Abstract
Endogenous and exogenous factors constantly challenge cellular DNA, generating cytotoxic and/or mutagenic DNA adducts. As a result, organisms have evolved different mechanisms to defend against the deleterious effects of DNA damage. Among these diverse repair pathways, direct DNA-repair systems provide cells with simple yet efficient solutions to reverse covalent DNA adducts. In this review, we focus on recent advances in the field of direct DNA repair, namely, photolyase-, alkyltransferase-, and dioxygenase-mediated repair processes. We present specific examples to describe new findings of known enzymes and appealing discoveries of new proteins. At the end of this article, we also briefly discuss the influence of direct DNA repair on other fields of biology and its implication on the discovery of new biology.
Simple direct repair mechanisms can fix DNA damage without breaking the DNA backbone. These essentially error-free processes include photolyase-, alkyltransferase-, and dioxygenase-mediated mechanisms.
Endogenous and environmental agents continuously threaten the genomic integrity of all living organisms. Replication of damaged DNA can lead to mutations that are tumorigenic, whereas DNA lesions that block replication or transcription can result in senescence and cell death. Therefore, cellular DNA must be promptly repaired. Well-known mechanisms include base-excision repair, nucleotide excision repair, mismatch repair, homologous recombination, and nonhomologous end joining. In addition, nature has also evolved several mechanisms in which the damage is directly reversed most often by a single repair protein without the incision of DNA backbone. Although such “direct repair” processes mediate the reversal of a relatively small set of DNA lesions, the simplicity and essentially error-free property of the direct reversal processes make them particularly attractive for a cell. Three major mechanisms of direct DNA repair have been identified to date: (i) photolyases reverse UV light-induced photolesions; (ii) O6-alkylguanine-DNA alkyltransferases (AGTs) reverse a set of O-alkylated DNA damage; and (iii) the AlkB family dioxygenases reverse N-alkylated base adducts (Fig. 1). This concise article intends to update knowledge since the publication of the second edition of DNA Repair and Mutagenesis (ASM) (Friedberg et al. 2006).
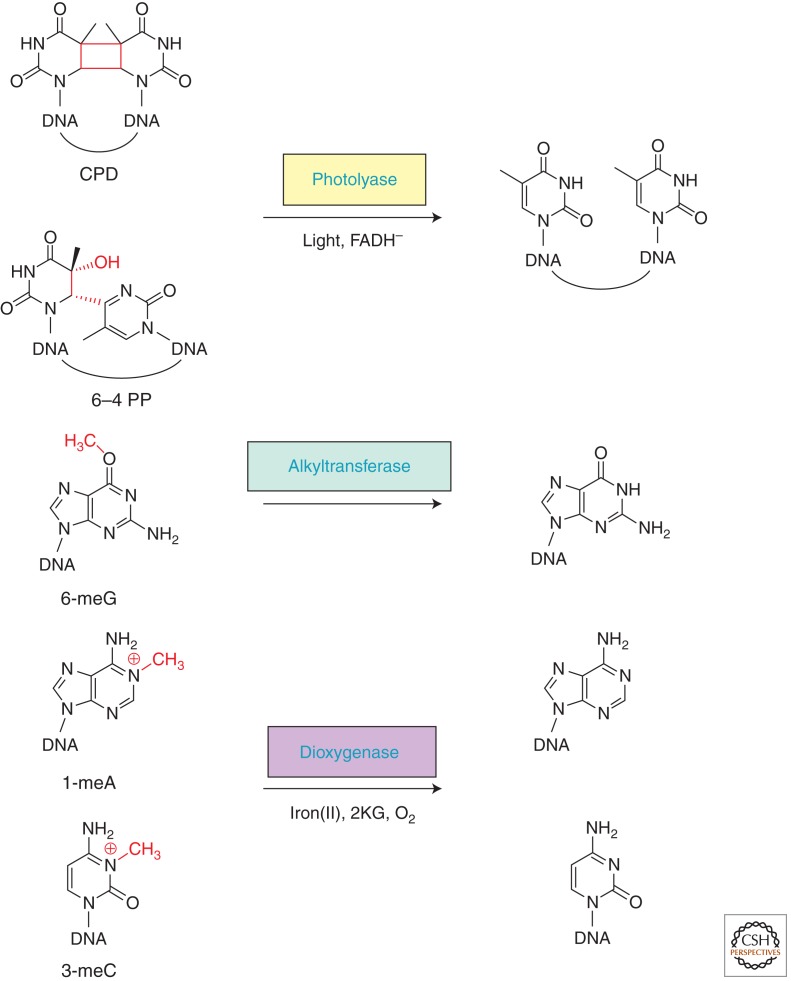
Figure 1.
Direct DNA-repair pathways: representative substrates, repair proteins and cofactors, and corresponding repair products.
PHOTOINDUCED REVERSAL OF UV RADIATION-CAUSED DNA DAMAGE BY PHOTOLYASES
UV radiation produces mainly two types of lesions in DNA: the cyclobutane pyrimidine dimers (CPDs) and the pyrimidine pyrimidones (6–4) photoproducts (6–4 PPs). Photolyases are specific to either CPD or 6–4 PP lesion (thus CPD photolyases and [6–4] photolyases, respectively) and use blue and near-UV light to reverse the UV-light-induced DNA damage. All photolyases bind the essential cofactor flavin adenine dinucleotide (FAD), and only the fully reduced FADH− is catalytically competent. Proposed mechanisms of photolyases involve an initial electron-transfer step from FADH− to the UV-induced lesions, a dimer-splitting process, and a final electron-transfer step from the pyrimidine monomer radical back to FADH·, thus regenerating FADH− (Fig. 2) (Muller and Carell 2009; Brettel and Byrdin 2010).
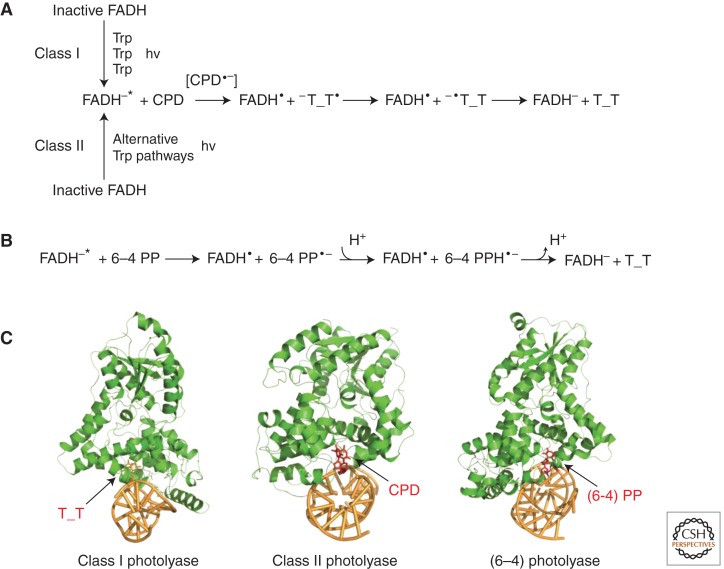
Figure 2.
Mechanisms and structures of representative photolyases from different classes. (A) Mechanisms of photorepair for CPD photolyases. The generation of fully reduced, active FADH−* species is different for Class I and Class II CPD photolyases. (B) Repair mechanism of (6–4) photolyases. (C) Representative complex structures of photolyases bound to double-stranded DNA (dsDNA) (PDB accession code: 1TEZ, 2XRZ, and 3CVU).
ADVANCES IN REACTION MECHANISMS OF CPD PHOTOLYASES
The advancing studies of the CPD repair mechanism that are based on direct measurements of Class I CPD photolyases have seen further development with ultrafast spectroscopy. For instance, using an Escherichia coli CPD photolyase, Zhong, Sancar, and colleagues observed electron transfer from the excited flavin cofactor to the CPD dimer in 170 ps and back electron transfer from the repaired thymines in 560 ps (Kao et al. 2005); further study by the investigators revealed that the CPD splits in two sequential steps within 90 ps (Liu et al. 2011). In another example by Brettel, Byrdin, and colleagues, splitting rates of the intradimer bond and electron return process of Aspergillus nidulans photolyase were estimated to be 0.2 and 1.5 ns, respectively (Thiagarajan et al. 2011). In both examples, the technology advance significantly enhanced our understanding of the spatiotemporal picture of CPD repair.
Class II photolyases have different amino acid sequences compared to those from Class I enzymes and are expected to differ in the pathway of electron transfer and the DNA-repair active site (Fig. 2A,C). The first crystal structure of a Class II photolyase (from Methanosarcina mazei), alone and in complex with CPD lesion-containing duplex DNA, revealed a larger lesion-binding site and an unusual (different from that of Class I) tryptophane dyad as the electron-transfer pathway to FAD (Kiontke et al. 2011). In addition, a distinct electron-transfer pathway (three Trp residues in this case) from the Class I pathway was also observed in Oriza sativa photolyase (Hitomi et al. 2012). Nevertheless, spectroscopic results for Arabidopsis thaliana and O. sativa photolyases showed photoreduction kinetics resembling those of Class I enzymes (Okafuji et al. 2010), indicating functional substitution between these different electron-transfer pathways.
ADVANCES IN THE STRUCTURAL AND FUNCTIONAL RELATIONSHIP OF (6–4) PHOTOLYASE
The reversal of 6–4 PP is thought to be more complex than that of CPD because 6–4 photolyase must catalyze not only covalent bond cleavage between two pyrmidine bases but also undergo a hydroxyl group transfer from the 5′ to the 3′ pyrimidine base. For this reason, an oxetane intermediate was assumed. However, Schlichting, Carell, and colleagues proposed a modified repair mechanism that lacked the strained oxetane intermediate (Maul et al. 2008). They crystallized Drosophila melanogaster (6–4) photolyase containing a 6–4 PP and initiated in situ photo repair (Fig. 2C). In the resulting structures, the positions of two conserved histidine residues essential for catalysis (His365 and His369) do not support a mechanism involving an oxetane structure. Instead, a mechanism that involves a water molecule formed in situ, which then attacks the acylimine, was postulated (Maul et al. 2008). More recent structural evidence from A. thaliana (6–4) photolyase revealed a narrower and deeper cavity for binding of 6–4 PP, also suggesting a different reaction mechanism compared to that of CPD photolyase (Fig. 2B) (Hitomi et al. 2009).
