DAergic projections to these brain regions post-TBI.
Environmental enrichment (EE) can be considered an animal model correlate of clinical rehabilitation, based on its ability to facilitate motor performance, spatial learning, and memory retention after both controlled cortical impact (CCI)24–30 and fluid percussion brain injury.31,32 EE is also effective in providing functional benefits after stroke,33,34 chemical toxin induced injury,35,36 and neurodegenerative diseases.37
One of the systems where EE appears to exert its effects is through DAergic signaling. EE increases DA levels in the nucleus accumbens,38 and increases motor activity when animals are challenged by DA receptor agonists.39 Animals exposed to EE exhibit increased resistance to DA-enhancing drugs such as cocaine40 and amphetamine.41 Also, there are relatively decreased levels of striatal and cortical DA transporter in female rats exposed to EE following TBI compared with TBI controls in standard (STD) environments.9 A better understanding of the effect that EE has on DAergic signaling post-TBI could assist in designing combined treatment strategies with pharmacologic manipulation to further improve the benefits of EE.
We have previously shown that TBI chronically alters DA signaling in the striatum and substantia nigra at 4 weeks after injury.9,10,42 Although various studies indicate that EE alters DA neurotransmission, the effect of EE following TBI in substantia nigra and VTA gene expression has not been reported. Furthermore, prior microarray analyses in TBI have focused on acute-to-subacute time points within the hippocampus and cortex, which may not successfully identify targets for rehabilitation strategies and do not directly examine the DAergic system. Therefore, to correlate with our previous reports of DA signaling changes at 4 weeks after TBI, we analyzed the effects of EE on substantia nigra and VTA genes. In this study, we utilized efficiency analysis43 to identify the optimal combination of methods (data transformation, normalization, and test for differential expression) for assessing differential gene expression for each of the three comparisons described in the Methods section. We report here, for the first time, the effects of a 4 week EE paradigm on substantia nigra and VTA gene expression after TBI, utilizing efficiency analysis of microarray data and employing the J5 test for differential expression.44
Methods
Animals and surgical procedures
Twenty-seven adult male Sprague–Dawley rats (Harlan, Indianapolis, IN) weighing 300–325 g were used. Of these, 12 were used for microarray and 15 were used for protein validation. They were initially housed with a 12 h light/dark cycle and were provided food and water ad libitum. All experiments were approved by the University of Pittsburgh Institutional Animal Care and Use Committee.
CCI device
The CCI injury device, as previously described,45–47 consists of a small (1.975 cm) bore, double-acting stroke-constrained pneumatic cylinder with a 5.0 cm stroke. The cylinder is rigidly mounted on a crossbar in an angled position. The lower rod end has an impact tip with a diameter of 6 mm and the upper end is attached to the transducer core of a linear velocity displacement transducer (LVDT). The impact tip is pneumatically driven at a pre-programmed velocity, depth, and duration of tissue deformation. The velocity of the cylinder is controlled by gas pressure and measured by LVDT.
The rats received inhaled anesthesia, which initially consisted of 4% isoflurane and 2:1 N2O/O2 for induction, and then was followed by 1–1.5% isoflurane for maintenance. The rats were subsequently placed in a stereotactic frame and a parasagittal 7 mm craniectomy (AP:+4.0 mm, L:+2.8 mm) was made between lambda and bregma exposing the dura mater over the right parietal cortex. After the craniectomy, a cortical injury was delivered to a depth of 2.7 mm at an impact velocity of 4 m/sec. The cortical injury was delivered at an ∼18 degree angle, such that the impactor was perpendicular with the dural plane. Sham animals underwent the same procedures, with the exception of the impact. Core body temperature was maintained at 37±0.5°C with a homeothermic blanket during surgery.
Housing conditions (environmental manipulation)
Following surgery, and after the effects of anesthesia abated (as evidenced by free movement in the holding cage), the rats were returned to the colony where those designated for enrichment were immediately placed in specifically designed 92×78×51 cm steel wire cages. The EE cages consisted of three levels with ladders so that the animals could ambulate from one level to another, and contained various toys (e.g., balls, blocks, and tubes), nesting materials (e.g., paper towels), and ad libitum food and water.25,28 To maintain novelty, the objects were rearranged every day and changed each time the cage was cleaned, which was twice per week. Rats in STD housing conditions were placed in typical laboratory steel wire mesh cages with only food and water.
Tissue preparation and hybridization
Four weeks after TBI or sham injury, the rats were killed and tissue from the substantia nigra and VTA was collected for analysis. Briefly, after deep anesthesia with pentobarbital (Nembutal, 80–100 mg/kg; Abbott Laboratories, North Chicago, IL), the rats were decapitated and the brains were quickly removed and dissected on a chilled ice plate. Total RNA was extracted from a region containing DAergic cell bodies (bilateral substantia nigra and VTAs). Poly-A RNA was isolated by two rounds of oligo-dT-conjugated latex bead selection (Oligotex; Qiagen, Chatsworth, CA). First-strand cDNA was synthesized from 2 g of pooled poly-A RNA using the Superscript Choice System (Gibco, Grand Island, NY) with an oligo-dT primer containing the T7 RNA polymerase promoter. After second-strand synthesis, the reaction products were extracted with phenol/chloroform/isoamyl alcohol and precipitated with ethanol, and then the cDNA pellet was resuspended in 3 L diethyl pyrocarbonate (DEPC)-treated water. In vitro transcription incorporating biotinylated rCTP and rUTP was performed on 1.5 L cDNA using Bioarray high-yield RNA transcript labeling reagents (Enzo Diagnostics, Syosset, NY) following the manufacturer's instructions. Oligonucleotide expression arrays (Affymetrix neurobiology array) containing 1322 gene sequences were used to determine the transcriptional profiles.
Microarray data analysis
Differentially expressed genes were categorized into the following groups selected through an extensive literature search: channels/transporters/receptors, proteolysis, metabolism, cytoskeleton, transcription/translation, secreted/extracellular, signal transduction, membrane associated, and miscellaneous. The four different conditions (n=3 animals in each) evaluated included enriched shams (ES), enriched injured (EI), non-enriched (i.e., STD housed) injured (NI), and non-enriched shams (NS). Three different comparisons were made to assess the effects of injury and EE on substantia nigra gene expression 1) NI versus NS, 2) ES versus NS, and 3) EI versus NI. For all comparisons, differentially expressed genes were identified using data analysis methods selected by efficiency analysis (EA)43 using AutoEA software.48 Efficiency analysis is an automated technique that uses data re-sampling to determine the optimal normalization, transformation, and feature selection (i.e., test for differential expression) that maximizes the reproducibility of a gene set. In doing so, one is able to identify the feature selection method that yields the most consistently repeatable results independent of gene identification, thus limiting external bias when choosing a method to determine expression significance. It is based upon the assumption that the most consistent test will provide the greatest amount of overlap at a fixed number of genes. The utility of this method has been previously demonstrated in other microarray studies.48–50
For comparison 1 (NI vs. NS), raw expression values were normalized through the additive global mean adjustment (GMA),44 and the J5 test44 was used at a threshold of 10.2 to generate the list of 86 differentially expressed genes. For comparison 2 (ES vs. NS), raw expression values were not normalized, and the J5 test44 was used at a threshold of 20.7 to generate the list of 26 differentially expressed genes. For comparison 3 (EI vs. NI), raw expression values were z-transformed within array, and the J5 test44 was used at a threshold of 14.0 to generate the list of 55 differentially expressed genes. Heat maps with hierarchical clustering were generated using Spotfire Decision Site software (TIBCO Spotfire, Somerville, MA).
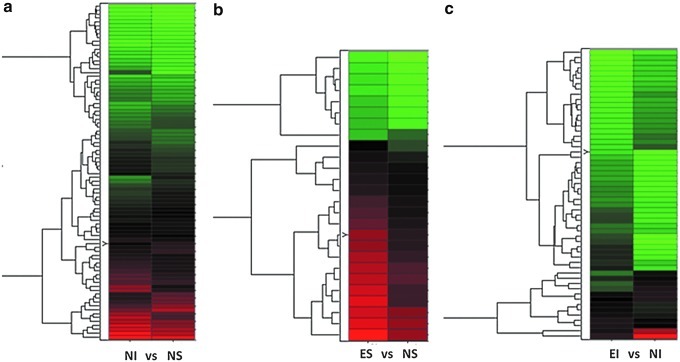
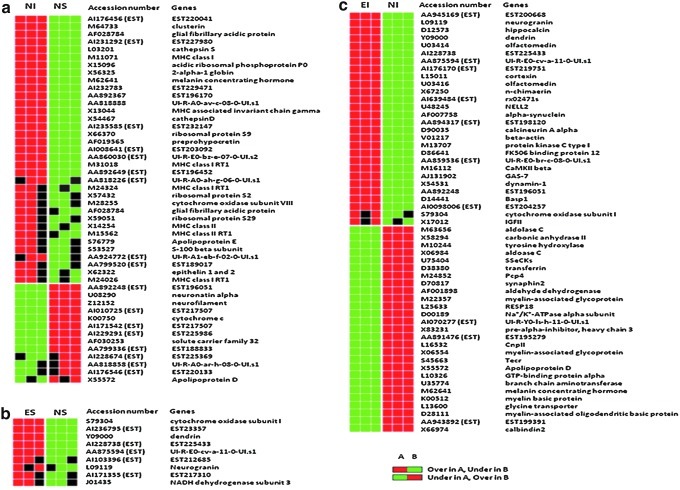
Gene expression pattern data (GEPD)44 grids were also used to prioritize the genes considered to be differentially expressed in most samples for each group-wise comparison (Fig. 1). The GEPD allows the direct visualization of the status of the genes in each sample in the microarray data. In this study, we focused on GEPD groups A and F, which included genes whose expression distributions tend to be unambiguously non-overlapping. Significantly impacted canonical Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways51 were determined for each comparison using impact analysis52 via pathway express.53
FIG. 1.