Zhong et al. used femtosecond spectroscopy to study the dynamics and mechanism of (6−4) photolyase from A. thaliana. The investigators observed a key cyclic proton transfer step between an active-site histidine residue and the substrate, which occurs in 425 ps and leads to 6–4 PP repair in tens of nanoseconds (Li et al. 2010). In addition, time-resolved experiments of Xenopus laevis (6−4) photolyase uncovered a drastic diffusion change, which was assigned to the rapid dissociation (time constant of ∼50 µs) of the protein from the repaired DNA product (Kondoh et al. 2011). Later on, 6–4 PP repair mediated by the same enzyme was investigated using Fourier transform infrared (FTIR) spectroscopy (Zhang et al. 2011). Differences in FTIR spectra revealed structural changes in the protein and DNA from binding to catalysis. Together, these data depict a general picture of how light is harvested and used to efficiently reverse 6–4 PP by (6−4) photolyase.
CRYPTOCHROME-DASH MEMBERS SHOW DNA-REPAIR ACTIVITY
The cryptochrome-DASH (CRY-DASH) subfamily is relatively closer to the animal cryptochrome subfamily, rather than the plant cryptochrome subfamily and the CPD photolyase subfamilies (Hitomi et al. 2000; Brudler et al. 2003). Therefore, CRY-DASH proteins were considered at first to be novel cryptochromes, rather than DNA-repair photolyases. However, weak CPD photorepair activities were detected for the Synechocystis protein and two vertebrate CRY-DASHs (Hitomi et al. 2000; Daiyasu et al. 2004). Crystal structure of the Synechocystis CRY-DASH further revealed high similarity to the structures of Class I CPD photolyases (Brudler et al. 2003). In 2008, progress from the Sancar laboratory provided an explanation of the observed weak DNA-repair activity. In actuality, CRY-DASH members robustly bind to and repair CPD in the context of single-stranded DNA (ssDNA), but not in double-stranded DNA (dsDNA) (Selby and Sancar 2006). Crystal structures of Arabidopsis cryptochrome 3 further indicate that the protein most likely lacks an efficient flipping mechanism to access CPD lesions within duplex DNA (Pokorny et al. 2008), and that the CPD-binding cavity in Arabidopsis cryptochrome 3 is less hydrophobic and thus less energetically favorable compared to that of Class I CPD photolyases (Huang et al. 2006). Together, this data reveals that CRY-DASH proteins are indeed light-driven DNA-repair enzymes, only less competent in base flipping and CPD binding.
DIRECT REVERSAL OF ALKYLATION DAMAGE BY ALKYLTRANSFERASES
Alkylating agents react with the hetereoatoms of DNA bases to generate a variety of cytotoxic and mutagenic covalent adducts ranging from simple methyl groups to bulky alkyl additions, thereby posing substantial threats to human health (Drablos et al. 2004; Shrivastav et al. 2010). On the other hand, certain alkylating agents are commonly used as chemotherapeutic drugs in cancer patients, with the goal of killing cancer cells. Such double-edged properties of alkylating agents thus impart great biological significance to the studies of the cellular pathways that determine the biological outcome of alkylating agents (Fu et al. 2012). In addition to the base-excision repair pathway that corrects many N-alkylated lesions, two direct DNA-repair pathways exist to combat the deleterious effects of alkylating agents: AGTs reverse O6-alkylated guanines and AlkB family dioxygenases reverse mainly N-alkylated lesions that block Watson–Crick pairings. Recent progress with regard to these two direct repair pathways is detailed below.
SUICIDAL REVERSAL OF ALKYLATION DAMAGE BY O6-ALKYLGUANINE-DNA ALKYLTRANSFERASES
O6-methyl guanine (6-meG) is mutagenic and carcinogenic as a result of its ability to cause G:C to A:T transitions during DNA replication. The first 6-meG alkyltransferse, identified in E. coli (named C-Ada), is the carboxy-terminal domain of a multifunctional repair protein (Demple et al. 1985; Sedgwick et al. 1988). The amino-terminal domain of E. coli Ada protein (N-Ada) mediates a direct removal of the methyl group from Sp-methylphosphotriester DNA backbone damage (Sedgwick et al. 1988; Myers et al. 1993; He et al. 2005). It uses a zinc-mediated thiol alkylation mechanism and signals the activation of the ada operon, which includes ada, alkA, alkB, and aidB (Teo et al. 1986; Lindahl et al. 1988; Sakumi and Sekiguchi 1989). Both Ada and AlkB are direct repair proteins. AlkA is a glycosylase conserved from bacterium to human (Wyatt et al. 1999; Hollis et al. 2000). The exact functional role of AidB is still unknown (Rohankhedkar et al. 2006; Bowles et al. 2008; Rippa et al. 2011).
The human homolog of C-Ada (hAGT) contains two domains, a zinc-bound amino-terminal domain and a carboxy-terminal domain that harbors the methyl recipient Cys145. The S-alkylated AGT cannot be restored and undergoes ubiquitin-mediated degradation (Srivenugopal et al. 1996). A recent discovery showed that the AGT homolog in Saccharomyces cerevisiae (Mgt1) is cotargeted for degradation by both the Ubr1/Rad6-dependent N-end rule pathway and the Ufd4/Ubc4-dependent ubiquitin-fusion degradation pathway, through a degron near its amino terminus (Hwang et al. 2009). Because Mgt1 and mammalian AGTs share high sequence homology, the next obvious question to be addressed is whether such a mechanism also applies to mammalian AGTs.
AGTs can recognize diverse O6-modified alkyl adducts. For instance, hAGT repairs not only 6-meG, but also many other bigger adducts including ethyl, 2-chloroethyl, and other aliphatic groups, and benzyl and pyridyloxobutyl adducts as well (Tubbs et al. 2007; Pegg 2011). Even O6-G-alkyl-O6-G interstrand cross-links could be repaired (Fang et al. 2008). As a result, hundreds of hAGT pseudosubstrates have been synthesized as inhibitors of hAGT, which are used in combination with therapeutic alkylating agents to improve the efficacy of cancer chemotherapy (Tubbs et al. 2007). One example is the clinical trial of O6-benzyl guanine (6-bzG) and temozolomide. Some responses were observed from patients with refractory central nervous system (CNS) tumors (Hammond et al. 2004; Warren et al. 2005; Quinn et al. 2009a,b). Another example is the combined use of O6-(4-bromothenyl) guanine (or PaTrin-2) and temozolomide in the treatment of refractory acute leukemia. Preclinical in vitro studies have shown that PaTrin-2 increases the inhibitory activity of temozolomide against human acute leukemia cells (Turriziani et al. 2006). Despite these initial positive feedbacks, it is clear that inhibitors with greater potency and higher tumor specificity are still needed (Zang et al. 2005; Guza et al. 2006; Pegg 2011).
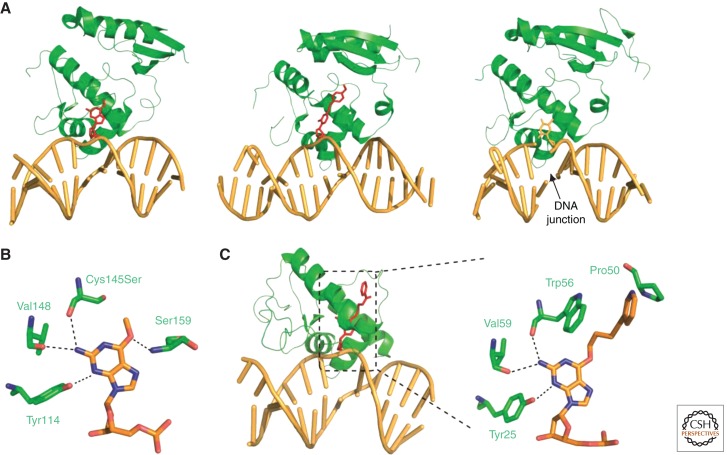
Several structures of hAGT/DNA complex are available for detailed examinations of substrate binding and nucleotide flipping. Tainer and colleagues reported the first structures of hAGT in complex with dsDNA, which revealed that hAGT uses a helix-turn-helix motif to mediate an unusual minor groove DNA binding (Fig. 3A,B) (Daniels et al. 2004). Our group also solved the structure of hAGT bound to a duplex DNA containing N4-p-xylylenediamine cytosine. The protein binds at two different sites: the modified cytosine and a partially flipped overhanging thymidine at the sticky ended DNA junction (Fig. 3A) (Duguid et al. 2005). The two different hAGT/DNA interactions suggest that hAGT may search for weakened and/or distorted base pairs to locate the lesion. Results from computational approaches have suggested a two-step base-flipping mechanism of hAGT, in which the existence of an extra-helical intermediate was postulated (Hu et al. 2008). It has also been revealed that while searching along a duplex DNA, hAGT shows 5′ to 3′ preference and binds DNA cooperatively (Daniels et al. 2004; Rasimas et al. 2007; Adams et al. 2009). Last, single-molecule spatial tracking measurements of C-Ada, the E. coli equivalence of hAGT, showed that sliding, with essentially no hopping, is the mechanism of C-Ada motion along stretched DNA (Lin et al. 2009).
Figure 3.
Overall structures and damage-binding pockets of hAGT and Schizosaccharomyces pombe Atl1. (A) Structures of hAGT/DNA bound to a 6-meG (1T38), an N4-p-xylylenediamine cytosine, and a partially flipped thymine (1YFH). (B) Active site of hAGT C145S with a bound 6-meG. (C) Overall and binding-site structure of Atl1 (3GYH).