Expression pattern grid. Differentially expressed genes are displayed for non-enriched (i.e., STD housed) injured (NI) versus non-enriched shams (NS) (a), enriched shams (ES) versus NS (b), and enriched injured (EI) versus NI (c) comparisons, with each box representing the gene expression level of individual rat's tissue sample. Some genes (e.g., myelin-associated glycoprotein) are repeated more than once, and expression sequence tags are labeled as EST next to accession numbers. When an individual sample expression value is greater than the 95th percentile compared with another group, it is represented as a red box. If the expression value is less than the 5th percentile, it is represented as a green box. If the expression value is within the 5th and 95th percentile, it is represented as a black box. The associated table (Table 2) demonstrates the J5 values for each significant gene and its associated category.
Quantitative real-time polymerase chain reaction (RT-PCR)
The validity of the inference of differential expression shown by the microarray analysis was verified by RT-PCR for the expression of tyrosine hydroxylase (TH). Because of the importance of TH to DA synthesis and previous examination of the expression of TH at chronic time points,42 we chose to evaluate TH expression utilizing RT-PCR. RT-PCR was performed using SYBR Green for detection of amplification using gene-specific primers designed with PrimerExpress (ABI) software. Primer sequences were as follows:
Rat 18S rRNA forward: TTGATTAAGTCCCTGCCCTTTG
Rat 18S rRNA reverse: GATCCGAGGGCCTCACTAAAC
Rat tyrosine hydroxylase forward: CAGAGCAGGATGCCAAGCA
Rat tyrosine hydroxylase reverse: TTTACAGCCCGAGACAAGGAG
PCR amplification and data collection were performed in triplicate for each sample on an ABI PRISM 7900HT Sequence Detection System (SDS). The reactions consisted of 1× SYBR Green Universal PCR Master Mix (ABI), 250 nM forward primer, 250 nM reverse primer and 2.5 μL cDNA (RT reaction) in a total volume of 25 μL. The thermal cycling conditions for PCR were 50°C for 2 min, 95°C for 12 min, 40 cycles of 95°C for 15 sec and 60°C for 1 min. The collected data were analyzed using SDS v2.0 software (ABI).
Western blot of cytochrome c oxidase
An additional experiment was performed to validate the differential expression of cytochrome c oxidase shown by the microarray analysis by Western blot. Anesthetized rats were killed after 4 weeks of STD or EE housing following surgery (n=3–4 for each group). Brains were dissected on a chilled ice plate and the substantia nigra/VTA region was frozen in liquid nitrogen and stored in −70°C until preparation. Tissues were prepared by sonicating them in a lysis buffer consisting of 0.1M NaCl, 0.01M Tris-HCl (pH 7.6), 0.001M ethylenediaminetetraacetic acid (EDTA) (pH 8.0), 1μg/mL phenylmethylsulfonyl fluoride, phosphatase inhibitor cocktails 1 and 2 (1:100, Sigma, St. Louis, MO), and a protease inhibitor cocktail (Complete Mini, Roche Applied Science, Mannheim, Germany). The sonicated tissues were centrifuged at 13,000×g for 30 min and supernatants were used for Western blot. Using a BCA protein assay kit (Pierce, Rockford, IL), samples containing 40 μg of protein were electrophoresed on 10% sodium dodecyl sulfate (SDS)-polyacrylamide gels, transferred to polyvinylidene fluoride membranes, and blocked by 5% bovine serum albumin (BSA) (Sigma, St. Louis, MO) in 0.05 M tris buffered saline (TBS) with 0.05% Tween-20 (TBST) for 1 h. The membranes were incubated with anti-cytochrome c oxidase (1:2,500, Cell Signaling) overnight, then washed with TBST and incubated for 1 h at room temperature with anti-mouse or anti-rabbit immunoglobulin G conjugated to peroxidase (1:20,000, Pierce). Membranes were exposed to chemiluminescence (Western Lighting, Perkins Elmer, Boston, MA) and cytochrome c oxidase was visualized by exposing the membranes to autoradiographic X-ray film from 10 sec to 1 min. Afterwards, membranes were stripped using 100 mM glycine pH 2.3 at 55°C for 1 h, incubated with anti-β-actin monoclonal antibody (1:10,000, Sigma-Aldrich) for 1 h, then incubated with anti-mouse immunoglobulin G conjugated to peroxidase. The same steps were taken as described to visualize β-actin blots. To measure the optical density of Western blots, ImageJ 1.44p PC software was used.
Results
Efficiency analysis of microarray data
The data quality for each of the individual comparisons was excellent, as determined using EA (Table 1), with high between-group correlations (r>0.95), low all-gene coefficients of variation (<0.03), and low confounding indices (<1.144). For each comparison, EA43 was used to identify the optimal methods for normalization and for identifying differentially expressed genes. The optimal method varied slightly among the comparisons (Table 1). Because EA indicated the optimal threshold to maximize reproducibility of the differentially expressed gene list, no false discovery control method was applied to adjust the threshold of significance.
Table 1.
Demonstrating the Optimal Method Determined by Efficiency Analysis
| Comparison | Optimal method | r | Cov | CI | # genes |
|---|---|---|---|---|---|
| NI vs. NS | J5 (GMA) | 0.992 | 0.0 | 1.003 | 86 |
| ES vs. NS | J5 (no normalization) | 0.991 | 0.029 | 1 | 26 |
| EI vs. NI | J5 (sqrt Z) | 0.965 | 0.008 | 1.031 | 55 |
NI, non-enriched (i.e., STD housed) injured; NS, non-enriched shams; ES, enriched shams; EI, enriched injured; GMA, global mean adjustment; sqrt, square root; cov, coefficient of variation.
Differential expression (microarray) and pathway analysis
Overall, the data analysis resulted in differential expression of 167 genes in three comparisons (Fig. 2). The NI versus NS comparison showed the highest number of differential expressions (n=86), which were mostly increases in the NI group compared with the NS group. There were fewer differential expressions in the ES versus NS comparison (n=26) and EI versus NI comparison (n=55). As shown in Figure 2, the number of transcripts showing higher expression in EI compared with NI was similar to that showing higher expression in NI compared with EI. Among these, only the genes that expressed the most consistent differences (two of three in each group showing either increased or decreased expression via the GEPD) were selected for further interpretation (Fig. 1). Genes with no known function (expressed sequence tags) and repeated genes in the microarray were excluded, leaving 69 genes with major differential expressions. These 69 genes were from the three comparisons: 1) NI versus NS, 2) ES versus NS, and 3) EI versus NI. There were differential expressions in 25, 4, and 40 genes for each of the comparisons, respectively (Fig. 1). For NI versus NS, TBI increased expression of 20 genes and decreased expression of 5 (Fig. 1a). As shown in Table 2, these included genes in the following categories: two channels/transporters/receptors, two proteolysis, two cytoskeleton, four transcrption/translation, six secreted/extracellular, one signal transduction, six membrane associated, and two metabolism. In the ES versus NS comparison, EE increased the expression of four genes and did not lead to any decreases (Fig. 1b). These included genes in the following categories: one membrane associated, two metabolism, and one signal transduction. For the EI versus NI comparison, EE in the setting of TBI increased the expression of 18 genes and decreased the expression of 22 genes (Fig. 1c). These included genes in the following categories: 6 membrane-associated, 12 signal transduction, 7 metabolism, 8 secreted/extracellular, 3 channels/transporters/receptors, 1 cytoskeleton, 1 transcription/translation, and 2 miscellaneous.
FIG. 2.
Hierarchical clustering of genes differentially expressed by efficiency analysis (EA) prior to further selection. Each row represents a gene and each column shows one of the four groups: non-enriched (i.e., STD housed) injured (NI), non-enriched shams (NS), enriched injured (EI), and enriched shams (ES). Three comparisons are made: NI versus NS (a), ES versus NS (b), and EI versus NI (c). Red indicates upregulated genes and green indicates downregulated genes. In comparisons with at least one injured group there was a large number of downregulated genes compared with upregulated ones.
Table 2.
Demonstrating Each Gene that Was Significantly Different for the Separate Comparisons, its Associated Accession Number, the Category it Belongs to, and the J5 Value
|
NS>NI | |||
|---|---|---|---|
| Accession | Gene | Category | J5 Value |
| U08290 | Neuronatin alpha | Membrane associated | 20.788 |
| K00750 | Cytochrome c | Metabolism | 12.796 |
| AF030253 | Solute carrier family 32, member 1 | Channels/transporters | 12.151 |
| X55572 | Apolipoprotein D | Secreted/extracellular | 11.716 |
| Z12152 | Neurofilament | Cytoskeleton | 11.536 |
| NS<NI | |||
|---|---|---|---|
| M64733 | Clusterin | Channels/transporters | −55.772 |
| AF028784 | Glial fibrillary acidic proteins alpha and delta | Cytoskeleton | −47.374 |
| S76779 | Apolipoprotein E | Secreted/extracellular | −30.681 |
| M28255 | Cytochrome c oxidase subunit VIII | Metabolism | −26.351 |
| L03201 | Cathepsin s | Proteolysis | −25.531 |
| M11071 | MHC class I | Membrane associated | −25.289 |
| X15096 | Acidic ribosomal phosphoprotein P0 | Transcription/translation | −22.521 |
| X56325 | Hemoglobin alpha 2 chain | Secreted/extracellular | −22.504 |
| X59051 | Ribosomal protein S29 | Transcription/translation | −21.907 |
| M62641 | Melanin concentrating hormone | Secreted/extracellular | −21.422 |
| X13044 | MHC-associated invariant chain gamma | Membrane associated | −18.411 |
| X54467 | Cathepsin D | Proteolysis | −17.564 |
| S53527 | S-100 beta subunit | Secreted/extracellular | −17.402 |
| X57432 | Ribosomal protein S2 | Transcription/translation | −13.452 |
| X66370 | Ribosomal protein S9 | Transcription/translation | −13.243 |
| AF019565 | Proprohypocretin | Secreted/extracellular | −13.113 |
| X14254 | MHC class II associated invariant chain | Membrane associated | −12.989 |
| M15562 | MHC class II RT1 | Membrane associated | −12.948 |
| M24026 | MHC class I RT1 | Membrane associated | −11.365 |
| ES>NS | |||
|---|---|---|---|
| S79304 | Cytochrome oxidase subunit I | Metabolism | 34.754 |
| Y09000 | Dendrin | Membrane associated | 25.951 |
| L09119 | Neurogranin | Signal transduction | 25.945 |
| J01435#4 | NADH dehydrogenase subunit 3 | Metabolism | 25.299 |
| EI>NI | |||
|---|---|---|---|
| L09119 | Neurogranin | Signal transduction | 39.017 |
| D12573 | Hippocalcin | Signal transduction | 38.083 |
| Y09000 | Dendrin | Membrane associated | 37.107 |
| U03414 | Neuronal olfactomedin | Secreted/extracellular | 34.408 |
| L15011 | Cortexin | Membrane associated | 22.785 |
| X67250 | n-chimaerin | Channels/transporters/receptors | 22.018 |
| U48245 | NELL2 | Secreted/extracellular | 21.028 |
| AF007758 | α-synuclein | Misc | 20.847 |
| D90035 | Calcineurin alpha | Signal transduction | 18.931 |
| V01217 | β-actin | Cytoskeleton | 17.841 |
| M13707 | PKC type 1 | Signal transduction | 17.259 |
| X17012 | IGF-2 | Secreted/extracellular | 16.852 |
| D86641 | FK506 binding protein 12 | Signal transduction | 16.78 |
| M16112 | CaMKIIβ | Signal transduction | 15.989 |
| AJ131902 | Gas7 | Transcription/translation | 15.492 |
| X54531 | Dynamin-1 | Signal transduction | 15.204 |
| S79304 | Cytochrome oxidase subunit I | Metabolism | 14.855 |
| D14441 | Basp1 | Signal transduction | 14.012 |
| EI<NI | |||
|---|---|---|---|
| K00512 | Myelin basic protein (Mbp) | Membrane associated | −39.44 |
| D38380 | Transferrin | Secreted/extracellular | −35.385 |
| D28111 | Myelin-associated oligodendrocytic basic protein | Membrane associated | −32.066 |
| M63656 | Aldolase C | Metabolism | −26.88 |
| M24852 | Purkinje cell protein 4 | Signal transduction | −22.492 |
| M10244 | Tyrosine hydroxylase | Metabolism | −21.464 |
| D00189 | Na/K ATPase alpha subunit | Channels/transporters/receptors | −20.845 |
| X55572 | Apolipoprotein D | Secreted/extracellular | −19.468 |
| X66974 | Calbindin2 | Signal transduction | −18.173 |
| X58294 | Carbonic anhydrase II | Metabolism | −17.451 |
| U75404 | Src suppressed c kinase substrate | Signal transduction | −17.024 |
| D70817 | Synaphin | Misc | −16.651 |
| AF001898 | Aldehyde dehydrogenase | Metabolism | −16.427 |
| M22357 | Myelin associated glycoprotein | Membrane associated | −16.397 |
| L25633 | Neuroendocrine-specific protein | Secreted/extracellular | −16.344 |
| X83231 | Inter-alpha trypsin inhibitor, heavy chain 3 | Secreted/extracellular | −15.67 |
| L16532 | 2’,3’-cyclic nucleotide 3’-phosphodiesterase | Membrane associated | −15.567 |
| S45663 | Trans-2,3-enoyl-CoA reductase | Metabolism | −15.413 |
| L10326 | GTP-binding protein alpha subunit | Signal transduction | −15.071 |
| U35774 | Branched chain aminotransferase 1 | Metabolism | −14.953 |
| M62641 | Pro-melanin-concentrating hormone | Secreted/extracellular | −14.932 |
| L13600 | Solute carrier family 6 | Channels/transporters/receptors | −14.569 |
ES, environmental enrichment sham; EI, environmental enrichment injured; NS, standard housing sham; NI, standard housing injured; MHC, major histocompatibility complex; PKC, protein kinase C; IGF-2, insulin-like growth factor 2; CaMKII, calcium/calmodulin dependent protein kinase II; Gas7, growth arrest-specific protein 7; Basp1, brain abundant membrane attached signal protein 1; GTP, Guanosine-5′-triphosphate.
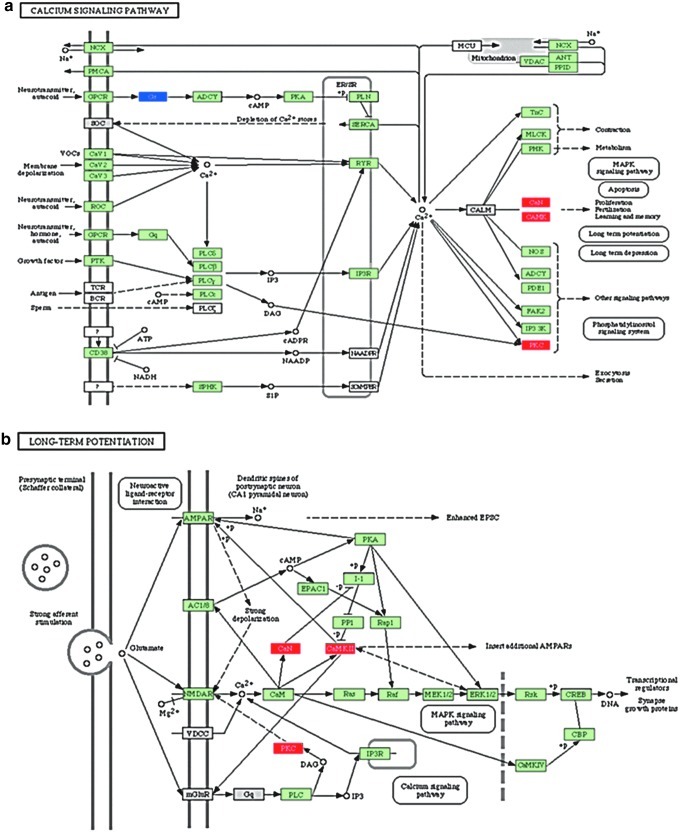
Pathway analysis identified interactions of various genes among multiple pathways, showing the functional complexity of these differentially expressed genes. For example, we found several genes in the calcium signaling pathway that were upregulated in the EI versus NI comparison to have various downstream functions affected for mitogen-activated protein kinase (MAPK) signaling, phospotidylinositol signaling, and apoptosis (Fig. 3a). These same genes are also activated as a part of long-term potentiation (LTP) (Fig. 3b).