REPAIR OF ALKYLATION DAMAGE BY ALKYLTRANSFERASE-LIKE PROTEINS (ATLs)
In silico analysis of genomic sequences suggests the presence of a family of AGT homologs (ATLs) with sequence similarity to the AGT catalytic domain, with the exception that the active cysteine residue is replaced by tryptophan, alanine, or other residues, all of which are not capable of nucleophilic attack (Margison et al. 2003, 2007). ATLs tightly bind ss/dsDNA that contain O6-alkyl guanine, but they display no alkyltransferase, demethylase, glycosylase, or endonuclease activity (Pearson et al. 2005, 2006; Chen et al. 2008; Morita et al. 2008). In fact, preincubation of alkylated oligonucleotides inhibits the repair activity of hAGT (Pearson et al. 2005, 2006). Yet it was clear that ATL must be playing a role in alkylation damage protection in vivo, because S. pombe and Thermus thermophilus were rendered more sensitive to alkylating reagents on inactivation of their ATL genes (Pearson et al. 2006; Morita et al. 2008). Through a series of genetic and biochemical experiments, ATL was discovered to be linked to the nucleotide excision repair (NER) pathway (Mazon et al. 2009; Tubbs et al. 2009). In a proposed model, ATLs first recognize the often bulky O6-alkyl lesion, then induce extensive DNA bending and switch from an open conformation to a closed state that is not seen in AGTs (Tubbs and Tainer 2010). Large DNA distortions are known to initiate NER recognition, and ATL in the closed state is also believed to facilitate recruitment of NER proteins. Crystal structures of ATLs from Vibrio parahaemolyticus and S. pombe are also in good agreement with the working model (Fig. 3C) (Tubbs et al. 2009; Aramini et al. 2010).
OXIDATIVE REVERSAL OF ALKYLATION DAMAGE BY AlkB FAMILY DIOXYGENASES
Oxidative DNA repair mediated by the AlkB family demethylases is one of the most exciting and rapidly growing areas in the field of DNA repair during the past decade since the initial ground-breaking discovery made independently by two groups in 2002 (Falnes et al. 2002; Trewick et al. 2002). Later on, AlkB was shown to repair RNA lesions as well (Aas et al. 2003; Ougland et al. 2004). So far, nine human homologs of E. coli AlkB have been identified (termed ALKBH1-8, plus FTO), representing a major class of DNA/RNA demethylases in human cells (Kurowski et al. 2003; Gerken et al. 2007; Sanchez-Pulido and Andrade-Navarro 2007). In the event of oxidative demethylation, the AlkB family proteins use an iron(II) site to activate the dioxygen molecule for oxidation of the aberrant alkyl groups. The hydroxylated alkyl groups, which are attached to the N1 position of adenine or N3 position of cytosine, then undergo facile C–N bond cleavage to yield the unmodified base and formaldehyde (Drablos et al. 2004; Sedgwick 2004; Falnes et al. 2007; Yi et al. 2009). Such an oxidation mechanism is shared by a variety of enzymes within the nonheme iron-containing protein family, which has also inspired the discovery of the JmjC-domain-containing histone demethylases that mediate epigenetic histone demethylation (Tsukada et al. 2006).
E. coli AlkB: UNIQUE BASE-FLIPPING, DIVERSE SUBSTRATES, AND OXIDATIVE DEMETHYLATION
Like photolyases and alkyltransferases, AlkB and related dioxygenases also need to flip the damaged base out of duplex DNA for repair reactions. AlkB has been shown to repair alkylation lesions in both ssDNA and dsDNA, with a preference toward single-stranded substrates. Structures of AlkB bound to a trimer d(T-1-meA-T) DNA provided the first glance of the folding and substrate binding of AlkB (Fig. 4A) (Yu et al. 2006); yet the characterization of the AlkB/dsDNA complex structure had been hampered by AlkB's weak and nonsequence-specific binding property (Dinglay et al. 2000; Mishina et al. 2004a). Utilizing a disulfide cross-linking technique pioneered by Verdine et al. to capture transient protein/DNA interactions (Huang et al. 1998; Verdine and Norman 2003), we stabilized the complex formed between AlkB and dsDNA (Fig. 4B) (Mishina and He 2003; Mishina et al. 2004b). We first engineered the cross-link into the active site of AlkB and determined the structure of the resulting complex (Yang et al. 2008). With the information revealed from the “active-site cross-linked” structure, a distal cross-link was designed to obtain a complex with an intact enzymatic pocket. Both of these structures show an unprecedented base-flipping mechanism. AlkB squeezes together the two bases flanking the flipped-out one to access the damaged base (Fig. 4B). In contrast to DNA distortion in the lesion-containing strand, AlkB has few interactions with the complementary strand, which merely accommodates DNA distortion of the lesion-containing strand through spontaneous conformational rearrangements. Thus, the complementary strand acts as a noncompetitive inhibitor, which explains the observed preference of AlkB toward ssDNA (Yang et al. 2008).
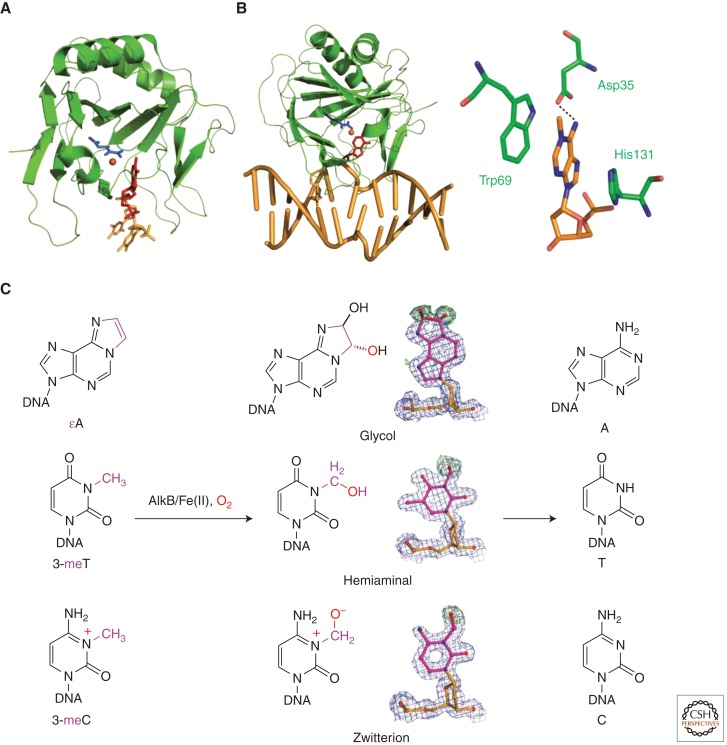
Figure 4.
Structure and mechanism of AlkB-mediated oxidative demethylation. (A) Structure of AlkB/d(T-1meA-T) (2FD8). (B) Structure of overall AlkB/DNA complex and the active site (3BIE). (C) Direct observation of oxidative-demethylation intermediates in crystallo.
AlkB recognizes diverse substrates: N1-methyl adenine (1-meA) and N3-methyl cytosine (3-meC) are the best substrates (Falnes et al. 2002; Trewick et al. 2002); N1-methyl guanine and N3-methyl thymine (3-meT) are repaired at lower rates (Delaney and Essigmann 2004; Falnes 2004; Koivisto et al. 2004). Furthermore, exocyclic DNA adducts such as 1,N6-ethenoadenine (εA), 1,N6-ethano adenine, and 3,N4-ethenocytosine are also substrates of AlkB (Delaney et al. 2005; Mishina et al. 2005; Frick et al. 2007). Mechanistically, such promiscuous substrate recognition has been extensively investigated with AlkB crystal structures containing different substrates (Yu et al. 2006; Yang et al. 2008; Yu and Hunt 2009; Holland and Hollis 2010; Yi et al. 2010). A “pinch” sequence (Thr51 to Tyr55) and a flexible loop (His72 to Tyr76) anchor the phosphate backbone; His131 and Trp69 stack against the base plane; Tyr78 and a “substrate recognition” loop (Lys134 to Glu136) form specific hydrogen bonds with the flipped bases (Yu and Hunt 2009; Yi et al. 2010). Overall, AlkB strictly controls the final location of substrates in the binding pocket so that the aberrant alkyl group is positioned precisely for efficient oxidative dealkylation.
Knowing how AlkB recognizes and accommodates different lesions, the next question concerns the mechanism of the oxidative repair. It was proposed that oxidative demethylation proceeds through hydroxylation at the aberrant methyl group followed by heterocleavage of the C–N bond; yet direct evidence supporting the process is lacking. To address this question, we took an in crystallo oxidation approach and captured different reaction intermediates (a zwitterionic structure, a hemiaminal, and a glycol) in the demethylation processes of 3-meC, 3-meT, and εA, respectively (Fig. 4C) (Yi et al. 2010). Essigmann and coworkers have also observed an epoxide intermediate in AlkB-mediated εA repair using mass spectrometry (Delaney et al. 2005). These observations provide direct support of the oxidative-demethylation mechanism. Together with solution observations (Bleijlevens et al. 2008, 2012), a more complete picture of cofactor binding, substrate flipping and processing, and product release is now emerging.
ALKBH1: AP LYASE OR DEMETHYLASE?
Among the nine homolog proteins, ALKBH1 displays the strongest homology with AlkB, although early studies failed to detect any demethylation activities (Duncan et al. 2002; Aas et al. 2003). A later study showed that, as a mitochondrial protein, ALKBH1is able to demethylate 3-meC in vitro (Westbye et al. 2008). Yet this is just the beginning of the enigma of ALKBH1. Two groups then independently showed that both human ALKBH1 and the S. pombe homolog Abh1 show AP-lyase activity (Muller et al. 2010; Korvald et al. 2012). The lyase activity of human ALKBH1 and Abh1 is not dependent on iron(II) or 2KG and is not affected by mutation of the putative metal-binding residues; furthermore, Abh1 failed to show demethylase activity against methylated DNA or etheno adducts, nor was the yeast abh1− mutant sensitive toward alkylating agents. But if the in vitro AP-lyase activity has any biological significance, it remains elusive. What does seem clear, however, is that ALKBH1 plays a role in placental trophoblast lineage differentiation and participates in transcriptional regulation (Pan et al. 2008; Nordstrand et al. 2010).