FIG. 3.
Selected pathways from enriched injured (EI) versus non-enriched (i.e., STD housed) injured (NI): demonstrating calcium signaling importance. Calcium signaling pathway (a). Upregulation of calcineurin (CaN), calcium/calmodulin dependent protein kinase II (CaMK), and protein kinase c (PKC) is shown. There are downstream contributions to multiple functions such as apoptosis, MAPK signaling, and phosphatidylinositol signaling. The same components are also affected in the long-term potentiation pathways (b).
Differential expression validation: quantitative PCR and Western blot
Increased TH expression occurs in the substantia nigra and striatum at 4 weeks post-TBI.42 TH acts as the rate-limiting enzyme for the conversion of the amino acid l-tyrosine to dihydroxyphenylalanine (DOPA), which is subsequently converted to DA. Given the importance of the TH to DA signaling, we examined its mRNA expression with PCR as well as the microarray analysis in the EI versus the NI group. The PCR analysis confirmed that in the EI group, there was a downregulation of TH relative to NI. RT-PCR of all rats in triplicate demonstrated that a greater number of cycles were needed to amplify the RNA of the EI group to reach threshold compared with the RNA of the NI group (16.96±0.090 vs. 11.04±0.091; p<0.001). In STD animals, TBI produced an increase in the protein expression of cytochrome c oxidase to 230±72% of sham levels. Protein expression levels of cytochrome c oxidase after EE increased to 211±29% of sham expression levels. EE increased the expression of cytochrome c oxidase in both the sham and injured groups by 386±53% and 153.0±41%, respectively, relative to their corresponding STD housed group.
Discussion
Prior studies investigating alterations in gene expression in the hippocampus and cortex have shown that TBI affects genes responsible for oxidative stress, metabolism, inflammation, cell structure, and cellular signaling.54–56 The majority of these studies have investigated acute and/or subacute expression changes after TBI in an effort to better understand primary injury and rapidly evolving secondary injuries such as apoptosis and inflammation.57,58 Here, we have identified alterations in gene expression post-TBI in substantia nigra/VTA tissue that assists in a better understanding of the chronic alterations in DAergic signaling that occur after TBI. Concurrent analysis of EE afforded the identification of important pathways of cellular function within the substantia nigra/VTA that may contribute to functional recovery. The findings from the substantia nigra tissue at 4 weeks post-injury demonstrate that TBI induces chronic alterations in areas of the brain distant from the site of impact similar to previously reported acute changes.59
NI versus NS
The first comparison performed was between TBI and sham animals in STD housing (i.e., not enriched). There was an upregulation of 20 genes and a downregulation of 5 genes in TBI rats compared with shams. A notable increase in mRNA expression was found in five major histocompatibility complex (MHC) proteins: MHC class II associated invariant chain, MHC class II RT1, MHC class I RT1, MHC class I, and MHC-associated invariant chain gamma. This change may indicate that there are chronic immunological modifications that could explain the sensitivity of the substantia nigra to toxic events post-TBI. However, there was no chronic upregulation in genes related to inflammatory cytokines such as IL-1, IL-6, IFN-γ, and TNFα.
Increased expression for three ribosomal proteins S9, S2, and S29 was also identified after TBI. As these proteins are subunits of ribosome, these increases may indicate an increase in protein synthesis to accommodate numerous cellular process involved in repair and remodeling of the neuronal network. The expression of 18s ribosomal gene, that was used as our quantitative PCR (qPCR) housekeeping gene, did not differ between our experimental groups.
Cytochrome c oxidase subunit VIII is part of complex IV in the electron transport chain of mitochondria. There is an upregulation of cytochrome c oxidase subunit VIII after TBI, indicating alterations in mitochondrial respiratory chain function, as it is a terminal enzyme of the respiratory chain regulating proton gradient across mitochondrial membrane. Cytochrome c oxidase protein levels were also found to be increased after TBI. Gene expression of cytochrome c, another essential component of the electron transport chain, is reduced following brain trauma. Because cytochrome c also functions as an apoptogenic factor, triggered by pro-apoptotic factors such as Bcl-2–associated X protein (Bax),60 downregulation of cytochrome c may indicate a change in apoptotic signaling within the substantia nigra. The effect of increased cytochrome c oxidase subunit VIII and decreased cytochrome c on mitochrondrial function is unclear, and requires future investigation, particularly given the importance of mitochondrial function to disease pathology within the DAergic system.61
ES versus NS
A second analysis involved comparison of STD and EE sham animals. There was an upregulation of four genes in the ES versus NS comparison. These included cytochrome oxidase subunit I, NADH dehydrogenase subunit 3, dendrin, and neurogranin. The increased expression of cytochrome oxidase subunit I and NADH dehydrogenase subunit 3 demonstrated alterations in substantia nigra mitochondrial function following EE. In EE shams, NADH dehydrogenase subunit 3, and mitochondrial complex I gene were upregulated. Similar to the previous explanations for cytochrome c oxidase, complex I and NADH dehydrogenase are important for energy metabolism, and increases in their expression may reflect increased metabolic activity induced by EE stimulation. Dendrin is a protein of unknown function, but its mRNA is localized in dendritic spines,62 where it interacts with cytoskeletal components in order to modulate structural changes.63 Neurogranin is a small protein expressed highly in dendritic spines, implicated in regulation of cytoskeletal architecture,64 and may also have an important role in learning and memory.65 These changes are consistent with increased energy demand and morphological changes of neural connections occurring in the setting of EE.
EI versus NI
In TBI animals, EE induced the upregulation of 18 genes and the downregulation of 22 genes. Three of the four upregulations in the ES versus NS comparison were repeated: cytochrome oxidase subunit I, neurogranin, and dendrin, demonstrating the robustness of neural processes induced by EE regardless of physiological or injury state. However, there were increases in 15 and decreases in 22 additional genes, demonstrating that functional improvement after EE in injured rats may result from activation of numerous pathways.
Alterations in genes involved with insulin-like growth factor 2 (IGF-2) and Src suppressed c kinase substrate (SSecKs)
Despite many studies reporting upregulation of neurotrophins such as brain-derived neurotrophic factor (BDNF), nerve growth factor (NGF), and neurotrophin-3 (NT-3) in regions such as the hippocampus, cerebral cortex, basal forebrain, and cerebellum in an EE paradigm,66,67 we did not see increases in expression of these factors in the substantia nigra. Instead, there was a genetic upregulation of IGF-2, which has been shown to induce memory consolidation and LTP.68
One interesting downregulation was in SSeCKS, a substrate of protein kinase C (PKC). Previous studies have shown that SSeCK localizes to focal contact sites in fibroblasts and binds to the cytoskeletal matrix.69 Moreover, astrocytes overexpressing SSeCKs inhibit angiogenesis, increase tight junction proteins found in endothelial cells, and reduce vascular endothelial growth factor (VEGF) expression.70 EE rats show increased VEGF expression and neurogenesis.71 Therefore, downregulation of SSeCKS may contribute to the cognitive improvements in animals exposed to EE, because of its effect on VEGF.
Upregulation of genes for differentiation, proliferation, and neurite outgrowth of neurons: olfactomedin, cortexin 1, NELL2, n-chimaerin, growth arrest-specific protein 7 (Gas7), and brain abundant membrane attached signal protein 1 (Basp1)
TBI rats exposed to EE also had upregulation of various genes that are highly expressed in the embryonic nervous system, such as olfactomedin, cortexin1, NELL2, and n-chimaerin. These genes have important roles in neural development, promoting differentiation, proliferation, and emigration of neurons.72–76
Similar to n-chimaerin, two other genes that can promote neurite outgrowth were also upregulated: Gas7 and Basp1. Gas7 has an important role in promoting neurite outgrowth.77–79 Similarly, Basp1 promotes neurite outgrowth80 and axonal regeneration after damage.81 Increased expression of genes implicated for differentiation, proliferation, and neurite outgrowth is consistent with the possibility that EE leads to the reversion to ontogenetic functions for reorganization.82,83
Upregulation of genes important for calcium signaling: PKC, calcium/calmodulin-dependent protein kinase II (CaMKII), calcineurin (CaN)
There were three genes directly regulating the calcium signaling pathway that were upregulated after EE in the setting of TBI compared with STD: PKC, CaMKII, and CaN α. Intracellular calcium can activate PKC and CaMKII, which leads to activation of pathways leading to LTP.