ALKBH2: A BONA FIDE HOUSEKEEPING DNA-REPAIR ENZYME
ALKBH2 demethylates both 1-meA and 3-meC in vitro (Duncan et al. 2002; Aas et al. 2003); mouse experiments have also established mAlkbh2 as the primary repair enzyme guarding the mammalian genome against these two lesions (Ringvoll et al. 2006). Moreover, ALKBH2 was shown to be the principle enzyme for εA repair in vivo as well (Duncan et al. 2002). Very recent data suggests that ALKBH2 protects against lethality and mutation in primary mouse embryonic fibroblasts (Nay et al. 2012), and that ALKBH2 may be involved in the molecular mechanism of gastric cancer through the inhibition of the proliferating gastric cancer cells (Gao et al. 2011). ALKBH2 ortholog in A. thaliana has also been shown to protect Arabidopsis against methylation DNA damage (Meza et al. 2012). Thus, unlike ALKBH1, ALKBH2 has been convincingly established as a bona fide DNA-repair enzyme that protects the genome against alkylation damage.
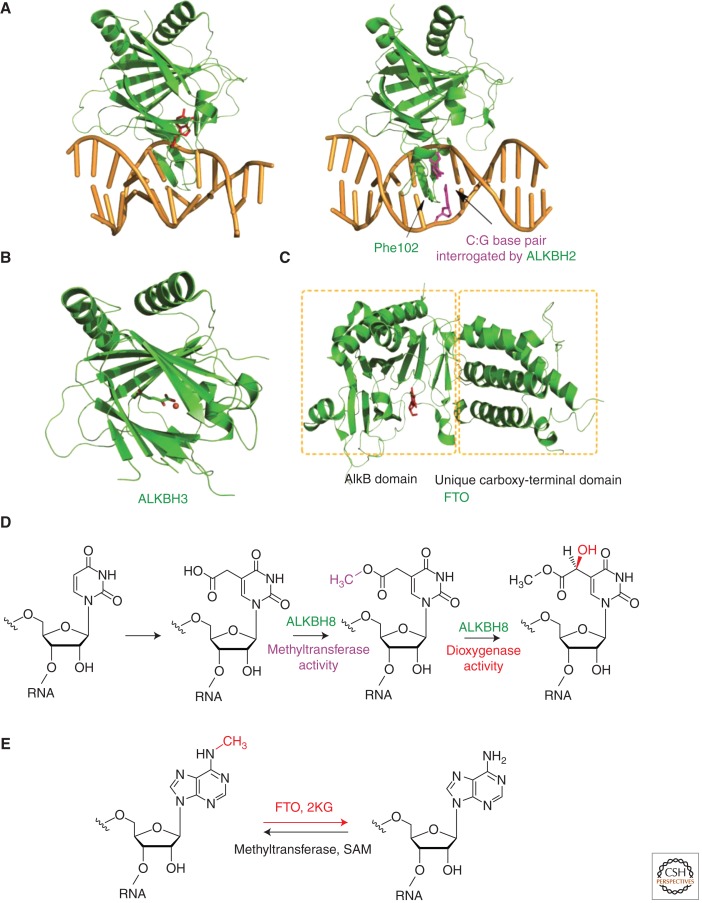
Crystallographic evidence has provided insights into the lesion-recognition mode and damage-searching mechanism of ALKBH2 (Fig. 5A). Using a disulfide cross-linking technique, we have succeeded in the structural characterization of ALKBH2 bound to duplex DNA containing different base lesions (Yang et al. 2008; Yi et al. 2012). ALKBH2 uses several active-site residues to recognize 1-meA and uses an aromatic finger residue Phe102 to facilitate base flipping (Yang et al. 2008). In addition, Phe102 can also probe the stability of base pairs when ALKBH2 is interrogating DNA for damage; the unique oxidation chemistry of ALKBH2 then ensures that only a cognate substrate will be modified (Yi et al. 2012). It remains to be seen if ALKBH2 could signal or recruit additional repair factors when it encounters noncognate damage (Gilljam et al. 2009).
Figure 5.
Structures and functions of AlkB homologs. (A) Structures of ALKBH2 with a bound 1-meA (3BTY) and a central C:G base pair (3RZG), which the protein is interrogating. (B) Structure of ALKBH3(2IUW). (C) Structure of FTO (3LFM). (D) ALKBH8 is a transfer RNA (tRNA) modification enzyme. (E) Demethylation of 6-meA by FTO enables reversible RNA modification.
ALKBH3: A DNA-REPAIR ENZYME THAT TARGETS 3-meC WITH ssDNA PREFERENCE
ALKBH3 demethylates both 1-meA and 3-meC in vitro, with a preference toward single-stranded nucleic acids (Duncan et al. 2002; Aas et al. 2003). Inside cells, ALKBH3 has significant roles in both prostate cancers and lung cancers (Liu et al. 2007; Tasaki et al. 2011), and it contributes to cell survival and invasion through discoidin receptor 1 (Shimada et al. 2008). In fact, studies have suggested ALKBH3 as a promising target molecule for developing therapeutic agents to treat castrate-resistant prostate cancer (Koike et al. 2012). To address the relevance of ALKBH3 in mammalian cells, Shi and colleagues revealed that ALKBH3 physically associates with ASCC3, a 3′ to 5′ DNA helicase (Dango et al. 2011). Such an association is critical to the ALKBH3-mediated repair process because ASCC3's DNA-unwinding activity presents the 3-meC substrate in ssDNA context, allowing ALKBH3 to access 3-meC within the double-stranded region of DNA. Thus, for tumors showing ALKBH3 and ASCC3 overexpression, therapeutic approaches that can couple 3-meC cytotoxicity to the disturbance of the activities, or association of these two proteins, may have therapeutic effects against these tumors.
Slupphaug, Tainer, and colleagues solved the crystal structure of the catalytic core of ALKBH3 and identified a flexible hairpin involved in base-flipping and ss/dsDNA discrimination (Fig. 5B) (Sundheim et al. 2006). Sequence alignments of ALKBH2 and ALKBH3 reveal a very hydrophobic Val101-Phe102-Gly103 motif for ALKBH2 and a heavily charged Arg122-Glu123-Asp124 sequence for ALKBH3 at the nucleotide-flipping region (Yi et al. 2009). Swapping the two sequences surprisingly switches the ss/dsDNA preference of the two proteins (Chen et al. 2010; Monsen et al. 2010). Yet a complex structure of ALKBH3/DNA would still be highly desirable to address the exact protein/DNA interactions in ALKBH3.
ALKBH5: AN OXYGENASE TARGETED BY HYPOXIA-INDUCING FACTOR 1α
Like other members of the ALKB family, ALKBH5 possesses the metal- and 2KG-binding motif. Pollard et al. showed that ALKBH5 is largely localized to the nucleus and is a 2KG oxygenase; however, the decarboxylation of 2KG by ALKBH5 is not stimulated by known AlkB substrates, hinting that ALKBH5 does not act on 1-meA-type DNA damage (Thalhammer et al. 2011). What makes ALKBH5 unique among the ALKBH genes is probably that it is a direct transcriptional target of hypoxia-inducible factor-1 and is induced by hypoxia in a range of cell types. Yet the biological function of ALKBH5 is currently unknown.
ALKBH8: A tRNA MODIFICATION ENZYME RATHER THAN A DNA-REPAIR PROTEIN
ALKBH8 is a multidomain protein that possesses an amino-terminal RNA recognition domain, the AlkB domain, and a carboxy-terminal methyltransferase domain. The methyltransferase domain has sequence homology with the S. cerevisiae tRNA methyltransferase Trm9, which catalyzes the methyl esterification of U34 of tRNAArg and tRNAGlu, forming 5-methoxycarbonylmethyluridine (mcm5U) and 5-methoxycarbonylmethyl-2-thiouridine, respectively. Two reports independently showed that ALKBH8 is a tRNA methyltransferase required for the biogenesis of mcm5U (Fu et al. 2010a; Songe-Moller et al. 2010). Soon after, we and others showed that the AlkB domain of ALKBH8 catalyzes the hydroxylation of mcm5U into (S)-5-methoxycarbonylhydroxymethyluridine, thereby firmly establishing ALKBH8 as a tRNA hypermodification enzyme (Fig. 5D) (Fu et al. 2010b; van den Born et al. 2011). The recently solved structure of ALKBH8, comprising the RNA recognition and AlkB domains, shows disordered loops flanking the active site in the AlkB domain, providing insights into the evolutionary diversification of AlkB domains (Pastore et al. 2012). Interestingly, ALKBH8 was shown to contribute to the progression of human bladder cancer; and silencing of ALKBH8 significantly suppressed invasion, angiogenesis, and growth of bladder cancers in vivo (Shimada et al. 2009). How the tRNA modification activity can attribute to this observed phenotype remains to be established.
FTO: LINKING OBESITY TO RNA DEMETHYLAION
A genome-wide association study found the FTO (fat-mass and obesity-associated) gene to be associated with body mass index (BMI) (Frayling et al. 2007). In this study, the authors found that a common variant in the FTO gene predisposes a person to diabetes through an effect on BMI. Adults who are homozygous for the risk allele weighed about 3 kg more compared to those without a risk allele. Two independent studies in close succession reported other intron 1 FTO single-nucleotide polymorphisms (SNPs) and extended the association to other obesity-related traits (Dina et al. 2007; Scuteri et al. 2007). Numerous subsequent studies also confirmed the association among 22 distinct populations, which have been summarized elsewhere (Tung and Yeo 2011).
In the mouse, FTO null individuals (FTO−/−) displayed decreased fat and lean body mass, increased metabolic rate, and elevated food intake, which shows that FTO is involved in energy homeostasis through control of energy expenditure (Fischer et al. 2009; Gao et al. 2010). Consistent with this, FTO was found to be highly expressed in the brain, particularly in the hypothalamus. Furthermore, nutritional status directly regulates the expression of FTO in a bidirectional manner. Food intake can also be bidirectionally influenced through manipulation of the FTO level in the arcuate nucleus of hypothalamus (Gerken et al. 2007; Tung et al. 2010). In humans, a catalytic incompetent mutation Arg316Gln renders the affected individuals a polymalformation syndrome (Boissel et al. 2009). Other loss-of-function mutations of FTO have also been identified, which appear to influence BMI as well (Meyre et al. 2010).
Shortly after FTO was reported, bioinformatics analysis revealed that FTO encodes an iron(II)- and 2KG-dependent dioxygenase, which is closely related to E. coli AlkB (Gerken et al. 2007; Sanchez-Pulido and Andrade-Navarro 2007). Crystal structure of FTO did show an AlkB-like domain and a unique carboxy-terminal domain (Fig. 5C) (Han et al. 2010). Biochemically, FTO was first shown to demethylate 3-meT in ssDNA and later 3-meU in ssRNA in vitro (Gerken et al. 2007; Jia et al. 2008). However, such demethylation activity is exceedingly low compared to other AlkB-family proteins. We discovered in 2011 that FTO shows efficient demethylation activity toward the abundant N6-methyl adenine (6-meA) residues in RNA, both in vitro and in vivo (Fig. 5E) (Jia et al. 2011). In addition, FTO partially colocalizes with nuclear speckles, which supports the notion that 6-meA in nuclear RNA is a physiological substrate. Future experiments demonstrating the specific messenger RNA (mRNA) targets of FTO is expected to link the demethylation activity to the observed phenotype of FTO.
CONCLUDING REMARKS AND OUTLOOK
The last decade has witnessed exciting advances and rapid growth in the field of direct DNA repair. Such rapid expansion of knowledge has not only enabled us to understand DNA-repair pathways in general and for therapeutic purposes (Rabik et al. 2006; Tubbs et al. 2007), but has also accelerated new discoveries in biology (Wu and Zhang 2011). For instance, the oxidative-demethylation mechanism of the AlkB family dioxygenases has inspired the discovery of histone lysine demethylases (Tsukada et al. 2006). The recently identified TET family proteins also share oxidative hydroxylation chemistry with AlkB. These proteins consecutively convert 5-methylcytosine, a hallmark of DNA epigenetics, to 5-hydroxymethylcytosine, 5-formylcytosine, and 5-carboxylcytosine (Tahiliani et al. 2009; Ito et al. 2010, 2011; He et al. 2011). Interestingly, the enzyme that recognizes and excises the oxidation end products of 5-formylcytosine and 5-carboxylcytosine is a DNA-repair glycosylase, thymine DNA glycosylase or TDG, which is a DNA mismatch repair protein (He et al. 2011; Maiti and Drohat 2011; Zhang et al. 2012). These advances are among the most exciting breakthroughs in the field of DNA epigenetics. In addition, the discovery that RNA methylation is reversed by FTO also brought up the notion of reversible RNA modification in biological regulation (Fig. 5E) (He 2010). Nevertheless, many puzzling questions remain. For example, what are enzymatic activities of ALKBH1, 4, 5, 6, and 7 and what are their biological roles? How is the demethylation activity of FTO related to its phenotypic observations? Hopefully, continued investigations will shine light on such questions that are related to or go beyond the field of DNA repair in the future.
Footnotes
Editors: Errol C. Friedberg, Stephen J. Elledge, Alan R. Lehmann, Tomas Lindahl, and Marco Muzi-Falconi
Additional Perspectives on DNA Repair, Mutagenesis, and Other Responses to DNA Damage available at www.cshperspectives.org
REFERENCES
- Aas PA, Otterlei M, Falnes PO, Vagbo CB, Skorpen F, Akbari M, Sundheim O, Bjoras M, Slupphaug G, Seeberg E, et al. 2003. Human and bacterial oxidative demethylases repair alkylation damage in both RNA and DNA. Nature 421: 859–863 [DOI] [PubMed] [Google Scholar]
- Adams CA, Melikishvili M, Rodgers DW, Rasimas JJ, Pegg AE, Fried MG 2009. Topologies of complexes containing O6-alkylguanine-DNA alkyltransferase and DNA. J Mol Biol 389: 248–263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aramini JM, Tubbs JL, Kanugula S, Rossi P, Ertekin A, Maglaqui M, Hamilton K, Ciccosanti CT, Jiang M, Xiao R, et al. 2010. Structural basis of O6-alkylguanine recognition by a bacterial alkyltransferase-like DNA repair protein. J Biol Chem 285: 13736–13741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleijlevens B, Shivarattan T, Flashman E, Yang Y, Simpson PJ, Koivisto P, Sedgwick B, Schofield CJ, Matthews SJ 2008. Dynamic states of the DNA repair enzyme AlkB regulate product release. EMBO Rep 9: 872–877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleijlevens B, Shivarattan T, van den Boom KS, de Haan A, van der Zwan G, Simpson PJ, Matthews SJ 2012. Changes in protein dynamics of the DNA repair dioxygenase AlkB upon binding of Fe2+ and 2-oxoglutarate. Biochemistry 51: 3334–3341 [DOI] [PubMed] [Google Scholar]
- Boissel S, Reish O, Proulx K, Kawagoe-Takaki H, Sedgwick B, Yeo GS, Meyre D, Golzio C, Molinari F, Kadhom N, et al. 2009. Loss-of-function mutation in the dioxygenase-encoding FTO gene causes severe growth retardation and multiple malformations. Am J Hum Genet 85: 106–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowles T, Metz AH, O'Quin J, Wawrzak Z, Eichman BF 2008. Structure and DNA binding of alkylation response protein AidB. Proc Natl Acad Sci 105: 15299–15304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brettel K, Byrdin M 2010. Reaction mechanisms of DNA photolyase. Curr Opin Struct Biol 20: 693–701 [DOI] [PubMed] [Google Scholar]
- Brudler R, Hitomi K, Daiyasu H, Toh H, Kucho K, Ishiura M, Kanehisa M, Roberts VA, Todo T, Tainer JA, et al. 2003. Identification of a new cryptochrome class. Structure, function, and evolution. Mol Cell 11: 59–67 [DOI] [PubMed] [Google Scholar]
- Chen CS, Korobkova E, Chen H, Zhu J, Jian X, Tao SC, He C, Zhu H 2008. A proteome chip approach reveals new DNA damage recognition activities in Escherichia coli. Nat Methods 5: 69–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B, Liu H, Sun X, Yang CG 2010. Mechanistic insight into the recognition of single-stranded and double-stranded DNA substrates by ABH2 and ABH3. Mol BioSyst 6: 2143–2149 [DOI] [PubMed] [Google Scholar]
- Daiyasu H, Ishikawa T, Kuma K, Iwai S, Todo T, Toh H 2004. Identification of cryptochrome DASH from vertebrates. Genes Cells 9: 479–495 [DOI] [PubMed] [Google Scholar]
- Dango S, Mosammaparast N, Sowa ME, Xiong LJ, Wu F, Park K, Rubin M, Gygi S, Harper JW, Shi Y 2011. DNA unwinding by ASCC3 helicase is coupled to ALKBH3-dependent DNA alkylation repair and cancer cell proliferation. Mol Cell 44: 373–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels DS, Woo TT, Luu KX, Noll DM, Clarke ND, Pegg AE, Tainer JA 2004. DNA binding and nucleotide flipping by the human DNA repair protein AGT. Nat Struct Mol Biol 11: 714–720 [DOI] [PubMed] [Google Scholar]
- Delaney JC, Essigmann JM 2004. Mutagenesis, genotoxicity, and repair of 1-methyladenine, 3-alkylcytosines, 1-methylguanine, and 3-methylthymine in alkB Escherichia coli. Proc Natl Acad Sci 101: 14051–14056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaney JC, Smeester L, Wong C, Frick LE, Taghizadeh K, Wishnok JS, Drennan CL, Samson LD, Essigmann JM 2005. AlkB reverses etheno DNA lesions caused by lipid oxidation in vitro and in vivo. Nat Struct Mol Biol 12: 855–860 [DOI] [PubMed] [Google Scholar]
- Demple B, Sedgwick B, Robins P, Totty N, Waterfield MD, Lindahl T 1985. Active site and complete sequence of the suicidal methyltransferase that counters alkylation mutagenesis. Proc Natl Acad Sci 82: 2688–2692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dina C, Meyre D, Gallina S, Durand E, Korner A, Jacobson P, Carlsson LM, Kiess W, Vatin V, Lecoeur C, et al. 2007. Variation in FTO contributes to childhood obesity and severe adult obesity. Nat Genet 39: 724–726 [DOI] [PubMed] [Google Scholar]
- Dinglay S, Trewick SC, Lindahl T, Sedgwick B 2000. Defective processing of methylated single-stranded DNA by E. coli AlkB mutants. Genes Dev 14: 2097–2105 [PMC free article] [PubMed] [Google Scholar]
- Drablos F, Feyzi E, Aas PA, Vaagbo CB, Kavli B, Bratlie MS, Pena-Diaz J, Otterlei M, Slupphaug G, Krokan HE 2004. Alkylation damage in DNA and RNA—repair mechanisms and medical significance. DNA Repair (Amst) 3: 1389–1407 [DOI] [PubMed] [Google Scholar]
- Duguid EM, Rice PA, He C 2005. The structure of the human AGT protein bound to DNA and its implications for damage detection. J Mol Biol 350: 657–666 [DOI] [PubMed] [Google Scholar]