LTP in midbrain DA synapses depends on glutamate receptors in the category of N-methyl-d-aspartate receptors (NMDA-Rs).84 NMDA-Rs induce intracellular calcium increase, and in addition to activation of diacylglycerol, can lead to PKC activation leading to LTP (Fig. 3). As shown, PKC can, in turn, enhance NMDA-R activation. Calcium influx also activates calmodulin, in turn activating CaMKII, which induces LTP by enhancing currents via α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid receptor (AMPA-R), a different category of glutamate receptors. This leads to protein synthesis and structural enlargement of the synapse. CaN, on the other hand, is activated by low intracellular increase in calcium, and induces long-term depression (LTD) by protein phosphatase 1 (PP1). Increased expression of PKC, CaMII, and CaN after exposure to EE in the setting of TBI shows that there is enhanced signaling in pathways leading to LTP as well as LTD. As both LTP and LTD are important for motor learning and memory, changes in related genes may contribute to the behavioral recovery after EE, as previously reported.26,28,47
Pharmacotherapies targeting calcium signaling pathways have shown some promise for treatment post TBI.85–87 These findings further define calcium signaling as an important potential target for TBI therapies and of potential importance for EE as well. Of particular interest is the fact that our data demonstrate calcium signaling alterations to be important even at a relatively chronic time point, suggesting that drug therapies for calcium targets could potentially play a role in both secondary injury at subacute time points and in chronic recovery post-TBI.
Alterations of genes important for dopamine signaling: α-synuclein, TH, aldehyde dehydrogenase 1 (ALDH1)
Injured rats exposed to EE showed upregulation of the α-synuclein gene. The function of α-synuclein has been implicated to function as a molecular chaperone.88 Another important function of α-synuclein is the regulation of DA synthesis by inhibition of TH89 and aromatic amino acid decarboxylase.90 Based on this finding, upregulation of α-synuclein in TBI rats exposed to EE may reduce TH function. In addition, the EI versus NI comparison demonstrated downregulation of TH mRNA, a major synthesizing enzyme for DA, which was confirmed with PCR analysis. The significant decrease in TH expression by PCR in the EI group can also be seen when compared with ES and NS. By increasing α-synuclein and decreasing TH expression, EE would, in theory, lead to a reduction of DAergic output from the substantia nigra/VTA, which has interesting implications for DAergic treatment strategies.
Along with the upregulation of genes that mediate proliferation of neurons and neurite outgrowth, mRNA changes for TH may indicate that DAergic neurons are undergoing morphological reorganization to make the DA signaling pathway more efficient. In addition to downregulation of TH, there is a downregulation of ALDH1, which is involved in the metabolism of DA. Although the metabolism of DA may be slowed, DA metabolites such as 3,4-dihydroxyphenyl acetaldehyde (DOPAL) may accumulate, which can induce oxidative stress and damage DA neurons.91,92 Because DOPAL accumulation can be detrimental to DA neurons, it is difficult to interpret if downregulation of ALDH1 gene is part of a pathological process or has a compensatory role. It is also possible that TH gene downregulation and α-synuclein gene upregulation may be compensatory events to counter the damaging effect of ALDH1 downregulation, which may be the primary change induced by EE. Future studies are needed to clarify the exact roles of multiple proteins regulating the DA signaling pathway.
A limitation of the current study is that we did not perform behavioral or histological outcome measures in the rats used in the two experiments presented here. The reason for this is based on the several previous studies from our laboratory consistently showing that EE enhances both motor and cognitive performance, attenuates hippocampal cell loss, and reduces cortical lesion volume relative to STD housing after a TBI with the same severity as that used in the current study.24–30 Hence, we opted to focus on gene expression.
Conclusion
In conclusion, we present here for the first time the effect of 4 weeks of EE on substantia nigra/VTA gene expression following TBI. Consistent with prior studies analyzing microarray data post-TBI, we have demonstrated alterations in signaling pathways important for inflammation, cell signaling, and cellular plasticity. Of particular interest were the changes in genes related to calcium signaling pathway and dopamine signaling pathway in the TBI animals exposed to EE. The calcium signaling genes have a wide overlap in many functions such as apoptosis, LTP/LTD, and MAPK signaling. These chronic changes in calcium signaling pathways indicate that this can be a viable target for pharmacological agents in future studies. Also, EE-induced downregulation of TH and upregulation of α-synuclein post-TBI is counter to the effect we would predict to be beneficial to the DAergic system. Furthermore, we also found that many neurotrophins such as BDNF, NGF, and NT-3 induced by EE in other brain regions are not altered in the substantia nigra/VTA at this 4 week time point. We demonstrated that transcriptional change induced by EE after TBI has a unique profile in the substantia nigra/VTA region. The changes we report here may be important to consider when evaluating pharmacological targets for post-TBI intervention. Future experimental analysis involving targeted therapeutic strategies is needed to verify the functional significance of the genetic alterations that we have outlined in this study, and to serve as a guide in developing novel therapeutic techniques.
Acknowledgments
This research was supported by National Institutes of Health (NIH) grants R21NS047919, P01NS030318, R01NS060672, NIH F30 grant 5F30NS067731-03, and the Pittsburgh Copeland Foundation (CED), and R01NS060005 (AEK).
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Capruso D.X. Levin H.S. Cognitive impairment following closed head injury. Neurol. Clin. 1992;10:879–893. [PubMed] [Google Scholar]
- 2.Fujimoto S.T. Longhi L. Saatman K.E. Conte V. Stocchetti N. McIntosh T.K. Motor and cognitive function evaluation following experimental traumatic brain injury. Neurosci. Biobehav. Rev. 2004;28:365–378. doi: 10.1016/j.neubiorev.2004.06.002. [DOI] [PubMed] [Google Scholar]
- 3.Levin H.S. Grossman R.G. Behavioral sequelae of closed head injury. A quantitative study. Arch. Neurol. 1978;35:720–727. doi: 10.1001/archneur.1978.00500350024005. [DOI] [PubMed] [Google Scholar]
- 4.Binder L.M. Persisting symptoms after mild head injury: a review of the postconcussive syndrome. J. Clin. Exp. Neuropsychol. 1986;8:323–346. doi: 10.1080/01688638608401325. [DOI] [PubMed] [Google Scholar]
- 5.Binder L.M. Neurobehavioral recovery after mild head injury. J. Neurosurg. 1987;67:785–787. doi: 10.3171/jns.1987.67.5.0785. [DOI] [PubMed] [Google Scholar]
- 6.Levin H.S. Goldstein F.C. High W.M., Jr. Williams D. Automatic and effortful processing after severe closed head injury. Brain Cogn. 1988;7:283–297. doi: 10.1016/0278-2626(88)90003-6. [DOI] [PubMed] [Google Scholar]
- 7.Levin H.S. High W.M. Goldstein F.C. Williams D.H. Sustained attention and information processing speed in chronic survivors of severe closed head injury. Scand. J. Rehabil. Med. Suppl. 1988;17:33–40. [PubMed] [Google Scholar]
- 8.Bales J.W. Wagner A.K. Kline A.E. Dixon C.E. Persistent cognitive dysfunction after traumatic brain injury: A dopamine hypothesis. Neurosci. Biobehav. Rev. 2009;33:981–1003. doi: 10.1016/j.neubiorev.2009.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wagner A.K. Chen X. Kline A.E. Li Y. Zafonte R.D. Dixon C.E. Gender and environmental enrichment impact dopamine transporter expression after experimental traumatic brain injury. Exp. Neurol. 2005;195:475–483. doi: 10.1016/j.expneurol.2005.06.009. [DOI] [PubMed] [Google Scholar]