- Duncan T, Trewick SC, Koivisto P, Bates PA, Lindahl T, Sedgwick B 2002. Reversal of DNA alkylation damage by two human dioxygenases. Proc Natl Acad Sci 99: 16660–16665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falnes PO 2004. Repair of 3-methylthymine and 1-methylguanine lesions by bacterial and human AlkB proteins. Nucleic Acids Res 32: 6260–6267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falnes PO, Johansen RF, Seeberg E 2002. AlkB-mediated oxidative demethylation reverses DNA damage in Escherichia coli. Nature 419: 178–182 [DOI] [PubMed] [Google Scholar]
- Falnes PO, Klungland A, Alseth I 2007. Repair of methyl lesions in DNA and RNA by oxidative demethylation. Neuroscience 145: 1222–1232 [DOI] [PubMed] [Google Scholar]
- Fang Q, Noronha AM, Murphy SP, Wilds CJ, Tubbs JL, Tainer JA, Chowdhury G, Guengerich FP, Pegg AE 2008. Repair of O6-G-alkyl-O6-G interstrand cross-links by human O6-alkylguanine-DNA alkyltransferase. Biochemistry 47: 10892–10903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer J, Koch L, Emmerling C, Vierkotten J, Peters T, Bruning JC, Ruther U 2009. Inactivation of the Fto gene protects from obesity. Nature 458: 894–898 [DOI] [PubMed] [Google Scholar]
- Frayling TM, Timpson NJ, Weedon MN, Zeggini E, Freathy RM, Lindgren CM, Perry JR, Elliott KS, Lango H, Rayner NW, et al. 2007. A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science 316: 889–894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frick LE, Delaney JC, Wong C, Drennan CL, Essigmann JM 2007. Alleviation of 1,N6-ethanoadenine genotoxicity by the Escherichia coli adaptive response protein AlkB. Proc Natl Acad Sci 104: 755–760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedberg EC, Friedberg EC 2006. DNA repair and mutagenesis. ASM, Washington, DC [Google Scholar]
- Fu D, Brophy JA, Chan CT, Atmore KA, Begley U, Paules RS, Dedon PC, Begley TJ, Samson LD 2010a. Human AlkB homolog ABH8 is a tRNA methyltransferase required for wobble uridine modification and DNA damage survival. Mol Cell Biol 30: 2449–2459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y, Dai Q, Zhang W, Ren J, Pan T, He C 2010b. The AlkB domain of mammalian ABH8 catalyzes hydroxylation of 5-methoxycarbonylmethyluridine at the wobble position of tRNA. Angew Chem Int Ed Engl 49: 8885–8888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu D, Calvo JA, Samson LD 2012. Balancing repair and tolerance of DNA damage caused by alkylating agents. Nat Rev Cancer 12: 104–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X, Shin YH, Li M, Wang F, Tong Q, Zhang P 2010. The fat mass and obesity associated gene FTO functions in the brain to regulate postnatal growth in mice. PLoS ONE 5: e14005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao W, Li L, Xu P, Fang J, Xiao S, Chen S 2011. Frequent down-regulation of hABH2 in gastric cancer and its involvement in growth of cancer cells. J Gastroenterol Hepatol 26: 577–584 [DOI] [PubMed] [Google Scholar]
- Gerken T, Girard CA, Tung YC, Webby CJ, Saudek V, Hewitson KS, Yeo GS, McDonough MA, Cunliffe S, McNeill LA, et al. 2007. The obesity-associated FTO gene encodes a 2-oxoglutarate-dependent nucleic acid demethylase. Science 318: 1469–1472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilljam KM, Feyzi E, Aas PA, Sousa MM, Muller R, Vagbo CB, Catterall TC, Liabakk NB, Slupphaug G, Drablos F, et al. 2009. Identification of a novel, widespread, and functionally important PCNA-binding motif. J Cell Biol 186: 645–654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guza R, Rajesh M, Fang Q, Pegg AE, Tretyakova N 2006. Kinetics of O6-methyl-2′-deoxyguanosine repair by O6-alkylguanine DNA alkyltransferase within K-ras gene-derived DNA sequences. Chem Res Toxicol 19: 531–538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond LA, Eckardt JR, Kuhn JG, Gerson SL, Johnson T, Smith L, Drengler RL, Campbell E, Weiss GR, Von Hoff DD, et al. 2004. A randomized phase I and pharmacological trial of sequences of 1,3-bis(2-chloroethyl)-1-nitrosourea and temozolomide in patients with advanced solid neoplasms. Clin Cancer Res 10: 1645–1656 [DOI] [PubMed] [Google Scholar]
- Han Z, Niu T, Chang J, Lei X, Zhao M, Wang Q, Cheng W, Wang J, Feng Y, Chai J 2010. Crystal structure of the FTO protein reveals basis for its substrate specificity. Nature 464: 1205–1209 [DOI] [PubMed] [Google Scholar]
- He C 2010. Grand challenge commentary: RNA epigenetics? Nat Chem Biol 6: 863–865 [DOI] [PubMed] [Google Scholar]
- He C, Hus JC, Sun LJ, Zhou P, Norman DP, Dotsch V, Wei H, Gross JD, Lane WS, Wagner G, et al. 2005. A methylation-dependent electrostatic switch controls DNA repair and transcriptional activation by E. coli ada. Mol Cell 20: 117–129 [DOI] [PubMed] [Google Scholar]
- He YF, Li BZ, Li Z, Liu P, Wang Y, Tang Q, Ding J, Jia Y, Chen Z, Li L, et al. 2011. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science 333: 1303–1307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hitomi K, Okamoto K, Daiyasu H, Miyashita H, Iwai S, Toh H, Ishiura M, Todo T 2000. Bacterial cryptochrome and photolyase: Characterization of two photolyase-like genes of Synechocystis sp. PCC6803. Nucleic Acids Res 28: 2353–2362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hitomi K, DiTacchio L, Arvai AS, Yamamoto J, Kim ST, Todo T, Tainer JA, Iwai S, Panda S, Getzoff ED 2009. Functional motifs in the (6–4) photolyase crystal structure make a comparative framework for DNA repair photolyases and clock cryptochromes. Proc Natl Acad Sci 106: 6962–6967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hitomi K, Arvai AS, Yamamoto J, Hitomi C, Teranishi M, Hirouchi T, Yamamoto K, Iwai S, Tainer JA, Hidema J, et al. 2012. Eukaryotic class II cyclobutane pyrimidine dimer photolyase structure reveals basis for improved ultraviolet tolerance in plants. J Biol Chem 287: 12060–12069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland PJ, Hollis T 2010. Structural and mutational analysis of Escherichia coli AlkB provides insight into substrate specificity and DNA damage searching. PLoS ONE 5: e8680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollis T, Lau A, Ellenberger T 2000. Structural studies of human alkyladenine glycosylase and E. coli 3-methyladenine glycosylase. Mutat Res 460: 201–210 [DOI] [PubMed] [Google Scholar]
- Hu J, Ma A, Dinner AR 2008. A two-step nucleotide-flipping mechanism enables kinetic discrimination of DNA lesions by AGT. Proc Natl Acad Sci 105: 4615–4620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H, Chopra R, Verdine GL, Harrison SC 1998. Structure of a covalently trapped catalytic complex of HIV-1 reverse transcriptase: Implications for drug resistance. Science 282: 1669–1675 [DOI] [PubMed] [Google Scholar]
- Huang Y, Baxter R, Smith BS, Partch CL, Colbert CL, Deisenhofer J 2006. Crystal structure of cryptochrome 3 from Arabidopsis thaliana and its implications for photolyase activity. Proc Natl Acad Sci 103: 17701–17706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang CS, Shemorry A, Varshavsky A 2009. Two proteolytic pathways regulate DNA repair by cotargeting the Mgt1 alkylguanine transferase. Proc Natl Acad Sci 106: 2142–2147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito S, D'Alessio AC, Taranova OV, Hong K, Sowers LC, Zhang Y 2010. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature 466: 1129–1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito S, Shen L, Dai Q, Wu SC, Collins LB, Swenberg JA, He C, Zhang Y 2011. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science 333: 1300–1303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia G, Yang CG, Yang S, Jian X, Yi C, Zhou Z, He C 2008. Oxidative demethylation of 3-methylthymine and 3-methyluracil in single-stranded DNA and RNA by mouse and human FTO. FEBS Lett 582: 3313–3319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia G, Fu Y, Zhao X, Dai Q, Zheng G, Yang Y, Yi C, Lindahl T, Pan T, Yang YG, et al. 2011. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat Chem Biol 7: 885–887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao YT, Saxena C, Wang L, Sancar A, Zhong D 2005. Direct observation of thymine dimer repair in DNA by photolyase. Proc Natl Acad Sci 102: 16128–16132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiontke S, Geisselbrecht Y, Pokorny R, Carell T, Batschauer A, Essen LO 2011. Crystal structures of an archaeal class II DNA photolyase and its complex with UV-damaged duplex DNA. EMBO J 30: 4437–4449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koike K, Ueda Y, Hase H, Kitae K, Fusamae Y, Masai S, Inagaki T, Saigo Y, Hirasawa S, Nakajima K, et al. 2012. Anti-tumor effect of AlkB homolog 3 knockdown in hormone-independent prostate cancer cells. Curr Cancer Drug Targets 12: 847–856 [DOI] [PubMed] [Google Scholar]
- Koivisto P, Robins P, Lindahl T, Sedgwick B 2004. Demethylation of 3-methylthymine in DNA by bacterial and human DNA dioxygenases. J Biol Chem 279: 40470–40474 [DOI] [PubMed] [Google Scholar]
- Kondoh M, Hitomi K, Yamamoto J, Todo T, Iwai S, Getzoff ED, Terazima M 2011. Light-induced conformational change and product release in DNA repair by (6–4) photolyase. J Am Chem Soc 133: 2183–2191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korvald H, Falnes PO, Laerdahl JK, Bjoras M, Alseth I 2012. The Schizosaccharomyces pombe AlkB homolog Abh1 exhibits AP lyase activity but no demethylase activity. DNA Repair (Amst) 11: 453–462 [DOI] [PubMed] [Google Scholar]