- 10.Shin S.S. Bray E.R. Zhang C.Q. Dixon C.E. Traumatic brain injury reduces striatal tyrosine hydroxylase activity and potassium-evoked dopamine release in rats. Brain Res. 2011;1369:208–215. doi: 10.1016/j.brainres.2010.10.096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wagner A.K. Sokoloski J.E. Chen X. Harun R. Clossin D.P. Khan A.S. Andes–Koback M. Michael A.C. Dixon C.E. Controlled cortical impact injury influences methylphenidate-induced changes in striatal dopamine neurotransmission. J. Neurochem. 2009;110:801–810. doi: 10.1111/j.1471-4159.2009.06155.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bales J.W. Yan H.Q. Ma X. Li Y. Samarasinghe R. Dixon C.E. The dopamine and cAMP regulated phosphoprotein, 32 kDa (DARPP-32) signaling pathway: a novel therapeutic target in traumatic brain injury. Exp. Neurol. 2011;229:300–307. doi: 10.1016/j.expneurol.2011.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dixon C.E. Kraus M.F. Kline A.E. Ma X. Yan H.Q. Griffith R.G. Wolfson B.M. Marion D.W. Amantadine improves water maze performance without affecting motor behavior following traumatic brain injury in rats. Restor. Neurol. Neurosci. 1999;14:285–294. [PubMed] [Google Scholar]
- 14.Kline A.E. Yan H.Q. Bao J. Marion D.W. Dixon C.E. Chronic methylphenidate treatment enhances water maze performance following traumatic brain injury in rats. Neurosci. Lett. 2000;280:163–166. doi: 10.1016/s0304-3940(00)00797-7. [DOI] [PubMed] [Google Scholar]
- 15.Kline A.E. Massucci J.L. Ma X. Zafonte R.D. Dixon C.E. Bromocriptine reduces lipid peroxidation and enhances spatial learning and hippocampal neuron survival in a rodent model of focal brain trauma. J. Neurotrauma. 2004;21:1712–1722. doi: 10.1089/neu.2004.21.1712. [DOI] [PubMed] [Google Scholar]
- 16.Gualtieri C.T. Evans R.W. Stimulant treatment for the neurobehavioural sequelae of traumatic brain injury. Brain Inj. 1988;2:273–290. doi: 10.3109/02699058809150898. [DOI] [PubMed] [Google Scholar]
- 17.Meythaler J.M. Brunner R.C. Johnson A. Novack T.A. Amantadine to improve neurorecovery in traumatic brain injury-associated diffuse axonal injury: a pilot double-blind randomized trial. J. Head Trauma Rehabil. 2002;17:300–313. doi: 10.1097/00001199-200208000-00004. [DOI] [PubMed] [Google Scholar]
- 18.Whyte J. Hart T. Vaccaro M. Grieb–Neff P. Risser A. Polansky M. Coslett H.B. Effects of methylphenidate on attention deficits after traumatic brain injury: a multidimensional, randomized, controlled trial. Am. J. Phys. Med. Rehabil. 2004;83:401–420. doi: 10.1097/01.phm.0000128789.75375.d3. [DOI] [PubMed] [Google Scholar]
- 19.Whyte J. Vaccaro M. Grieb–Neff P. Hart T. Polansky M. Coslett H.B. The effects of bromocriptine on attention deficits after traumatic brain injury: a placebo-controlled pilot study. Am. J. Phys. Med. Rehabil. 2008;87:85–99. doi: 10.1097/PHM.0b013e3181619609. [DOI] [PubMed] [Google Scholar]
- 20.Kim J. Whyte J. Patel S. Europa E. Wang J. Coslett H.B. Detre J.A. Methylphenidate modulates sustained attention and cortical activation in survivors of traumatic brain injury: a perfusion fMRI study. Psychopharmacology (Berl) 2011;222:47–57. doi: 10.1007/s00213-011-2622-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Giacino J.T. Whyte J. Bagiella E. Kalmar K. Childs N. Khademi A. Eifert B. Long D. Katz D.I. Cho S. Yablon S.A. Luther M. Hammond F.M. Nordenbo A. Novak P. Mercer W. Maurer–Karattup P. Sherer M. Placebo-controlled trial of amantadine for severe traumatic brain injury. N. Engl. J. Med. 2012;366:819–826. doi: 10.1056/NEJMoa1102609. [DOI] [PubMed] [Google Scholar]
- 22.Shiel A. Burn J. P. Henry D. Clark J. Wilson B.A. Burnett M.E. McLellan D.L. The effects of increased rehabilitation therapy after brain injury: results of a prospective controlled trial. Clin. Rehabil. 2001;15:501–514. doi: 10.1191/026921501680425225. [DOI] [PubMed] [Google Scholar]
- 23.Zhu X.L. Poon W.S. Chan C.C. Chan S.S. Does intensive rehabilitation improve the functional outcome of patients with traumatic brain injury (TBI)? A randomized controlled trial. Brain Inj. 2007;21:681–690. doi: 10.1080/02699050701468941. [DOI] [PubMed] [Google Scholar]
- 24.Wagner A.K. Kline A.E. Sokoloski J. Zafonte R.D. Capulong E. Dixon C.E. Intervention with environmental enrichment after experimental brain trauma enhances cognitive recovery in male but not female rats. Neurosci. Lett. 2002;334:165–168. doi: 10.1016/s0304-3940(02)01103-5. [DOI] [PubMed] [Google Scholar]
- 25.Kline A.E. Wagner A.K. Westergom B.P. Malena R.R. Zafonte R.D. Olsen A.S. Sozda C.N. Luthra P. Panda M. Cheng J.P. Aslam H.A. Acute treatment with the 5-HT1A receptor agonist 8-OH-DPAT and chronic environmental enrichment confer neurobehavioral benefit after experimental brain trauma. Behav. Brain Res. 2007;177:186–194. doi: 10.1016/j.bbr.2006.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kline A.E. McAloon R.L. Henderson K.A. Bansal U.K. Ganti B.M. Ahmed R.H. Gibbs R.B. Sozda C.N. Evaluation of a combined therapeutic regimen of 8–OH-DPAT and environmental enrichment after experimental traumatic brain injury. J. Neurotrauma. 2010;27:2021–2032. doi: 10.1089/neu.2010.1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hoffman A.N. Malena R.R. Westergom B.P. Luthra P. Cheng J.P. Aslam H.A. Zafonte R.D. Kline A.E. Environmental enrichment-mediated functional improvement after experimental traumatic brain injury is contingent on task-specific neurobehavioral experience. Neurosci. Lett. 2008;431:226–230. doi: 10.1016/j.neulet.2007.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sozda C.N. Hoffman A.N. Olsen A.S. Cheng J.P. Zafonte R.D. Kline A.E. Empirical comparison of typical and atypical environmental enrichment paradigms on functional and histological outcome after experimental traumatic brain injury. J. Neurotrauma. 2010;27:1047–1057. doi: 10.1089/neu.2010.1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.de Witt B.W. Ehrenberg K.M. McAloon R.L. Panos A.H. Shaw K.E. Raghavan P.V. Skidmore E.R. Kline A.E. Abbreviated environmental enrichment enhances neurobehavioral recovery comparably to continuous exposure after traumatic brain injury. Neurorehabil. Neural Repair. 2011;25:343–350. doi: 10.1177/1545968310390520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Matter A.M. Folweiler K.A. Curatolo L.M. Kline A.E. Temporal effects of environmental enrichment-mediated functional improvement after experimental traumatic brain injury in rats. Neurorehabil. Neural Repair. 2011;25:558–564. doi: 10.1177/1545968310397206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Passineau M.J. Green E.J. Dietrich W.D. Therapeutic effects of environmental enrichment on cognitive function and tissue integrity following severe traumatic brain injury in rats. Exp. Neurol. 2001;168:373–384. doi: 10.1006/exnr.2000.7623. [DOI] [PubMed] [Google Scholar]
- 32.Hicks R.R. Zhang L. Atkinson A. Stevenon M. Veneracion M. Seroogy K.B. Environmental enrichment attenuates cognitive deficits, but does not alter neurotrophin gene expression in the hippocampus following lateral fluid percussion brain injury. Neuroscience. 2002;112:631–637. doi: 10.1016/s0306-4522(02)00104-5. [DOI] [PubMed] [Google Scholar]
- 33.Johansson B.B. Functional outcome in rats transferred to an enriched environment 15 days after focal brain ischemia. Stroke. 1996;27:324–326. doi: 10.1161/01.str.27.2.324. [DOI] [PubMed] [Google Scholar]
- 34.Dahlqvist P. Ronnback A. Bergstrom S. A. Soderstrom I. Olsson T. Environmental enrichment reverses learning impairment in the Morris water maze after focal cerebral ischemia in rats. Eur. J. Neurosci. 2004;19:2288–2298. doi: 10.1111/j.0953-816X.2004.03248.x. [DOI] [PubMed] [Google Scholar]
- 35.Saari M.J. Fong S. Shivji A. Armstrong J.N. Enriched housing masks deficits in place navigation induced by neonatal monosodium glutamate. Neurotoxicol. Teratol. 1990;12:29–32. doi: 10.1016/0892-0362(90)90109-p. [DOI] [PubMed] [Google Scholar]
- 36.Jadavji N.M. Kolb B. Metz G.A. Enriched environment improves motor function in intact and unilateral dopamine-depleted rats. Neuroscience. 2006;140:1127–1138. doi: 10.1016/j.neuroscience.2006.03.027. [DOI] [PubMed] [Google Scholar]
- 37.Jankowsky J.L. Melnikova T. Fadale D.J. Xu G.M. Slunt H.H. Gonzales V. Younkin L.H. Younkin S.G. Borchelt D.R. Savonenko A.V. Environmental enrichment mitigates cognitive deficits in a mouse model of Alzheimer's disease. J. Neurosci. 2005;25:5217–5224. doi: 10.1523/JNEUROSCI.5080-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Segovia G. Del Arco A. De Blas M. Garrido P. Mora F. Environmental enrichment increases the in vivo extracellular concentration of dopamine in the nucleus accumbens: a microdialysis study. J. Neural Transm. 2010;117:1123–1130. doi: 10.1007/s00702-010-0447-y. [DOI] [PubMed] [Google Scholar]
- 39.Hoffmann L.C. Schutte S.R. Koch M. Schwabe K. Effect of “enriched environment” during development on adult rat behavior and response to the dopamine receptor agonist apomorphine. Neuroscience. 2009;158:1589–1598. doi: 10.1016/j.neuroscience.2008.11.035. [DOI] [PubMed] [Google Scholar]
- 40.Solinas M. Thiriet N. El Rawas R. Lardeux V. Jaber M. Environmental enrichment during early stages of life reduces the behavioral, neurochemical, and molecular effects of cocaine. Neuropsychopharmacology. 2009;34:1102–1111. doi: 10.1038/npp.2008.51. [DOI] [PubMed] [Google Scholar]
- 41.Bardo M.T. Bowling S.L. Rowlett J.K. Manderscheid P. Buxton S.T. Dwoskin L.P. Environmental enrichment attenuates locomotor sensitization, but not in vitro dopamine release, induced by amphetamine. Pharmacol. Biochem. Behav. 1995;51:397–405. doi: 10.1016/0091-3057(94)00413-d. [DOI] [PubMed] [Google Scholar]
- 42.Yan H.Q. Ma X. Chen X. Li Y. Shao L. Dixon C.E. Delayed increase of tyrosine hydroxylase expression in rat nigrostriatal system after traumatic brain injury. Brain Res. 2007;1134:171–179. doi: 10.1016/j.brainres.2006.11.087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jordan R. Patel S. Hu H. Lyons–Weiler J. Efficiency analysis of competing tests for finding differentially expressed genes in lung adenocarcinoma. Cancer Inform. 2008;6:389–421. doi: 10.4137/cin.s791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Patel S. Lyons–Weiler J. caGEDA: a web application for the integrated analysis of global gene expression patterns in cancer. Appl. Bioinformatics. 2004;3:49–62. doi: 10.2165/00822942-200403010-00007. [DOI] [PubMed] [Google Scholar]
- 45.Dixon C.E. Clifton G.L. Lighthall J.W. Yaghmai A.A. Hayes R.L. A controlled cortical impact model of traumatic brain injury in the rat. J. Neurosci. Methods. 1991;39:253–262. doi: 10.1016/0165-0270(91)90104-8. [DOI] [PubMed] [Google Scholar]
- 46.Yan H.Q. Yu J. Kline A.E. Letart P. Jenkins L.W. Marion D.W. Dixon C.E. Evaluation of combined fibroblast growth factor-2 and moderate hypothermia therapy in traumatically brain injured rats. Brain Res. 2000;887:134–143. doi: 10.1016/s0006-8993(00)03002-x. [DOI] [PubMed] [Google Scholar]
- 47.Kline A.E. Olsen A.S. Sozda C.N. Hoffman A.N. Cheng J.P. Evaluation of a combined treatment paradigm consisting of environmental enrichment and the 5-HT1A receptor agonist buspirone after experimental traumatic brain injury. J. Neurotrauma. 2012;29:1960–1969. doi: 10.1089/neu.2012.2385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hsu H. Shi H. Jordan R.M. Lyons–Weiler J. Pilewski J.M. Feghali–Bostwick C.A. Lung tissues in patients with systemic sclerosis have gene expression patterns unique to pulmonary fibrosis and pulmonary hypertension. Arthritis Rheum. 2011;63:783–794. doi: 10.1002/art.30159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Founds S.A. Conley Y.P. Lyons–Weiler J.F. Jeyabalan A. Hogge W.A. Conrad K.P. Altered global gene expression in first trimester placentas of women destined to develop preeclampsia. Placenta. 2009;30:15–24. doi: 10.1016/j.placenta.2008.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Montecalvo A. Larregina A.T. Shufesky W.J. Stolz D.B. Sullivan M.L. Karlsson J.M. Baty C.J. Gibson G.A. Erdos G. Wang Z. Milosevic J. Tkacheva O.A. Divito S.J. Jordan R. Lyons–Weiler J. Watkins S.C. Morelli A.E. Mechanism of transfer of functional microRNAs between mouse dendritic cells via exosomes. Blood. 2012;119:756–766. doi: 10.1182/blood-2011-02-338004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ogata H. Goto S. Sato K. Fujibuchi W. Bono H. Kanehisa M. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 1999;27:29–34. doi: 10.1093/nar/27.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Draghici S. Khatri P. Tarca A. L. Amin K. Done A. Voichita C. Georgescu C. Romero R. A systems biology approach for pathway level analysis. Genome Res. 2007;17:1537–1545. doi: 10.1101/gr.6202607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Khatri P. Voichita C. Kattan K. Ansari N. Khatri A. Georgescu C. Tarca A.L. Draghici S. Onto-Tools: new additions and improvements in 2006. Nucleic Acids Res. 2007;35(Web Server issue):W206–W211. doi: 10.1093/nar/gkm327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Marciano P.G. Eberwine J.H. Ragupathi R. Saatman K.E. Meaney D.F. McIntosh T.K. Expression profiling following traumatic brain injury: a review. Neurochem. Res. 2002;27:1147–1155. doi: 10.1023/a:1020973308941. [DOI] [PubMed] [Google Scholar]
- 55.Matzilevich D.A. Rall J.M. Moore A.N. Grill R.J. Dash P.K. High-density microarray analysis of hippocampal gene expression following experimental brain injury. J. Neurosci. Res. 2002;67:646–663. doi: 10.1002/jnr.10157. [DOI] [PubMed] [Google Scholar]
- 56.Li H.H. Lee S.M. Cai Y. Sutton R.L. Hovda D.A. Differential gene expression in hippocampus following experimental brain trauma reveals distinct features of moderate and severe injuries. J. Neurotrauma. 2004;21:1141–1153. doi: 10.1089/neu.2004.21.1141. [DOI] [PubMed] [Google Scholar]
- 57.Rall J.M. Matzilevich D.A. Dash P.K. Comparative analysis of mRNA levels in the frontal cortex and the hippocampus in the basal state and in response to experimental brain injury. Neuropathol. Appl. Neurobiol. 2003;29:118–131. doi: 10.1046/j.1365-2990.2003.00439.x. [DOI] [PubMed] [Google Scholar]
- 58.Redell J.B. Liu Y. Dash P.K. Traumatic brain injury alters expression of hippocampal microRNAs: potential regulators of multiple pathophysiological processes. J. Neurosci. Res. 2009;87:1435–1448. doi: 10.1002/jnr.21945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hall E.D. Sullivan P.G. Gibson T.R. Pavel K.M. Thompson B.M. Scheff S.W. Spatial and temporal characteristics of neurodegeneration after controlled cortical impact in mice: more than a focal brain injury. J. Neurotrauma. 2005;22:252–265. doi: 10.1089/neu.2005.22.252. [DOI] [PubMed] [Google Scholar]
- 60.Kadenbach B. Arnold S. Lee I. Huttemann M. The possible role of cytochrome c oxidase in stress-induced apoptosis and degenerative diseases. Biochim. Biophys. Acta. 2004;1655:400–408. doi: 10.1016/j.bbabio.2003.06.005. [DOI] [PubMed] [Google Scholar]
- 61.de Castro I.P. Martins L.M. Loh S.H. Mitochondrial quality control and Parkinson's disease: a pathway unfolds. Mol. Neurobiol. 2011;43:80–86. doi: 10.1007/s12035-010-8150-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Herb A. Wisden W. Catania M.V. Marechal D. Dresse A. Seeburg P.H. Prominent dendritic localization in forebrain neurons of a novel mRNA and its product, dendrin. Mol. Cell Neurosci. 1997;8:367–374. doi: 10.1006/mcne.1996.0594. [DOI] [PubMed] [Google Scholar]