- Kurowski MA, Bhagwat AS, Papaj G, Bujnicki JM 2003. Phylogenomic identification of five new human homologs of the DNA repair enzyme AlkB. BMC Genomics 4: 48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Liu Z, Tan C, Guo X, Wang L, Sancar A, Zhong D 2010. Dynamics and mechanism of repair of ultraviolet-induced (6–4) photoproduct by photolyase. Nature 466: 887–890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, Zhao T, Jian X, Farooqui Z, Qu X, He C, Dinner AR, Scherer NF 2009. Using the bias from flow to elucidate single DNA repair protein sliding and interactions with DNA. Biophys J 96: 1911–1917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindahl T, Sedgwick B, Sekiguchi M, Nakabeppu Y 1988. Regulation and expression of the adaptive response to alkylating agents. Annu Rev Biochem 57: 133–157 [DOI] [PubMed] [Google Scholar]
- Liu BQ, Wu YD, Li PH, Wei JX, Zhang T, Liu RL 2007. Prostate cancer antigen-1 as a potential novel marker for prostate cancer. Asian J Androl 9: 821–826 [DOI] [PubMed] [Google Scholar]
- Liu Z, Tan C, Guo X, Kao YT, Li J, Wang L, Sancar A, Zhong D 2011. Dynamics and mechanism of cyclobutane pyrimidine dimer repair by DNA photolyase. Proc Natl Acad Sci 108: 14831–14836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiti A, Drohat AC 2011. Thymine DNA glycosylase can rapidly excise 5-formylcytosine and 5-carboxylcytosine: Potential implications for active demethylation of CpG sites. J Biol Chem 286: 35334–35338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margison GP, Povey AC, Kaina B, Santibanez Koref MF 2003. Variability and regulation of O6-alkylguanine-DNA alkyltransferase. Carcinogenesis 24: 625–635 [DOI] [PubMed] [Google Scholar]
- Margison GP, Butt A, Pearson SJ, Wharton S, Watson AJ, Marriott A, Caetano CM, Hollins JJ, Rukazenkova N, Begum G, et al. 2007. Alkyltransferase-like proteins. DNA Repair (Amst) 6: 1222–1228 [DOI] [PubMed] [Google Scholar]
- Maul MJ, Barends TR, Glas AF, Cryle MJ, Domratcheva T, Schneider S, Schlichting I, Carell T 2008. Crystal structure and mechanism of a DNA (6–4) photolyase. Angew Chem Int Ed Engl 47: 10076–10080 [DOI] [PubMed] [Google Scholar]
- Mazon G, Philippin G, Cadet J, Gasparutto D, Fuchs RP 2009. The alkyltransferase-like ybaZ gene product enhances nucleotide excision repair of O6-alkylguanine adducts in E. coli. DNA Repair (Amst) 8: 697–703 [DOI] [PubMed] [Google Scholar]
- Meyre D, Proulx K, Kawagoe-Takaki H, Vatin V, Gutierrez-Aguilar R, Lyon D, Ma M, Choquet H, Horber F, Van Hul W, et al. 2010. Prevalence of loss-of-function FTO mutations in lean and obese individuals. Diabetes 59: 311–318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meza TJ, Moen MN, Vagbo CB, Krokan HE, Klungland A, Grini PE, Falnes PO 2012. The DNA dioxygenase ALKBH2 protects Arabidopsis thaliana against methylation damage. Nucleic Acids Res 40: 6620–6631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishina Y, He C 2003. Probing the structure and function of the Escherichia coli DNA alkylation repair AlkB protein through chemical cross-linking. J Am Chem Soc 125: 8730–8731 [DOI] [PubMed] [Google Scholar]
- Mishina Y, Chen LX, He C 2004a. Preparation and characterization of the native iron(II)-containing DNA repair AlkB protein directly from Escherichia coli. J Am Chem Soc 126: 16930–16936 [DOI] [PubMed] [Google Scholar]
- Mishina Y, Lee CH, He C 2004b. Interaction of human and bacterial AlkB proteins with DNA as probed through chemical cross-linking studies. Nucleic Acids Res 32: 1548–1554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishina Y, Yang CG, He C 2005. Direct repair of the exocyclic DNA adduct 1,N6-ethenoadenine by the DNA repair AlkB proteins. J Am Chem Soc 127: 14594–14595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monsen VT, Sundheim O, Aas PA, Westbye MP, Sousa MM, Slupphaug G, Krokan HE 2010. Divergent ss-hairpins determine double-strand versus single-strand substrate recognition of human AlkB-homologues 2 and 3. Nucleic Acids Res 38: 6447–6455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita R, Nakagawa N, Kuramitsu S, Masui R 2008. An O6-methylguanine-DNA methyltransferase-like protein from Thermus thermophilus interacts with a nucleotide excision repair protein. J Biochem 144: 267–277 [DOI] [PubMed] [Google Scholar]
- Muller M, Carell T 2009. Structural biology of DNA photolyases and cryptochromes. Curr Opin Struct Biol 19: 277–285 [DOI] [PubMed] [Google Scholar]
- Muller TA, Meek K, Hausinger RP 2010. Human AlkB homologue 1 (ABH1) exhibits DNA lyase activity at abasic sites. DNA Repair (Amst) 9: 58–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers LC, Terranova MP, Ferentz AE, Wagner G, Verdine GL 1993. Repair of DNA methylphosphotriesters through a metalloactivated cysteine nucleophile. Science 261: 1164–1167 [DOI] [PubMed] [Google Scholar]
- Nay SL, Lee DH, Bates SE, O'Connor TR 2012. Alkbh2 protects against lethality and mutation in primary mouse embryonic fibroblasts. DNA Repair (Amst) 11: 502–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordstrand LM, Svard J, Larsen E, Nilsen A, Ougland R, Furu K, Lien GF, Rognes T, Namekawa SH, Lee JT, et al. 2010. Mice lacking Alkbh1 display sex-ratio distortion and unilateral eye defects. PLoS ONE 5: e13827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okafuji A, Biskup T, Hitomi K, Getzoff ED, Kaiser G, Batschauer A, Bacher A, Hidema J, Teranishi M, Yamamoto K, et al. 2010. Light-induced activation of class II cyclobutane pyrimidine dimer photolyases. DNA Repair (Amst) 9: 495–505 [DOI] [PubMed] [Google Scholar]
- Ougland R, Zhang CM, Liiv A, Johansen RF, Seeberg E, Hou YM, Remme J, Falnes PO 2004. AlkB restores the biological function of mRNA and tRNA inactivated by chemical methylation. Mol Cell 16: 107–116 [DOI] [PubMed] [Google Scholar]
- Pan Z, Sikandar S, Witherspoon M, Dizon D, Nguyen T, Benirschke K, Wiley C, Vrana P, Lipkin SM 2008. Impaired placental trophoblast lineage differentiation in Alkbh1−/− mice. Dev Dyn 237: 316–327 [DOI] [PubMed] [Google Scholar]
- Pastore C, Topalidou I, Forouhar F, Yan AC, Levy M, Hunt JF 2012. Crystal structure and RNA binding properties of the RNA recognition motif (RRM) and AlkB domains in human AlkB homolog 8 (ABH8), an enzyme catalyzing tRNA hypermodification. J Biol Chem 287: 2130–2143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson SJ, Ferguson J, Santibanez-Koref M, Margison GP 2005. Inhibition of O6-methylguanine-DNA methyltransferase by an alkyltransferase-like protein from Escherichia coli. Nucleic Acids Res 33: 3837–3844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson SJ, Wharton S, Watson AJ, Begum G, Butt A, Glynn N, Williams DM, Shibata T, Santibanez-Koref MF, Margison GP 2006. A novel DNA damage recognition protein in Schizosaccharomyces pombe. Nucleic Acids Res 34: 2347–2354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pegg AE 2011. Multifaceted roles of alkyltransferase and related proteins in DNA repair, DNA damage, resistance to chemotherapy, and research tools. Chem Res Toxicol 24: 618–639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pokorny R, Klar T, Hennecke U, Carell T, Batschauer A, Essen LO 2008. Recognition and repair of UV lesions in loop structures of duplex DNA by DASH-type cryptochrome. Proc Natl Acad Sci 105: 21023–21027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinn JA, Jiang SX, Reardon DA, Desjardins A, Vredenburgh JJ, Rich JN, Gururangan S, Friedman AH, Bigner DD, Sampson JH, et al. 2009a. Phase II trial of temozolomide plus O6-benzylguanine in adults with recurrent, temozolomide-resistant malignant glioma. J Clin Oncol 27: 1262–1267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinn JA, Jiang SX, Reardon DA, Desjardins A, Vredenburgh JJ, Rich JN, Gururangan S, Friedman AH, Bigner DD, Sampson JH, et al. 2009b. Phase I trial of temozolomide plus O6-benzylguanine 5-day regimen with recurrent malignant glioma. Neuro Oncol 11: 556–561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabik CA, Njoku MC, Dolan ME 2006. Inactivation of O6-alkylguanine DNA alkyltransferase as a means to enhance chemotherapy. Cancer Treat Rev 32: 261–276 [DOI] [PubMed] [Google Scholar]
- Rasimas JJ, Kar SR, Pegg AE, Fried MG 2007. Interactions of human O6-alkylguanine-DNA alkyltransferase (AGT) with short single-stranded DNAs. J Biol Chem 282: 3357–3366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ringvoll J, Nordstrand LM, Vagbo CB, Talstad V, Reite K, Aas PA, Lauritzen KH, Liabakk NB, Bjork A, Doughty RW, et al. 2006. Repair deficient mice reveal mABH2 as the primary oxidative demethylase for repairing 1meA and 3meC lesions in DNA. EMBO J 25: 2189–2198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rippa V, Duilio A, di Pasquale P, Amoresano A, Landini P, Volkert MR 2011. Preferential DNA damage prevention by the E. coli AidB gene: A new mechanism for the protection of specific genes. DNA Repair (Amst) 10: 934–941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohankhedkar MS, Mulrooney SB, Wedemeyer WJ, Hausinger RP 2006. The AidB component of the Escherichia coli adaptive response to alkylating agents is a flavin-containing, DNA-binding protein. J Bacteriol 188: 223–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakumi K, Sekiguchi M 1989. Regulation of expression of the ada gene controlling the adaptive response. Interactions with the ada promoter of the Ada protein and RNA polymerase. J Mol Biol 205: 373–385 [DOI] [PubMed] [Google Scholar]
- Sanchez-Pulido L, Andrade-Navarro MA 2007. The FTO (fat mass and obesity associated) gene codes for a novel member of the non-heme dioxygenase superfamily. BMC Biochem 8: 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scuteri A, Sanna S, Chen WM, Uda M, Albai G, Strait J, Najjar S, Nagaraja R, Orru M, Usala G, et al. 2007. Genome-wide association scan shows genetic variants in the FTO gene are associated with obesity-related traits. PLoS Genet 3: e115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedgwick B 2004. Repairing DNA-methylation damage. Nat Rev Mol Cell Biol 5: 148–157 [DOI] [PubMed] [Google Scholar]
- Sedgwick B, Robins P, Totty N, Lindahl T 1988. Functional domains and methyl acceptor sites of the Escherichia coli ada protein. J Biol Chem 263: 4430–4433 [PubMed] [Google Scholar]
- Selby CP, Sancar A 2006. A cryptochrome/photolyase class of enzymes with single-stranded DNA-specific photolyase activity. Proc Natl Acad Sci 103: 17696–17700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada K, Nakamura M, Ishida E, Higuchi T, Yamamoto H, Tsujikawa K, Konishi N 2008. Prostate cancer antigen-1 contributes to cell survival and invasion though discoidin receptor 1 in human prostate cancer. Cancer Sci 99: 39–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada K, Nakamura M, Anai S, De Velasco M, Tanaka M, Tsujikawa K, Ouji Y, Konishi N 2009. A novel human AlkB homologue, ALKBH8, contributes to human bladder cancer progression. Cancer Res 69: 3157–3164 [DOI] [PubMed] [Google Scholar]
- Shrivastav N, Li D, Essigmann JM 2010. Chemical biology of mutagenesis and DNA repair: Cellular responses to DNA alkylation. Carcinogenesis 31: 59–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Songe-Moller L, van den Born E, Leihne V, Vagbo CB, Kristoffersen T, Krokan HE, Kirpekar F, Falnes PO, Klungland A 2010. Mammalian ALKBH8 possesses tRNA methyltransferase activity required for the biogenesis of multiple wobble uridine modifications implicated in translational decoding. Mol Cell Biol 30: 1814–1827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivenugopal KS, Yuan XH, Friedman HS, Ali-Osman F 1996. Ubiquitination-dependent proteolysis of O6-methylguanine-DNA methyltransferase in human and murine tumor cells following inactivation with O6-benzylguanine or 1,3-bis(2-chloroethyl)-1-nitrosourea. Biochemistry 35: 1328–1334 [DOI] [PubMed] [Google Scholar]
- Sundheim O, Vagbo CB, Bjoras M, Sousa MM, Talstad V, Aas PA, Drablos F, Krokan HE, Tainer JA, Slupphaug G 2006. Human ABH3 structure and key residues for oxidative demethylation to reverse DNA/RNA damage. EMBO J 25: 3389–3397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L, et al. 2009. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science 324: 930–935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tasaki M, Shimada K, Kimura H, Tsujikawa K, Konishi N 2011. ALKBH3, a human AlkB homologue, contributes to cell survival in human non-small-cell lung cancer. Br J Cancer 104: 700–706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teo I, Sedgwick B, Kilpatrick MW, McCarthy TV, Lindahl T 1986. The intracellular signal for induction of resistance to alkylating agents in E. coli. Cell 45: 315–324 [DOI] [PubMed] [Google Scholar]
- Thalhammer A, Bencokova Z, Poole R, Loenarz C, Adam J, O'Flaherty L, Schodel J, Mole D, Giaslakiotis K, Schofield CJ, et al. 2011. Human AlkB homologue 5 is a nuclear 2-oxoglutarate dependent oxygenase and a direct target of hypoxia-inducible factor 1α (HIF-1α). PLoS ONE 6: e16210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiagarajan V, Byrdin M, Eker AP, Muller P, Brettel K 2011. Kinetics of cyclobutane thymine dimer splitting by DNA photolyase directly monitored in the UV. Proc Natl Acad Sci 108: 9402–9407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trewick SC, Henshaw TF, Hausinger RP, Lindahl T, Sedgwick B 2002. Oxidative demethylation by Escherichia coli AlkB directly reverts DNA base damage. Nature 419: 174–178 [DOI] [PubMed] [Google Scholar]
- Tsukada Y, Fang J, Erdjument-Bromage H, Warren ME, Borchers CH, Tempst P, Zhang Y 2006. Histone demethylation by a family of JmjC domain-containing proteins. Nature 439: 811–816 [DOI] [PubMed] [Google Scholar]
- Tubbs JL, Tainer JA 2010. Alkyltransferase-like proteins: Molecular switches between DNA repair pathways. Cell Mol Life Sci 67: 3749–3762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tubbs JL, Pegg AE, Tainer JA 2007. DNA binding, nucleotide flipping, and the helix-turn-helix motif in base repair by O6-alkylguanine-DNA alkyltransferase and its implications for cancer chemotherapy. DNA Repair (Amst) 6: 1100–1115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tubbs JL, Latypov V, Kanugula S, Butt A, Melikishvili M, Kraehenbuehl R, Fleck O, Marriott A, Watson AJ, Verbeek B, et al. 2009. Flipping of alkylated DNA damage bridges base and nucleotide excision repair. Nature 459: 808–813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tung YC, Yeo GS 2011. From GWAS to biology: Lessons from FTO. Ann NY Acad Sci 1220: 162–171 [DOI] [PubMed] [Google Scholar]
- Tung YC, Ayuso E, Shan X, Bosch F, O'Rahilly S, Coll AP, Yeo GS 2010. Hypothalamic-specific manipulation of Fto, the ortholog of the human obesity gene FTO, affects food intake in rats. PLoS ONE 5: e8771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turriziani M, Caporaso P, Bonmassar L, Buccisano F, Amadori S, Venditti A, Cantonetti M, D'Atri S, Bonmassar E 2006. O6-(4-bromothenyl)guanine (PaTrin-2), a novel inhibitor of O6-alkylguanine DNA alkyl-transferase, increases the inhibitory activity of temozolomide against human acute leukaemia cells in vitro. Pharmacol Res 53: 317–323 [DOI] [PubMed] [Google Scholar]
- van den Born E, Vagbo CB, Songe-Moller L, Leihne V, Lien GF, Leszczynska G, Malkiewicz A, Krokan HE, Kirpekar F, Klungland A, et al. 2011. ALKBH8-mediated formation of a novel diastereomeric pair of wobble nucleosides in mammalian tRNA. Nat Commun 2: 172. [DOI] [PubMed] [Google Scholar]
- Verdine GL, Norman DP 2003. Covalent trapping of protein-DNA complexes. Annu Rev Biochem 72: 337–366 [DOI] [PubMed] [Google Scholar]
- Warren KE, Aikin AA, Libucha M, Widemann BC, Fox E, Packer RJ, Balis FM 2005. Phase I study of O6-benzylguanine and temozolomide administered daily for 5 days to pediatric patients with solid tumors. J Clin Oncol 23: 7646–7653 [DOI] [PubMed] [Google Scholar]
- Westbye MP, Feyzi E, Aas PA, Vagbo CB, Talstad VA, Kavli B, Hagen L, Sundheim O, Akbari M, Liabakk NB, et al. 2008. Human AlkB homolog 1 is a mitochondrial protein that demethylates 3-methylcytosine in DNA and RNA. J Biol Chem 283: 25046–25056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Zhang Y 2011. Mechanisms and functions of Tet protein-mediated 5-methylcytosine oxidation. Genes Dev 25: 2436–2452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyatt MD, Allan JM, Lau AY, Ellenberger TE, Samson LD 1999. 3-methyladenine DNA glycosylases: Structure, function, and biological importance. Bioessays 21: 668–676 [DOI] [PubMed] [Google Scholar]
- Yang CG, Yi C, Duguid EM, Sullivan CT, Jian X, Rice PA, He C 2008. Crystal structures of DNA/RNA repair enzymes AlkB and ABH2 bound to dsDNA. Nature 452: 961–965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi C, Yang CG, He C 2009. A non-heme iron-mediated chemical demethylation in DNA and RNA. Acc Chem Res 42: 519–529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi C, Jia G, Hou G, Dai Q, Zhang W, Zheng G, Jian X, Yang CG, Cui Q, He C 2010. Iron-catalysed oxidation intermediates captured in a DNA repair dioxygenase. Nature 468: 330–333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi C, Chen B, Qi B, Zhang W, Jia G, Zhang L, Li CJ, Dinner AR, Yang CG, He C 2012. Duplex interrogation by a direct DNA repair protein in search of base damage. Nat Struct Mol Biol 19: 671–676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu B, Hunt JF 2009. Enzymological and structural studies of the mechanism of promiscuous substrate recognition by the oxidative DNA repair enzyme AlkB. Proc Natl Acad Sci 106: 14315–14320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu B, Edstrom WC, Benach J, Hamuro Y, Weber PC, Gibney BR, Hunt JF 2006. Crystal structures of catalytic complexes of the oxidative DNA/RNA repair enzyme AlkB. Nature 439: 879–884 [DOI] [PubMed] [Google Scholar]
- Zang H, Fang Q, Pegg AE, Guengerich FP 2005. Kinetic analysis of steps in the repair of damaged DNA by human O6-alkylguanine-DNA alkyltransferase. J Biol Chem 280: 30873–30881 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Iwata T, Yamamoto J, Hitomi K, Iwai S, Todo T, Getzoff ED, Kandori H 2011. FTIR study of light-dependent activation and DNA repair processes of (6–4) photolyase. Biochemistry 50: 3591–3598 [DOI] [PubMed] [Google Scholar]
- Zhang L, Lu X, Lu J, Liang H, Dai Q, Xu GL, Luo C, Jiang H, He C 2012. Thymine DNA glycosylase specifically recognizes 5-carboxylcytosine-modified DNA. Nat Chem Biol 8: 328–330 [DOI] [PMC free article] [PubMed] [Google Scholar]