- 63.Kremerskothen J. Kindler S. Finger I. Veltel S. Barnekow A. Postsynaptic recruitment of Dendrin depends on both dendritic mRNA transport and synaptic anchoring. J. Neurochem. 2006;96:1659–1666. doi: 10.1111/j.1471-4159.2006.03679.x. [DOI] [PubMed] [Google Scholar]
- 64.Diez–Guerra F.J. Neurogranin, a link between calcium/calmodulin and protein kinase C signaling in synaptic plasticity. IUBMB Life. 2010;62:597–606. doi: 10.1002/iub.357. [DOI] [PubMed] [Google Scholar]
- 65.Chen S.J. Sweatt J.D. Klann E. Enhanced phosphorylation of the postsynaptic protein kinase C substrate RC3/neurogranin during long-term potentiation. Brain Res. 1997;749:181–187. doi: 10.1016/s0006-8993(96)01159-6. [DOI] [PubMed] [Google Scholar]
- 66.Ickes B.R. Pham T.M. Sanders L.A. Albeck D.S. Mohammed A.H. Granholm A.C. Long-term environmental enrichment leads to regional increases in neurotrophin levels in rat brain. Exp. Neurol. 2000;164:45–52. doi: 10.1006/exnr.2000.7415. [DOI] [PubMed] [Google Scholar]
- 67.Gelfo F. Cutuli D. Foti F. Laricchiuta D. De Bartolo P. Caltagirone C. Petrosini L. Angelucci F. Enriched environment improves motor function and increases neurotrophins in hemicerebellar lesioned rats. Neurorehabil. Neural Repair. 2011;25:243–252. doi: 10.1177/1545968310380926. [DOI] [PubMed] [Google Scholar]
- 68.Chen D.Y. Stern S.A. Garcia–Osta A. Saunier–Rebori B. Pollonini G. Bambah–Mukku D. Blitzer R.D. Alberini C.M. A critical role for IGF-II in memory consolidation and enhancement. Nature. 2011;469:491–497. doi: 10.1038/nature09667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lin X. Tombler E. Nelson P.J. Ross M. Gelman I.H. A novel src- and ras-suppressed protein kinase C substrate associated with cytoskeletal architecture. J. Biol. Chem. 1996;271:28,430–28,438. doi: 10.1074/jbc.271.45.28430. [DOI] [PubMed] [Google Scholar]
- 70.Lee S.W. Kim W.J. Choi Y.K. Song H.S. Son M.J. Gelman I.H. Kim Y.J. Kim K.W. SSeCKS regulates angiogenesis and tight junction formation in blood–brain barrier. Nat. Med. 2003;9:900–906. doi: 10.1038/nm889. [DOI] [PubMed] [Google Scholar]
- 71.During M.J. Cao L. VEGF, a mediator of the effect of experience on hippocampal neurogenesis. Curr. Alzheimer Res. 2006;3:29–33. doi: 10.2174/156720506775697133. [DOI] [PubMed] [Google Scholar]
- 72.Coulter P.M., 2nd Bautista E.A. Margulies J.E. Watson J.B. Identification of cortexin: a novel, neuron-specific, 82-residue membrane protein enriched in rodent cerebral cortex. J. Neurochem. 1993;61:756–759. doi: 10.1111/j.1471-4159.1993.tb02183.x. [DOI] [PubMed] [Google Scholar]
- 73.Kuroda S. Oyasu M. Kawakami M. Kanayama N. Tanizawa K. Saito N. Abe T. Matsuhashi S. Ting K. Biochemical characterization and expression analysis of neural thrombospondin-1-like proteins NELL1 and NELL2. Biochem. Biophys. Res. Commun. 1999;265:79–86. doi: 10.1006/bbrc.1999.1638. [DOI] [PubMed] [Google Scholar]
- 74.Barembaum M. Moreno T.A. LaBonne C. Sechrist J. Bronner–Fraser M. Noelin-1 is a secreted glycoprotein involved in generation of the neural crest. Nat. Cell Biol. 2000;2:219–225. doi: 10.1038/35008643. [DOI] [PubMed] [Google Scholar]
- 75.Moreno T.A. Bronner–Fraser M. The secreted glycoprotein Noelin-1 promotes neurogenesis in Xenopus. Dev. Biol. 2001;240:340–360. doi: 10.1006/dbio.2001.0472. [DOI] [PubMed] [Google Scholar]
- 76.Tomarev S.I. Nakaya N. Olfactomedin domain-containing proteins: possible mechanisms of action and functions in normal development and pathology. Mol. Neurobiol. 2009;40:122–138. doi: 10.1007/s12035-009-8076-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ju Y.T. Chang A.C. She B.R. Tsaur M.L. Hwang H.M. Chao C.C. Cohen S.N. Lin–Chao S. gas7: A gene expressed preferentially in growth-arrested fibroblasts and terminally differentiated Purkinje neurons affects neurite formation. Proc. Natl. Acad. Sci. U.S.A. 1998;95:11,423–11,428. doi: 10.1073/pnas.95.19.11423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chao C.C. Su L.J. Sun N.K. Ju Y.T. Lih J.C. Lin–Chao S. Involvement of Gas7 in nerve growth factor-independent and dependent cell processes in PC12 cells. J. Neurosci. Res. 2003;74:248–254. doi: 10.1002/jnr.10763. [DOI] [PubMed] [Google Scholar]
- 79.Chang P.Y. Kuo J.T. Lin–Chao S. Chao C.C. Identification of rat Gas7 isoforms differentially expressed in brain and regulated following kainate-induced neuronal injury. J. Neurosci. Res. 2005;79:788–797. doi: 10.1002/jnr.20409. [DOI] [PubMed] [Google Scholar]
- 80.Frey D. Laux T. Xu L. Schneider C. Caroni P. Shared and unique roles of CAP23 and GAP43 in actin regulation, neurite outgrowth, and anatomical plasticity. J. Cell Biol. 2000;149:1443–1454. doi: 10.1083/jcb.149.7.1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bomze H.M. Bulsara K.R. Iskandar B.J. Caroni P. Skene J.H. Spinal axon regeneration evoked by replacing two growth cone proteins in adult neurons. Nat. Neurosci. 2001;4:38–43. doi: 10.1038/82881. [DOI] [PubMed] [Google Scholar]
- 82.Cramer S.C. Chopp M. Recovery recapitulates ontogeny. Trends Neurosci. 2000;23:265–271. doi: 10.1016/s0166-2236(00)01562-9. [DOI] [PubMed] [Google Scholar]
- 83.Caleo M. Tropea D. Rossi C. Gianfranceschi L. Maffei L. Environmental enrichment promotes fiber sprouting after deafferentation of the superior colliculus in the adult rat brain. Exp. Neurol. 2009;216:515–519. doi: 10.1016/j.expneurol.2008.12.016. [DOI] [PubMed] [Google Scholar]
- 84.Overton P.G. Richards C.D. Berry M.S. Clark D. Long-term potentiation at excitatory amino acid synapses on midbrain dopamine neurons. Neuroreport. 1999;10:221–226. doi: 10.1097/00001756-199902050-00004. [DOI] [PubMed] [Google Scholar]
- 85.Singleton R.H. Stone J.R. Okonkwo D.O. Pellicane A.J. Povlishock J.T. The immunophilin ligand FK506 attenuates axonal injury in an impact-acceleration model of traumatic brain injury. J. Neurotrauma. 2001;18:607–614. doi: 10.1089/089771501750291846. [DOI] [PubMed] [Google Scholar]
- 86.Okonkwo D.O. Buki A. Siman R. Povlishock J.T. Cyclosporin A limits calcium-induced axonal damage following traumatic brain injury. Neuroreport. 1999;10:353–358. doi: 10.1097/00001756-199902050-00026. [DOI] [PubMed] [Google Scholar]
- 87.Okonkwo D.O. Melon D.E. Pellicane A.J. Mutlu L.K. Rubin D.G. Stone J.R. Helm G.A. Dose-response of cyclosporin A in attenuating traumatic axonal injury in rat. Neuroreport. 2003;14:463–466. doi: 10.1097/00001756-200303030-00033. [DOI] [PubMed] [Google Scholar]
- 88.Burre J. Sharma M. Tsetsenis T. Buchman V. Etherton M.R. Sudhof T.C. Alpha-synuclein promotes SNARE-complex assembly in vivo and in vitro. Science. 2010;329:1663–1667. doi: 10.1126/science.1195227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Alerte T.N. Akinfolarin A.A. Friedrich E.E. Mader S.A. Hong C.S. Perez R.G. Alpha-synuclein aggregation alters tyrosine hydroxylase phosphorylation and immunoreactivity: lessons from viral transduction of knockout mice. Neurosci. Lett. 2008;435:24–29. doi: 10.1016/j.neulet.2008.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Tehranian R. Montoya S.E. Van Laar A.D. Hastings T.G. Perez R.G. Alpha-synuclein inhibits aromatic amino acid decarboxylase activity in dopaminergic cells. J. Neurochem. 2006;99:1188–1196. doi: 10.1111/j.1471-4159.2006.04146.x. [DOI] [PubMed] [Google Scholar]
- 91.Li S.W. Lin T.S. Minteer S. Burke W.J. 3,4-Dihydroxyphenylacetaldehyde and hydrogen peroxide generate a hydroxyl radical: possible role in Parkinson's disease pathogenesis. Brain Res. Mol. Brain Res. 2001;93:1–7. doi: 10.1016/s0169-328x(01)00120-6. [DOI] [PubMed] [Google Scholar]
- 92.Burke W.J. Li S.W. Williams E.A. Nonneman R. Zahm D.S. 3,4-Dihydroxyphenylacetaldehyde is the toxic dopamine metabolite in vivo: implications for Parkinson's disease pathogenesis. Brain Res. 2003;989:205–213. doi: 10.1016/s0006-8993(03)03354-7. [DOI] [PubMed] [Google Scholar]